Abstract
We have used membrane surface charge to modulate the structural dynamics of an integral membrane protein, phospholamban (PLB), and thereby its functional inhibition of the sarcoplasmic reticulum Ca-ATPase (SERCA). It was previously shown by EPR, in vesicles of neutral lipids, that the PLB cytoplasmic domain is in equilibrium between an ordered T state and a dynamically disordered R state, and that phosphorylation of PLB increases the R state and relieves SERCA inhibition, suggesting that R is less inhibitory. Here we sought to control the T/R equilibrium by an alternative means – varying the lipid headgroup charge, thus perturbing the electrostatic interaction of PLB’s cationic cytoplasmic domain with the membrane surface. We resolved the T and R states not only by EPR in the absence of SERCA, but also by time-resolved fluorescence resonance energy transfer (TR-FRET) from SERCA to PLB, thus probing directly the SERCA-PLB complex. Compared to neutral lipids, anionic lipids increased both the T population and SERCA inhibition, while cationic lipids had the opposite effects. In contrast to conventional models, decreased inhibition was not accompanied by decreased binding. We conclude that PLB binds to SERCA in two distinct structural states of the cytoplasmic domain, an inhibitory T state that interacts strongly with the membrane surface, and a less inhibitory R state that interacts more strongly with the anionic SERCA cytoplasmic domain. Modulating membrane surface charge provides an effective way of investigating the correlation between structural dynamics and function of integral membrane proteins.
Keywords: phospholamban, SERCA, FRET, EPR, electrostatic interactions
Introduction
The functions of integral membrane proteins depend on the interplay of protein structure and dynamics with the lipid environment1. In the present study, we use the lipid environment as a tool to perturb the system, followed by measurement of structure, dynamics, and function, to elucidate mechanistic principles. We vary membrane surface electrostatics by manipulating lipid headgroup charge, which has been shown to be a powerful approach in the analysis of peripheral membrane proteins2.
The sarcoplasmic reticulum Ca-ATPase (SERCA) actively transports Ca from the cytoplasm to the SR lumen and initiates muscle relaxation. In the cardiomyocyte, an integral membrane protein phospholamban (PLB)3 regulates SERCA activity by decreasing SERCA’s apparent Ca affinity4. This inhibition can be relieved by elevated Ca or by phosphorylation of PLB in response to β-adrenergic stimulation5. Decreasing this inhibitory regulation relieves cardiomyopathy, so elucidating the interaction mechanism between SERCA and PLB is essential for understanding cardiac pathology and for devising new cardiac therapies6.
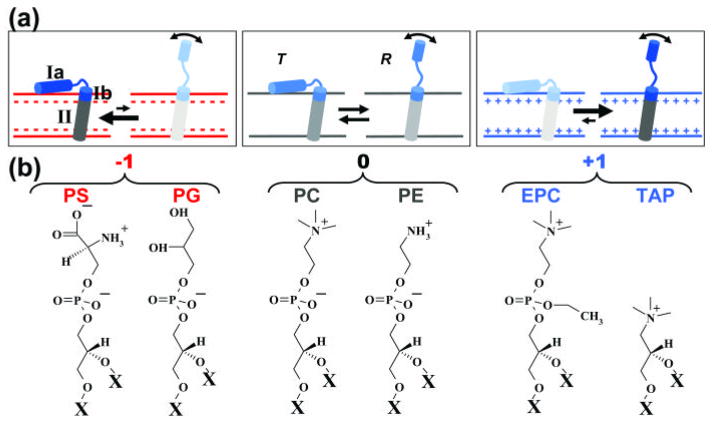
PLB exists in equilibrium between monomeric and pentameric forms, but the monomer is the principle species that binds to and inhibits SERCA 7, so we used the monomeric AFA-PLB mutant (C36A/C41F/C46A) 8; 9 throughout this study. The high-resolution structural dynamics of free PLB monomer in a lipid bilayer has been determined using nuclear magnetic resonance (NMR) and electron paramagnetic resonance (EPR) 8; 9; 10; 11. PLB consists of an N-terminal cytoplasmic helix, a loop, and a transmembrane helix (Fig. 1). The top of the TM helix (domain Ib) is hydrophilic, directly interacts with lipid headgroups, and is more dynamic than the rest of the TM helix (domain II) 9; 11; 12. EPR of TOAC, a spin label attached rigidly to the peptide backbone, shows that the cytoplasmic domain of PLB (Ia and Ib) is in equilibrium between an ordered T state and a dynamically disordered (partially unfolded) R state (sometimes called “excited state”), while the transmembrane helix is quite stable 9. The cytoplasmic domain is associated with the membrane surface in T but dissociated in R 9. Phosphorylation of PLB induces a shift in the T/R equilibrium toward R, suggesting that R is less inhibitory than T 10; 13.
Fig. 1.
Tuning the T/R equilibrium using lipid headgroup charge. Here and in subsequent figures, red indicates negative charge, blue positive. (a) The cationic cytoplasmic domain (Ia and Ib) of monomeric PLB is in equilibrium between an ordered T state and a dynamically disordered R state, while domain II is stable 9. (b) Structures of lipid headgroups and their net charges. All lipids have the same fatty acid chain, X = C18:1 (oleic acid).
Numerous high-resolution structures of SERCA in its enzymatic cycle have been obtained from X-ray diffraction 14; 15, but there is no high-resolution structure of the SERCA-PLB complex. Based on crosslinking, mutagenesis and structures of free SERCA and free PLB, a docking model has been constructed, in which the cytoplasmic domain of PLB extends above the membrane surface and interacts with the cytoplasmic domain of SERCA16. Conventional models hypothesize that dissociation of this inhibitory SERCA-PLB complex is necessary for the relief of SERCA inhibition, either by high Ca, phosphorylation of PLB, mutagenesis of PLB, or addition of a PLB antibody17; 18, but EPR and NMR studies suggest that PLB remains bound to SERCA in both T (inhibitory) and R (less- inhibitory) states10; 19; 20. However, none of these spectroscopic studies probed specifically the bound SERCA-PLB complex.
To help resolve this controversy, in the present study we have probed directly the structure of the SERCA-PLB complex, and we systematically tuned the structural dynamics of the cationic cytoplasmic domain of PLB by adjusting membrane surface charge using charged lipids. We first used EPR 10; 21 of TOAC-PLB in the absence of SERCA, to show that we can control the T/R equilibrium using lipid headgroup charge. We then used time-resolved fluorescence resonance energy transfer (TR-FRET) 22 to directly measure SERCA-PLB binding and simultaneously resolve the T and R structural states of the SERCA-PLB complex. We performed ATPase assays to determine the correlation of these observations with PLB inhibitory function. With this combined approach we constructed a revised model for the structural and functional regulation of the SERCA-PLB complex. This approach has implications far beyond SERCA, demonstrating that variation of membrane surface electrostatics, in conjunction with high-resolution spectroscopy, is a potentially powerful approach to systematically tune the structural and functional dynamics of integral membrane proteins.
Results
We used lipid headgroup charge as a means of perturbing electrostatically the structural equilibria of the SERCA-PLB system. The advantage of this approach is that it does not alter the native chemical compositions of the proteins, compared with conventional modifications such as mutagenesis, phosphorylation, and crosslinking. All lipids used have the same unsaturated fatty acid chains, di(C18:1 ), but varying headgroups and charges: phosphatidyl choline (PC, 0), phosphatidyl ethanolamine (PE, 0), phosphatidyl glycerol (PG, −1), phosphatidyl serine (PS, −1), ethyl-phosphocholine (EPC, +1), and trimethyl-ammonium-propane (TAP, +1) (Fig. 1). We hypothesized that the principal effect of this variation of membrane surface charge would be to perturb the equilibrium between the T state (membrane bound and highly ordered) and the R state (dissociated from the membrane and highly disordered). If our hypothesis is true, negatively charged lipids should increase the T state population and SERCA inhibition (Fig. 1, top left), while positively charged lipids should have the opposite effects (Fig. 1, top right). In this work, membranes were composed of PC, PE, and L at molar ratios 4/1/1, where L is PS, PG, PC, EPC, or TAP (Fig. 1, bottom). PC and PE are in all samples, because they are the major constituent lipids in cardiac SR and are important for SERCA activity and PLB-dependent regulation in reconstituted membranes 23; 24. The molar ratios of SERCA/PLB/lipid, when one or both proteins were present, were 1/10/700, which results in a functional regulation of SERCA by PLB that matches that in the native environment 25; 26.
EPR shows that the PLB structural distribution depends on lipid headgroup charge
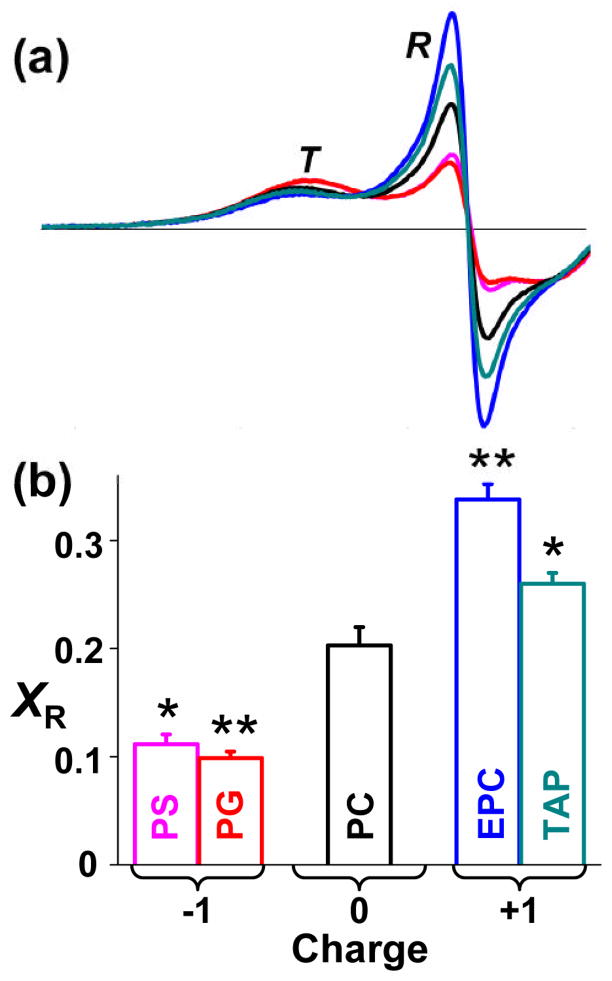
We have previously shown that the TOAC spin label, rigidly coupled to the peptide backbone at position 11 on PLB, clearly resolves the T and R states 9; 10; 21. Therefore, we used this EPR approach to determine the effect of lipid headgroup charge on the T/R equilibrium (Fig. 2). The T state is dynamically restricted, resulting in a broad peak at lower field, while the dynamically disordered R state results in a sharp peak at higher field (Fig. 2a). The positions and shapes of the two components did not change significantly with lipid headgroup charge, indicating that only the populations of the two states were affected. The mole fraction of PLB in the R state (XR) was determined by digital analysis of EPR spectra as described previously 21 (Fig. 2b). XR is about 0.20 in zwitterionic PC, while anionic PS and PG decrease XR substantially and cationic EPC and TAP increase XR (Fig. 2). These results strongly support the hypothesis in Fig. 1: the T/R equilibrium is influenced by the electrostatic interaction between the cationic cytoplasmic domain of PLB and the membrane surface charge; anionic lipid headgroups attract the cationic PLB cytoplasmic domain to the membrane surface, stabilizing the membrane-associated T state, while cationic lipid headgroups repel the cationic PLB cytoplasmic domain from the membrane surface, stabilizing the R state.
Fig. 2.
Effect of lipid headgroup charge on EPR of 11-TOAC spin-labeled PLB in lipid bilayers, with lipid composition and abbreviations as defined in Fig. 1 (color scheme indicated in Fig. 1b). (a) Low-field portion (3305 to 3341 G) of the spectrum resolves two distinct dynamic states of the PLB cytoplasmic domain, an ordered T state and a dynamically disordered R state. (b) Mole fraction of R state (XR). Mean ± SEM (n = 3). Students t-test, compared with PC: * P < 0.05, ** P < 0.01.
TR-FRET from SERCA to PLB resolves two structural states of the SERCA-PLB complex
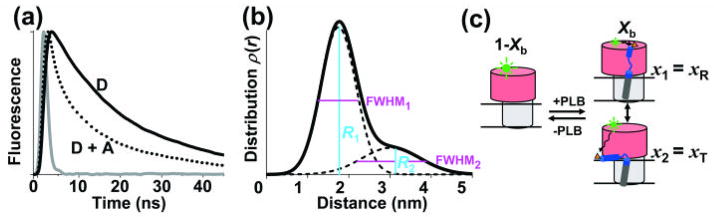
To further resolve the structure of the SERCA-PLB complex, we performed TR-FRET to measure the distance between IAEDANS labeled SERCA (AEDANS-SERCA, donor) and Dabcyl labeled PLB (Dabcyl-PLB, acceptor). The Förster distance (R0) between this pair is 3.2 nm 25. IAEDANS labels SERCA specifically at Cys 674, with the molar ratio of bound dye to SERCA = 1.02 ± 0.05 27. Synthetic AFA-PLB was labeled by attaching Fmoc-Lys(Dabcyl)-OH to the N-terminus at the last step of synthesis 26. The time-resolved fluorescence of AEDANS-SERCA without (donor only, FD(t)) or with Dabcyl-AFA-PLB (donor plus acceptor, FD+A(t)) was measured by direct waveform recording using a high-performance time-resolved fluorescence instrument28 (see Methods) (Fig. 3a), then analyzed (Eq. S1–S7) using non- linear fitting software. Conventional steady-state FRET only measures the ensemble-averaged interprobe distance. The principal advantage of TR-FRET is that it resolves directly the fraction Xb of the donor that has acceptor bound, thus reporting directly the structure of the SERCA-PLB complex (Eq. S4 – Eq. S7). Within this bound complex, TR-FRET also resolves multiple structural states, as defined by Gaussian interprobe distance distributions, each characterized by the center (Ri), width (FWHMi), and mole fraction (xi) (Eq. S4 – Eq. S7) 22; 29. For the bound complex in PC, two Gaussian components are necessary and sufficient to fit FDA(t) (Fig. 3b, Fig. S2). The shorter interprobe distance (R1) is 1.75 ± 0.03 nm, with width FWHM1 = 0.99 ± 0.07 nm and mole fraction x1 = 0.77 ± 0.02. The longer interprobe distance (R2) is 3.03 ± 0.02 nm, with FWHM2 being 1.67 ± 0.24 nm, and x2 = 1− x1 = 0.23 ± 0.02. The fraction of SERCA bound to PLB (Eq. S6), is Xb = 0.82 ± 0.01 in PC, indicating that 82% of SERCA is bound to PLB. The se results support the model that the complex between SERCA and PLB is in equilibrium between two structural states (Fig. 3), as is PLB in the absence of SERCA (Fig. 2).
Fig. 3.
TR-FRET from AEDANS-SERCA (donor) to Dabcyl-PLB (acceptor) in PC. (a) Example of TR fluorescence, measured by direct waveform recording in PC membranes. D = donor-only, D+A = donor plus acceptor. Gray = instrument response function (IRF). (b) SERCA-PLB interprobe distance distribution determined from data in a, containing two Gaussian components (solid curve = sum of two dashed curves), centered at R1 and R2. (c) TR-FRET data resolves free SERCA (left, mole fraction 1-Xb) from PLB-bound SERCA-PLB (right, Xb). Two structural states of the bound SERCA-PLB complex are resolved (b), consistent with the model shown here. x1 and x2 are the mole fractions of the two states, corresponding to bound T and R states, as shown below.
Ensemble average FRET shows that the average interprobe distance between SERCA and PLB is affected by lipid headgroup charge
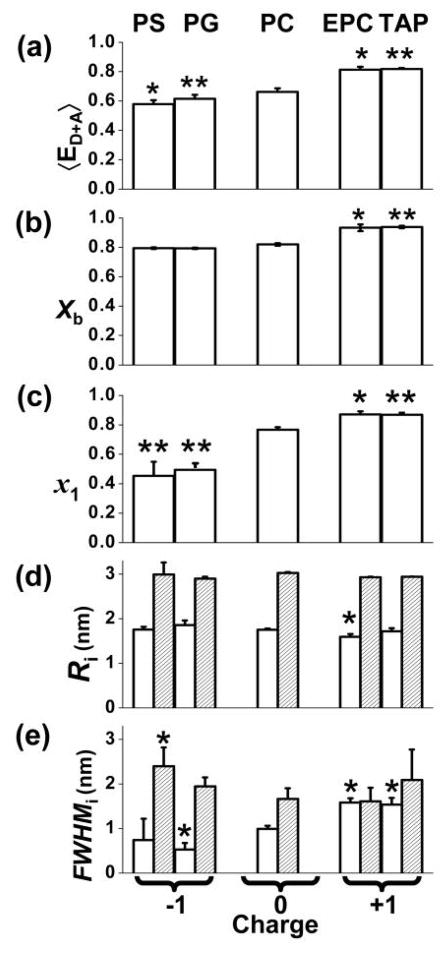
Ensemble-average FRET efficiency 〈ED+A〉 (Fig. 4a) was calculated using the average lifetime (Eq. S3), which is equivalent to (but more precise than) FRET efficiency measured by fluorescence intensity under steady illumination30. Compared to zwitterionic PC, anionic PS and PG decreased FRET, suggesting an increase in the average interprobe distance, while cationic EPC and TAP increased the ensemble-average FRET, suggesting a decreased average interprobe distance (Fig. 4a). These results suggest that the proximity between SERCA and PLB is affected by the electrostatic interaction between the lipid headgroup and the PLB cytoplasmic domain. However, this ensemble average measurement has no structural resolution, so it can not distinguish a change in binding from a change in the structure of the complex. Only TR-FRET can resolve the ambiguity.
Fig. 4.
Effects of lipid charge on SERCA-PLB structural distribution, determined from TR-FRET. Two-Gaussian components distance distributions were determined as in (Fig. 3). Students t-test, compared with PC: * P < 0.05, ** P < 0.01. (a) Ensemble average FRET. (b) Mole fraction of SERCA bound to PLB. (c) Mole fraction of the short distance state (x1 = xR). (d) Centers R1 (open) and R2 (shaded) of the two distance distributions. (e) Widths.
Lipid headgroup charge modulates the distribution of the two structural states of the SERCA-PLB complex
TR-FRET waveforms were fitted as described in Fig. 3. In all cases, across the five different lipid compositions, two structural states were found to be necessary and sufficient to fit the data. The independently determined parameters (Fig. 4b–e) were the fraction of SERCA bound to PLB (Xb) and mole fractions of bound states (x1 and x2) (Fig. 3c), along with the structural characteristics of each bound state (R1, FWHM1, R2, FWHM2) (Eq. S1–S7) (Fig. 3b). Membrane surface charge only slightly affects the binding (Xb) between SERCA and PLB (Fig. 4b). In PS, PG and PC, Xb is ~ 0.8, while in EPC and TAP, Xb is ~ 0.9. Thus, most SERCA has PLB bound, and these small effects can not explain the substantial dependence of 〈ED+A〉 on charge (Fig. 4a). The two structural states have consistent properties, justifying their being treated as “states.” The central interprobe distances of the two SERCA-PLB structural states were found to be quite invariant, with a short distance R1 ~ 1.8 nm, and a long distance R2 ~ 3.0 nm (Fig. 4d). Some of the widths (FWHMi, defining the structural heterogeneity) of the distance distributions are slightly dependent on lipid headgroup charge (Fig. 4e).
The most prominent effect of membrane surface charge is to shift the equilibrium between the two structural states (Fig. 4c). Compared to zwitterionic PC (x1 = 0.77 ± 0.02), anionic PS and PG decrease x1 to 0.45 ± 0.10 and 0.49 ± 0.05 respectively. Cationic EPC and TAP increase x1 to 0.87 ± 0.02 and 0.87 ± 0.01 respectively. Thus in both isolated PLB (EPR in Fig. 2) and in the SERCA-PLB complex (TR-FRET in Fig. 4), the electrostatic interaction between the cytoplasmic domain of PLB and the membrane surface charge shifts the structural equilibrium between two structural states. Based on a comparison between EPR and TR-FRET data, it appears that population 1, the short distance state detected by TR-FRET, corresponds to the R state, while population 2 corresponds to the T state, as depicted in Fig. 3c. Negative surface charge attracts the positively charged PLB cytoplasmic domain and thus increases the fraction (x2 = xT ) of the membrane-associated bound T state (long interprobe distance), while positively charged headgroups have the opposite effect and thus increase the bound R state fraction (x1 = xR).
SERCA activity in the absence of PLB depends on lipid headgroup charge
To establish control values, the ATPase activity of SERCA alone was measured at different pCa, and the pCa-dependence was fitted using Eq. 1. There were no significant effects on the Vmax (activity at saturating Ca), but there were significant effects of headgroup charge on pKCa, defining the apparent Ca affinity (Table 1). Compared to PC (zwitterionic), PS and PG (anionic) increase pKCa, while EPC and TAP (cationic) decrease it.
Table 1.
Lipid charge effects on SERCA activity (Eq. 1). (Mean ± SEM. n ≥ 4)
| Lipid (charge) | pKCa | Vmax |
|---|---|---|
| PS (−) | 6.81 ± 0.01 | 1.65 ± 0.03 |
| PG (−) | 6.73 ± 0.02 | 1.55 ± 0.12 |
| PC (0) | 6.68 ± 0.02 | 1.65 ± 0.19 |
| EPC (+) | 6.53 ± 0.03 | 1.46 ± 0.10 |
| TAP (+) | 6.51 ± 0.02 | 1.66 ± 0.19 |
Lipid headgroup charge affects SERCA inhibition by PLB
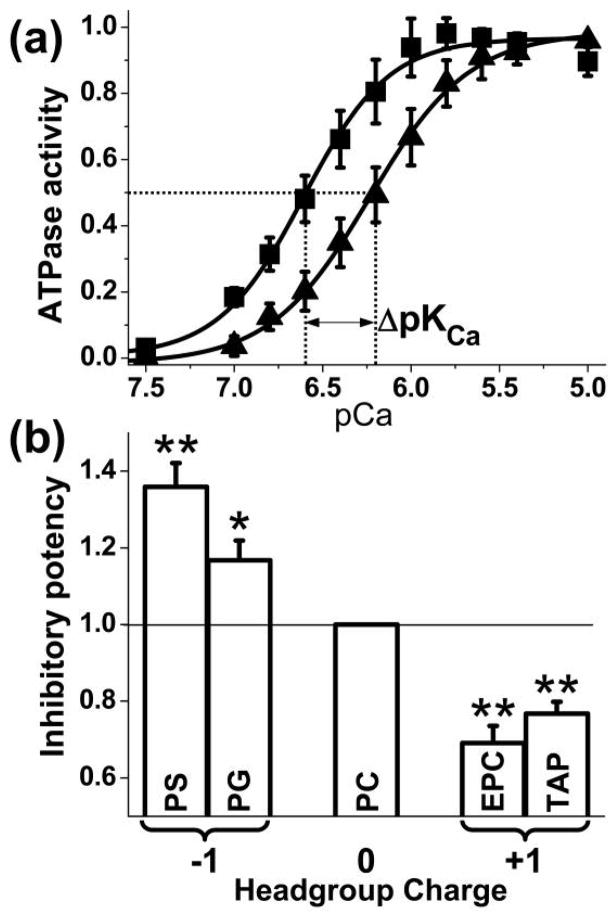
The inhibitory function of PLB is defined by its shift of the apparent SERCA Ca affinity, ΔpKCa (Fig. 5a, Eq. 2). In order to compare the inhibitory potency of PLB in various lipid environments, ΔpKCa was normalized to the value obtained with PC (Fig. 5b). The results show that PLB is more inhibitory in the presence of anionic lipids PS and PG, and less inhibitory in the presence of cationic lipids EPC and TAP. Previous results showed that PLB phosphorylation, which decreases inhibition of SERCA, increases the population in the dynamically disordered R state 10. The results of Fig. 5b show that membrane surface charge modulates PLB’s effect on SERCA by an analogous mechanism, confirming the conclusion above that the R state corresponds to the structural state having the shorter interprobe distance (population 1 in Fig. 3 and Fig. 4).
Fig. 5.
Effects of lipid headgroup charge on SERCA inhibition. (a) Activity of SERCA in PC without (¦) and with (?) AFA-PLB. Solid curves are the best fit using Eq. 1. Error bars are SEM (n = 4). (b) Inhibitory potency, defined as ΔpKCa normalized to the value for PC (mean ± SEM. n = 4–6). Students t-test, compared with PC: * P < 0.05, ** P < 0.01.
Discussion
Bimodal structure of the SERCA-PLB complex resolved by TR-FRET
Using EPR, we demonstrated that anionic lipids attract the cationic cytoplasmic domain of PLB to the membrane surface and thus increase the population of the membrane-associated T state, while cationic lipids do the opposite, increasing the R state population (Fig. 1). We then used TR-FRET to resolve two distinct structural states of the bound SERCA-PLB complex (Fig. 3) and observed a similar effect, with the R state assigned to the population having the shorter interprobe distance (Fig. 4). Membrane surface charge shifts the equilibrium between the T/R states with little or no affect on the two structural states themselves (Fig. 4d). This supports a model in which the cytoplasmic domain of the SERCA-bound T state is membrane associated, while that of the SERCA-bound R state loses contact with the membrane surface and contacts the SERCA cytoplasmic domain (Fig. 3c).
Mechanism of SERCA regulation
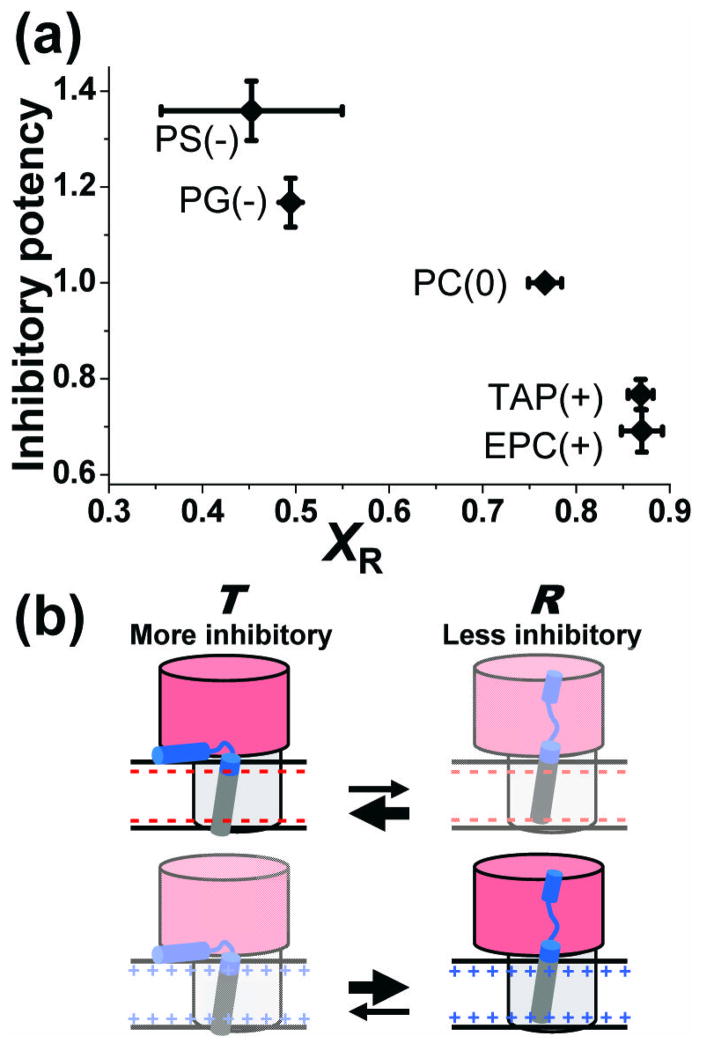
As we systematically varied the membrane surface charge, we observed a strong correlation between the population of the R state and PLB inhibitory function (Fig. 6a). This finding is consistent with previous results showing that phosphorylation of PLB, which decreases SERCA inhibition, also increases xR 10, as do some loss-of- inhibition mutations in PLB 31. In the current study, we show that varying the membrane surface charge serves not only to relieve inhibition (positive charge increases xR), but also to increase inhibition (negative charge decreases xR). Thus we obtain convincing evidence that this correlation between structural dynamics and function holds even in the absence of PLB covalent modification (e.g., phosphorylation or mutation): it is primarily the T/R equilibrium that determines SERCA function (Fig. 6b).
Fig. 6. Correlation of inhibitory potency with the T/R equilibrium.
(a) As the fraction of SERCA-PLB complex in the R state (xR) increases, in response to increasing membrane surface charge, the inhibitory potency of PLB (defined in Fig. 5) decreases. (b) Model consistent with the data. Both T and R states bind to SERCA. The membrane-associated T state is more inhibitory than the extended R state. Negative surface charge shifts the equilibrium toward T, increasing inhibition (top), while positive surface charge does the opposite (bottom).
Structural dynamics, not SERCA affinity, determines the inhibitory potency of PLB
TR-FRET clearly resolves free SERCA from the bound SERCA-PLB complex (Fig. 3c), and shows that relief of inhibition does not arise from a change in Xb, the fraction of SERCA bound to PLB (Fig. 4d). In both neutral (PC) or anionic lipids (PS, PG) (Fig. 4b), 80% of the SERCA is bound to PLB (Xb ~ 0.8), but PLB is more inhibitory in anionic lipids (Fig. 5b). Cationic lipids (EPC and TAP) actually increase slightly the fraction of SERCA bound to PLB (Xb ~ 0.9, Fig. 4b), but PLB inhibitory function decreases (Fig. 5b). This is opposite from the effect expected if relief of inhibition were due to dissociation of the complex. Therefore, the functional effects are due to structural changes within the bound SERCA-PLB complex (Fig. 6b), not to changes in SERCA-PLB affinity. It has been shown that the transmembrane helix of PLB without the cytoplasmic domain is sufficient to inhibit SERCA activity 32, so the role of the cytoplasmic domain is to relieve this inhibition when it is in the R state. Future studies must investigate how the dynamic disorder of the R state propagates allosterically to the transmembrane domain, interrupting the inhibitory interaction of the transmembrane domain with SERCA. This will have important implications for therapeutic engineering in this system, since it offers the hope of designing mutant proteins or drugs that stabilize the non- inhibitory R state and thus relieve SERCA inhibition, without the need to dissociate PLB from SERCA 26; 31.
Relationship to previous work
The fraction xR is much greater when PLB is bound to SERCA (Fig. 3, Fig. 4c) than when it is free (Fig. 2); this is consistent with previous studies by EPR 10; 19 and crosslinking 16, all of which suggest that SERCA decreases PLB’s interaction with the membrane surface. This is presumably due in part to the negative charge of the SERCA cytoplasmic domain (indicated by red color in Fig. 3c and Fig. 6b), which attracts the positively charged PLB cytoplasmic domain. Thus a negatively charged membrane surface competes most effectively for PLB binding (Fig. 4c). Previous NMR results suggested that several residues around K3 on PLB are in contact with SERCA 20. NMR also provides more detailed structural insight into the interactions between PLB and the membrane surface, involving both hydrophobic and hydrophilic side chains 33; 34. Previous NMR results showed that the anionic PG increases the population of the restricted T state of PLB in the absence of SERCA34, consistent with the EPR results here (Fig. 2).
Implications for SERCA regulation in native SR
The primary purpose of our manipulation of the lipid environment in this study was to perturb the structural and functional dynamics of SERCA-PLB through a mechanism distinct from PLB phosphorylation. However, it is also important to ask how these results relate to conditions in native cardiac SR. Our lipid headgroup composition is similar to that of cardiac SR, where PC is predominant (53%), followed by PE (27%) and PS (10%)35. However, most (76%) of the negatively charged PS headgroups face the lumen35, so it is unlikely that they interact significantly with the cytosolic domain of PLB. Thus, since virtually all of the lipids facing the cytosol are neutral, it is likely that the conditions in cardiac SR are best mimicked by our sample in which all lipids are neutral, where 77% of the SERCA-PLB complex is in the less inhibitory R state (Fig. 6a). These results suggest that in cardiac SR, a minor population of PLB in the T state is enough to inhibit SERCA substantially. At first glance, this seems surprising, but it means that phosphorylation of PLB need only shift about 23% of PLB to the R state to maximally activate SERCA. This is analogous to the poised equilibrium in the regulatory light chain of smooth muscle myosin, where phosphorylation causes just a 22% shift in the dynamic structural equilibrium but results in profound activation22.
Lipid headgroup charge as a research tool to tune the structural and functional dynamics of integral membrane proteins
It has been an effective strategy to vary the physical properties of the hydrophobic core of the membrane, such as the hydrophobic thickness and fluidity36; 37, to investigate the structural and functional dynamics of integral membrane proteins. The composition of lipid headgroups is also critical for the function of membrane proteins2, including SERCA23; 38. Researchers have varied the zwitterionic and anionic lipid compositions to mimic native membrane environments39; 40. Cationic lipids, on the other hand, do not occur naturally and have been used primarily as tools in liposomal transfection41 and lipid transfer42. The present study introduces them as agents to perturb the structure and function of membrane proteins. Using lipids with anionic, zwitterionic, and cationic headgroups, we controlled the surface electrostatics and tuned successfully the structural dynamics of an integral membrane protein complex. Compared to altering the protein structure through direct chemical modifications such as mutagenesis, phosphorylation, lipidation, methylation, and crosslinking, this method preserves the chemical integrity of the proteins involved. Although the present study focuses on charge variation, it is clear from the data that charge is not the only headgroup property that affects structural dynamics and function in the SERCA-PLB system – the two anionic lipids do not cause identical effects; neither do the two cationic lipids. Nevertheless, despite substantial variation in headgroup size and shape, the correlations of structure and function with charge are clear (Fig. 6).
Conclusion
We have systematically controlled the structural dynamics of an integral membrane protein complex, SERCA-PLB, by varying the membrane surface charge. Both in the absence of SERCA (measured by EPR of spin-labeled PLB) and bound to SERCA (measured by TR-FRET from SERCA to PLB), we found that PLB is in equilibrium between a dynamically disordered and extended R state, and an ordered membrane-associated T state. Compared with uncharged lipids, negative membrane surface charge shifts PLB toward the T state and increases SERCA inhibition, while positive charge shifts PLB toward the R state and decreases SERCA inhibition. TR-FRET measures the T/R structural equilibrium directly in the SERCA-PLB complex, while simultaneously and independently measuring SERCA-PLB binding. The results show that the observed functional effects are not caused by changes in the affinity of PLB for SERCA. Thus the correlation between SERCA function and PLB’s T/R structural equilibrium is established directly in the SERCA-PLB complex, independently of PLB phosphorylation or mutation. This work resolves crucial questions about the SERCA-PLB regulatory mechanism and, more generally, demonstrates the utility of lipid headgroup charge as an effective research tool to control membrane protein structural dynamics by perturbing electrostatics without changing the chemical composition of the protein.
Materials and Methods
Sample preparation and assays
SERCA was purified from rabbit skeletal muscle using reactive red in 0.1% octaethylene glycol monododecyl ether (C12E8) 25. Purified SERCA was labeled with AEDANS as previously described 25 and flash frozen in sucrose buffer (300 mM sucrose, 20 mM 3-(N-morpholino) propanesulfonic acid (MOPS), pH 7.0, 4° C). The dye-to-protein ratio was determined by measuring the absorbance at 334 nm (e = 6100 M−1cm−1) in a denaturing buffer (0.1 M NaOH, 1% sodium dodecyl sulfate). Solid-phase peptide synthesis and HPLC purification were used to prepare AFA-PLB, as previously reported 9; 10. Fluorescence Labeling at the N-terminus was accomplished by incorporation of Fmoc-Lys(Dabcyl)-OH during peptide synthesis. Fmoc-TOAC-OH (2,2,6,6-tetramethylpiperidine-1oxyl-4-amino-4-carboxylic acid) was incorporated in the AFA-PLB sequence at position 11, as previously reported 43. Characterization was accomplished by mass spectrometry (MALDI-TOF) and Edman protein sequencing. PLB concentrations were measured with the BC A assay (Pierce) and by amino acid analysis. Functional reconstitution of SERCA and/or PLB was performed as described previously 10; 44, adapted for systematic variation of lipid composition. The final molar ratio of SERCA/PLB/lipid was 1/10/700, with either one or both proteins present. The molar lipid composition was PC/PE/L = 4/1/1, where L = PS (−), PG (−), PC (0), EPC (+) and TAP (+).
Ca-ATPase activity was measured at 25°C as a function of pCa using an enzyme- linked ATPase assay in a microplate reader 25. The data were fitted by
| Eq. 1 |
where Vmax is the maximum ATPase rate, pKCa is the apparent Ca affinity, and nH is the Hill coefficient. The inhibitory potency of PLB was defined as the decrease in the apparent Ca affinity of SERCA:
| Eq. 2 |
EPR
EPR spectra were acquired with a Bruker EleXsys E500 spectrometer equipped with a 4122 SHQ cavity. A quartz dewar and Bruker N2 temperature controller were used to maintain the samples at 25 ± 0.1°C. Spectra were acquired using 12.6 mW microwave power, 100kHz modulation frequency with 1 G peak-to-peak amplitude, and a 120 G sweep width. Mole fractions of populations, resolved by rotational dynamics, were determined by fitting the spectra to numerical simulations 10.
FRET
Fluorescence waveforms were acquired using a high-performance time-resolved fluorescence (HPTRF) spectrometer constructed in this laboratory28, which uses direct waveform recording (DWR) rather than the conventional method of time-correlated single-photon counting (TCSPC). As shown previously, when identical samples are studied, this DWR instrument offers 105 times higher throughput than TCSPC, while providing at least comparable performance in signal/noise, accuracy, and resolution of distinct components28. AEDANS-SERCA was excited using a passively Q-switched microchip YAG laser (NanoUV-355; JDS Uniphase), at 355 nm with a pulse repetition frequency of 10 kHz. The high energy (1 μJ / pulse) narrow (~ 1 ns full width at half maximum) laser pulses are highly uniform in shape and intensity. Emitted photons pass through a polarizer set to the magic angle (54.7°), followed by an interference bandpass filter (Semrock 470/22 nm), detection with a photomultiplier tube (PMT) module (H5773-20, Hamamatsu), and digitization (Acqiris DC252, time resolution 0.125 ns ). TR-FRET waveforms were analyzed as described previously22 and described in SI. For all FRET samples analyzed, two Gaussian distance distributions were necessary and sufficient to fit the data (Fig. S2).
Supplementary Material
HIGHLIGHTS.
We varied lipid charge to modulate regulation of the calcium pump (SERCA) by PLB.
Time-resolved FRET shows that the SERCA-PLB complex has 2 structural states, R and T.
Membrane surface charge affects the R/T structural distribution, not binding.
The R state of the SERCA-PLB complex is more active than the T state.
SERCA-PLB function is regulated by the R/T structural distribution, not binding.
Acknowledgments
Fluorescence and EPR experiments were performed at the Biophysical Spectroscopy Center, University of Minnesota. Computational resources were provided by the Minnesota Supercomputing Institute. We thank Razvan Cornea and Gianluigi Veglia for helpful discussions. Special thanks to Octavian Cornea for assistance with manuscript preparation and submission. This study was supported by NIH grants to DDT (R01 GM27906, P30 AR0507220). JL was supported by a University of Minnesota Doctoral Dissertation Fellowship, and ZMJ was supported by NIH training grant T32 AR007612)
Abbreviations used
- PLB
phospholamban
- SERCA
sarcoplasmic reticulum Ca-ATPase
- EPR
electron paramagnetic resonance
- TR-FRET
time-resolved fluorescence resonance energy transfer
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPE
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DOPS
1,2-dioleoyl-sn-glycero-3-phospho-L-serine
- DOPG
1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- DOECP
1,2-dioleoyl-sn-glycero-3-ethylphosphocholine
- DOTAP
1,2-dioleoyl-3-trimethylammonium-propane
- TOAC
2,2,6,6-tetramethyl-piperidine-1-oxyl-4-amino-4-carboxylic acid
- AEDANS
5-((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid
- Dabcyl
4-((4-(dimethylamino)phenyl)azo)-benzoic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sachs JN, Engelman DM. Introduction to the membrane protein reviews: the interplay of structure, dynamics, and environment in membrane protein function. Annu Rev Biochem. 2006;75:707–12. doi: 10.1146/annurev.biochem.75.110105.142336. [DOI] [PubMed] [Google Scholar]
- 2.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–11. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 3.Tada M, Inui M. Regulation of calcium transport by the ATPase-phospholamban system. J Mol Cell Cardiol. 1983;15:565–75. doi: 10.1016/0022-2828(83)90267-5. [DOI] [PubMed] [Google Scholar]
- 4.Cantilina T, Sagara Y, Inesi G, Jones LR. Comparative studies of cardiac and skeletal sarcoplasmic reticulum ATPases. Effect of a phospholamban antibody on enzyme activation by Ca2+ J Biol Chem. 1993;268:17018–25. [PubMed] [Google Scholar]
- 5.Simmerman HK, Jones LR. Phospholamban: protein structure, mechanism of action, and role in cardiac function. Physiol Rev. 1998;78:921–47. doi: 10.1152/physrev.1998.78.4.921. [DOI] [PubMed] [Google Scholar]
- 6.MacLennan DH, Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol. 2003;4:566–77. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 7.Cornea RL, Jones LR, Autry JM, Thomas DD. Mutation and phosphorylation change the oligomeric structure of phospholamban in lipid bilayers. Biochemistry. 1997;36:2960–7. doi: 10.1021/bi961955q. [DOI] [PubMed] [Google Scholar]
- 8.Zamoon J, Mascioni A, Thomas DD, Veglia G. NMR solution structure and topological orientation of monomeric phospholamban in dodecylphosphocholine micelles. Biophys J. 2003;85:2589–98. doi: 10.1016/s0006-3495(03)74681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karim CB, Kirby TL, Zhang Z, Nesmelov Y, Thomas DD. Phospholamban structural dynamics in lipid bilayers probed by a spin label rigidly coupled to the peptide backbone. Proc Natl Acad Sci U S A. 2004;101:14437–42. doi: 10.1073/pnas.0402801101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karim CB, Zhang Z, Howard EC, Torgersen KD, Thomas DD. Phosphorylation-dependent Conformational Switch in Spin-labeled Phospholamban Bound to SERCA. J Mol Biol. 2006;358:1032–40. doi: 10.1016/j.jmb.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 11.Traaseth NJ, Shi L, Verardi R, Mullen DG, Barany G, Veglia G. Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach. Proc Natl Acad Sci U S A. 2009;106:10165–70. doi: 10.1073/pnas.0904290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metcalfe EE, Zamoon J, Thomas DD, Veglia G. 1H/15N Heteronuclear NMR spectroscopy shows four dynamic domains for phospholamban reconstituted in dodecylphosphocholine micelles. Biophys J. 2004;87:1205–14. doi: 10.1529/biophysj.103.038844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metcalfe EE, Traaseth NJ, Veglia G. Serine 16 phosphorylation induces an order-to-disorder transition in monomeric phospholamban. Biochemistry. 2005;44:4386–96. doi: 10.1021/bi047571e. [DOI] [PubMed] [Google Scholar]
- 14.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature. 2000;405:647–55. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 15.Moller JV, Olesen C, Winther AM, Nissen P. The sarcoplasmic Ca2+-ATPase: design of a perfect chemi-osmotic pump. Q Rev Biophys. 2010;43:501–66. doi: 10.1017/S003358351000017X. [DOI] [PubMed] [Google Scholar]
- 16.Toyoshima C, Asahi M, Sugita Y, Khanna R, Tsuda T, MacLennan DH. Modeling of the inhibitory interaction of phospholamban with the Ca2+ ATPase. Proc Natl Acad Sci U S A. 2003;100:467–72. doi: 10.1073/pnas.0237326100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asahi M, McKenna E, Kurzydlowski K, Tada M, MacLennan DH. Physical interactions between phospholamban and sarco(endo)plasmic reticulum Ca2+-ATPases are dissociated by elevated Ca2+, but not by phospholamban phosphorylation, vanadate, or thapsigargin, and are enhanced by ATP. J Biol Chem. 2000;275:15034–8. doi: 10.1074/jbc.275.20.15034. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Akin BL, Jones LR. Mechanism of reversal of phospholamban inhibition of the cardiac Ca2+-ATPase by protein kinase A and by anti-phospholamban monoclonal antibody 2D12. J Biol Chem. 2007;282:20968–76. doi: 10.1074/jbc.M703516200. [DOI] [PubMed] [Google Scholar]
- 19.Kirby TL, Karim CB, Thomas DD. Electron paramagnetic resonance reveals a large-scale conformational change in the cytoplasmic domain of phospholamban upon binding to the sarcoplasmic reticulum Ca-ATPase. Biochemistry. 2004;43:5842–52. doi: 10.1021/bi035749b. [DOI] [PubMed] [Google Scholar]
- 20.Zamoon J, Nitu F, Karim C, Thomas DD, Veglia G. Mapping the interaction surface of a membrane protein: unveiling the conformational switch of phospholamban in calcium pump regulation. Proc Natl Acad Sci U S A. 2005;102:4747–52. doi: 10.1073/pnas.0406039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nesmelov YE, Karim CB, Song L, Fajer PG, Thomas DD. Rotational dynamics of phospholamban determined by multifrequency electron paramagnetic resonance. Biophys J. 2007;93:2805–12. doi: 10.1529/biophysj.107.108910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kast D, Espinoza-Fonseca LM, Yi C, Thomas DD. Phosphorylation-induced structural changes in smooth muscle myosin regulatory light chain. Proc Natl Acad Sci U S A. 2010;107:8207–12. doi: 10.1073/pnas.1001941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter GW, Negash S, Squier TC. Phosphatidylethanolamine modulates Ca-ATPase function and dynamics. Biochemistry. 1999;38:1356–64. doi: 10.1021/bi9822224. [DOI] [PubMed] [Google Scholar]
- 24.Reddy LG, Cornea RL, Winters DL, McKenna E, Thomas DD. Defining the molecular components of calcium transport regulation in a reconstituted membrane system. Biochemistry. 2003;42:4585–92. doi: 10.1021/bi026995a. [DOI] [PubMed] [Google Scholar]
- 25.Mueller B, Karim CB, Negrashov IV, Kutchai H, Thomas DD. Direct detection of phospholamban and sarcoplasmic reticulum Ca-ATPase interaction in membranes using fluorescence resonance energy transfer. Biochemistry. 2004;43:8754–65. doi: 10.1021/bi049732k. [DOI] [PubMed] [Google Scholar]
- 26.Lockamy EL, Cornea RL, Karim CB, Thomas DD. Functional and physical competition between phospholamban and its mutants provides insight into the molecular mechanism of gene therapy for heart failure. Biochem Biophys Res Commun. 2011;408:388–92. doi: 10.1016/j.bbrc.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birmachu W, Nisswandt FL, Thomas DD. Conformational transitions in the calcium adenosinetriphosphatase studied by time -resolved fluorescence resonance energy transfer. Biochemistry. 1989;28:3940–7. doi: 10.1021/bi00435a047. [DOI] [PubMed] [Google Scholar]
- 28.Muretta JM, Kyrychenko A, Ladokhin AS, Kast DJ, Gillispie GD, Thomas DD. High-performance time -resolved fluorescence by direct waveform recording. Rev Sci Instrum. 2010;81:103101. doi: 10.1063/1.3480647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agafonov RV, Negrashov IV, Tkachev YV, Blakely SE, Titus MA, Thomas DD, Nesmelov YE. Structural dynamics of the myosin relay helix by time -resolved EPR and FRET. Proc Natl Acad Sci U S A. 2009;106:21625–30. doi: 10.1073/pnas.0909757106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2. Kluwer Academic/Plenum Press; New York: 1999. [Google Scholar]
- 31.Ha KN, Traaseth NJ, Verardi R, Zamoon J, Cembran A, Karim CB, Thomas DD, Veglia G. Controlling the inhibition of the sarcoplasmic Ca2+-ATPase by tuning phospholamban structural dynamics. J Biol Chem. 2007;282:37205–14. doi: 10.1074/jbc.M704056200. [DOI] [PubMed] [Google Scholar]
- 32.Karim CB, Marquardt CG, Stamm JD, Barany G, Thomas DD. Synthetic null-cysteine phospholamban analogue and the corresponding transmembrane domain inhibit the Ca-ATPase. Biochemistry. 2000;39:10892–7. doi: 10.1021/bi0003543. [DOI] [PubMed] [Google Scholar]
- 33.Gustavsson M, Traaseth NJ, Karim CB, Lockamy EL, Thomas DD, Veglia G. Lipid-Mediated Folding/Unfolding of Phospholamban as a Regulatory Mechanism for the Sarcoplasmic Reticulum Ca(2+)-ATPase. J Mol Biol. 2011 doi: 10.1016/j.jmb.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gustavsson M, Traaseth NJ, Veglia G. Probing ground and excited states of phospholamban in model and native lipid membranes by magic angle spinning NMR spectroscopy. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbamem.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bick RJ, Buja LM, Van Winkle WB, Taffet GE. Membrane asymmetry in isolated canine cardiac sarcoplasmic reticulum: comparison with skeletal muscle sarcoplasmic reticulum. J Membr Biol. 1998;164:169–75. doi: 10.1007/s002329900402. [DOI] [PubMed] [Google Scholar]
- 36.Cornea RL, Thomas DD. Effects of membrane thickness on the molecular dynamics and enzymatic activity of reconstituted Ca-ATPase. Biochemistry. 1994;33:2912–20. doi: 10.1021/bi00176a022. [DOI] [PubMed] [Google Scholar]
- 37.Sonntag Y, Musgaard M, Olesen C, Schiott B, Moller JV, Nissen P, Thogersen L. Mutual adaptation of a membrane protein and its lipid bilayer during conformational changes. Nat Commun. 2011;2:304. doi: 10.1038/ncomms1307. [DOI] [PubMed] [Google Scholar]
- 38.Hughes E, Clayton JC, Middleton DA. Cytoplasmic residues of phospholamban interact with membrane surfaces in the presence of SERCA: a new role for phospholipids in the regulation of cardiac calcium cycling? Biochim Biophys Acta. 2009;1788:559–66. doi: 10.1016/j.bbamem.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 39.Lee AG. Lipid-protein interactions in biological membranes: a structural perspective. Biochim Biophys Acta. 2003;1612:1–40. doi: 10.1016/s0005-2736(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 40.Gustavsson MB, Traaseth NJ, Veglia G. Activating and Deactivating Roles of Lipid Bilayers on the Ca2+-Atpase/Phospholamban Complex. Biochemistry. 2011 doi: 10.1021/bi200759y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo X, Huang L. Recent Advances in Nonviral Vectors for Gene Delivery. Acc Chem Res. 2011 doi: 10.1021/ar200151m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kunze A, Svedhem S, Kasemo B. Lipid transfer between charged supported lipid bilayers and oppositely charged vesicles. Langmuir. 2009;25:5146–58. doi: 10.1021/la802758h. [DOI] [PubMed] [Google Scholar]
- 43.Karim CB, Zhang Z, Thomas DD. Synthesis of TOAC spin-labeled proteins and reconstitution in lipid membranes. Nat Protoc. 2007;2:42–9. doi: 10.1038/nprot.2007.2. [DOI] [PubMed] [Google Scholar]
- 44.Reddy LG, Autry JM, Jones LR, Thomas DD. Co-reconstitution of phospholamban mutants with the Ca-ATPase reveals dependence of inhibitory function on phospholamban structure. J Biol Chem. 1999;274:7649–55. doi: 10.1074/jbc.274.12.7649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.