SUMMARY
The association between hyper-inflammatory states and numerous diseases is widely recognized, but our understanding of the molecular strategies that have evolved to prevent uncontrolled activation of inflammatory responses remains incomplete. Here, we report a critical, non-transcriptional role of GPS2 as a guardian against hyperstimulation of the TNFα-induced gene program. GPS2 cytoplasmic actions are required to specifically modulate RIP1 ubiquitylation and JNK activation by inhibiting TRAF2/Ubc13 enzymatic activity. In vivo relevance of GPS2 anti-inflammatory role is confirmed by inhibition of TNFα target genes in macrophages and by improved insulin signaling in the adipose tissue of aP2-GPS2 transgenic mice. As the non-transcriptional role is complemented by GPS2 functioning as positive and negative cofactor for nuclear receptors, in vivo overexpression also results in elevated circulating level of Resistin and development of hepatic steatosis. Together, these studies define GPS2 as a molecular guardian required for precise control of inflammatory responses involved in immunity and homeostasis.
INTRODUCTION
Preventing the hyper-stimulation of inflammatory responses is a key requirement for normal homeostasis, which can be achieved by different strategies, including regulating the production of inflammatory cytokines, restricting the response to basal stimuli and ensuring the proper termination of the activated signaling cascade. Excessive or prolonged activation of pro-inflammatory signaling pathways has been implicated in the pathogenesis of several human diseases including autoimmune disorders, neurodegenerative diseases and cancer (Amor et al., 2010; Grivennikov et al., 2010). Chronic inflammation has also been linked with obesity, insulin resistance and development of Type 2 Diabetes and convincing evidences have indicated that local production of pro-inflammatory cytokines, including TNFα, leads to the disruption of the insulin response in adipose tissue and peripheral organs (Hotamisligil, 2006; Lee and Pratley, 2005). Pro-inflammatory signals, together with elevated levels of FFA and ROS, activate a series of stress-induced serine kinases, including JNK, and IKKβ, which play a key role in the development of insulin resistance as indicated by the improvement in insulin sensitivity associated with their chemical inhibition or with the genetic disruption of JNK (Hirosumi et al., 2002; Sabio and Davis, 2010).
Activation of JNK and IKKβ kinases in response to inflammatory stimuli is achieved via the coordinated actions of a large number of signaling proteins and enzymatic activities. In the case of TNFα, trimerization of liganded TNFR1 leads to the formation of a membrane-associated complex including TRADD, TRAF2, RIP1 and cIAP1/2, with one TRAF2 trimer required for the recruitment of each cIAP2 molecule (Chen and Goeddel, 2002; Zheng et al., 2010). Overall, the enzymatic activities of TRAF2, cIAP1 and Ubc13 are together required for the polyubiquitination of RIP1, which was initially thought to act as a general scaffold for the assembly of TAK1/TAB1/TAB2 and IKKα/IKKβ/NEMO complexes. However, this simple model has been recently revised, and in the emerging picture, there is a significant separation between the activation of JNK and IKKβ in terms of the adaptor proteins, the enzymatic activities and the type of ubiquitin chains utilized in the activation cascade (Bianchi and Meier, 2009; Deribe et al., 2010; Liu and Chen, 2010). Because of the importance of avoiding uncontrolled stimulation of inflammatory responses, each of the signaling steps has to be tightly regulated. While some inhibitory strategies have been uncovered, including factors such as A20 (TNFAIP3) and CYLD that negatively regulate the ubiquitylation status of key signaling molecules (Bhoj and Chen, 2009; Sun, 2008), the full extent of the cell's ability to control inflammatory responses at the molecular level is still incompletely understood.
G-protein pathway suppressor 2 (GPS2) is a small, ubiquitous protein that was originally identified while screening for suppressors of Ras activation in the yeast pheromone response pathway (Spain et al., 1996). While GPS2 role in transcriptional regulation has been indicated by several studies reporting its interactions with a series of transcriptional regulators (Jakobsson et al., 2009; Lee et al., 2006; Pan et al., 2008; Peng et al., 2000; Peng et al., 2001; Sanyal et al., 2007), a clear understanding of GPS2 function is still strikingly incomplete. Intriguingly, in many cases GPS2 was suggested to act as a coactivator, whereas biochemically it was identified as an intrinsic component of a major transcriptional repressor complexes, the NCoR/SMRT nuclear receptor corepressor complex (Zhang et al., 2002).
Here, we have further contributed to the understanding of the positive and negative transcriptional roles of GPS2, while also uncovering a critical non-nuclear role for GPS2 in inhibiting the pro-inflammatory TNFα pathway. This reflects an unexpected, non-transcriptional function, with GPS2 being required for inhibiting the enzymatic activity of key components of the ubiquitin conjugating machinery in the TNFR1 signaling cascade. Consistent with these findings, overexpression of GPS2 impaired the activation of pro-inflammatory responses, both in vitro and in vivo in adipose tissue and macrophages, by specifically inhibiting JNK activation.
RESULTS
GPS2 interacts with the TRAF/Ubc13 K63 ubiquitylation machinery
While previous studies identified GPS2 as a transcriptional cofactor and indicated that it could mediate both gene repression and activation, GPS2 initial discovery a yeast genetic screen for revertants of Ras activation and its ability to inhibit JNK activation (Spain et al., 1996; Zhang et al., 2002), suggested the additional possibility of a critical, non-transcriptional role in the regulation of signaling pathways.
To investigate a potential role of GPS2 in the regulation of pro-inflammatory signals, we first examined transcriptional activation of a LacZ reporter gene driven by the TNFα-responsive Cox2 promoter, finding that knockdown of GPS2 by siRNA microinjection was sufficient to induce gene activation (Fig 1A). However, basal activation of the Cox2 promoter in cells microinjected with siGPS2 was rescued by JNK specific inhibitor SP600125 (Fig 1A), suggesting that the transcriptional output of GPS2 downregulation might be dependent on the global activation of JNK in the cell. Also, siRNA-dependent downregulation of GPS2 proved to be sufficient to induce basal activation of JNK in the cytoplasm, and to promote a stronger activation in response to TNFα, as indicated by western blotting with an antibody specific for phospho-JNK (Fig 1B), whereas overexpression of HA-GPS2 inhibited the cytoplasmic activation of JNK upon TNFα treatment (Fig 1C). Importantly, the hypothesis of a non-transcriptional role for GPS2 in the regulation of JNK activation was consistent with GPS2 localization both to the cell nucleus and cytoplasm. (Supplement Fig S1).
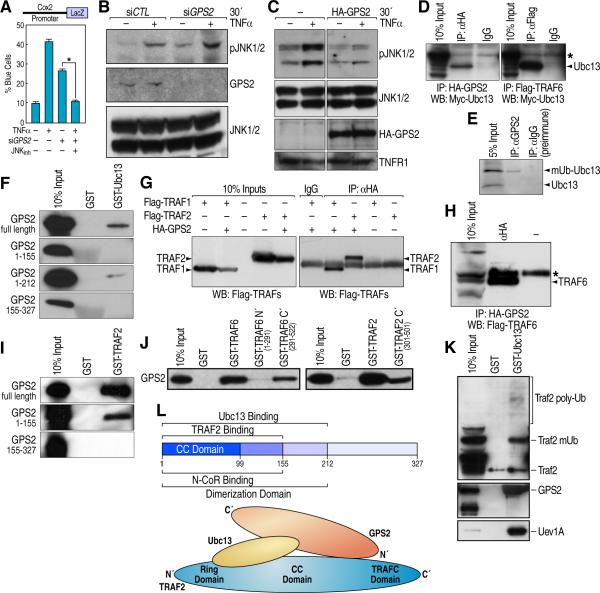
Figure 1. GPS2 interacts with the K63 ubiquitylation machinery.
(A) Single cell microinjection of Cox2-LacZ reporter with GPS2 siRNA and JNK inhibitor SP600125. All graphic data is +/- SD, * indicates P<0.05 (B, C) Western blot (WB) for P-JNK in 293T cytoplasmic extracts. (D) Co-immunoprecipitation (co-IP) of HA-GPS2/Myc-Ubc13 in 293T cells (left panel), and Flag-TRAF6/Myc-Ubc13 as positive control (right panel). (E) Endogenous co-IP between GPS2 and Ubc13 in 293T cells using a protein A/G column with chemically crosslinked GPS2 antibody. (F) Mapping of direct interaction between GPS2 and Ubc13 by GST pull-down. (G) Co-IP of HA-GPS2 with Flag-TRAF1 and Flag-TRAF2. (H) Co-IP of HA-GPS2 with Flag-TRAF6. (I) GST pull down mapping of GPS2-TRAF2 interaction. (J) Interaction between GPS2 and the TRAFC domain of TRAF6 (upper panel) and TRAF2 (bottom panel) (K) GST-Ubc13 affinity column loaded with 293T cellular extracts: Ubc13 interaction with Uev1a, Traf2 and GPS2. (L) Schematic model of the interaction surfaces between GPS2, Ubc13 and Traf2. * indicates an aspecific band.
To identify GPS2 interacting partners that could provide insights into GPS2 regulation of JNK activity, we performed a yeast-two-hybrid assay and discovered an unexpected interaction between GPS2, used as bait, and the E2 ubiquitinconjugating enzyme Ubc13 responsible for making K63-branched ubiquitin chains in response to the activation of various inflammatory receptors, including TNFα receptor (Fukushima et al., 2007). Direct interaction between GPS2 and Ubc13 was confirmed by coimmunoprecipitation in 293T cells (Fig 1D, 1E) and by GST pull down (Fig 1F). Interestingly, the endogenous co-IP revealed a preferential binding between GPS2 and the activated form of Ubc13 bound to ubiquitin (Fig 1E). Mapping identified a direct binding between Ubc13 and a central fragment of GPS2 corresponding to aa 155-212 (Fig 1F). Unexpectedly, strong, direct interaction was also observed between GPS2 and members of the TRAF family, which include several E3 ligases working together with Ubc13 in the activation of the inflammatory signaling cascades (Wertz and Dixit, 2009). Coimmunoprecipitation assays showed interaction between GPS2, TRAF1, TRAF2 and TRAF6 (Fig 1G and 1H), and in GST pull downs deletion mutants indicated that the N’ terminal fragment of GPS2 (1-155) interacted directly with the TRAFC domain of TRAF2 and TRAF6 (Fig 1I, 1J). To verify whether Ubc13 could interact with both GPS2 and the E3 ligase at the same time we made a GSTUbc13 affinity column and loaded it with 293T cellular extracts. As indicated by immunoblots in Fig 1K, Ubc13 could easily pull down together GPS2, TRAF2 and the E2 ubiquitin conjugating cofactor Uev1A. A summary of the interactions identified and the protein surfaces required for such interactions is provided in Fig 1L.
GPS2 regulates the activation of TNFR1 in response to TNFα
In mammalian cells, TNFα binds to two receptors: TNFR1, which is ubiquitously expressed, and TNFR2, expression of which is restricted to cells of the immune system. Depending on the complexes assembled upon ligand binding, TNFR1 can activate pro-survival as well as pro-apoptotic pathways, with pro-survival pathways being activated by the assembling of a membrane complex, including the ligand-bound receptor, the adaptor protein TRADD, the ubiquitylation machinery composed by the E2 conjugating enzyme Ubc13, the E3 ligases TRAF2 and cIAP1/2 and their target protein RIP1 (Silke and Brink, 2009; Wertz and Dixit, 2009). Because of the unexpected interaction uncovered between GPS2, Ubc13 and TRAF2, we elected to investigate whether the anti-inflammatory role of GPS2 might be mediated by its intervention in these early steps of TNFα signaling.
First, we investigated GPS2 recruitment to the TNFR1 complex, confirming that endogenous GPS2 would bind to the receptor within minutes of TNFα treatment, in an NCoR-independent manner, whereas transiently overexpressed HA-GPS2 would bind to the receptor even in the unstimulated cells (Supplemental Fig S2A). Because TNFR1-bound GPS2 was rapidly poly-ubiquitylated upon TNFα stimulation (Supplemental Fig S2B), we tested whether GPS2 could be modified by Ubc13/TRAF2 using an in vitro ubiquitylation assay. While GPS2 was not poly-ubiquitylated in this assay (Supplemental Fig S2C), we found that TRAF2/Ubc13-dependent poly-ubiquitylation was greatly inhibited by GPS2 (Fig 2A). In particular the inhibition seemed to be specific for TRAF2-dependent ubiquitylation as the short ubiquitin chains assembled by the E2 conjugating complex alone were not affected (Fig 2A), and no changes were observed in the interaction between Ubc13 and its coenzyme Uev1A (Supplemental Fig S2D, S2E). Regulation of TRAF2 enzymatic activity by GPS2 was also observed within the activated TNFR1 complex, with TRAF2 auto-ubiquitylation being highly impaired by GPS2 overexpression, or increased with GPS2 downregulation (Fig 2B). TRAF2 recruitment to the receptor, on the contrary, was not affected either by GPS2 overexpression or downregulation (Fig 2C), suggesting that GPS2 does not affect complex formation, but is important to keep an inhibitory control over TRAF2 enzymatic activity.
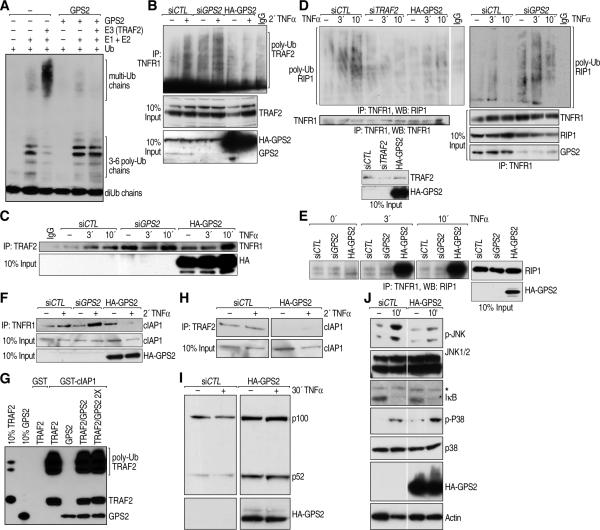
Figure 2. TNFα-dependent activation of TNFR1 is regulated by GPS2.
(A) GPS2 inhibits TRAF2 enzymatic activity in in vitro ubiquitination assay with recombinant E1, recombinant E2 (Ubc13/Uev1a) and bacterially expressed and purified E3 (TRAF2). (B) Polyubiquitylation of TRAF2 bound to TNFR1 in TNFα-stimulated 293T with siGPS2/HAGPS2. (C) No changes are observed in TRAF2 binding to TNFR1 when GPS2 is down- or up-regulated. (D) Poly-ubiquitylated RIP1 as immunoprecipitated with TNFR1 from cells transfected with siCTL, siTRAF2, HA-GPS2 (left panel) or siGPS2 (right panel). (E) Accumulation of unmodified RIP1 bound to TNFR1 with GPS2 overexpression. (F) CIAP1 binding to TNFR1 is regulated GPS2 in co-IP. (G) GST pull down for cIAP1-TRAF2 interaction in presence of increasing amount of GPS2. (H) Co-IP between TRAF2 and cIAP1 in 293T cytoplasmatic extracts. (I) Increase of p100/p52 processing in GPS2 overexpressing cells by WB. (J) JNK, NFkB, p38 activation measured by P-JNK, IkB and P-p38 blot. * indicates an aspecific band
Next, we investigated the TNFα-dependent poly-ubiquitylation of RIP1, a key target of TRAF2 activity, which is required for the activation of JNK. While RIP1 poly-ubiquitylation was observed within minutes of TNFα stimulation in the control sample, RIP1 modification was almost completely inhibited by GPS2 overexpression, similarly to when TRAF2 itself is deleted (Fig 2D). Conversely, RIP1 poly-ubiquitylation was increased in cells transfected with siGPS2 (Fig 2D). Consistent with the observation that unmodified RIP1 is associated with proapoptotic cellular responses (Vandenabeele et al.), we found that the accumulation of the unmodified form of RIP1 protein observed with GPS2 overexpression (Fig 2E) correlates with an increase formation of the RIP1/FADD/CASP8 complex (Supplemental Fig S2F). Consequently, TNFα-induced apoptosis was highly increased in cells overexpressing GPS2, as indicated by the presence of markers of mature apoptosis including cleaved PARP1 and cleaved Caspase 3 (Supplemental Fig S2G).
Recent studies performed to clarify the enzymology of RIP1 post-translational modifications have indicated that RIP1 may not be directly poly-ubiquitylated by TRAF2, but rather by another E3 ligase which is recruited in a TRAF2-dependent manner, namely cIAP1 (Bertrand et al., 2008; Mahoney et al., 2008; Varfolomeev et al., 2008; Vince et al., 2009). In agreement with the lack of RIP1 polyubiquitylation, cIAP1 recruitment was also abrogated in presence of GPS2 overexpression (Fig 2F). Conversely, GPS2 downregulation by siRNA induced a significant increase in the amount of cIAP1 recruited upon TNFα treatment (Fig 2F). Because these data implicated an inhibitory role for GPS2 on TRAF2 post-translational modification that affected the correct recruitment of cIAP1, we further investigated the interaction between TRAF2 and cIAP1 by GST pull down. As indicated in Fig 2G, a strong direct interaction was observed between in vitro translated TRAF2 and GST-cIAP1, with a clear enrichment in the poly-ubiquitylated fraction of TRAF2 bound to cIAP1. Under these conditions, we also observed direct interaction between cIAP1 and GPS2, finding that increasing the amount of GPS2 in the reaction did not displace the binding of TRAF2 to cIAP1 (Fig 2G). Together these results indicate that GPS2 is not an allosteric inhibitor of the formation of this complex, but rather plays a role in modulating the posttranslational modifications of TRAF2. Indeed, the interaction between TRAF2 and cIAP1 was less easily detected using GST-TRAF2 expressed in bacteria and therefore not modified by ubiquitin (Data not shown). Interestingly, the ability of GPS2 to regulate TRAF2-cIAP1 interaction was not restricted at the receptor level, as we could observe a global decrease in cIAP1 interaction with TRAF2 by co-immunoprecipitation in cytoplasmic extracts (Fig 2H). Because a cytosolic cIAP1/2-TRAF2-TRAF3 complex is known to be required for maintaining negative regulation of the NFκB non-canonical pathway (Vallabhapurapu et al., 2008; Zarnegar et al., 2008), we investigated the role of GPS2 in the regulation of this pathway as well. In keeping with its ability to abrogate TRAF2-cIAP1 interaction, we found that GPS2 overexpression was sufficient to produce basal activation of the non-canonical pathway, as indicated by processing of the p100/p52 subunits (Fig 2I).
GPS2 inhibitory role is specific for JNK activation
A key remaining question was whether the entire TNFα signaling pathway was dependent on GPS2 regulation. Interestingly, numerous reports have suggested that the Ubc13/TRAF2 complex and the K63-ubiquitylation activity are required for JNK activation, but dispensable for the other arms of TNFα signaling (Habelhah et al., 2004; Yamamoto et al., 2006; Zhang et al., 2010). And, the participation of RIP1 itself to TNFα-induced activation of NFκB has been questioned (Wong et al., 2009). To investigate the breadth of the effect of GPS2 overexpression, we looked at the activation of various downstream effectors of TNFα signaling. As expected, JNK activation was inhibited, as indicated by a decrease in P-JNK1/2 (Fig 2J), but neither downregulation of the IKK pathway, as indicated by IκB protein degradation, nor loss of activation of p38, as indicated by immunoblotting for P-p38, were observed (Fig 2J). Thus, these results confirmed a very specific role for GPS2 in modulating the machinery responsible for the activation of JNK and its downstream signaling events.
Hyperactivation of a pro-inflammatory transcriptional program in absence of GPS2
To further test the relevance of this non-transcriptional role of GPS2 for the downstream effect of TNFα signaling, we investigated the activation of the pro-inflammatory gene transcription program in response to GPS2 depletion or overexpression. We first examined the regulation of two well-known TNFα targets, IL8 and TNFα. While basal repression was not affected by GPS2 downregulation, a significant increase in gene activation could be observed in response to TNFα treatment (Fig 3A). The specificity of this effect was confirmed by rescuing the downregulation of human GPS2 with the overexpression of its murine isoform (Supplemental Fig S3A). To broaden our investigation, we performed RNA-sequencing in 293T cells, untreated or treated with TNFα for 6h, and expression of genes upregulated in response to TNFα were then compared between siCTL- and siGPS2-transfected cells. As shown in Fig 3B, a large number of genes were found further upregulated with siGPS2, and GO term analysis of these genes indicated significant enrichment in categories, like cytokine activity, cell adhesion and motility, inflammatory and immune responses, that are characteristic of TNFαresponse (Table S1). Thus, this genome wide analysis confirmed that loss of GPS2 causes a large increase in the pro-inflammatory response to TNFα. Conversely, overexpression of GPS2 in transient transfection was sufficient to significantly inhibit the activation of several TNFα target genes, including VCAM1, IL8 and NFKBIA. The inhibition was also observed in the cases of CCL20 and CXCL10, which had not shown a significant increase in activation in the RNA-seq with siGPS2, possibly because they were already maximally upregulated (Fig 3C and Table S1). Interestingly, the expression of GPS2 itself showed the expected kinetics for an endogenous inhibitor of TNFα signaling, with GPS2 expression being downregulated upon treatment with TNFα (Supplemental Fig S3B).
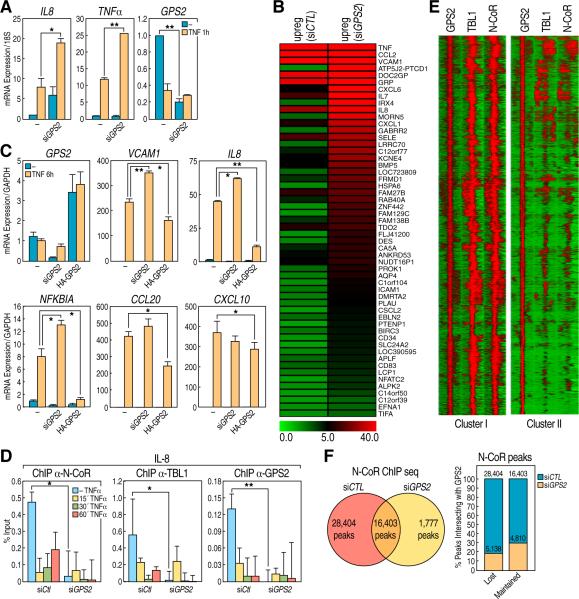
Figure 3. Role of GPS2 in the regulation of pro-inflammatory gene network program.
(A) qPCR analysis of IL8, TNFα and GPS2 genes expression in 293T cells treated with TNFα. (B) RNAseq in 293T cells transfected with siCTL or siGPS2. Heat map indicates the fold changes upon 6h TNFα treatment for all the genes that are further upregulated when comparing siGPS2 and siCTL (Ratio between RPKM values >1.4) (C) qPCR analysis of GPS2, VCAM1, IL8, NFKBIA, CCL20, CXCL10 genes expression upon GPS2 downregulation or overexpression. (D) ChIP for NCoR, TBL1 and GPS2 on the IL8 promoter. (E) NCoR, GPS2, TBL1 ChIPseq. Heat map representations of the location of NCoR and TBL1 peaks within a +/-1kb region surrounding the center of GPS2 peaks with overlapping peaks for NCoR/TBL1/GPS2 (Cluster I) or GPS2 peaks alone (Cluster II, see Venn diagram in Supplemental Fig S4). (F) NCoR ChIPseq: Venn diagram showing overlapping between the NCoR peaks in 293T cells with siGPS2 or siCTL (left); graph showing the percentage of NCoR peaks that overlaps with GPS2 peaks (right). The NCoR peaks are divided between peaks that are lost or maintained with siGPS2 and in both cases the overlapping is about 20-30%. Yellow bars represent the percentage of NCoR peaks overlapping with GPS2 binding, blue bars represent the percentage of NCoR peaks that do not overlap with GPS2. All graphic data is +/- SD, * if P<0.05, ** if P<0.01.
Being GPS2 a component of the NCoR/SMRT corepressor complex, this increase in gene expression could be interpreted as a result of derepression alone or as a combined effect of transcriptional and non-transcriptional roles of GPS2. To distinguish between these two possibilities, we first tested whether removal of GPS2 from downregulated promoters could induce gene activation based on dismissal of the NCoR/SMRT co-repressors. Indeed, ChIP analysis of the IL8 promoter indicated that both NCoR and TBL1 were dismissed in cells transfected with siGPS2 (Fig 3D). However, this could still alternatively represent either the cause or a consequence of gene activation. To formally adjudicate this issue, we performed ChIP-Sequencing analysis for determining the genome-wide localization of GPS2, either alone or with the other components of the complex. The objective was to distinguish between the alternative possibilities that local actions of GPS2 were exclusively responsible for keeping the genes from being upregulated, or that GPS2 non-transcriptional role was also important in determining the transcriptional regulation of genes that were not directly repressed by GPS2 and NCoR. ChIP-Seq experiments performed for GPS2, NCoR and TBL1 revealed, as might have been expected, a high level of co-localization among these factors, confirming that a large transcriptional program is regulated by the NCoR/TBL1/GPS2 complex (Fig 3E and Supplement Fig S4). However, there was an equally large number of locations where GPS2 was recruited independently of the NCoR complex (Fig 3E and Supplemental Fig S4), possibly via some of the transcription factors that can directly bind to GPS2, including p53 and the nuclear receptors LXR, HNF4 and FXR (Peng et al., 2001; Sanyal et al., 2007). Indeed, the differential motif analysis showed a more significant enrichment in the NR motifs (including LXRE, PPARE, RXRE, ERE, and ARE) for the GPS2 single peaks, while the common peaks were more significantly enriched in other motifs including those for NRF1, Sp1, ETS, CTCF, and YY1. Consistent with this motif analysis, we also found that the overlapping NCoR/TBL1/GPS2 peaks were enriched in promoter regions (almost 40% versus the 15-20% of the single components), while intergenic and intronic regions were enriched among GPS2-alone peaks (Supplemental Fig S4). ChIP-Seq analysis for NCoR binding in presence of siGPS2 was performed in the same cells, indicating a dramatic loss of NCoR binding with a decrease in more than 50% of peaks (Fig 3F). Surprisingly, when the NCoR peaks, divided among peaks that were “lost” or “maintained” without GPS2, were overlapped with the GPS2 binding locations, we found that only ~20% of the “lost” NCoR peaks were originally marked by the presence of GPS2. An even slightly higher ratio, (~30%) was observed for the NCoR peaks that were maintained with siGPS2 (Fig 3F). While these results together do not exclude the importance of GPS2 as a transcriptional coregulator, they do indicate that the strong upregulation of pro-inflammatory genes observed in presence of siGPS2 is not caused solely by a local effect on regulated promoters, but rather by the removal of a global inhibitory effect.
GPS2 anti-inflammatory role is required for PPARγ-dependent differentiation of adipocytes
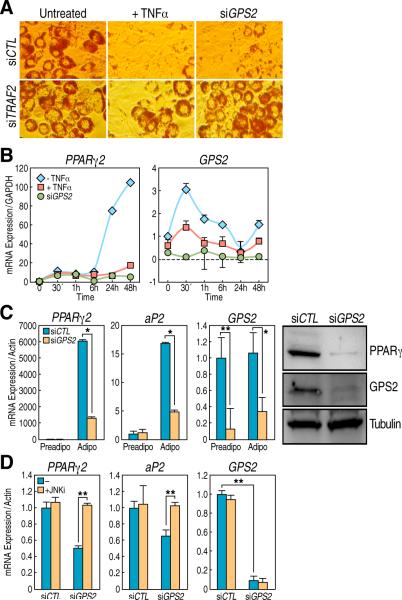
The nuclear receptor corepressors NCoR and SMRT have been previously associated with the transcriptional repression mediated by PPARγ and an increase in adipogenic differentiation was observed in 3T3-L1 cells depleted of NCoR/SMRT (Yu et al., 2005). However, when we tested the physiological relevance of GPS2 in the same model of differentiation, we found that GPS2-depleted cells were unable to fully differentiate into adipocytes (Fig 4A). Hence, the adipogenic conversion of 3T3-L1 provided a powerful experimental system in which we could differentiate between the transcriptional and the nontranscriptional role of GPS2. Because inflammatory signals are known to exert critical negative regulation on the differentiation process during the early stages of adipogenesis (Xu et al., 1999; Ruan et al., 2002; Cawthorn and Sethi, 2008), we reasoned that the adipogenic differention in 3T3-L1 cells treated with siGPS2 may be blocked because the TNFα signaling pathway was improperly activated. Indeed, the downregulation of TRAF2, a key mediator of TNFR1 signaling, was sufficient to rescue the adipogenic differentiation of GPS2-depleted cells, as indicated by fat droplets accumulation and Red Oil staining (Fig 4A). Also, when comparing the effects of TNFα treatment with the effect of depleting cells of GPS2 using specific siRNA, we generally observed a very similar phenotype in respect to the misregulation of key early genes such as CHOP and c-Myc (data not shown). Similarly, in both cells transfected with siGPS2 and treated with TNFα, we could observe a pronounced inhibition of the expression of PPARγ2 itself (Fig 4B, 4C), which prevented further differentiation of cells depleted of GPS2, thus impairing expression of markers of mature adipogenic differentiation, including aP2/FABP4 (Fig 4C). Interestingly, GPS2 expression was found to be increased during the first few hours of differentiation, prior to PPARγ2 expression, and the increase in GPS2 expression was abrogated when cells were stimulated with TNFα (Fig 4B). Finally, we tested whether we could rescue the inhibition of PPARγ2 expression in 3T3-L1 cells treated with siGPS2 by specifically inhibiting JNK. As shown in Fig 4D, inhibition of JNK activity with the specific inhibitor SP600125 was sufficient to fully restore PPARγ expression after three days of differentiation. Thus, these results independently confirm that the non-transcriptional role of GPS2 is required during the early stages of differentiation to prevent undesired activation of inflammatory signals and to guarantee the correct upregulation of PPARγ, linking the effects on JNK activation to gene transcriptional events.
Figure 4. GPS2 is required for PPARg-dependent adipogenesis.
(A) Oil Red O staining of 3T3-L1 after 7 days of differentiation: downregulation of TRAF2 rescues the siGPS2 phenotype. (B) qPCR analysis of PPARγ2 and GPS2 genes expression in 3T3-L1 cells at various stages of differentiation. (C) Downregulation of PPARγ by GPS2: qPCR analysis of PPARγ2 and aP2 expression (left), PPARγ and GPS2 protein level (right) (D) Loss of PPARγ2 and aP2 upregulation upon siGPS2 is rescued by JNK inhibitor SP600125. All graphic data is +/- SD, * if P<0.05, ** if P<0.01.
The anti-inflammatory role of GPS2 is important to protect aP2-GPS2 transgenic mice from HFD-induced development of insulin resistance in adipose tissue
The next question was whether the overexpression of GPS2 could keep pro-inflammatory pathways under negative regulation in vivo, in a situation where having a reduced inflammatory response could be beneficial for the organism. To this end, we generated aP2-GPS2 transgenic mice in which HA-GPS2 expression in adipose tissue is driven by the aP2/FABP4 enhancer region (Ross et al., 1990)(Fig 5A, 5B). Initial examination of these mice indicated that a significant reduction in the activation of JNK1/2 could be seen in the adipose tissue (Fig 5C), thus confirming the inhibitory role of GPS2. In addition, aP2-GPS2 mice consistently exhibited a slightly higher expression of PPARγ2 and some of its target genes such as aP2 and Adiponectin (Supplemental Fig S5A), consistent with the observations made in preadipocytes, where GPs2 downregulation caused a reduction in the expression level of PPARγ2, and with the fact that inflammatory stimuli can transcriptionally downregulate PPARγ2 (Zhang et al., 1996). The increase in PPARγ expression was very limited though, and did not translate in a significant increase in the size of the adipose mass nor in total body weight (Supplemental Fig S5B and data not shown).
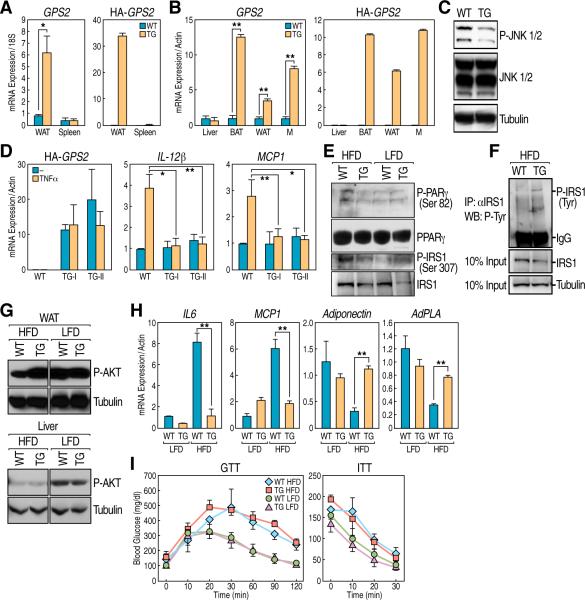
Figure 5. GPS2 anti-inflammatory role in the aP2-GPS2 mice.
(A) qPCRs for endogenous GPS2 and for HA tagged overexpressed form confirming expression in the WAT of ap2-GPS2 mice. Spleen is shown as a negative control. (B) qPCR confirming GPS2 is also overexpressed in BAT and macrophages (M) but not in liver. (C) P-JNK activation in the adipose tissue is inhibited in transgenic mice. (D) qPCR in bone marrow-derived macrophages: loss of IL12b and mcp1 upregulation by TNFα. (E) WB showing a decrease in serine phosphorylation of IRS1 and PPARγ. (F) Increase in tyrosine phosphorylation of IRS1 as indicated by immunoprecipitation of IRS1 followed by blot against P-Tyr. (G) Marked increase in AKT phosphorylation in the WAT from obese transgenic mice, no changes in the liver. (H) qPCR analysis of inflammatory cytokines, Adiponectin and AdPLA in the WAT of obese wt and transgenic mice. (I) Glucose Tolerant Test (GTT, left panel) and Insulin Tolerant Test (ITT, right panel) showing no difference in serum glucose levels, after glucose or insulin peritoneal injection, between HFD-fed transgenic and wt mice. All graphic data is +/- SD, * if P<0.05, ** if P<0.01, *** if P<0.001.
As expected, because of the aP2 expression pattern, we found GPS2 being overexpressed not only in the white and brown adipose tissue (WAT and BAT), but also in macrophages (Fig 5B). We, therefore, took advantage of the macrophage system and showed that GPS2 overexpression was sufficient to abrogate the activation of two major TNFα targets, IL-12β (IL12p40) and Mcp-1 (CCL2)(Ghisletti et al., 2009). This was equally observed in bone marrow-derived macrophages (Fig 5D) and in thioglycollate-induced macrophages (data not shown), indicating that GPS2 plays an important inhibitory role on the activation of the TNFα signaling pathway in vivo. Interestingly, a strong inhibition was also observed in the macrophages response to LPS stimulation (Supplemental Fig S5C), suggesting that GPS2 may regulate Ubc13/TRAF enzymatic activity during the activation of TLR receptors as well.
Next, we investigated whether GPS2 could play a protective role against the activation of pro-inflammatory pathways in a condition of diet-induced obesity, which are known to significantly contribute to the development of insulin resistance. For this purpose, wild-type and transgenic littermates were compared after >12 weeks of high-fat diet (HFD), whereas control animals from the same litter were kept on a low-fat diet (LFD). Consistent with the inhibition of JNK activity, serine phosphorylation of IRS1 was found significantly decreased and phosphorylation of PPARγ2 on Ser82 was also diminished in the WAT of aP2-GPS2 mice (Fig 5E). As a result, a significant improvement in the activation of the insulin pathway was observed in the adipose tissue of the HFD-fed transgenic mice, with a significant increase in IRS1 phosphorylation on Tyrosine residues (Fig 5F), and the activation of AKT being rescued to the same level of the control mice in LFD (Fig 5G). Conversely, no changes were observed in the liver, where GPS2 was not overexpressed (Fig 5G). In addition, the expression of pro-inflammatory cytokines characteristically associated with obesity-induced insulin resistance, such as IL6 and Mcp1, was markedly decreased in the obese transgenic mice when compared with their wild-type counterparts (Fig 5H). Conversely, the expression level of adiponectin, an insulin sensitizer adipokine secreted by the adipose tissue, was repressed in the wild-type mice in HFD, but rescued in the GPS2 transgenic mice to levels similar to the lean mice (Fig 5H). The insulin-induced activation of the adipose-specific phospholipase AdPLA, a key enzyme to inhibit lipolysis in response to feeding (Jaworski et al., 2009), was also rescued in the obese transgenic mice (Fig 5H). Together these results indicate that GPS2 overexpression in the fat and macrophages of diet-induced obese mice impairs pro-inflammatory responses, resulting into an improvement in activation of the insulin signaling pathway in the adipose tissue to levels comparable to lean mice. However, the aP2-GPS2 mice did not show the expected improvement in global insulin sensitivity, as indicated by glucose tolerance and insulin tolerance tests (Fig 5I).
Resistin overexpression and hepatic steatosis in aP2-GPS2 transgenic mice
In addition to this rather surprising result, GPS2 transgenic animals exhibited fatty livers, not only in diet-induced obese animal, but even when the mice were kept under a low fat diet. (Fig 6A). In HFD-fed mice, where the HFD alone is sufficient to induce mild steatosis in the wild type mice, GPS2 overexpression also strongly augmented the fatty liver phenotype (Fig 6A). Because the non-alcoholic fatty liver is often characterized by the development of hepatic inflammation, while GPS2 overexpression had proven to have an anti-inflammatory effect we investigated whether the hepatic steatosis would progress to steatohepatitis in the GPS2 transgenic mice, finding a significant decrease in the expression of the markers of macrophages activation, f4/80 and CD14 (Fig 6B). This suggested that GPS2 overexpression was driving two opposite effects – an increase in hepatic steatosis and overall insulin resistance, while still maintaining repression of the inflammatory responses. As GPS2 expression was specifically driven in the adipose and macrophage tissues, and not in the liver, we investigated the possibility that adipokines secreted from the WAT were responsible for the hepatic phenotype. In this regard, resistin was originally identified as an adipocyte-specific secreted factor associated with obesity and insulin resistance (Steppan et al., 2001) and found to be downregulated by TZDs and by TNFα(Fasshauer et al., 2001; Steppan et al., 2001). Moreover, elevated levels of resistin are associated with fatty liver, apoB secretion and increase in hepatocyte lipid content, while loss of resistin ameliorates hepatic steatosis in obese mice (Costandi et al., 2011; Singhal et al., 2008). Interestingly, we found extremely elevated levels of resistin in the plasma of GPS2 transgenic mice compared to their wild-type littermates (Fig 6C) and a significant increase in the expression of resistin in the adipose tissue, the main secreting organ in mice (Fig 6D). Consistent with the resistin upregulation and with the lack of improvement in glucose tolerance, we also observed downregulation of GLUT4 expression in the muscle of the aP2-GPS2 mice (Fig 6E).
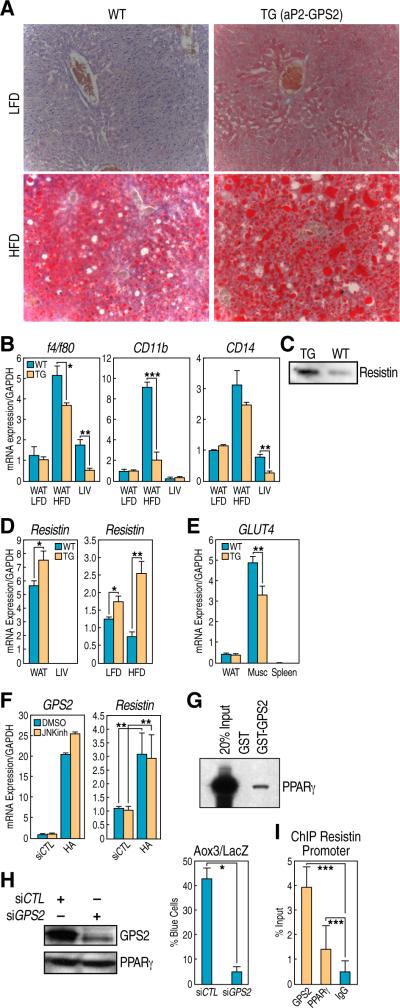
Figure 6. Hepatic steatosis and elevated Resistin levels in aP2-GPS2 mice.
(A) Oil Red O staining on liver sections: increase in lipid droplets accumulation in aP2-GPS2 mice. (B) qPCR analysis of macrophages markers f4/80, CD11b and CD14 in adipose and liver tissue. (C) Elevated level of Resistin in the plasma of GPS2 transgenic mice by WB (D) Increase in Resistin gene expression by qPCR in WAT. (E) Down-regulation of GLUT4 expression by qPCR in muscle. (F) qPCR analysis in 3T3-L1 cells: Resistin up-regulation by HA-GPS2 is independent of JNK. (G) Interaction between GPS2 and PPARγ by GST pull down. (H) PPARγ protein level is unchaged when GPS2 is downregulated after differentiation (right); PPARγ-dependent Aox3-LacZ reporter assay in single cell microinjection. (I) ChIP with GPS2 and PPARγ on Resistin promoter. All graphic data is +/- SD, * if P<0.05, ** if P<0.01, *** if P<0.001.
To confirm that GPS2 was primarily responsible for the increase in resistin expression, we tested in vitro, in 3T3-L1 cells, the effect of transient overexpression of GPS2. As indicated in Fig 6F, we observed a significant increase in resistin expression at the mRNA level, which was not dependent on the inhibition of JNK activity. Because this result suggested that GPS2 overexpression was not modulating resistin expression by inhibiting the TNFα/JNK pathway, we considered the alternative hypothesis of a transcriptional effect. In addition to its participation in the NCoR corepressor complex, GPS2 has also been recently described as a transcriptional coactivator for a series a nuclear receptors including FXR, SHP, HNF4 and LXR (Jakobsson et al., 2009; Sanyal et al., 2007). Similarly, we found that GPS2 could also directly interact with PPARγ (Fig 6G) and participate in mediating its transcriptional activity, as indicated by the loss in the activation of the PPARγ-dependent Aox3 promoter on GPS2 knock-down. Importantly, in this case GPS2 was removed by specific siRNA transfection only after several days of differentiation to ensure that PPARγ expression would not be affected (Fig 6H). Although PPARγ can repress resistin when activated by TDZs in mature adipocytes, the fat-specific expression of resistin during adipogenesis is positively regulated by PPARγ together with C/EBP factors (Tomaru et al., 2009), suggesting that overexpression of GPS2 may directly upregulate resistin expression by participating to a transcriptional activating complex with PPARγ and other transcription factors. Indeed, ChIP analysis performed in differentiating 3T3-L1 on the resistin promoter, where previous genome-wide analysis had indicated the presence of a strong PPARγ binding site (Lefterova et al., 2010; Mikkelsen et al.), confirmed GPS2 recruitement together with PPARγ (Fig 6I). Together these results indicate that GPS2 overexpression in the aP2-GPS2 mice causes a significant increase in Resistin expression, possibly because of a direct effect on PPARγ-mediated transcriptional regulation in the adipose tissue, which explain the development of fatty liver and the lack of improvement in systemic insulin sensitivity. In conclusion, GPS2 is required to mediate both non-transcriptional and transcriptional effects that are important to regulate inflammatory and metabolic responses, as indicated by the complex phenotype of the aP2-GPS2 mice.
DISCUSSION
GPS2 non-transcriptional anti-inflammatory role
The importance of maintaining a strict control on the inflammatory responses is emphasized by the large numbers of studies that, over the past 20 years, have provided convincing linkage between inflammation and numerous chronic illnesses, including atherosclerosis, cancer, diabetes, and neurodegenerative disease (Amor et al., 2010; Grivennikov et al., 2010; Hotamisligil, 2006; Lee and Pratley, 2005). In this manuscript, we have described an unexpected cytoplasmic function of the transcriptional cofactor GPS2 in the regulation of TNFR1 signaling and provided the initial evidence that GPS2 is a key component, both in vivo and in vitro, of the endogenous intracellular mechanisms responsible for preventing the uncontrolled activation of inflammatory gene programs. Intriguingly, a growing number of cofactors have lately been reported of playing non-transcriptional functions that, while mechanistically distinct, complement their roles in transcriptional regulation. This is consistent with the idea that cells can exploit the specific abilities of each factor by using them in different contexts. In the transmission of inflammatory signaling, this strategy has been adopted several times: Ubc13, for example, is an essential component of inflammatory receptors signaling, but its ubiquitin-conjugating enzymatic activity is also required in the nucleus, where Ubc13 functions as a chromatin modifying factor for DNA damage-induced ubiquitylation of histone H2A (Huen et al., 2007; Mailand et al., 2007). A similar dual strategy for GPs2 has emerged here, with this small protein being required for modulating the TNFR1 signaling complex at the plasma membrane, while also acting as a transcriptional cofactor in the nucleus.
Recent reports have indicated that the activation of the different signaling pathways activated in response to TNFR1 stimulation are distinctly regulated by specific ubiquitylation events and by the formation of different types of ubiquitin chains. The presence of Ubc13, the assembly of K63-ubiquitin chains and the integrity of the TRAF2 RING domain are all required for the activation of JNK, but they seem dispensable for the activation of IKK and NFκB, for which the formation of K48- and linear-ubiquitin chains is instead required (Bianchi and Meier, 2009; Habelhah, 2010). Our data are consistent with this specificity, with GPS2 inhibition of TRAF2/Ubc13 enzymatic activity translating into a significant decrease of JNK activation without affecting TNFα-induced stimulation of the NFκB canonical pathway, which may be important to avoid the deleterious effects of prolonged JNK activation, while ensuring that the prosurvival NFκB signaling pathway is not affected.
A remaining question is how the endogenous GPS2 is regulated during normal inflammatory responses. Transcriptional downregulation is certainly used to modulate the levels of GPS2 expression upon TNFα-induced activation (as shown in Fig 4B and Supplemental Fig S3B), but an additional level of regulation must exist to rapidly eliminate the inhibitory checkpoint when the inflammatory signal is first received from the cell, as the signaling complexes need to be available for a fast response. Interestingly, upon TNFα stimulation, GPS2 is itself post-translationally regulated by ubiquitylation and we would speculate that this step may be important to release the inhibition on TRAF2/Ubc13. Indeed, we found that only a very limited fraction of the overexpressed GPS2 protein is modified by ubiquitylation, suggesting that its strong inhibitory effect could be due to the inability of the cell to regulate its activity as for the endogenous protein. Further studies will be required to completely understand how the post-translational modifications of GPS2 can affect the regulation of TNFR signaling.
In Vivo Roles of GPS2
To determine the physiological importance of the anti-inflammatory role of GPS2 in vivo we generated transgenic mice overexpressing GPS2 in macrophages and adipose tissues. The strong inhibitory effect of GPS2 overexpression on pro-inflammatory pathways was recapitulated in vivo in macrophages where a limited increase in GPS2 expression level was sufficient to induce a significant impairment in the cell ability to respond to TNFα and LPS stimulation. Also, in the white adipose tissue of GPS2 transgenic mice we observed a significant improvement in the ability to respond to insulin stimulation, as indicated by the activation of different mediators of insulin signaling, by a decrease in the production of cytokines, and by significant increase in the expression of Adiponectin in the obese aP2-GPS2 mice. Significantly, overexpressing GPS2 was not sufficient to recapitulate the phenotypes observed in other mice models, in which directly suppressing JNK expression or activity in the adipose tissue fully rescued HFD-induced insulin resistance and glucose intolerance at the whole body level (Sabio et al., 2008; Zhang et al., 2011). However, we believe that this “apparent” contradiction reflects the complementary effects of GPS2 overexpression at the transcriptional and the non-transcriptional level. Thus, while the anti-inflammatory role of GPS2 in WAT and macrophages would dictate a positive effect on insulin sensitivity, the profound increase in circulating levels of Resistin in plasma, due to higher Resistin gene expression, correlates with the hepatic phenotype and with a decrease in glucose uptake that are normally observed in mouse models of insulin resistance. In this light, it is intriguing that while the aP2-GPS2 mice presented hepatic steatosis even under low dietary fat conditions, they did not develop steatohepatitis, as usually observed in the progression of the non-alcoholic liver disease (NAFLD). As JNK activity in hematopoietic cells is required for the progression to steatohepatitis and fibrosis (Kodama et al., 2009), we are tempted to speculate that considering the interplay between GPS2 anti-inflammatory role in macrophages and its ability to regulate resistin expression in WAT is critical to fully understand the resulting phenotype.
Finally, whereas the major finding in this manuscript is the identification of an unexpected cytosolic role for GPS2 in the regulation of ubiquitin signaling, the results of this study does not diminish the importance of the roles played by GPS2 as a nuclear transcriptional cofactor and actually provide an in vivo validation of their relevance by identifying Resistin as a direct target of GPS2 transcriptional regulation in the adipose tissue.
GPS2 as a transcriptional cofactor
From a biochemical point of view, we have mapped GPS2 chromatin binding genome-wide and revealed a large number of regulatory regions where GPS2 is recruited to chromatin either as a component of the NCoR corepressor complex or by itself. This is consistent with the emerging picture that GPS2 can work as a coactivator as well as a corepressor for nuclear receptors and other transcription factors. In the case of nuclear receptors, GPS2 was reported to be recruited either indirectly via the corepressors or directly by the receptor (Jakobsson et al., 2009; Sanyal et al., 2007). Our data indicate that both strategies are important in the regulation of PPARγ transcriptional activity, even though the most striking effect in this study was the almost complete loss of PPARγ expression upon GPS2 downregulation due to early activation of the TNFα signaling pathway. Thus, we believe that the regulation of PPARγ exemplifies how cells can use the same factor, in this case GPS2, to regulate a biological output by acting at different levels and in different cellular compartments. GPS2 cytosolic actions are indeed important in differentiating adipocytes to keep the TNFα pro-inflammatory pathway under negative regulation, therefore allowing PPARγ expression and avoiding inhibition of PPARγ transcriptional activity by JNK-dependent phosphorylation, while in the nucleus GPS2 modifies PPARγ transcriptional regulation acting both as a corepressor within the NCoR complex, and also as a dedicated coactivator.
In conclusion, this study has uncovered an unexpected and hopefully important strategy that serves as a guardian function to prevent cells from unleashing a hyper-inflammatory response to TNFα signaling. The pivotal role of GPS2 acting at the level of the plasma membrane to control a central inflammatory signaling pathway, in concert with its nuclear roles, uncovers a bi-functional strategy for normal cellular and metabolic regulation, with intriguing implications for metabolic diseases.
EXPERIMENTAL PROCEDURES
Generation of transgenic mice and metabolic studies
GPS2 was expressed under regulation of the aP2 promoter region (Dr. Kahn, Addgene plasmid 11424). In the diet-induced obesity studies pairs of same sex mice were fed high fat diet (HFD, 45% of calories from fat, D12451; Research Diets) or low fat diet (LFD, 15% of calories from fat, D12450B; Research Diets) up to 20 weeks. To measure insulin sensitivity, mice were injected with insulin (Humulin-R, 0.75u/kg body weight) and tissues harvested 30 min later for protein and RNA extraction.
In vitro ubiquitination and protein interaction assays
Ubiquitination assays were carried out with bacterially purified TRAF2 and GPS2, recombinant E1 and E2. See Supplemental Information for details on protein purification and immunoprecipitation techniques.
RNA-Sequencing and ChIP-sequencing
For preparation of RNA-seq samples 293T cells were subjected to standard RNA isolation and libraries were constructed with RNA-Seq Sample Preparation kit (Illumina) prior to deep sequencing on a Solexa Genome Analyzer II. Image analysis and base calling were done by using Illumina Pipeline, using Ibis for improved base calling (Kircher et al., 2009). Similarly, for ChIP-Seq sample preparation 293T cells were subjected to standard ChIP followed by library preparation and sequencing. Description of bioinformatic analysis is in the Supplemental Information.
Supplementary Material
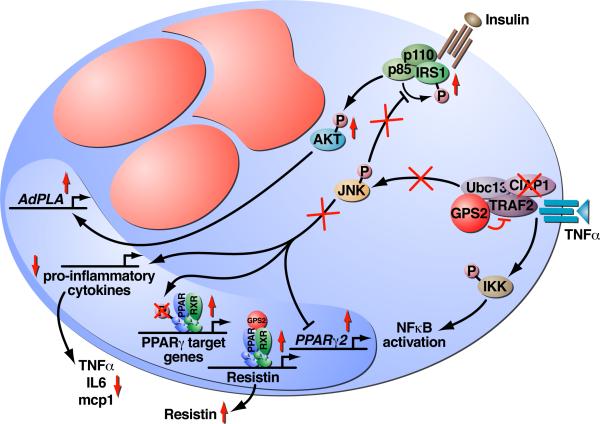
Figure 7. Model of GPS2 functions in the inhibition of JNK activity.
Schematic of an adipocyte cell with large red lipid droplets, indicating that overexpressed GPS2 binds to TRAF2 and Ubc13 at the level of the TNFR1 membrane complex and inhibits their enzymatic activity. Recruitment of cIAP1 is also inhibited, resulting in specific downregulation of JNK activity while the canonical NFκB pathway is not affected. Inhibition of JNK activation ameliorates insulin signaling, with IRS1 being correctly phosphorylated on Tyr residues in response to insulin stimulation and with the activation of the AKT kinase leading to correct upregulation of insulin target genes (i.e. AdPLA). Conversely, inhibition of the inflammatory response leads to downregulation of the expression of pro-inflammatory cytokines. GPS2 anti-inflammatory role contributes also to license PPARγ transcriptional activity by preventing PPARγ2 downregulation and inhibiting JNK-dependent phosphorylation of PPARγ, whereas GPS2 recruitment to regulatory units is important for PPARγ-mediated transcriptional activation.
ACKNOWLEGDMENTS
We are grateful to J. Wang and C. Nelson for excellent technical assistance; Dr. W. Huang and Dr. K. Saijo for macrophages protocols; Dr. A. Aggarwaal for insightful discussions; J. Hightower for figure preparation. M.G.R. is an HHMI investigator, V.P. is supported by NIDDK (grant ROODK078756), M.D.C. is supported by the Susan G. Komen Foundation. Research supported by DK074868, DK039949, HL065445, NS047101, DK018477 to MGR and CKG.
Footnotes
Accession Numbers
Datasets for RNA-seq and ChIP-seq experiments are available at:
REFERENCES
- Amor S, Puentes F, Baker D, van der Valk P. Inflammation in neurodegenerative diseases. Immunology. 2010;129:154–169. doi: 10.1111/j.1365-2567.2009.03225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ, Barker PA. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- Bianchi K, Meier P. A tangled web of ubiquitin chains: breaking news in TNF-R1 signaling. Mol Cell. 2009;36:736–742. doi: 10.1016/j.molcel.2009.11.029. [DOI] [PubMed] [Google Scholar]
- Cawthorn WP, Sethi JK. TNF-alpha and adipocyte biology. FEBS Lett. 2008;582:117–131. doi: 10.1016/j.febslet.2007.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- Costandi J, Melone M, Zhao A, Rashid S. Human resistin stimulates hepatic overproduction of atherogenic ApoB-containing lipoprotein particles by enhancing ApoB stability and impairing intracellular insulin signaling. Circ Res. 2011;108:727–742. doi: 10.1161/CIRCRESAHA.110.238949. [DOI] [PubMed] [Google Scholar]
- Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17:666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- Fasshauer M, Klein J, Neumann S, Eszlinger M, Paschke R. Tumor necrosis factor alpha is a negative regulator of resistin gene expression and secretion in 3T3-L1 adipocytes. Biochem Biophys Res Commun. 2001;288:1027–1031. doi: 10.1006/bbrc.2001.5874. [DOI] [PubMed] [Google Scholar]
- Fukushima T, Matsuzawa S, Kress CL, Bruey JM, Krajewska M, Lefebvre S, Zapata JM, Ronai Z, Reed JC. Ubiquitin-conjugating enzyme Ubc13 is a critical component of TNF receptor-associated factor (TRAF)-mediated inflammatory responses. Proc Natl Acad Sci U S A. 2007;104:6371–6376. doi: 10.1073/pnas.0700548104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habelhah H. Emerging complexity of protein ubiquitination in the NF-kappaB pathway. Genes Cancer. 2010;1:735–747. doi: 10.1177/1947601910382900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habelhah H, Takahashi S, Cho SG, Kadoya T, Watanabe T, Ronai Z. Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-kappaB. EMBO J. 2004;23:322–332. doi: 10.1038/sj.emboj.7600044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, Karin M, Hotamisligil GS. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Hu E, Kim JB, Sarraf P, Spiegelman BM. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARgamma. Science. 1996;274:2100–2103. doi: 10.1126/science.274.5295.2100. [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson T, Venteclef N, Toresson G, Damdimopoulos AE, Ehrlund A, Lou X, Sanyal S, Steffensen KR, Gustafsson JA, Treuter E. GPS2 is required for cholesterol efflux by triggering histone demethylation, LXR recruitment, and coregulator assembly at the ABCG1 locus. Mol Cell. 2009;34:510–518. doi: 10.1016/j.molcel.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Jaworski K, Ahmadian M, Duncan RE, Sarkadi-Nagy E, Varady KA, Hellerstein MK, Lee HY, Samuel VT, Shulman GI, Kim KH, et al. AdPLA ablation increases lipolysis and prevents obesity induced by high-fat feeding or leptin deficiency. Nat Med. 2009;15:159–168. doi: 10.1038/nm.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Stenzel U, Kelso J. Improved base calling for the Illumina Genome Analyzer using machine learning strategies. Genome Biol. 2009;10:R83. doi: 10.1186/gb-2009-10-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y, Kisseleva T, Iwaisako K, Miura K, Taura K, De Minicis S, Osterreicher CH, Schnabl B, Seki E, Brenner DA. c-Jun N-terminal kinase-1 from hematopoietic cells mediates progression from hepatic steatosis to steatohepatitis and fibrosis in mice. Gastroenterology. 2009;137:1467–1477. e1465. doi: 10.1053/j.gastro.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Yi W, Griswold MD, Zhu F, Her C. Formation of hMSH4-hMSH5 heterocomplex is a prerequisite for subsequent GPS2 recruitment. DNA Repair (Amst) 2006;5:32–42. doi: 10.1016/j.dnarep.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Lee YH, Pratley RE. The evolving role of inflammation in obesity and the metabolic syndrome. Curr Diab Rep. 2005;5:70–75. doi: 10.1007/s11892-005-0071-7. [DOI] [PubMed] [Google Scholar]
- Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G, Manduchi E, Grant GR, Lazar MA. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol Cell Biol. 2010;30:2078–2089. doi: 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chen ZJ. Expanding role of ubiquitination in NF-kappaB signaling. Cell Res. 2010;21:6–21. doi: 10.1038/cr.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney DJ, Cheung HH, Mrad RL, Plenchette S, Simard C, Enwere E, Arora V, Mak TW, Lacasse EC, Waring J, et al. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci U S A. 2008;105:11778–11783. doi: 10.1073/pnas.0711122105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YC, Breiding DE, Sverdrup F, Richard J, Androphy EJ. AMF-1/Gps2 binds p300 and enhances its interaction with papillomavirus E2 proteins. J Virol. 2000;74:5872–5879. doi: 10.1128/jvi.74.13.5872-5879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YC, Kuo F, Breiding DE, Wang YF, Mansur CP, Androphy EJ. AMF1 (GPS2) modulates p53 transactivation. Mol Cell Biol. 2001;21:5913–5924. doi: 10.1128/MCB.21.17.5913-5924.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Rangwala SM, Rhoades B, Shapiro JS, Rich AS, Kim JK, Shulman GI, Kaestner KH, Lazar MA. Genetic modulation of PPARgamma phosphorylation regulates insulin sensitivity. Dev Cell. 2003;5:657–663. doi: 10.1016/s1534-5807(03)00274-0. [DOI] [PubMed] [Google Scholar]
- Ross SR, Graves RA, Greenstein A, Platt KA, Shyu HL, Mellovitz B, Spiegelman BM. A fat-specific enhancer is the primary determinant of gene expression for adipocyte P2 in vivo. Proc Natl Acad Sci U S A. 1990;87:9590–9594. doi: 10.1073/pnas.87.24.9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H, Hacohen N, Golub TR, Van Parijs L, Lodish HF. Tumor necrosis factor-alpha suppresses adipocyte-specific genes and activates expression of preadipocyte genes in 3T3-L1 adipocytes: nuclear factor-kappaB activation by TNF-alpha is obligatory. Diabetes. 2002;51:1319–1336. doi: 10.2337/diabetes.51.5.1319. [DOI] [PubMed] [Google Scholar]
- Sabio G, Das M, Mora A, Zhang Z, Jun JY, Ko HJ, Barrett T, Kim JK, Davis RJ. A stress signaling pathway in adipose tissue regulates hepatic insulin resistance. Science. 2008;322:1539–1543. doi: 10.1126/science.1160794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabio G, Davis RJ. cJun NH2-terminal kinase 1 (JNK1): roles in metabolic regulation of insulin resistance. Trends Biochem Sci. 2010;35:490–496. doi: 10.1016/j.tibs.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S, Bavner A, Haroniti A, Nilsson LM, Lundasen T, Rehnmark S, Witt MR, Einarsson C, Talianidis I, Gustafsson JA, et al. Involvement of corepressor complex subunit GPS2 in transcriptional pathways governing human bile acid biosynthesis. Proc Natl Acad Sci U S A. 2007;104:15665–15670. doi: 10.1073/pnas.0706736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silke J, Brink R. Regulation of TNFRSF and innate immune signalling complexes by TRAFs and cIAPs. Cell Death Differ. 2009;17:35–45. doi: 10.1038/cdd.2009.114. [DOI] [PubMed] [Google Scholar]
- Singhal NS, Patel RT, Qi Y, Lee YS, Ahima RS. Loss of resistin ameliorates hyperlipidemia and hepatic steatosis in leptin-deficient mice. Am J Physiol Endocrinol Metab. 2008;295:E331–338. doi: 10.1152/ajpendo.00577.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain BH, Bowdish KS, Pacal AR, Staub SF, Koo D, Chang CY, Xie W, Colicelli J. Two human cDNAs, including a homolog of Arabidopsis FUS6 (COP11), suppress G-protein- and mitogen-activated protein kinase-mediated signal transduction in yeast and mammalian cells. Mol Cell Biol. 1996;16:6698–6706. doi: 10.1128/mcb.16.12.6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppan CM, Bailey ST, Bhat S, Brown EJ, Banerjee RR, Wright CM, Patel HR, Ahima RS, Lazar MA. The hormone resistin links obesity to diabetes. Nature. 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- Sun SC. Deubiquitylation and regulation of the immune response. Nat Rev Immunol. 2008;8:501–511. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaru T, Steger DJ, Lefterova MI, Schupp M, Lazar MA. Adipocyte-specific expression of murine resistin is mediated by synergism between peroxisome proliferator-activated receptor gamma and CCAAT/enhancer-binding proteins. J Biol Chem. 2009;284:6116–6125. doi: 10.1074/jbc.M808407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng PH, Keats JJ, Wang H, Vignali DA, Bergsagel PL, Karin M. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol. 2008;9:1364–1370. doi: 10.1038/ni.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele P, Declercq W, Van Herreweghe F, Vanden Berghe T. The role of the kinases RIP1 and RIP3 in TNF-induced necrosis. Sci Signal. 3:re4. doi: 10.1126/scisignal.3115re4. [DOI] [PubMed] [Google Scholar]
- Varfolomeev E, Goncharov T, Fedorova AV, Dynek JN, Zobel K, Deshayes K, Fairbrother WJ, Vucic D. c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. J Biol Chem. 2008;283:24295–24299. doi: 10.1074/jbc.C800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vince JE, Pantaki D, Feltham R, Mace PD, Cordier SM, Schmukle AC, Davidson AJ, Callus BA, Wong WW, Gentle IE, et al. TRAF2 must bind to cellular inhibitors of apoptosis for tumor necrosis factor (tnf) to efficiently activate nf-{kappa}b and to prevent tnf-induced apoptosis. J Biol Chem. 2009;284:35906–35915. doi: 10.1074/jbc.M109.072256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WW, Gentle IE, Nachbur U, Anderton H, Vaux DL, Silke J. RIPK1 is not essential for TNFR1-induced activation of NF-kappaB. Cell Death Differ. 2009;17:482–487. doi: 10.1038/cdd.2009.178. [DOI] [PubMed] [Google Scholar]
- Xu H, Sethi JK, Hotamisligil GS. Transmembrane tumor necrosis factor (TNF)-alpha inhibits adipocyte differentiation by selectively activating TNF receptor 1. J Biol Chem. 1999;274:26287–26295. doi: 10.1074/jbc.274.37.26287. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Okamoto T, Takeda K, Sato S, Sanjo H, Uematsu S, Saitoh T, Yamamoto N, Sakurai H, Ishii KJ, et al. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat Immunol. 2006;7:962–970. doi: 10.1038/ni1367. [DOI] [PubMed] [Google Scholar]
- Yu C, Markan K, Temple KA, Deplewski D, Brady MJ, Cohen RN. The nuclear receptor corepressors NCoR and SMRT decrease peroxisome proliferator-activated receptor gamma transcriptional activity and repress 3T3-L1 adipogenesis. J Biol Chem. 2005;280:13600–13605. doi: 10.1074/jbc.M409468200. [DOI] [PubMed] [Google Scholar]
- Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J, Shiba T, Yang X, Yeh WC, Mak TW, et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. 2008;9:1371–1378. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Berger J, Hu E, Szalkowski D, White-Carrington S, Spiegelman BM, Moller DE. Negative regulation of peroxisome proliferator-activated receptor-gamma gene expression contributes to the antiadipogenic effects of tumor necrosis factor-alpha. Mol Endocrinol. 1996;10:1457–1466. doi: 10.1210/mend.10.11.8923470. [DOI] [PubMed] [Google Scholar]
- Zhang D, Harry GJ, Blackshear PJ, Zeldin DC. G-protein pathway suppressor 2 (GPS2) interacts with the regulatory factor X4 variant 3 (RFX4_v3) and functions as a transcriptional co-activator. J Biol Chem. 2008;283:8580–8590. doi: 10.1074/jbc.M708209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- Zhang L, Blackwell K, Shi Z, Habelhah H. The RING domain of TRAF2 plays an essential role in the inhibition of TNFalpha-induced cell death but not in the activation of NF-kappaB. J Mol Biol. 2010;396:528–539. doi: 10.1016/j.jmb.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xu A, Chung SK, Cresser JH, Sweeney G, Wong RL, Lin A, Lam KS. Selective Inactivation of c-Jun NH2-Terminal Kinase in Adipose Tissue Protects Against Diet-Induced Obesity and Improves Insulin Sensitivity in Both Liver and Skeletal Muscle in Mice. Diabetes. 2011;60:486–495. doi: 10.2337/db10-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Kabaleeswaran V, Wang Y, Cheng G, Wu H. Crystal structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 complexes: affinity, specificity, and regulation. Mol Cell. 2010;38:101–113. doi: 10.1016/j.molcel.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.