Abstract
Endocytosis, the internalization and transport of extracellular cargo, is an essential cellular process. The ultimate step in endocytosis is the intracellular degradation of extracellular cargo for use by the cell. While live cell imaging and single particle tracking have been well-utilized to study the internalization and transport of cargo, the final degradation step has required separate biochemical assays. We describe the use of self-quenched endocytic cargo to image the intracellular transport and degradation of endocytic cargo directly in live cells. We first outline the fluorescent labeling and quantification of two common endocytic cargos; a protein, bovine serum albumin, and a lipid nanoparticle, low-density lipoprotein. In vitro measurements confirm that self-quenching is a function of the number of fluorophores bound to the protein or particle and that recovery of the fluorescent signal occurs in response to enzymatic degradation. We then use confocal fluorescence microscopy and flow cytometry to demonstrate the use of self-quenched bovine serum albumin with standard fluorescence techniques. Using live cell imaging and single particle tracking, we find that the degradation of bovine serum albumin occurs in an endo-lysosomal vesicle that is positive for LAMP1.
Keywords: albumin, endocytosis, fluorescence microscopy, live cell imaging, self-quenching, single particle tracking
The ability to characterize the intracellular transport of extracellular cargo is essential to understanding human health and disease [1–5]. Extracellular cargo, including nutrients, toxins, and nanoparticles, bind to the plasma membrane of the cell and are internalized into membrane-bound endosomes. These endocytic vesicles are then transported by motor proteins along the cytoskeleton of the cell. The final step in this process is the enzyme-mediated degradation of cargo within an endo-lysosomal vesicle. Degradation of extracellular cargo is the step by which extracellular cargo is processed for use by the cell. Understanding endocytic transport and degradation requires spatial information, to determine which population of endocytic vesicles is involved in transport, and temporal information, to follow the motion of the extracellular cargo during transport. For this reason, fluorescence microscopy, combined with single particle tracking, has been well-utilized for the study of endocytic transport [6–15]. While imaging endocytic transport is well-established, current fluorescence microscopy approaches lack the ability to probe the degradation of cargo within the cell. For example, it is possible to use live cell imaging to track cargo and relevant proteins during binding, internalization, and transport, but degradation must be probed with biochemical assays such as western blotting. This results in a discontinuity in endocytic transport experiments in which one method, microscopy, is used to probe the initial internalization and transport, and a second method must be used to probe the final step in the endocytic process. Our goal is to develop and utilize a method that allows the complete endocytic pathway, from binding to degradation of cargo, to be probed with a single technique.
Degradation of cargo within an endocytic vesicle is carried out by lysosomal enzymes such as the cathepsins [16]. Current methods to probe enzyme activity include the use of self-quenched, fluorogenic, or FRET-based substrates that report on the activity of a specific enzyme [17–21]. Commercially available substrates (Anaspec, Sigma-Aldrich, others) are designed to probe the activity of the lysosomal enzymes. For intracellular studies of endocytosis, we sought to develop a method that would build on the advantages of single particle tracking, the ability to follow particle motion and identify relevant proteins, but that would also allow us to measure a change that indicates where and when degradation of endocytic cargo occurs. Additionally, we would like a method that can be used across multiple scales - from subcellular measurements with fluorescence microscopy to the analysis of tens of thousands of cells with flow cytometry - without any modification to existing experimental platforms.
Self-quenched substrates, in which a high local concentration of fluorophores bound to a substrate results in limited fluorescence emission, are good candidates for the imaging of endocytic transport and degradation. Self-quenching, in comparison to FRET, requires only a single color fluorophore making it compatible with multicolor imaging and simple optical setups. Self-quenching of fluorophores bound to substrates has been used previously for lysosomal enzyme activity assays including solution-phase measurements [22, 23], static imaging [24, 25], in vivo imaging [26], and flow cytometry [24, 27, 28]. While single particle tracking has not been used previously in combination with enzyme assays, it has been used to observe the effect of pH change on viral fusion during endocytic transport [7, 29].
To illustrate the use of self-quenched substrates to probe endocytic transport and degradation, we first fluorescently labeled two common endocytic cargos in a self-quenched configuration. The first cargo was a single protein molecule, bovine serum albumin (BSA), covalently labeled with an amine-reactive fluorophore. The second cargo was a single particle, low density lipoprotein (LDL), labeled with a lipophilic fluorophore. We first describe the labeling scheme used for BSA and LDL and the in vitro measurements used to quantify self-quenching. The results described below confirm that self-quenching is a function of the number of fluorophores and that the recovery of the fluorescent signal occurs in response to the enzymatic degradation of proteins. We then use this assay to characterize the intracellular degradation of BSA, probed with cellular imaging, flow cytometry, and single particle tracking.
Materials and methods
Cell culture
BS-C-1 cells (ATCC, Manassas, VA) were maintained in a 37°C, 5% carbon dioxide environment in Minimum Essential Medium (MEM, Invitrogen, Carlsbad, CA) with 10% (v/v) fetal bovine serum (FBS, Invitrogen). Cells were passaged every 3 days. For fluorescence imaging, cells were cultured in 35 mm glass-bottom cell culture dishes (MatTek, Ashland, MA). Nuclei were stained with 27.25 µM DAPI (4',6-diamidino-2-phenylindole, D3571, Invitrogen) in full growth medium for 30 minutes prior to experiments.
Labeling of BSA and LDL with fluorescent dyes
BSA (BP1600, Fisher Bioreagents) was labeled with AlexaFluor647 carboxylic acid succinimidyl ester (AF647, A20006, Invitrogen) according to the manufacturer’s instructions. To obtain different labeling ratios, 136 µM BSA was incubated with 91 mM, 68 mM or 0.91 mM AF647 for 1 hr resulting in 2.48, 0.93 or 0.07 AF647 per BSA, respectively. The labeling ratio of 2.70 AF647 per BSA was obtained by allowing 136 µM BSA to incubate with 91 mM AF647 for 16 hours. In all cases, the reaction was stopped with 1.5 M hydroxylamine and free AF647 was separated from BSA using a NAP5 size exclusion column (17–0853-02, GE Healthcare, Buckinghamshire, UK). Final concentrations of BSA and AF647 were measured with a UV-Vis spectrophotometer (DU800, Beckman Coulter, Fullerton, CA, USA).
Human LDL (BT-903, Biomedical Technologies, Stoughton, MA) was labeled with 1,1’-dioctadecyl-3,3,3’,3’-tetramethylindodicarbocyanine perchlorate (DiD, D-307, Invitrogen) at a concentration of 1.8 mM for a ratio of 200 DiD molecules per LDL particle. Lower labeling ratios were achieved by diluting the DiD stock solution by 25, 50 and 500-fold. LDL and DiD were mixed every 10 min for 1 hr before removal of excess DiD on a NAP5 size exclusion column. The ratio of DiD molecules per LDL particle was measured using a UV-Vis spectrophotometer.
Gel electrophoresis
Samples were prepared in Laemmli SDS sample buffer (BP-110R, Boston BioProducts). Each sample (10–15 µg) was loaded onto a precast polyacrylamide gel (4–20%, 161–1105, Bio-Rad, Hercules, CA) and run at 130 V for 1 hr. Gels were stained with SimplyBlue SafeStain (LC6060, Invitrogen) for 1 hr and destained in water for at least 2 hr.
Pepstatin treatment
Pepstatin A methyl ester (pepstatin, 516485, EMD Chemicals), a cell permeable derivative of pepstatin A, was used to inhibit the activity of acid proteases. The activity of pepsin in solution was inhibited by the addition of 7 nM pepstatin for 5 min prior to the addition of BSA. Cells were incubated in serum-deficient medium (MEM with 0.2% FBS) for 18 hr before incubation with 10 µM pepstatin. Cells were pre-treated with pepstatin for 2 hrs and it remained present throughout the course of each experiment.
Confocal fluorescence microscopy
Images were collected with a FluoView 1000 laser scanning confocal microscope (Olympus, Center Valley, PA) using a 1.42 N.A., 60x, oil immersion objective. DAPI was excited with a 405 nm diode laser and AF647 was excited by a 635 nm diode laser. For DAPI, a 430–470 nm band pass filter was used to filter emission and for AF647 a 655–755 nm band pass filter was used. For all images, the pinhole was set to obtain a 1 µm thick optical slice.
Single particle tracking fluorescence microscopy
For two-color, single particle tracking experiments, an inverted microscope (Olympus IX71, Center Valley, PA) in an epi-fluorescent configuration with a 1.45 N A, 60x, oil immersion objective (Olympus) was used. Excitation was supplied by two lasers: an argon ion laser tuned to 514 nm (35-LAP-431-208, Melles Griot, Carlsbad, CA) and a red diode (635-25C, Coherent, Santa Clara, CA). Excitation beams were overlapped using a dichroic mirror (Z514BCM, Chroma, Rockingham, VT) and focused on the back focal plane of the microscope objective. A shutter (Uniblitz, Rochester, NY) limited exposure of the cells to the lasers. Cells were illuminated by both laser lines using a dichroic mirror (Z514/633RPC, Chroma). Emission was separated into two channels based on wavelength using a 620 nm long pass filter (620DCXR, Chroma). Excitation light was filtered out of the emission by the appropriate filters: EYFP - HQ580/50 (Chroma), AF647 - HQ680/60 (Chroma). A second 620 nm long pass filter (Chroma) was used to image both emission paths side by side on a single CDD camera (DU-888, Andor, South Windsor, CT). Images were recorded at a rate of 0.5 frames/s with a 250 ms exposure. Experiments were conducted at 37°C.
Image analysis
Image J (http://rsb.info.nih.gov/ij/) was used for tracking and quantifying colocalization. Particle tracking was performed with the Image J plugin, ‘‘Manual Tracking’’ (http://rsb.info.nih.gov/ij/plugins/track/track.html). The intensity of BSA-AF647-containing endosomes, referred to as particles, was determined using ImageJ with the MOSAIC plugin [30]. The MOSAIC Particle Tracker 2D/3D feature was used to detect the center of each particle in the image. Coordinates and images were imported into MATLAB and a custom written function which determined the average intensity within a 2 pixel radius of each particle was applied. The average background was determined in ImageJ for an area within the cell that did not contain particles (typical area 30–60 µm2). The background was then subtracted from the particle intensities and any remaining values that were below zero were eliminated from further analysis as false-positive detections. The particle intensities (3039 particles in the control cells, 3259 in pepstatin-treated cells values) were then compared using a Wilcoxon signed-rank test for a p-value of 0.005. Images for publication were background subtracted and intensities were adjusted equally within each data set.
Flow cytometry
Cells were cooled to 4°C for 15 minutes before incubation with BSA-AF647 for 15 minutes. Unbound BSA-AF647 was removed by washing 2 times with 2 mL of full growth medium. After a 1 hr incubation at 37°C, cells were trypsinized, pelleted (9000 rcf for 5 min), and washed with Leibovitz’s L-15 medium (21083, Invitrogen). Flow cytometry was carried out on a BD LSR-II flow cytometer (Becton Dickinson, Franklin Lakes, NJ). For each experiment, 30,000 cells were sampled.
Results and discussion
Fluorescent labeling of BSA and LDL
We illustrate two labeling schemes; covalent attachment of fluorophores to an individual protein molecule, BSA, and the use of a lipophilic fluorophore to label a naturally occurring lipoprotein nanoparticle, LDL. Specifically, BSA was labeled with amine-reactive AlexaFluor647 (AF647) according to the manufacturer’s instructions as described in the Materials and Methods. BSA-fluorescein has been used previously as a self-quenched substrate [22]. Fluorophores such as AF647 that emit in the red region of the visible spectrum are advantageous for multicolor imaging experiments in combination with variants of green fluorescent protein that emit in the blue-green region. LDL was labeled with a lipophilic fluorophore, DiD, that inserts into the lipid component of LDL. The ratio of fluorophores to BSA or LDL was calculated based on their absorption measured with UV-Vis spectrophotometry as described in the Materials and Methods.
Dequenching increases with the number of fluorophores
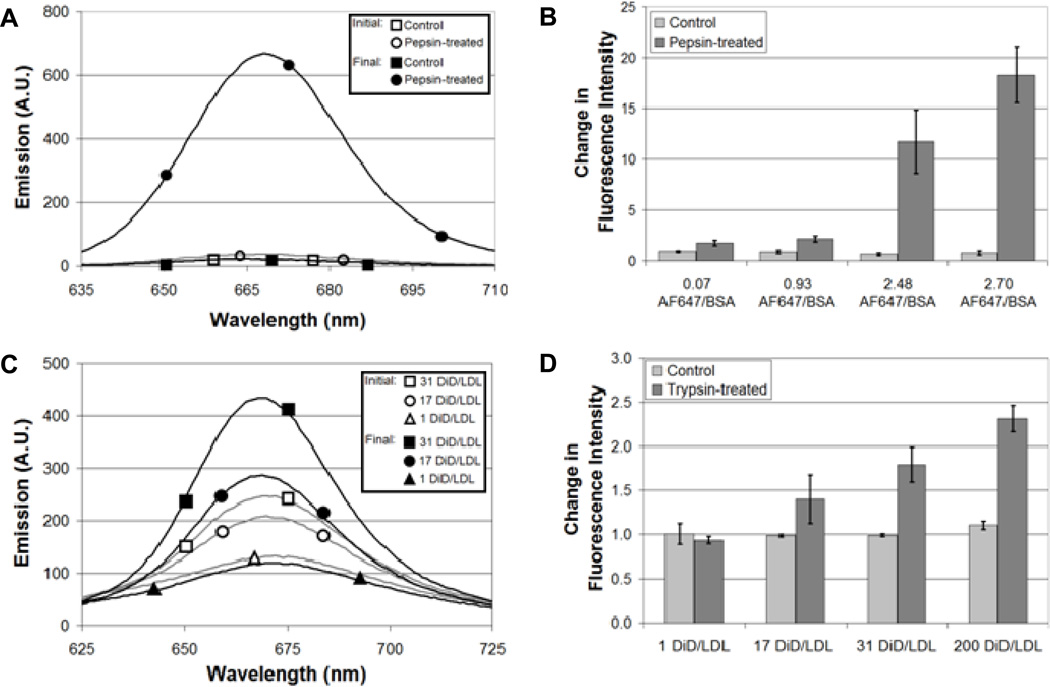
The number of fluorophores covalently bound to BSA or inserted into the lipids of LDL was controlled by the concentration of fluorophores added to BSA or LDL during labeling. Dequenching, defined as an increase in fluorescence in response to a decreased local concentration of fluorophores, was measured for increasing numbers of fluorophores bound to BSA or LDL. The measurements were carried out in solution using an enzyme to degrade BSA or LDL and thereby reduce the local concentration of fluorophores (Fig. 1). BSA-AF647 was incubated with 0.1 mg/mL pepsin, an acid protease known to degrade BSA [31, 32], for 1 hr at 37°C in a TCA solution at pH 1.2. As a control, BSA-AF647 was incubated in the same TCA solution at pH 1.2 to ensure the low pH did not lead to a change in emission. LDL-DiD was incubated with trypsin, a protease shown previously to degrade LDL [14], in PBS for 2 hr at 37°C. As a control, LDL-DiD was incubated in PBS at 37°C in the absence of trypsin. Data were analyzed identically for BSA and LDL. The emission of either BSA-AF647 or LDL-DiD was measured with a fluorimeter (RF-5301PC, Shimadzu, Japan) before and after incubation with the enzyme (Fig. 1A and C). Emission measurements were normalized by the absorption of BSA-AF647 or LDL-DiD before and after incubation although there was little change in absorption (Supplementary Material, Fig. S1). Dequenching was measured as the change in fluorescence intensity following enzymatic treatment (Fig. 1B and D). No change in emission would be indicated by a value of 1. For low numbers of fluorophores, little change was observed after incubation with pepsin or trypsin. For example, BSA labeled with an average of 0.93 AF647 molecules per BSA increased emission by a factor of 1.75 after incubation with pepsin (Fig. 1B). An identically labeled sample that was not exposed to pepsin had a change in fluorescence intensity of 0.97, essentially no change in intensity. The low level of dequenching observed for 0.93 AF647 molecules per BSA is expected for a Poisson distribution of fluorophores bound to BSA. LDL labeled with 1 DiD molecule per LDL particle showed no change in emission (0.94) after incubation with trypsin (Fig. 1D). An identically labeled sample that was not exposed to trypsin also showed no change in emission (1.01). In comparison, increasing the number of fluorophores resulted in a greater change in fluorescence intensity. BSA labeled with an average of 2.70 AF647 molecules increased emission by a factor of 18.25 and LDL labeled with an average of 200 DiD molecules increased emission by a factor of 2.32. Unless noted otherwise, a labeling ratio of 2.48 AF647 molecules per BSA molecule, with a dequenching value of 11.73, was used for the experiments described below.
Fig. 1.
Dequenching of BSA and LDL as function of fluorophore number. (A) Emission spectra (625 nm excitation) of BSA-AF647 labeled with an average of 2.70 AF647 molecules per BSA molecule in solution (TCA, pH 1.2). Final spectra were measured after a 1 hr incubation at 37°C. Absorption spectra are included in Supplementary Material, Fig. S1. (B) Fluorescence emission increased with increasing numbers of AF647 conjugated to BSA. A value of 1 indicates no change in emission. (C) Emission spectra (600 nm excitation) of LDL-DiD in a PBS solution. Final spectra were measured after a 2 hr incubation at 37°C in the presence of trypsin. Absorption spectra showed little change after the 1 hr incubation.14 (D) Fluorescence emission increased with increasing numbers of DiD inserted into the lipid component of LDL. Error bars represent the standard deviation of 3 experiments.
Dequenching is specific to enzyme activity
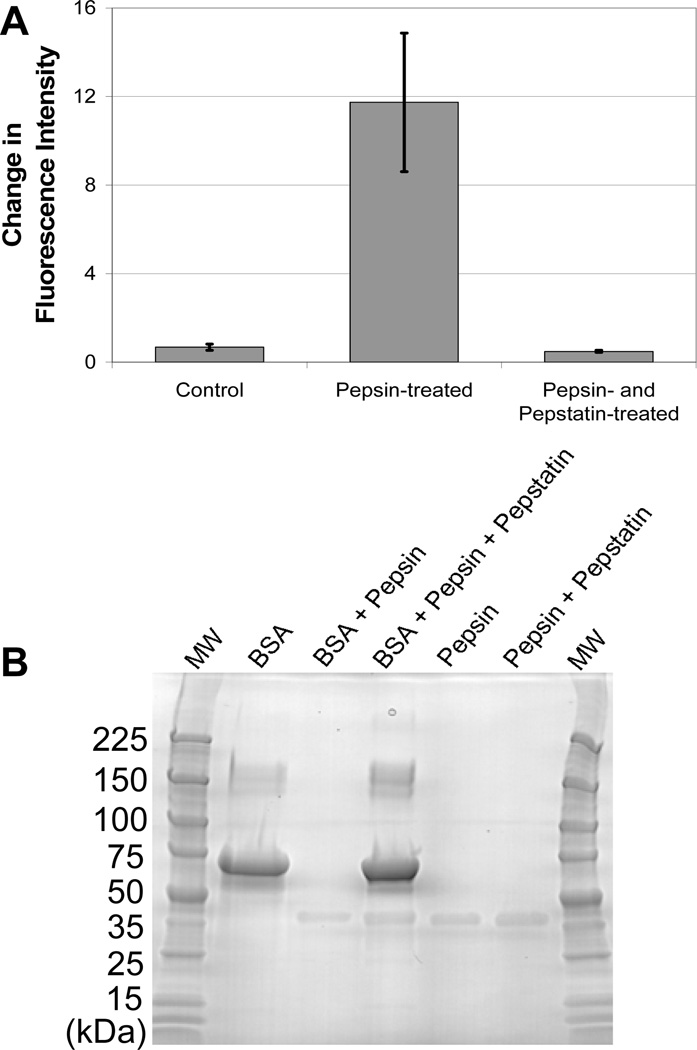
Control experiments described above demonstrate that dequenching does not occur in response to pH or temperature, but instead requires the presence of a proteolytic enzyme. To confirm that enzymatic activity was specific, dequenching was measured in the presence of an enzyme inhibitor. The enzymatic activity of pepsin and other acid proteases is inhibited by pepstatin [33, 34]. AF647-BSA was incubated with both pepsin and pepstatin for 1 hr as described in the Materials and Methods. Treatment of AF647-BSA with pepsin results in a dequenching value of 11.7. In comparison, the addition of pepstatin reduced dequenching to 0.5 (Fig. 2A). Gel electrophoresis confirms that the degradation of BSA by pepsin is inhibited by pepstatin (Fig. 2B).
Fig. 2.
Dequenching of BSA is specific to protease activity. (A) Dequenching was not observed when BSA-AF647 was incubated with a combination of pepsin and pepstatin, a pepsin inhibitor. (B) Gel electrophoresis (4–20% gradient, polyarcrylamide, SimplyBlue SafeStain) shows that the degradation of BSA (67 kDa) by pepsin is inhibited by pepstatin. Pepsin (35 kDa) is visible on the gel while pepstatin (0.7 kDa) and the degraded BSA fragments are too small to observe.
Self-quenching as an intracellular assay of enzyme activity
Dequenching provides a method to probe protein or particle degradation in solution, as described above, and also in live cells. Upon enzymatic degradation and the separation of the fluorophores, the fluorescent signal increases, indicating that degradation has occurred. This approach has the advantage of an increase in signal as an indicator of a cellular event, as compared to decreases in signal which are difficult to decouple from photobleaching. Cellular experiments can be carried out on the subcellular level, probing the degradation of cargo in individual endocytic vesicles, or on the cellular level using either fluorescence microscopy to image individual cells or flow cytometry to measure dequenching in tens of thousands of cells.
Intracellular degradation of BSA is inhibited by pepstatin: Cellular imaging and flow cytometry
Use of a self-quenched substrate such as BSA-AF647, in combination with an enzyme inhibitor, makes it possible to determine which lysosomal enzyme, or combination of enzymes, is responsible for the degradation of endocytic cargo. Traditionally these measurements have been carried out with biochemical assays such as western blotting. Probing degradation directly in the cell makes it possible to determine at which intracellular site a specific enzyme is involved in degradation and removes the need for antibodies or other indirect probe of protein degradation. The intracellular degradation of albumin by cathepsin D has been determined previously [35, 36], making BSA a good substrate to test the use of self-quenching in intracellular measurements. Cathepsin D, like pepsin, is an acid protease that is inhibited by pepstatin.
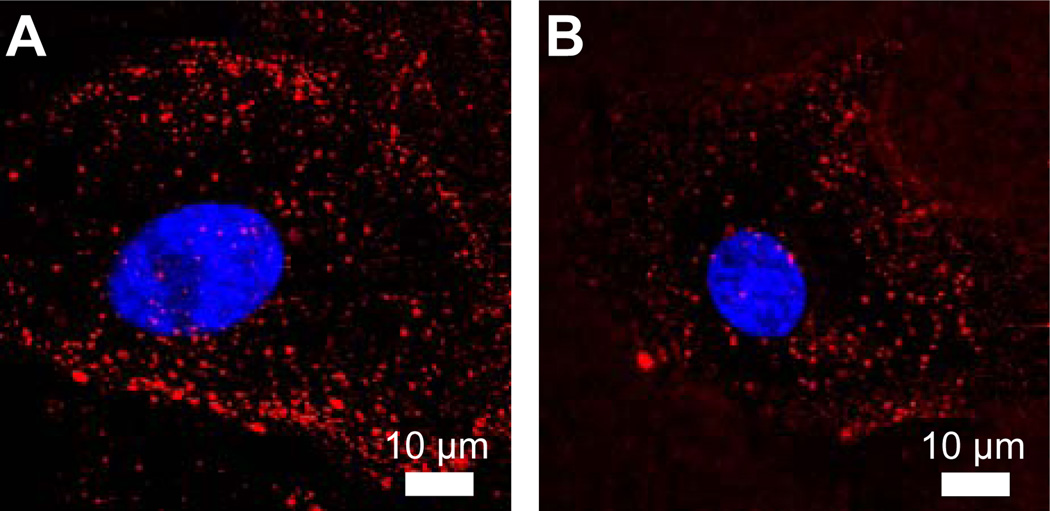
Confocal fluorescence microscopy was used to image cells incubated with 80 µg/mL BSA-AF647 for 1 hr at 37°C. Fluorescence microscopy images show a punctate BSA-AF647 signal indicative of BSA-AF647 within endocytic vesicles (Fig. 3A). In comparison, cells incubated with pepstatin, which inhibits cathepsin D, show the same punctate staining, but weaker fluorescence (Fig. 3B). An analysis of the intensity of each endocytic vesicle, described in the Materials and Methods, in control and pepstatin-treated cells showed that, on average, the intensity of the BSA-AF647-containing vesicles was 1.5-fold greater in control cells than in pepstatin-treated cells. The intensity analysis was carried out for >3000 endosomes for 7 cells of each condition, p = 0.005. The reduced degradation of BSA-AF647 in pepstatin-treated cells is in good agreement with previous inhibition assays [28].
Fig. 3.
Cellular imaging of BSA dequenching. (A) A representative confocal fluorescence microscopy image of a cell following a 1 hr incubation with BSA-AF647. (B) The same incubation and imaging parameters were used to image cells treated with pepstatin, an inhibitor of acid proteases. Nuclei were stained with DAPI (blue). Unmerged and brightfield images are included in Supplementary Material (Fig. S2).
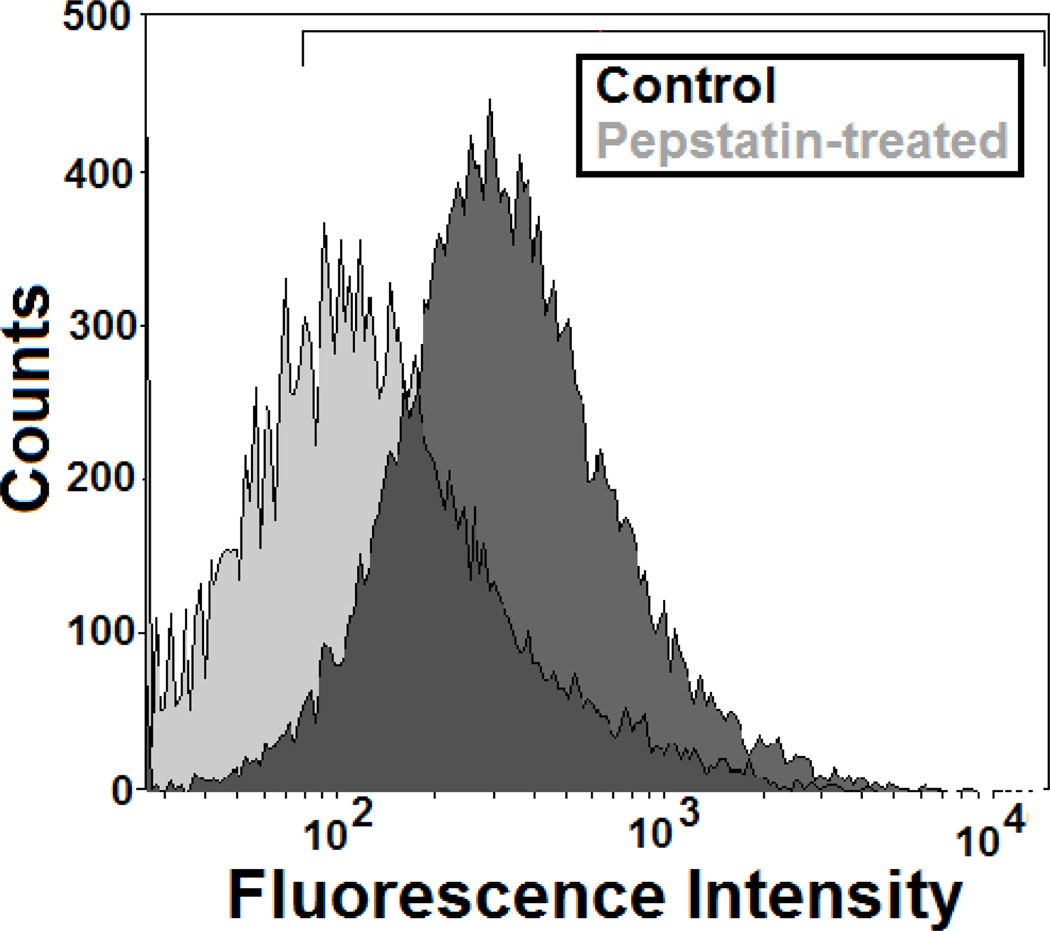
Cellular imaging has the advantage of providing spatial information, but is relatively low throughput as individual cells or organelles must be imaged and analyzed. In comparison, flow cytometry measures the fluorescence intensity of individual cells at relatively high speeds (~10,000 cells/min). While spatial information is lost, there are clear advantages to acquiring large amounts of data rapidly. Flow cytometry (BD LSR-II) was used to measure the emission of cells following a 1 hr incubation with 80 µg/mL BSA-AF647 in either untreated or pepstatin-treated cells (Fig. 4). Analysis of flow cytometry data shows that untreated, control cells show greater fluorescence intensity, normalized to 90%, as compared to pepstatin-treated cells, 44%. These results are in good agreement with previous flow cytometry measurements showing pepstatin resulted in a 30–50% inhibition of BSA degradation in macrophages [28].
Fig. 4.
Flow cytometry was used to measure the emission of cells incubated with BSA-AF647. Treatment of cells with pepstatin decreased the BSA-AF674 signal. A representative scatter plot is shown in Supplementary Material, Fig. S3.
BSA is degraded in LAMP1-positive vesicles
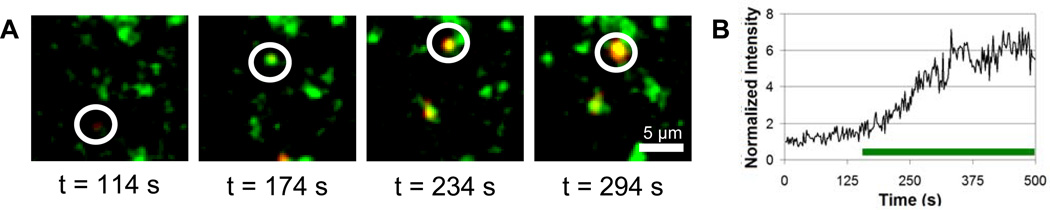
Single particle tracking is an especially powerful method for the study of endocytic transport. The use of a self-quenched endocytic cargo, used in combination with single particle tracking, allows us to follow intracellular transport and determine the intracellular site of degradation directly. To utilize this approach for the study of BSA degradation, LAMP1, a late endosomal and lysosomal protein [15, 37–42], was labeled with EYFP and used to generate BSC-1 monkey kidney cells that stably express LAMP1-EYFP [14, 43]. BSA-AF647 was incubated with cells at a concentration of 80 µg/mL for 30 min in full growth medium. These conditions allow BSA-AF647 to be internalized into endocytic vesicles [44, 45]. Two-color live cell imaging, using a custom built microscope with laser excitation, was used to image LAMP1-EYFP and BSA-AF647 simultaneously in real time in live cells. Data were collected as a series of images (0.5 Hz) and analyzed to detect increases in the BSA-AF647 signal (Fig. 5). BSA-AF647 was considered to dequench if the emission intensity, localized within an endo-lysosomal vesicle, increased by at least a factor of 2 within 100 s. Endo-lysosomal vesicles containing BSA-AF647 that underwent dequenching were then scored for the presence of LAMP1-EYFP. BSA-AF647 and LAMP1-EYFP were considered colocalized if they overlapped and moved through the cell together for at least 10 seconds. Of 10 BSA-AF647 dequenching events, all were localized in a LAMP1-positive vesicle. Two-color imaging with LAMP1-EYFP provides the first confirmation that BSA is degraded in a LAMP1-positive vesicle.
Fig. 5.
Two-color single particle tracking of BSA-AF647 and LAMP1-EYFP. (A) Snapshots illustrate the dequenching of BSA-AF647 (red) in a LAMP1-positive endo-lysosomal vesicle (green). Images were recorded at a rate of 0.5 Hz. (B) Intensity of the BSA-AF647 as a function of time. The horizontal bar under the intensity trace indicates the period of time during which BSA-AF647 was colocalized with a LAMP1-vesicle.
Conclusions
Self-quenched substrates allow us to combine single particle tracking fluorescence microscopy with the measurement of enzyme activity. We apply this method to the study of endocytosis in which cellular transport is followed by enzymatic degradation. We first describe the fluorescent labeling scheme and in vitro characterization for the self-quenching of BSA and LDL, two common endocytic cargos. We then use self-quenched BSA, in combination with static cellular imaging and flow cytometry, to illustrate the suitability of self-quenched substrates for use with existing fluorescence microscopy and flow cytometry platforms. Using two-color single particle tracking fluorescence microscopy, we find that the degradation of BSA occurs in an endo-lysosomal vesicle that is positive for LAMP1. BSA, or the human analog, is essential for human health [46, 47]. It is also increasingly important in nanobiotechnology for nanoparticle and drug delivery [48–50]. Determining the intracellular site and mechanism of BSA degradation is essential for the design of BSA-mediated delivery systems. While the use of standard fluorophores makes it possible to probe cellular location as a function of time, the change in intensity of self-quenched cargo is an indicator for the site at which cargo undergoes degradation providing a valuable new method for the study of endocytosis.
Supplementary Material
Acknowledgments
This research was supported by a NIH Director’s New Innovator Award (1DP2OD006470) to C.K.P. The authors thank Craig Szymanski and Jairo Zapata for assistance with experiments and data analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular Biology of the Cell. New York: Garland Publishing; 1994. [Google Scholar]
- 2.Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 3.Gruenberg J. The endocytic pathway: a mosaic of domains. Nat. Rev. Mol. Cell Bio. 2001;2:721–730. doi: 10.1038/35096054. [DOI] [PubMed] [Google Scholar]
- 4.Mellman I. Endocytosis and molecular sorting. Annu. Rev. Cell Dev. Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 5.Marsh M, Helenius A. Virus entry: Open sesame. Cell. 2006;124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandenburg B, Zhuang XW. Virus trafficking - learning from single-virus tracking. Nature Reviews Microbiology. 2007;5:197–208. doi: 10.1038/nrmicro1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakadamyali M, Rust MJ, Babcock HP, Zhuang XW. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lakadamyali M, Rust MJ, Zhuang X. Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell. 2006;124:997–1009. doi: 10.1016/j.cell.2005.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Payne CK, Jones SA, Chen C, Zhuang XW. Internalization and trafficking of cell surface proteoglycans and proteoglycan-binding ligands. Traffic. 2007;8:389–401. doi: 10.1111/j.1600-0854.2007.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 11.Hess GT, Humphries WH, Fay NC, Payne CK. Cellular binding, motion, and internalization of synthetic gene delivery polymers. BBA-Mol. Cell Res. 2007;1773:1583–1588. doi: 10.1016/j.bbamcr.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Payne CK. Imaging gene delivery with fluorescence microscopy. Nanomedicine. 2007;2:847–860. doi: 10.2217/17435889.2.6.847. [DOI] [PubMed] [Google Scholar]
- 13.Ewers H, Smith AE, Sbalzarini IF, Lilie H, Koumoutsakos P, Helenius A. Single-particle tracking of murine polyoma virus-like particles on live cells and artificial membranes. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15110–15115. doi: 10.1073/pnas.0504407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphries WH, Fay NC, Payne CK. Intracellular degradation of low-density lipoprotein probed with two-color fluorescence microscopy. Integr. Biol. 2010;2:536–544. doi: 10.1039/c0ib00035c. [DOI] [PubMed] [Google Scholar]
- 15.Szymanksi CJ, Humphries WH, Payne CK. Single particle tracking as a method to resolve differences in highly colocalized proteins. Analyst. 2011;136:3527–3533. doi: 10.1039/c0an00855a. [DOI] [PubMed] [Google Scholar]
- 16.Voet D, Voet JG. Biochemistry. New Jersey: John Wiley & Sons; 2004. [DOI] [PubMed] [Google Scholar]
- 17.Miller JN. Fluorescence energy transfer methods in bioanalysis. Analyst. 2005;130:265–270. doi: 10.1039/b314346p. [DOI] [PubMed] [Google Scholar]
- 18.Fischer R, Bachle D, Fotin-Mleczek M, Jung G, Kalbacher H, Brock R. A targeted protease substrate for a quantitative determination of protease activities in the endolysosomal pathway. ChemBioChem. 2006;7:1428–1434. doi: 10.1002/cbic.200600209. [DOI] [PubMed] [Google Scholar]
- 19.Reymond JL, Fluxa VS, Maillard N. Enzyme assays. Chem. Commun. 2009:34–46. doi: 10.1039/b813732c. [DOI] [PubMed] [Google Scholar]
- 20.Goddard JP, Reymond JL. Recent advances in enzyme assays. Trends Biotechnol. 2004;22:363–370. doi: 10.1016/j.tibtech.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Boonacker E, Van Noorden CJF. Enzyme cytochemical techniques for metabolic mapping in living cells, with special reference to proteolysis. J. Histochem. Cytochem. 2001;49:1473–1486. doi: 10.1177/002215540104901201. [DOI] [PubMed] [Google Scholar]
- 22.Voss EW, Workman CJ, Mummert ME. Detection of protease activity using a fluorescence-enhancement globular substrate. Biotechniques. 1996;20:286–291. doi: 10.2144/96202rr06. [DOI] [PubMed] [Google Scholar]
- 23.Jones LJ, Upson RH, Haugland RP, PanchukVoloshina N, Zhou MJ. Quenched BODIPY dye-labeled casein substrates for the assay of protease activity by direct fluorescence measurement. Anal. Biochem. 1997;251:144–152. doi: 10.1006/abio.1997.2259. [DOI] [PubMed] [Google Scholar]
- 24.Reis RCM, Sorgine MHF, Coelho-Sampaio T. A novel methodology for the investigation of Intracellular proteolytic processing in intact cells. Eur. J. Cell Biol. 1998;75:192–197. doi: 10.1016/S0171-9335(98)80061-7. [DOI] [PubMed] [Google Scholar]
- 25.French T, So PTC, Weaver DJ, CoelhoSampaio T, Gratton E, Voss EW, Carrero J. Two-photon fluorescence lifetime imaging microscopy of macrophage-mediated antigen processing. J Microsc. 1997;185:339–353. doi: 10.1046/j.1365-2818.1997.d01-632.x. [DOI] [PubMed] [Google Scholar]
- 26.Weissleder R, Tung CH, Mahmood U, Bogdanov A. In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat. Biotechnol. 1999;17:375–378. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 27.Weaver DJ, Durack G, Voss EW. Analysis of the intracellular processing of proteins: Application of fluorescence polarization and a novel fluorescent probe. Cytometry. 1997;28:25–35. [PubMed] [Google Scholar]
- 28.Weaver DJ, Cherukuri A, Carrero J, CoelhoSampaio T, Durack G, Voss EW. Macrophage mediated processing of an exogenous antigenic fluorescent probe: Time-dependent elucidation of the processing pathway. Biol. Cell. 1996;87:95–104. [PubMed] [Google Scholar]
- 29.van der Schaar HM, Rust MJ, Chen C, van der Ende-Metselaar H, Wilschut J, Zhuang XW, Smit JM. Dissecting the Cell Entry Pathway of Dengue Virus by Single-Particle Tracking in Living Cells. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sbalzarini IF, Koumoutsakos P. Feature Point Tracking and Trajectory Analysis for Video Imaging in Cell Biology. J. Struct. Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Weber G, Young LB. Fragmentation of boine serum albumin by pepsin. 1. Origin of acid expansion of albumin molecule. J. Biol. Chem. 1964;239:1415–1423. [PubMed] [Google Scholar]
- 32.Weber G, Young LB. Fragmentation of bovine serum albumin by pepsin. 2. Isolation, amino acid composition, and physical properties of fragments. J. Biol. Chem. 1964;239:1424–1431. [PubMed] [Google Scholar]
- 33.Kageyama T. Pepsinogens, progastricsins, and prochymosins: structure, function, evolution, and development. Cell. Mol. Life Sci. 2002;59:288–306. doi: 10.1007/s00018-002-8423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rich DH, Sun ETO. Mechanism of inhibition of pepsin by pepstatin-effect of inhibitor structure on dissociation constant and and time-dependent inhibition. Biochem. Pharmacol. 1980;29:2205–2212. doi: 10.1016/0006-2952(80)90199-9. [DOI] [PubMed] [Google Scholar]
- 35.Mego JL. Role of thiols, pH and cathepsin D in the lysosomal catabolism of serum albumin. Biochem. J. 1984;218:775–783. doi: 10.1042/bj2180775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baricos WH, Zhou YW, Fuerst RS, Barrett AJ, Shah SV. The role of aspartic and cysteine proteinases in albumin degradation by rat kidney cortical lysosomes. Arch. Biochem. Biophys. 1987;256:687–691. doi: 10.1016/0003-9861(87)90625-4. [DOI] [PubMed] [Google Scholar]
- 37.Chen JW, Murphy TL, Willingham MC, Pastan I, August JT. Identification of two lysosomal membrane glycoproteins. J Cell Biol. 1985;101:85–95. doi: 10.1083/jcb.101.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis V, Green SA, Marsh M, Vihko P, Helenius A, Mellman I. Glycoproteins of the lysosomal membrane. J Cell Biol. 1985;100:1839–1847. doi: 10.1083/jcb.100.6.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lippincott-Schwartz J, Fambrough DM. Lysosomal membrane dynamics: Structure and interoganellar movement of a major lysosomal membrane glycoprotein. J Cell Biol. 1986;102:1593–1605. doi: 10.1083/jcb.102.5.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clague MJ. Molecular aspects of the endocytic pathway. Biochem. J. 1998;336:271–282. doi: 10.1042/bj3360271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffiths G, Hoflack B, Simons K, Mellman I, Kornfeld S. The mannose 6-phosphate receptor and the biogenesis of lysosomes. Cell. 1988;52:329–341. doi: 10.1016/s0092-8674(88)80026-6. [DOI] [PubMed] [Google Scholar]
- 42.Geuze HJ, Stoorvogel W, Strous GJ, Slot JW, Bleekemolen JE, Mellman I. Sorting of mannose 6-phosphate receptors and lysosomal membrane-proteins in endocytic vesicles. J Cell Biol. 1988;107:2491–2501. doi: 10.1083/jcb.107.6.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherer NM, Lehmann MJ, Jimenez-Soto LF, Ingmundson A, Horner SM, Cicchetti G, Allen PG, Pypaert M, Cunningham JM, Mothes W. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic. 2003;4:785–801. doi: 10.1034/j.1600-0854.2003.00135.x. [DOI] [PubMed] [Google Scholar]
- 44.Tagawa M, Yumoto R, Oda K, Nagai J, Takano M. Low-Affinity Transport of FITC-Albumin in Alveolar Type II Epithelial Cell Line RLE-6TN. Drug Metab. Pharmacok. 2008;23:318–327. doi: 10.2133/dmpk.23.318. [DOI] [PubMed] [Google Scholar]
- 45.Yumoto R, Nishikawa H, Okamoto M, Katayama H, Nagai J, Takano M. Clathrin-mediated endocytosis of FITC-albumin in alveolar type II epithelial cell line RLE-6TN. Am. J. Physiol.-Lung C. 2006;290:L946–L955. doi: 10.1152/ajplung.00173.2005. [DOI] [PubMed] [Google Scholar]
- 46.Anderson NL, Anderson AG. The human plasma proteome. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 47.Osicka TM, Houlihan CA, Chan JG, Jerums G, Comper WD. Albuminuria in patients with type 1 diabetes is directly linked to changes in the lysosome-mediated degradation of albumin during renal passage. Diabetes. 2000;49:1579–1584. doi: 10.2337/diabetes.49.9.1579. [DOI] [PubMed] [Google Scholar]
- 48.Hanaki K, Momo A, Oku T, Komoto A, Maenosono S, Yamaguchi Y, Yamamoto K. Semiconductor quantum dot/albumin complex is a long-life and highly photostable endosome marker. Biochem. Biophys. Res. Commun. 2003;302:496–501. doi: 10.1016/s0006-291x(03)00211-0. [DOI] [PubMed] [Google Scholar]
- 49.Kratz F. Albumin as a drug carrier: Design of prodrugs, drug conjugates and nanoparticles. J. Control. Release. 2008;132:171–183. doi: 10.1016/j.jconrel.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 50.Doorley GW, Payne CK. Cellular binding of nanoparticles in the presence of serum proteins. Chem. Commun. 2011;47:466–468. doi: 10.1039/c0cc02618b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.