Abstract
Differentiation of insulin producing beta-cells is a genetically well defined process that involves functions of various conserved transcription factors. Still, the transcriptional mechanisms underlying specification and determination of beta-cell fate are poorly defined. Here we provide the description of a beta-cell progenitor specific enhancer as a model to study initial steps of beta-cell differentiation. We show that evolutionary non-conserved upstream sequences of the zebrafish hb9 gene are required and sufficient for regulating expression in beta-cells prior to the onset of insulin expression. This enhancer contains binding sites for paired-box transcription factors and two E-boxes that in EMSA studies show interaction with Pax6b and NeuroD, respectively. We show that Pax6b is a potent activator of endodermal hb9 expression and that this activation depends on the beta-cell enhancer. Using genetic approaches we show that pax6b is crucial for maintenance but not induction of pancreatic hb9 transcription. As loss of Pax6b or Hb9 independently results in the loss of insulin expression, the data reveal a novel cross-talk between the two essential regulators of early beta-cell differentiation. While we find that the known pancreatic E-box binding proteins NeuroD and Ngn3 are not required for hb9 expression we also show that removal of both E-boxes selectively eliminates pancreatic specific reporter expression. The data provide evidence for an Ngn3 independent pathway of beta-cell specification that requires function of currently not specified E-box binding factors.
Highlights
► Beta-cell progenitor enhancer localizes to non-conserved hb9 promoter sequences. ► E-boxes are required for hb9 induction. ► Induction is independent of Ngn3 and NeuroD. ► Pax6b activates enhancer. ► Pax6b is required for hb9 maintenance.
Introduction
The Hb9/Hlxb9/Mnx1 gene encodes a homeobox transcription factor that in vertebrates has conserved functions in motoneuron differentiation and pancreas development (Broihier and Skeath, 2002; Ferrier et al., 2001; Grapin-Botton et al., 2001; Tanabe et al., 1998; Wendik et al., 2004). Furthermore, in humans, the heterozygous loss of HLXB9 function is the major cause for sacral agenesis in patients with Currarino syndrome (Ross et al., 1998). Consistent with these diverse functions, Hb9 genes in all vertebrates show a complex pattern of expression that is conserved in motoneurons, pancreatic endoderm, differentiating beta-cells and different mesodermal structures including the posterior notochord (Harrison et al., 1999; Li and Edlund, 2001; Li et al., 1999; Ross et al., 1998; Saha et al., 1997; Tanabe et al., 1998; Thaler et al., 1999; Wendik et al., 2004). While Hb9 genes have been subject to various promoter studies (Lee et al., 2004; Nakano et al., 2005; Woolfe et al., 2005), not much is known about the molecular mechanism underlying their complex regulation. Previous studies focused on highly conserved non-coding sequence motifs identified in comparative genome promoter studies. The conservation is most striking in two 5 prime sequences, a distal 320 bp motif termed the A-box and a more proximal motif of about 150 bp termed the B-box (Nakano et al., 2005) (Fig. 1A). As these motifs show 74–94% identity in diverse species such as zebrafish and human it was suggested that they might have functions in gene regulation. Consistent with this notion, studies in mouse showed that the B-box and a second more proximal, mammalian-specific conserved motif termed MNE are sufficient and required for targeting reporter expression to motoneurons (Lee and Pfaff, 2003; Lee et al., 2004; Nakano et al., 2005). However, sequence motifs related to MNE have not been detected outside of mammals (Lee and Pfaff, 2003; Lee et al., 2004; Nakano et al., 2005) and mechanisms underlying hb9 regulation in non-mammalian organisms and in endodermal and mesodermal structures remain to be investigated.
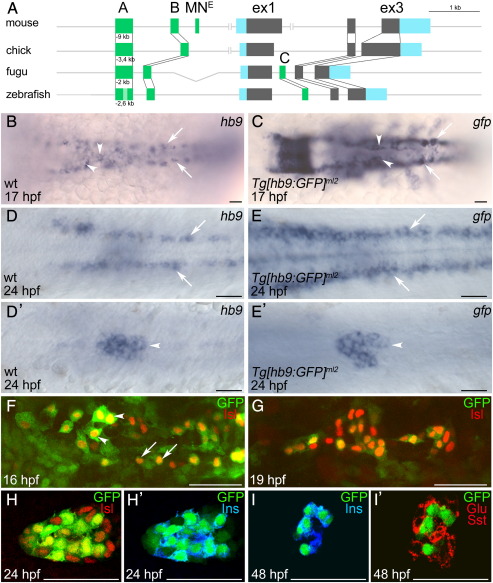
Fig. 1.
Motoneuron and beta-cell specific GFP expression in Tg[hb9:GFP]ml2 embryos. (A) Schematic representation of the hb9 gene from different species. Highlighted are conserved non-coding regions (dark green), and exons (coding sequences in grey, untranslated regions in blue). (B–I′) In situ hybridization analysis comparing mRNA expression of hb9 (B, D, D′) and gfp (C, E, E′) in Tg[hb9:GFP]ml2 embryos at 17 hpf (B, C) and 24 hpf (D, E) reveals similar pattern in motoneurons and ventral interneurons (B–E, white arrows) and pancreas (B, C, white arrowheads). Asterisk in C marks ectopic gfp expression in rhombomere. D′, E′ show different focal planes of the embryos shown in D, E. (F–I) Confocal image projections of Tg[hb9:GFP]ml2 embryos labelled with GFP (green) and islet antibodies (red) at 16 hpf (F), 19 hpf (G) and 24 hpf (H) show overlapping expression of GFP and islet proteins in motoneurons (arrows) and in a subset of endocrine cells (arrowheads). (H–I′) Confocal image stacks of Tg[hb9:GFP]ml2 embryos labelled with antibodies for GFP and pancreatic hormones at 24 hpf (H′) and 48 hpf (I, I′) show that GFP overlaps with insulin (blue in H′, I) but not with glucagon (Glu) and somatostatin (Sst) (Glu + Sst in red, I′). Embryos are shown in dorsal (B–E) or ventral (F–I) view with anterior to the left. Scale bars in all figures correspond to 50 μm.
Here we use the zebrafish as a model to study hb9 regulatory elements with a focus on pancreas-specific regulation. Hb9 genes in zebrafish and mouse are expressed during two distinct phases of pancreas development (Harrison et al., 1999; Li et al., 1999; Wendik et al., 2004). Before onset of pancreatic organ morphogenesis, Hb9 is expressed in the entire prospective pancreatic endoderm. With the onset of organ morphogenesis, expression is first downregulated and later reactivated specifically in differentiating beta-cells. Knock-out studies in mouse revealed independent requirements for the early and late pancreatic Hb9 expression in initiating pancreas morphogenesis and in regulating beta-cell maturation, respectively. In particular, one of the pancreatic anlagen, the dorsal bud, is missing in the mutants and the second, ventral bud, is formed but fails to establish mature beta-cells as revealed by reduced expression of the early beta-cell differentiation marker insulin and loss of the late differentiation marker Glut2 (Harrison et al., 1999; Li et al., 1999). In zebrafish, morpholino knock down of hb9 results in a strong reduction or even loss of embryonic insulin expression while early pancreas morphogenesis appears unaffected (Wendik et al., 2004). The data suggest that only the later beta-cell specific expression of hb9 is associated with an evolutionary conserved function in beta-cell differentiation and maturation. Consistent with the requirement for hb9 upstream of insulin, the onset of hb9 expression in beta-cells precedes that of insulin (Wendik et al., 2004). This makes hb9 one of the earliest specific markers for differentiating beta-cells and suggests that hb9 expression is linked with initial steps of beta-cell specification.
Beta-cell formation requires activities of a conserved network of cross-regulating transcription factors which interact during pancreas development to establish the characteristic expression programmes for the distinct pancreatic cell types (Edlund, 2002; Habener et al., 2005; Kinkel and Prince, 2009; Schwitzgebel, 2001; Wilson et al., 2003). Genetic studies in mouse identified more than a dozen transcription factors that contribute to this network and that are required for proper differentiation of beta-cells. So far only a few of these factors have been studied in the context of zebrafish pancreas development. Among these, the homeobox-containing proteins Nkx6.1 and Nkx6.2 are required for defining a beta-cell progenitor pool (Binot et al., 2010), while Nkx2.2a and Pax6b are essential for beta-cell fate determination (Pauls et al., 2007; Verbruggen et al., 2010). Ngn3/Neurog3, a basic helix–loop–helix (bHLH) factor that in mouse is a key regulator of endocrine cell differentiation was reported to be expressed in the larval but not the embryonic zebrafish pancreas (Apelqvist et al., 1999; Gradwohl et al., 2000; Jenny et al., 2002; Kinkel and Prince, 2009; Moro et al., 2009; Rukstalis and Habener, 2009; Schwitzgebel et al., 2000; Zecchin et al., 2007). The bHLH factor NeuroD/Beta2, that is expressed, similar to the situation in mouse (Huang et al., 2002; Itkin-Ansari et al., 2005; Naya et al., 1997), in all endocrine precursors, was found to be missing in nkx6.1/nkx6.2 double morphants (Binot et al., 2010) and reduced in nkx2.2a single morphant zebrafish embryos (Pauls et al., 2007). Notably, reduction of neuroD expression in these morphants correlates with the loss of insulin expression, which is consistent with a possible function of NeuroD in beta-cell fate determination downstream of the nkx genes.
We have previously generated a hb9:GFP transgenic zebrafish line Tg[hb9:GFP]ml2 (also known as Tg[mnx1:GFP]ml2) in which 3.1 kb of hb9 genomic sequences, including the A- and B-box, are sufficient to recapitulate several aspects of hb9 expression (Flanagan-Steet et al., 2005; Kimmel and Meyer, 2010). In contrast to the currently available Hb9 mouse promoter lines (Arber et al., 1999; Nakano et al., 2005; Thaler et al., 1999; Wichterle et al., 2002), these fish also show reporter expression in the pancreas, specifically recapitulating the functionally conserved hb9 expression in beta-cells. Here we provide the first characterization of an hb9 beta-cell enhancer element and of upstream transcription factors mediating the beta-cell specific expression. We show that in zebrafish the hb9 beta-cell enhancer element localizes to 478 bp of non-conserved distal sequences and we provide molecular and genetic evidence for a direct binding of the endocrine progenitor factors NeuroD and Pax6b on this element. Our data reveal distinct functions of the 478 bp motive in establishing and maintaining beta-cell specific expression, further indicating that the hb9 pancreas enhancer represents a promising model for detailed analyses of the molecular regulation of beta-cell fate specification and determination.
Materials and methods
Transgenic lines and fish maintenance
The Tg[ins:dsRed] transgenic line (Shin et al., 2008) was used for injection of DNA constructs. AB strain and Tg[hb9:GFP]ml2 transgenic line were used for mRNA and morpholino injections. The pax6bsa0086 allele (Verbruggen et al., 2010) was obtained from the Sanger Center.
DNA constructs and promoter analysis
The zebrafish hb9 promoter fragments were initially isolated from PAC clones that were identified by hybridizing with a radioactive labelled hb9 cDNA. Southern analysis of the PAC DNA revealed the presence of the 5′ UTR of hb9 on a 5.3 kb EcoRI fragment. This fragment was sub-cloned into pBluescript and sequenced on an ABIPrism 310 Genetic Analyzer (ABI Prism® BigDye™ Terminator, Applied Biosystems). Sequencing of the amplified hb9 promoter revealed an 8 bp deletion at − 1558 bp and 2 substitutions at − 1046 and − 717. Conserved promoter regions of the hb9 gene were identified by comparing the zebrafish, mouse, human, chick and Fugu locus by BLAST searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and ClustalW2 Multiple Sequence Alignment (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Potential binding sites were identified by Match-1.0 Public, Patch 1.0 and P-Match-Public 1.0 Public programmes (www.gene-regulation.com).
hb9pG1 vector was generated by cloning the 3.1 kb hb9 promoter into the pG1 vector (Gilmour et al., 2002) by using EcoRI/BamHI restriction enzymes. The hb9-deletion constructs were generated based on hb9pG1 vector by using the indicated restriction enzymes. hb9NheI:GFP_pG1 was generated by introducing a NheI restriction site into the PE/A-box boundary with the Site-Directed Quick-Change Mutagenesis Kit (Stratagene). From this constructs hb9∆NheI:GFP and hb9∆A:GFP were generated by cutting with EcoRI/NheI and NheI/EagI respectively and blunting with T4 Polymerase (Fermentas, EP0061) and religation. For generation of hb9∆1:GFP and hb9∆2:GFP constructs PCR was used to amplify the desired elements and PCR-fragments were cloned upstream of A-Box into hb9NheI:GFP_pG1. hb9∆1:GFP was generated with hb9_EcoRI_F (GAATTCCTGAGGAAAACGCTTTCG) and NheI_R (TCGATCATGCTAGCACAAATGGGCCGTGC) primers, hb9∆2:GFP was generated with SalI_F (GTCGACATTTAAATTAGCCTGGCATCTGG) and dr_hb9_NheI_R (GCTAGCCAGTTTGCTTTGATAAGCC) primers. Quick-Change Mutagenesis Kit (Stratagene) was used for the generation of hb9Em:GFP. The following primers were used: NrD_mut_F (CATTTAAATTAGCCTGGAGATCTGACATCGTCAATCACG), NrD_mut_R (CGTGATTGACGATGTCAGATCTCCAGGCTAATTTAAATG), E47mut_F (GTGTTCGGTTAAGTAAACTAGTGTTTTCGGGCTGGTTC) and E47mut_R (GAACCAGCCCGAAAACACTAGTTTACTTAACCGAACAC).
For generation of hb9PE229:gfp DNA the PE229 fragment was PCR-amplified by using SalI_F and NheI_R primers and cloned into hb9pG1, linearized with SalI/StuI. For generation of hb9∆AB:GFP plasmid hb9NheI:GFP_pG1 was digested with NheI/StuI and religated. For cloning of reporter vectors PE+ hsp:GFP, A+hsp:GFP and PE+ A+hsp:GFP zebrafish hsp70 promoter was first amplified by PCR with the following primers: hsp70_StuI_F (AGGCCTTCAGGGGTGTCGCTTGGT) and hsp70_BamHI_R (GGATCCTTGTACAAACTTGGCAGGAA). PE+ hsp:GFP construct was generated by cloning hsp70 PCR fragment into hb9NheI:GFP_pG1 after cutting of last with NheI/BamHI and blunt of NheI-site. For generation of A+hsp:GFP hsp70 was cloned into hb9∆NheI:GFP_pG1 after serial cutting with AvrII/BamHI and blunt of AvrII site. For generation of PE+ A+hsp:GFP reporter construct hsp-PCR fragment was cloned into hb9pG1 after digest with AvrII/BamHI. Before injection promoters, except vectors showed on Fig. 3, A, were cloned into pT2KXIG (Kawakami, 2004) vector by restriction with XhoI/BamHI (vector) and SalI/BamHI (promoter).
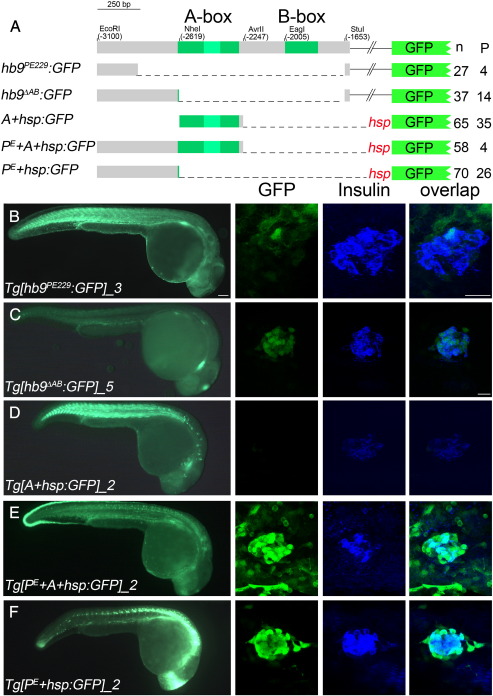
Fig. 3.
478 bp of proximal hb9 promoter sequences are sufficient for beta-cell specific regulation. (A) Overview of transient GFP reporter experiments with a scheme of deletion constructs on the left and corresponding numbers of GFP positive embryos (n) and embryos expressing GFP in the pancreas (P) on the right side. (B–F) GFP expression in representative transgenic lines at 28 hpf in whole embryos (left panel) and in fixed tissue (right panel) stained with antibodies for GFP (green) and insulin (blue). The right panel shows confocal image projections and the corresponding overlays (right-most panel). Scale bar corresponds to 100 μm (whole embryos) and 20 μm (confocal image stacks). For more details see also Supplementary Table 2.
Generation of transgenic zebrafish
Tg[hb9:GFP]ml2 transgenic line was generated as described by Flanagan-Steet et al. (2005). For microinjection of hb9:GFP, hb9∆A:GFP, hb9∆NheI:GFP, hb9∆1:GFP, hb9∆2:GFP, hb9PE229:GFP, hb9∆AB:GFP, PE+ hsp:GFP, A+hsp:GFP, PE+ A+hsp:GFP and hb9Em:GFP constructs the intact Tol2 vectors, containing corresponding hb9 promoter were injected into 1-cell stage Tg[ins:dsRed] zebrafish embryos together with transposase mRNA. GFP-positive embryos were raised to adulthood and screened for transgene transmission to their progeny.
Microinjection of DNA, mRNA and morpholinos
For microinjection of constructs shown on Fig. 3, A, DNA were linearized with NotI and purified DNA were injected into one-cell stage zebrafish embryos. Knock-down experiments were performed with published splicing morpholinos against pax6b (Mo2Pax6b (in this text called MOpax6b), 5′-TTGATTTGCACTCACGCTCGGTATG) (Verbruggen et al., 2010), ATG morpholino for neuroD (nrdMO) (in this text called MOneuroD: TGACTTCGTCATGTCGGAACTCTAG) (Sarrazin et al., 2006), and ATG morpholino for ngn3 (MOngn3: GGATCTTGGAGTCATTCTCTTGCAA). Morpholino was injected into 1-cell stage AB-strain or Tg[hb9:GFP]ml2 zebrafish embryos.
For induction experiments single-cell injection was performed at 16–32 cell stage in AB-strain, Tg[hb9:GFP]ml2 and Tg[hb9∆NheI:GFP]. The following mRNA were injected: Casanova (cas_pCS2 +), nucRFP (h2b-RFP Red_pCS2+; obtained from W. Driever), Pax6b-HA (Pax6b-HA_pCS2 +) and NeuroD-Flag (NeuroD-Flag_pCS2 +). For mRNA synthesis vectors were restricted with NotI and mRNA was produced by using Sp6 mMessage mMachine Kit, Ambion.
EMSA
The following oligos were used for EMSA (only forward primers are listed, reverse primers present the same sequence but reverse and complement):
E1 (CATTTAAATTAGCCTGGCATCTGGACATCGTCAATCACG)
E1m (CATTTAAATTAGCCTGGAGATCTGACATCGTCAATCACG)
E2 (GTGTTCGGTTAAGTAACACCTGGTTTTCGGGCTGGTTC)
E2m (GTGTTCGGTTAAGTAAACTAGTGTTTTCGGGCTGGTTC)
P1 + 2 + 3 (GCATCTGGACATCGTCAATCACGGGCTCGTGACACCGAG)
P4 + 5 + 6 (AGAGGGGACTCGTGGGTCATCTCGTTTAAAGCGCATTTTAAATTAC)
P7 + 8 (TGTGTTCGGTTAAGTAACACCTGGTTTTCGGGCTGGTTCATCAGT)
P9 + 10 (TGAATCACTTAAGAGATAATCAGGCTTATCAAAGC)
P11 (ACTTTGTATTATGCAATGTTTTCAAAA)
P12 (GAAAGGAAAAAGCATGAAGCACTGAATC)
P613 (GTGTCCTTTAATGACTTTTATAGTTGG)
P14 (CCCATTTGTGCAATTATGATCGAAAATATTTA).
EMSA was performed according to a published protocol with some modifications (Hartl et al., 2006). Oligos were end-labelled using Adenosine 5′-triphosphate ([γ-32P], PerkinElmer) and polynucleotide kinase (Fermentas, EK0031). Pax6b-HA protein and NeuroD-Flag/E47 complex were synthesised in vitro by TNT Quick-coupled Transcription/Translation kit (Promega) using the following plasmids: E47_pcDNA3 (obtained from S. Pfaff), Pax6b-HA_pCS2+, and NeuroD-Flag_pCS2+. After oligo labelling and protein synthesis the binding reaction was performed using 25,000 cpm for each oligo, 2 μl protein in the presence of 0.1 mg/ml of poly(dI-dC) and 1 × binding buffer (5 × buffer: 60 mM HEPES pH 7.9, 20 mM Tris–HCl pH 7.9, 300 mM KCl, 60% (v/v) glycerol, 5 mM DTT and 5 mM EDTA). The reaction was incubated for 45 min at room temperature. For competition assays, the unlabelled competitors were added at a 100-fold higher concentration and incubated at room temperature for 15 min, then the labelled probes were added and incubated for an additional 40 min. Binding was analysed by running a native 5% polyacrylamide gel, followed by overnight exposure of film (GE Healthcare, Amersham HyperfilmTM MP) at − 80 °C before development.
Whole-mount in situ hybridization and immunohistochemistry
Whole mount in situ hybridization using antisense RNA-probes labelled with digoxigenin (DIG) was performed as previously described (Hauptmann and Gerster, 2000). The following probes were used: insulin (obtained from W. Driever), hb9 (Wendik et al., 2004) and gfp (pG1) (Gilmour et al., 2002). RNA probes were prepared by using T7 or Sp6 RNA-Polymerases and DIG-Labelling Mix (Roche) according to the supplied protocol. All probes were detected using Anti-digoxygenin-AP FAB fragments (Roche).
For immunohistochemistry embryos were fixed for 1 h at RT in 4% PFA and 1% DMSO. After fixation embryos were blocked with blocking buffer for 1 h at RT and incubated with primary antibodies overnight at + 4 °C. The following day embryos were washed 3 times for 15 min in 1 × PBST and incubated in blocking buffer for 1 h at RT. Embryos were then incubated with secondary antibodies overnight at + 4 °C. Blocking and staining solutions contained 1% sheep serum, 1% BSA, 1% triton X-100, and 1% DMSO in PBS. Primary antibodies: anti-GFP (1: 200, rabbit IgG, Torrey Pines Biolabs), Islet-I Homeobox (1: 200, mouse IgG, 39.4D5, DSHB) and polyclonal guinea pig anti-insulin (1: 200, Dako), polyclonal rabbit anti-human glucagon (1:200, Dako) and polyclonal rabbit anti-somatostatin (1:200, Dako); secondary antibodies: Alexa Fluor 488 goat anti-rabbit IgG, Alexa Fluor 633 goat anti-guinea pig IgG (1: 1000, Invitrogen) and mouse Goat IgG, Cy3-conjugated (1:1000, Chemicon).
Imaging
Confocal image stacks were generated with Zeiss LSM Exciter5 microscope equipped with the following lasers: 25 mW Argon laser (468, 488, 514 nm), 1 mW 543 nm HeNe-laser, 5 mW 633 nm HeNe-laser and a 25 mW 405 nm diode-laser. Fluorescence detection was performed with the following dichroic beam splitters and filters: HFT405/488/543/633, NFT545, BP 505–530 (GFP and Alexa 488), LP 560 (Cy3) and LP 650 (Alexa 633). A Leica DM 6000 microscope was used for DIC images of whole mount in situ staining. AdobePhotoshop was used for image arrangement and adjustment.
Results
3.1 kb of 5′ flanking region of the zebrafish hb9 gene contains motoneuron and beta-cell specific regulatory elements
Tg[hb9:GFP]ml2 transgenic fish that contain 3.1 kb of 5 prime upstream non-coding region of the zebrafish hb9 gene fused to the gfp cDNA show GFP expression in motoneurons and in the pancreas. In order to clarify whether the reporter expression recapitulates the endogenous hb9 expression, we compared hb9 and gfp mRNA expression at different stages of development (Fig. 1, Supplemental Fig. 1). Whole mount mRNA in situ hybridisation analyses of Tg[hb9:GFP]ml2 embryos between 10 and 24 h post fertilization (hpf) revealed similarities but also differences between hb9 and gfp expression. In particular we found that gfp expression in Tg[hb9:GFP]ml2 does not recapitulate hb9 expression in early endoderm, notochord, hypochord and swim bladder and that gfp is expressed ectopically in various tissues including hindbrain, eyes and anterior somites (Supplemental Fig. 1). The differences in expression suggest that additional enhancer and silencer elements outside the 3.1 kb fragment must contribute to the complex hb9 expression. However, consistent with the expected overlapping expression in motoneurons and beta-cells, we observed similar patterns of robust gfp and hb9 expression in the spinal cord and the primary pancreatic islet (Figs. 1B–E′). Immunofluorescence analyses of 12–24 somite stage Tg[hb9:GFP]ml2 embryos (16–24 hpf) with an anti-islet antibody, a marker for primary neurons and pancreatic endocrine cells, confirm that GFP is expressed in spinal motoneurons (Fig. 1F) and in a subpopulation of the early endocrine cells (Figs. 1F, G). To further define which endocrine cell types are expressing GFP, we stained 24 hpf and 48 hpf Tg[hb9:GFP]ml2 embryos with antibodies against insulin and with a mixture of the alpha and delta-cell markers anti-glucagon and anti-somatostatin (Figs. 1H–I′). Overlapping expression of GFP and insulin but not GFP and glucagon/somatostatin at 48 hpf shows that pancreatic expression in Tg[hb9:GFP]ml2 is specific to beta-cells (Figs. 1I, I′). In summary, this shows that cis-regulatory enhancer elements controlling the highly conserved hb9 expression in motoneurons and beta-cells are present within the 3.1 kb of the 5 prime upstream non-coding region.
In sequence alignments of 5 prime Hb9 regions from zebrafish, fugu (Takifugu rubripes), chicken, mouse and human, the A-box and B-box are found 2.6 and 2 kb upstream of ATG, respectively (Fig. 1A). To localize the specific regulatory elements required for pancreatic and motoneuron gene expression, we generated hb9:GFP-reporter constructs lacking the A and/or B box and flanking genomic regions (Fig. 3A, Supplemental Fig. 2A), and analysed their activities in embryos injected with the corresponding DNA. Such transient reporter assay results in mosaic F0 embryos in which only a small portion of the cells can show reporter expression due to the mosaic distribution of the injected DNA. To distinguish beta-cell specific expression from potential nonspecific expression in the surrounding cells, the DNA constructs were injected into Tg[ins:dsRed] transgenic embryos in which beta-cells express dsRED, and analysed for overlapping expression of GFP and dsRED. In independent control injections with the 3.1 kb hb9:GFP construct, we observed GFP/dsRED double labelled cells in 25–40% of the injected embryos (Supplemental Figs. 2B–D). Motoneuron specific expression, as judged by the presence of GFP labelled peripheral axonal projections, was observed in 80–100% of the embryos. No GFP expression could be observed after injection of constructs that were either missing the A- and B-Box-containing distal half or that were missing the proximal part between the B-box and HindIII restriction site (Fig. 2A, Supplemental Fig. 2A). Most importantly, GFP expression in motoneurons was observed following injection of all constructs containing proximal sequences together with the B-box, while beta-cell specific GFP expression was only observed with constructs that contained the most distal part including the A-box (Fig. 2A, Supplemental Fig. 2A).
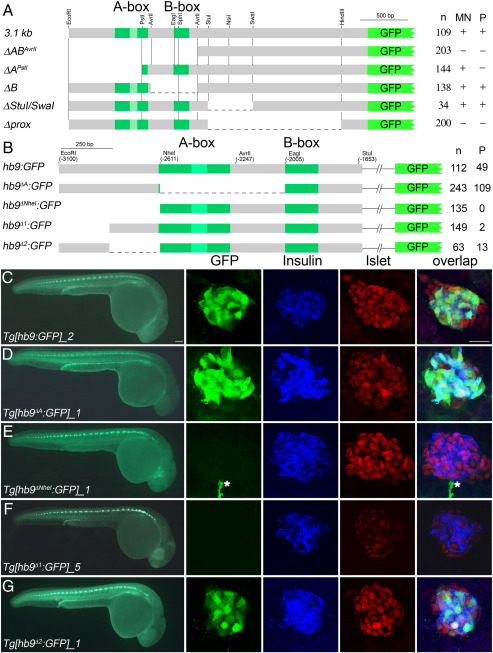
Fig. 2.
Pancreas specific GFP expression in Tg[hb9:GFP] embryos requires distal non-conserved hb9 promoter sequences. (A) Scheme of non-Tol2 hb9-reporter constructs (left) and results of the transient reporter assay (right, indicated are the numbers (n) of embryos showing presence (+) or absence (−) of GFP signal in spinal cord (SC) and pancreas (P)). (B) Scheme of Tol2 hb9-reporter constructs (left) and results of transient reporter assays (right, indicated are the numbers of GFP positive embryos (n) and of embryos with GFP positive beta-cell (P)). (C–G) GFP expression in representative transgenic lines at 28 hpf in whole embryos (left panel) and in fixed tissue (right panel) stained with antibodies for GFP (green), insulin (blue) and islet (red). The right panel shows confocal image projections and the corresponding overlays (white asterisk in E marks a motor axon). Pancreatic GFP expression is found for reporter constructs lacking either the A-box (Tg[hb9∆A:GFP], C) or the 249 bp flanking the A-box (Tg[hb9∆2:GFP], F), but not for constructs lacking the most distal 229 bp (Tg[hb9∆NheI:GFP], D; Tg[hb9∆1:GFP], E). Scale bar corresponds to 100 μm (whole embryos) and 20 μm (confocal image stacks). For further details on the GFP expression in all transgenic reporter lines see Supplementary Table 1.
In summary, the distal sequences that include both conserved motifs can direct tissue specific hb9 expression in the pancreas and motoneurons and the proximal non-conserved sequences seem to be important either for basal expression or to enhance expression levels. Furthermore, our analyses suggest a conserved role for the B-box in regulating motoneuron specific expression and provide a first hint for the presence of an hb9 beta-cell specific enhancer distal to the B-box.
478 bp of non-conserved sequences are required and sufficient for beta-cell specific expression
To better characterize the beta-cell enhancer we next generated constructs in which distal sequences were deleted (Fig. 2B). For these experiments, we used Tol2-mediated transgenesis in order to increase the likelihood of having beta-cell expression in our transient assays and to enhance recovery of stable transgenic lines. Surprisingly, loss of the A-box (hb9ΔA:GFP) did not lead to loss of reporter expression in beta-cells as identified by insulin antibody staining. Analyses of transiently expressing embryos revealed a similar frequency of pancreas specific GFP expression for hb9∆A:GFP and hb9:GFP constructs (Fig. 2B). To confirm the transient expression data, we isolated a total of 12 independent stable integrations for Tg[hb9∆A:GFP] and 2 independent alleles for Tg[hb9:GFP] (Figs. 2C, D, Supplemental Table 1). From those, nine of the Tg[hb9∆A:GFP] and both Tg[hb9:GFP] showed robust GFP expression in beta-cells and in motoneurons (Figs. 2C, D). The data suggest that the A-box is not required for pancreas and for motoneuron specific reporter expression. Notably, these transgenic lines showed a highly variable reporter expression in gut endoderm, epidermis, somites and nervous system, indicating that our reporter constructs are strongly influenced by positional effects (Supplemental Table 1).
The lack of beta-cell specific GFP expression in ΔAPstI:GFP transient line (Fig. 2A) but not in hb9∆A:GFP (Fig. 2B) suggests that the non-conserved distal part of the 3.1 kb fragment must mediate beta-cell specific expression. Consistent with this notion, we found that the loss of the distal 478 bp in hb9∆NheI:GFP injected embryos abolishes beta-cell specific expression in transient reporter assays and also in Tg[hb9∆NheI:GFP] stable transgenic allele (Figs. 2B, E, Supplemental Table 1). We therefore conclude that the non-conserved distal promoter sequences contain essential elements for pancreas specific expression.
To define the position of these elements more precisely, we generated constructs lacking either the distal or proximal halves of the non-conserved sequences between − 3097 and − 2868 bp (hb9∆1:GFP) and between − 2868 and − 2619 (hb9∆2:GFP), respectively (Fig. 2B). In transient reporter assays, hb9∆1:GFP DNA failed to induce pancreatic GFP expression (1.3% of the embryos showed overlapping GFP and dsRED expression, n = 149, Fig. 2B) while the hb9∆2:GFP (overlapping expression in 21% of embryos, n = 63, Fig. 3B) induced pancreas specific GFP expression, although the frequency was reduced as compared to the hb9:GFP control construct (43%, n = 112, Fig. 2). Results with stable transgenic lines were consistent with the transient assays as 6 out of 7 Tg[hb9∆1:GFP] alleles were negative for GFP expression in the pancreas while 7 out of 10 Tg[hb9∆2:GFP] alleles showed specific expression in beta-cells (Figs. 2F, G, Supplemental Table 1).
To determine if the distal sequences are sufficient to activate gfp expression in beta-cells, we generated two constructs in which the most distal 229 bp (hb9PE229:GFP) or 478 bp (hb9∆AB:GFP) were directly fused to the proximal part of the hb9 promoter (Fig. 3A). Both constructs induced pancreas specific expression in transient assays, indicating that the distal sequences contain the pancreas enhancer. However, in stable transgenic lines only Tg[hb9∆AB:GFP] alleles showed beta-cell specific GFP expression (Fig. 3C, Supplemental Table 2). While four of the nine Tg[hb9PE229:GFP] alleles showed weak expression in the pancreas, immunofluorescence analyses revealed that in three alleles GFP expression did not overlap with insulin (Fig. 3B, Supplemental Table 2). These data suggest that the most distal 229 bp are required but not sufficient for beta-cell specific gene expression. They further suggest that the 478 bp of non-conserved distal hb9 sequences, that we hereafter refer to as PE, contain multiple elements that together regulate beta-cell specific expression.
We next asked if the distal 478 bp contain all sequence motifs sufficient for beta-cell specific expression. To answer this question, we generated constructs in which the 478 bp PE fragment was cloned in front of the zebrafish hsp70 promoter linked to a gfp cDNA (PE+ hsp:GFP, Fig. 3A). Based on the weak beta-cell specific reporter expression in Tg[hb9∆AB:GFP] embryos we further asked if the A-box contributes to pancreas expression. To test this possibility we generated constructs in which hsp:GFP was combined with either the A-box only (A+hsp:GFP) or with PE and A-box (PE+ A+hsp:GFP, Fig. 3A). In transient assays 54% of the PE+ hsp:GFP (n = 65), 37% of the PE+ A+hsp:GFP (n = 70) but only 7% of the A+hsp:GFP (n = 58) injected embryos showed beta-cell specific expression (Fig. 3A). Consistent with an A-box independent beta-cell specific regulation by the PE, pancreatic GFP expression in three out of four Tg[PE+ hsp:GFP] lines was very similar to that in the two Tg[PE+ A+hsp:GFP] alleles while it was missing in all three Tg[A+hsp:GFP] alleles (Figs. 3D–F, Supplemental Table 2). Overall, the data show that the PE, and in particular the distal half of the PE, plays a critical role in beta-cell regulation and that the PE alone is sufficient for regulating beta-cell specific expression.
Direct binding of Pax6b and NeuroD to multiple sites within the hb9 PE
Bioinformatic analyses of the PE sequence revealed several potential binding sites for transcription factors with well described function in early endocrine cell differentiation, including consensus sequences for Pax6 (Epstein et al., 1994; Zhou et al., 2002), E-box-binding proteins such as Ngn3/NeuroD, and RAR (Mangelsdorf and Evans, 1995), among others. For our initial studies we focused on the endocrine progenitor factors Pax6b and NeuroD, for which 14 and 2 potential binding sites were found in the PE, respectively (Fig. 4A). To determine if these consensus sites are recognized by the corresponding transcription factors, we first tested direct binding in electrophoretic mobility shift assay (EMSA) with in vitro translated proteins and radioactively labelled synthetic double-stranded oligonucleotides. To confirm the specificity of the observed interaction, we performed competition assays with unlabeled self-competitors and also analysed whether the translated proteins recognized mutated consensus motifs.
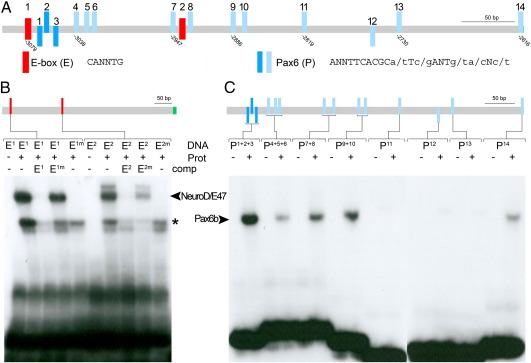
Fig. 4.
In vitro binding of NeuroD and Pax6b to PE specific sequence motives. (A) Schematic representation of potential binding sites for the transcription factors NeuroD (E-box in red) and Pax6 (blue) within the hb9 pancreas enhancer as revealed by bioinformatic sequence analyses. Pax6 sites with a core match between 0.76 and 0.9 are shown in light blue, and sites with a core match between 0.9 and 1 are shown in dark blue (core = TCACG). Orientation of Pax6 sites is indicated by position relative to the line (above: positive DNA strand; below: negative DNA). (B) EMSA studies show binding of in vitro synthesized NeuroD/E47 dimers (Prot) to oligonucleotides (DNA) spanning the E-boxes (E1, E2) but not to corresponding oligonucleotides with mutated E boxes (E1m and E2m). Competitor assays with unlabelled oligonucleotides (comp, as indicated) revealed efficient competition by E1 and E2 but not by E1m and E2m. Notably, unlabelled E2m reduced binding of labelled E2 to NeuroD/E47, suggesting that E2 binds NeuroD/E47 with lower affinity as compared to E1. Bands indicated by the asterisk presumably correspond to monomeric DNA–protein complexes. (C) EMSA studies with in vitro synthesized Pax6b protein (Prot) and oligonucleotides spanning individual or multiple potential Pax6b sites (as indicated). EMSA signals could be detected for P1 + 2 + 3, P4 + 5 + 6, P7 + 8, P9 + 10, P14 but not for P11, P12 and P13 (positions of protein-bound DNAs are indicated by arrowhead).
To test NeuroD binding activities, the potential binding sites (E-box motifs E1 and E2) were incubated with a mixture of NeuroD and its dimerization partner E47 (Fig. 4B). We found that NeuroD/E47 heterodimers bind to both E-boxes and that this interaction is specific, as NeuroD/E47 bound neither of the mutated E-boxes and unlabeled self-competitors efficiently prevented interaction with the labelled probes.
A slightly different strategy was used to test the 14 Pax6 paired box consensus sites (termed P1–P14). As ten of these sites are separated from their next neighbours by less than 12 nucleotides, we not only used DNA oligos containing individual paired box consensus sites but in some cases also oligos with additional neighbouring consensus sites (Fig. 4C). The EMSA studies revealed strongest signals for the oligonucleotides containing the most distal binding sites (P1 + 2 + 3) and slightly weaker signals for P4 + 5 + 6, P7 + 8, P9 + 10 and P14. The potential binding sites P11, P12 and P13 were not recognized by Pax6b protein (Fig. 4C). Notably, all Pax6b-interacting oligonucleotides showed a similar shift independent of whether one, two or three potential Pax6 binding sites were present on the oligonucleotides. We therefore reasoned that a single Pax6b protein binds each oligonucleotide. EMSA studies with mutated versions of P1 + 2 + 3 and P9 + 10 reveal different binding affinities for the neighbouring Pax6 consensus motifs, which could explain these results (Supplemental Fig. 3). In particular we find that P3 and P9 are dispensable for efficient binding to Pax6b while intact P1 + 2 and P10 sites are required for efficient competition with labelled P1 + 2 + 3 and P9 + 10, respectively (Supplemental Fig. 3). As steric hindrance might prevent simultaneous binding of two Pax6b factors on neighbouring P1 and P2 binding sites, the data suggest efficient interaction of Pax6b with either P1 or P2 and with P10.
In summary, our EMSA data show that at least two transcription factors with functions in endocrine progenitors, Pax6b and NeuroD, can directly bind to the binding sites in the hb9 PE. Furthermore, they reveal that the sequences with highest in vitro binding affinity for these proteins are localized in the distal part of PE, which is most critical for beta-cell specific expression as based on our promoter analyses.
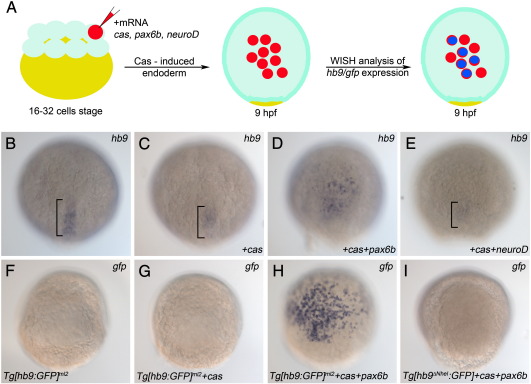
Ectopic induction of hb9 expression by Pax6b but not by NeuroD
Next we tested if overexpression of Pax6b and NeuroD encoding mRNA is sufficient to induce expression of either hb9 mRNA in wild type embryos or gfp mRNA in our transgenic reporter lines. In order to target the injected mRNA specifically to endoderm we took advantage of a previously described approach for cell-autonomous induction of endoderm by Casanova/Sox32 (Kikuchi et al., 2001; Stafford et al., 2006). In these experiments, a mixture of mRNAs encoding Cas together with Pax6b or NeuroD and the lineage tracer nucRFP (data not shown) was injected into a single-cell of 16–32 cell stage embryos (Fig. 5A). Due to Cas activity, the progeny of the injected cells adopted an endodermal fate which can then be analysed for hb9 or gfp mRNA expression. Based on the short half-life of injected mRNA we choose 9 hpf for the fixation of embryos. Our analyses revealed ectopic activation of hb9 in 63% (n = 11, Figs. 5B–D, Supplemental Table 3) of the pax6b mRNA injected embryos while neuroD mRNA had no effect (n = 13, Fig. 5E, Supplemental Table 3). To test if the Pax6b responsive elements are present on the 3.1 kb promoter fragment, we next injected the cas/pax6b mixture into Tg[hb9:GFP]ml2 transgenic embryos. These embryos showed robust expression of GFP protein as well as gfp mRNA, while non-injected embryos as well as embryos injected only with cas mRNA were gfp negative (Figs. 5F–H, Supplemental Table 3). To clarify whether Pax6b responsiveness requires the PE, we next injected pax6b/cas mRNA in Tg[hb9∆NheI:GFP] transgenic embryos, in which the PE is missing. No gfp expression could be observed in these embryos (Fig. 5I). Altogether, these data show that Pax6b is sufficient to activate endoderm specific hb9 expression and that this activation is mediated through the PE.
Fig. 5.
Pax6b but not NeuroD is sufficient to induce endodermal hb9 expression. (A) Schematic of the induction experiment: One cell of a 16–32 cell embryo stage was injected with mRNA encoding either Cas alone or a mixture of mRNAs encoding Cas together with NeuroD or Pax6b. Injected embryos were fixed 8 h later (9 hpf) and analysed by mRNA in situ hybridisation. (B–E) Expression of hb9 mRNA in uninjected control embryos (B) and in embryos injected with cas (C), cas/paxb6 (D) and cas/neuroD (E) mRNA. Note that only cas/pax6b mRNA injected embryos (D) show ectopic hb9 signals. Signals in posterior notochord (bracket in B, C, E) represent endogenous hb9 expression, while reduced signals in the cas (C), and cas/neuroD (E) injected embryos correlate with the developmental delay caused by the injection. (F–I) Expression of gfp mRNA in transgenic reporter lines in which gfp is controlled by PE-containing (Tg[hb9:GFP]ml2 in F–H) or PE-deleted (Tg[hb9∆NheI:GFP] in I) hb9 promoter-sequences. Uninjected (F) and cas-mRNA injected Tg[hb9:GFP]ml2 embryos (G) show no gfp signals while injection of cas/pax6b mRNA results in a robust gfp induction (H). Lack of gfp signals in cas/pax6b injected Tg[hb9∆NheI:GFP] shows that gfp induction by Pax6b is PE-dependent.
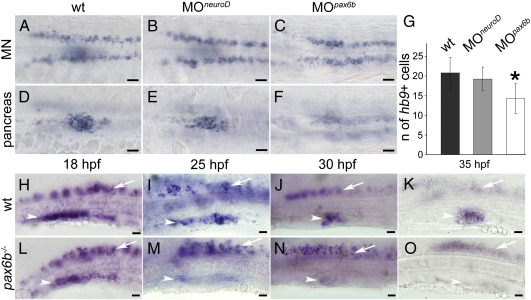
Pax6b is required for maintenance of hb9 expression in beta-cells
To test whether NeuroD and Pax6b are required for endogenous regulation of pancreas-specific hb9 expression we performed knock-down analyses with published morpholinos. We used a pax6b splicing morpholino which has been shown to cause loss of insulin expression (Verbruggen et al., 2010) and a neuroD morpholino that was used to study neuroD function of the lateral line organ (Sarrazin et al., 2006). Analyses of 24 hpf embryos injected with neuroD morpholino revealed no difference in the staining intensities or the number of hb9 expressing pancreatic cells between neuroD morphants (19.2 cells/embryo, n = 5) and wild type (wt) embryos (20.8 cells/embryo, n = 5) (Figs. 6A–D, G). Together with the gain-of-function analyses, this suggests that NeuroD is neither required nor sufficient for early hb9 regulation.
Fig. 6.
hb9 expression is initiated but not maintained in Pax6b deficient embryos. (A–F) Expression of hb9 mRNA at 24 hpf in motoneurons (MN in A, C, E) and in the pancreatic islet (pancreas in B, D, F) of uninjected wild type (wt) embryos (A, B) and embryos injected with neuroD (C, D) or pax6b morpholinos (E, F). Note that pancreatic hb9 expression is reduced in pax6b but not in neuroD morphants while motoneuron specific expression in the same embryos is not affected. Embryos are shown in dorsal view with anterior to the left. Scale bars in all figures correspond to 20 μm. (G) Number of hb9 positive cells in the pancreas of the wild type (wt), neuroD (MOneuroD) and pax6b (MOpax6b) morphants. (p-value for neuroD morphants 0.19429, p-value for pax6b morphants 0.00467). (H–O) hb9 expression in wild type embryos (H, I, J, K) and pax6b mutants (L, M, N, O) at 18 hpf (H, L), 25 hpf (I, M), 30 hpf (J, N) and 35 hpf (K, O). Note that hb9 expression in pancreas (white arrowhead) decreases during development in mutants, whereas expression in motoneurons (white arrow) is not affected. Embryos are shown in lateral view with anterior to the left. Scale bars in all figures correspond to 20 μm.
In contrast, pax6b morphants showed a reduced number of hb9 expressing pancreatic cells (14.3 cells/embryo, n = 9) and expression levels per cell appear much weaker as revealed by comparing staining intensities in the spinal cord and the pancreas of morphants and wild type embryos (Figs. 6A, B, D, F, G). We further noted that pax6b morpholino had only a minor effect on the pancreatic GFP protein expression levels in 24 hpf injected Tg[hb9:GFP]ml2 embryos (data not shown). GFP is a very stable protein that remains in the cells even when mRNA expression has stopped. We therefore reasoned that the difference between the GFP protein and hb9 mRNA expression levels in the beta-cells of 24 hpf pax6b morphants could indicate a defect in maintenance rather than induction of hb9 mRNA expression. To test this, we followed hb9 expression and gfp mRNA in pax6b−/− mutants from 18 hpf until 36 hpf (Figs. 6H–O, Supplemental Fig. 4). At 18 hpf, hb9 expression in pax6b mutants is similar to that in wild type (Figs. 6H, L), whereas between 25 hpf and 30 hpf absence of Pax6b leads to the progressive reduction of hb9 expression in the pancreas (Figs. 6I, J, M, N). At 35 hpf no hb9 expression could be detected in the endocrine pancreas (Figs. 6K, O). Importantly, at all stages analysed, motoneuron hb9 expression in pax6b −/− mutants was not affected (Figs. 6H–O), revealing the specific role for Pax6b in regulation of hb9 in the endocrine pancreas. Taken together, our results show that Pax6b is required for maintenance but not initiation of hb9 expression.
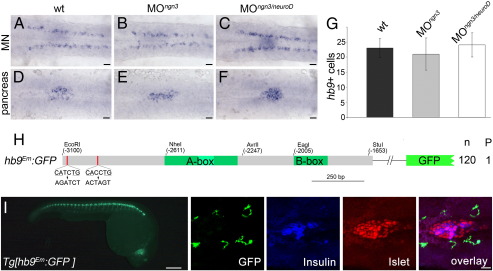
Induction of pancreatic hb9 expression requires E-box elements
While our studies suggest that NeuroD is not required for hb9 expression, they do not exclude the option that other bHLH proteins regulate hb9 transcription through interaction with the two E-box elements. A promising candidate for such factors is Ngn3 even though ngn3 expression has not been reported in the early zebrafish pancreas (Kinkel and Prince, 2009; Zecchin et al., 2007). As expression levels of ngn3 may be below detection limit, we used a genetic approach to determine ngn3 functions in embryonic hb9 transcription. However, comparison of pancreatic hb9 or gfp expression revealed no significant differences between uninjected embryos, and embryos that were injected with ngn3 morpholinos alone or with a combination of neuroD and ngn3 morpholinos (Figs. 7A, D, C, F, G, data not shown). This shows that neither ngn3 nor neuroD is required for early pancreatic hb9 expression.
Fig. 7.
E-box binding proteins but not Ngn3 and NeuroD are required for beta-cell specific hb9 expression. (A–F) Similar expression of hb9 mRNA at 24 hpf in motoneurons (MN in A–C) and in the pancreatic islet (pancreas in D–F) of uninjected wild type (wt) embryos (A, D) and embryos injected with ngn3 (B, E) or a combination of ngn3 and neuroD morpholinos (C, F). Different focus planes of the same embryos are shown in dorsal view with anterior to the left. (G) Countings of hb9 positive cells in the pancreas of the wild type (wt, n = 21), ngn3 (MOngn3, n = 32) and ngn3/neuroD (MOngn3/neuroD, n = 22) morphants at 24 hpf. (H) Schematic drawing of the hbEm:GFP construct in which both E-boxes (red line) were mutated by site-directed mutagenesis (origin and mutated sequences are shown with consensus sequences being underlined). Numbers on the right summarize the results of transient reporter assays in which a single embryo out of 120 GFP positive embryos (n) showed reporter expression within the pancreas (P). (I) GFP expression in Tg[hb9Em:GFP] embryos at 28 hpf in whole embryos and in embryos stained with antibodies for GFP (green, expression corresponds to motor axons), insulin (blue) and islet (red). The right images shown are confocal image projections with an overlay at the right. Scale bar corresponds to 100 μm (live embryo in I) and 20 μm (A–F, confocal images in I).
To test more directly for a function of the E-boxes we asked if they are required for beta-cell specific reporter expression. Injection of the hb9Em:GFP reporter construct in which both E-boxes were mutated by site-directed mutagenesis resulted in only 1 out of 120 embryos (0.8%) showing GFP expression in the pancreas. Further, analyses of two hb9Em:GFP transgenic fishlines revealed a complete absence of GFP expression in the endocrine pancreas (Fig. 7I). As motoneuron specific expression in these embryos was similar to that of Tg[hb9:GFP]ml2 embryos, our data suggest a specific requirement of one or several unspecified E-box-binding factors during initiation of beta-cell specific hb9 expression.
Discussion
Vertebrate hb9 genes show conserved expression and function in early beta-cell maturation. While hb9 genes have been subject to several promoter analyses, the enhancer elements regulating pancreas specific expression have not been identified. By using transient and stable transgenic reporter assays, we now show that in zebrafish, a non-conserved 5 prime upstream region of 478 bp is sufficient and required for targeting expression specifically to beta-cell progenitors. We further identify NeuroD and Pax6b as transcription factors that directly bind in vitro to this enhancer element and we present in vivo gain and loss-of-function analyses that suggest essential function of currently not specified E-box-binding factors and of Pax6b during initiation and maintenance of beta-cell specific expression of hb9, respectively.
hb9 regulation by conserved and non-conserved enhancer elements
Hb9 genes throughout vertebrates show complex embryonic expression in various tissues of endodermal, ectodermal and mesodermal origin (Arber et al., 1999; Broihier and Skeath, 2002; Ferrier et al., 2001; Grapin-Botton et al., 2001; Ross et al., 1998; Tanabe et al., 1998; Thaler et al., 1999; Wendik et al., 2004). Here we show that 3.1 kb of zebrafish hb9 upstream genomic DNA is sufficient to recapitulate the conserved expression of hb9 in motoneurons and in differentiating beta-cells. These sequences include two highly conserved non-coding motifs, the A- and B-box. Our data show that in zebrafish, the B-box mediates reporter expression in motoneurons and some ventral interneurons while the A-box is dispensable for embryonic hb9 regulation. Similar conclusions have been drawn from the analyses of the A- and B-boxes of the mouse hb9 promoter, in which three copies of the B-box fused to a minimal promoter were sufficient to regulate expression in motoneurons, while a corresponding construct with three copies of the A-box failed to induce any specific reporter expression (Nakano et al., 2005). Interestingly, motoneuron specific hb9 regulation in mouse also involves a second mammal-specific element termed MNE, which has a distinct mode of molecular activity as compared to the B-box (Lee and Pfaff, 2003; Lee et al., 2004; Nakano et al., 2005). While the B-Box is directly regulated by Hox/Pbx factors to mediate anterior–posterior positional information (Nakano et al., 2005), the MNE is regulated by basic helix–loop–helix (bHLH) and Lim-homeodomain (Lim-HD) transcription factors that integrate signals of dorsal–ventral patterning and cell differentiation (Lee and Pfaff, 2003; Lee et al., 2004; Lee et al., 2008). The highly conserved function of Lim-HD, bHLH and Hb9 proteins in motoneuron differentiation implies that molecular interactions between these factors are not restricted to mammals (Briscoe et al., 2000; Lee and Pfaff, 2003; Lee et al., 2004; Shirasaki and Pfaff, 2002; Zhou and Anderson, 2002), even though MNE related sequences are not detectable in non-mammalian system. Our transient reporter assays reveal that a critical region of 300 bp, including the B-box, is required for motoneuron specific expression in zebrafish (compare ΔASphI and ΔABAvrII in Fig. 2, Supplemental Fig. 1). While low sequence conservation may prevent bioinformatics-based detection of MNE related sequences, it is still possible that this critical region similarly receives combined regulatory input from bHLH, Lim-HD and Hox/Pbx factors. Alternatively, in non-mammalian organisms, the regulatory function of the MNE may be located to the enhancer of the hb9 paralog mnr2, which is missing in mammals (Wendik et al., 2004). Preliminary sequence analyses of the two mnr2 genes in zebrafish did not provide evidence for MNE related sequences and it will be interesting to determine how conserved factors which regulate the MNE, such as the bHLH proteins Olig2, Ngn2, NeuroD and the lim-HD factors Isl1/2 and Lhx3 (Lee and Pfaff, 2003), contribute to motoneuron specific hb9/mnr2 regulation in non-mammalian organisms.
Recent high throughput analyses of cis-regulatory elements came to the conclusion that a lot of enhancer elements are not conserved between vertebrates and this includes both activator and repressor elements (Birney et al., 2007; McGaughey et al., 2009; Roh et al., 2007). Consistent with this, we were unable to recapitulate all sites of endogenous hb9 mRNA expression with our transgenic hb9-reporter lines and most transgenic lines, including the initially reported Tg[hb9:GFP]ml2 and Tg[hb9:GFP]ml3 (Flanagan-Steet et al., 2005; Kimmel and Meyer, 2010) show ectopic reporter expression in hb9-negative tissues such as hindbrain and eye. This shows that hb9 is regulated by additional activating and repressing elements outside of the analysed 3.1 kb region. As these more distal sequences lack homology to corresponding regions of other vertebrate hb9 genes, this suggests functions for additional non-conserved element in hb9 regulation. Among those, the as-yet unidentified enhancer regulating expression in caudal structures and notochord is of major interest when considering the connection of human HLXB9 and Currarino syndrome. In these patients, reduced HLXB9 function resulting from loss of one HLXB9 allele is thought to cause formation of caudal teratomas, which in consequence lead to sacral agenesis (Ross et al., 1998).
Beta-cell specific regulation by the non-conserved PE
Previous analyses of the mouse hb9 gene failed to detect a beta-cell enhancer in a 9 kb fragment upstream of the hb9 gene (Arber et al., 1999; Nakano et al., 2005; Wichterle et al., 2002). Notably, this fragment contained the A-box at the most distal position. In our studies we localize the zebrafish hb9 beta-cell enhancer to a 478 bp sequence directly distal to the A-box, thus to a region that was not analysed in mouse. Our transient and transgenic reporter analyses demonstrate that these 478 bp, termed PE, are required and sufficient for targeting expression to beta-cell progenitors. While sequence alignments of the PE reveal no obvious homologies to the Medaka hb9 gene, we also found that the zebrafish hb9:GFP construct is sufficient to regulate beta-cell specific expression in hb9:gfp transgenic Medaka embryos (D. Meyer, unpublished data). This demonstrates that the regulatory activity of this early beta-cell enhancer is conserved, at least in fish. The lack of pancreas specific GFP-expression in Tg[hb9Δ1:GFP] but not in Tg[hb9Δ2:GFP] further suggests that the distal part of the PE contains essential elements while the proximal part is dispensable for beta-cell expression. Consistent with these data, the majority of the EMSA-confirmed Pax6b binding sites and both E-boxes localize to the distal half of the PE. Nevertheless, the distal part without the A- and B-box containing sequences is not sufficient to trigger beta-cell specific reporter expression. Possibly, the proximal half of the PE and the flanking conserved sequences both contain functionally redundant regulatory elements that interact with the distal elements.
Molecular regulation of the PE
Our studies revealed specific requirements of E-box-binding factors and Pax6 during initiation and maintenance of beta-cell specific hb9 expression. In particular, we show that the PE contains two E-boxes and multiple Pax6-binding sites that interact in vitro with NeuroD/E47 and Pax6b proteins, respectively. From the 14 potential Pax6-type paired box binding sites revealed by bioinformatic analyses, at least five are recognized by Pax6b protein in vitro. Interestingly, four of these Pax6b-interacting sequences contain two or three nearly adjacent Pax6 consensus motifs and one of these double Pax6 consensus motifs has an intervening E-box. The functional relevance of these clusters is unclear, in particular as the presence of multiple Pax6 consensus motifs did not result in binding of multiple Pax6b proteins.
In order to determine the biological role of the E-boxes and the Pax6 binding sites, we tested if loss or gain of protein function affects hb9 expression. We show that the E-boxes are essential to initiate beta-cell specific reporter expression in hb9:GFP transgenic embryos and that this induction is initiated independent of the E-box-binding proteins NeuroD and Ngn3. Consistent with the previously reported lack of ngn3 mRNA expression in the early zebrafish pancreas (Kinkel and Prince, 2009; Moro et al., 2009), we also find that ngn3 is dispensable for early beta-cell development in zebrafish. The results reveal major difference between mouse and zebrafish ngn3 functions in pancreas development and they imply that in zebrafish at least one additional E-box binding factor acts upstream or in parallel to NeuroD during beta-cell differentiation. Notably, in human only a few of the homozygous NGN3 missense mutations are associated with neonatal diabetes as an indicator for missing beta-cells (Rubio-Cabezas et al., 2011). While the non-diabetes phenotypes could result from hypomorphic NGN3 alleles it is also possible that differentiation of beta-cells in human may require additional to-be identified E-box-binding factors similar to the situation in zebrafish.
Our data further suggest that the paired-box binding sites within the PE mediate Pax6b-dependent hb9 transcriptional activation. We show that injection of pax6b mRNA results in ectopic induction of hb9 mRNA and of gfp mRNA in hb9:GFP transgenes, and that this induction is PE dependent. The data strongly suggest a direct regulation of the PE by Pax6b, while they also reveal that Pax6b is not required for the initiation but for the maintenance of pancreatic hb9 expression. Interestingly, almost no beta-cells are observed in pax6b-mutants showing that besides Hb9, other factors are probably acting downstream from Pax6b to drive the differentiation of beta-cells.
Conclusion
Currently, the vertebrate insulin promoter is the major model for studying the molecular basis of beta-cell specific transcription. Our studies now introduce the hb9-PE element as a novel model not only for studies of the regulation of early beta-cell specific transcription, but also for studying the molecular control of beta-cell differentiation. Our studies further revealed interactions of two classes of transcription factors with this novel beta-cell specific enhancer. We show that a currently unspecified combination of E-box-binding proteins is required to initiate PE based expression and that Pax6b is a potent activator of this enhancer that is required to maintain hb9 expression. Our analyses suggest that these genes have independent function during initial steps of beta-cell differentiation in zebrafish. As Pax6b and Hb9 are both required to initiate expression of insulin this raises the question whether the observed function of Pax6b in maintaining endocrine hb9 expression represents a conserved cross-regulation between two beta-cell fate-regulating transcription factors. Previously reported interactions between Pax6 and various co-factors in non-pancreatic tissues provide interesting opportunities to address questions on the molecular regulation of beta-cell restriction of transcription in the context of early induction in beta-cell progenitors or maintenance after maturation. Reported interactions of Pax6 with members of the Sox and Ets family of transcription factors and with the chromatin remodelling factor Brg1 provide interesting novel links for future studies on the genetic and epigenetic regulation of beta-cell restricted expression (Cvekl et al., 2004; Kamachi et al., 2001; Yang et al., 2006).
Acknowledgments
We thank Robin Kimmel and Elisabeth Ott for critical reading of the manuscript and all members of the institute for suggestions and fruitful discussions. Special thanks go to Frederic Biemar for help during the initial cloning of the hb9 promoter, to Wolfgang Driever for general support during the starting phase of this project and for providing Tg[ins:dsRed] fish and insulin in situ probes, to Marianne L. Voz for providing ngn3 morpholino and to Francesco Argenton and Samuel L. Pfaff for providing expression plasmid for NeuroD and E47, respectively. This project was supported by grant from the University of Freiburg (D.M.), FWF P-20492 (D.M.), Belgian State (SSTC, IUAP/PAI) and the European Union (B.P.).
Footnotes
Supplementary data to this article can be found online at doi:10.1016/j.ydbio.2012.03.001.
Contributor Information
Valeriya Arkhipova, Email: A.Valeriya@gmx.de.
Björn Wendik, Email: bjoern.wendik@perkinelmer.com.
Nathalie Devos, Email: nathalie.i.devos@gskbio.com.
Olivier Ek, Email: o.ek@student.ulg.ac.be.
Bernard Peers, Email: bpeers@ulg.ac.be.
Dirk Meyer, Email: dirk.meyer@uibk.ac.at.
Appendix A. Supplementary data
Supplementary materials.
References
- Apelqvist A., Li H., Sommer L., Beatus P., Anderson D.J., Honjo T., Hrabe de Angelis M., Lendahl U., Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Arber S., Han B., Mendelsohn M., Smith M., Jessell T.M., Sockanathan S. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- Binot A.C., Manfroid I., Flasse L., Winandy M., Motte P., Martial J.A., Peers B., Voz M.L. Nkx6.1 and nkx6.2 regulate alpha- and beta-cell formation in zebrafish by acting on pancreatic endocrine progenitor cells. Dev. Biol. 2010;340:397–407. doi: 10.1016/j.ydbio.2010.01.025. [DOI] [PubMed] [Google Scholar]
- Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., Kuehn M.S., Taylor C.M., Neph S., Koch C.M., Asthana S., Malhotra A., Adzhubei I., Greenbaum J.A., Andrews R.M., Flicek P., Boyle P.J., Cao H., Carter N.P., Clelland G.K., Davis S., Day N., Dhami P., Dillon S.C., Dorschner M.O., Fiegler H., Giresi P.G., Goldy J., Hawrylycz M., Haydock A., Humbert R., James K.D., Johnson B.E., Johnson E.M., Frum T.T., Rosenzweig E.R., Karnani N., Lee K., Lefebvre G.C., Navas P.A., Neri F., Parker S.C., Sabo P.J., Sandstrom R., Shafer A., Vetrie D., Weaver M., Wilcox S., Yu M., Collins F.S., Dekker J., Lieb J.D., Tullius T.D., Crawford G.E., Sunyaev S., Noble W.S., Dunham I., Denoeud F., Reymond A., Kapranov P., Rozowsky J., Zheng D., Castelo R., Frankish A., Harrow J., Ghosh S., Sandelin A., Hofacker I.L., Baertsch R., Keefe D., Dike S., Cheng J., Hirsch H.A., Sekinger E.A., Lagarde J., Abril J.F., Shahab A., Flamm C., Fried C., Hackermuller J., Hertel J., Lindemeyer M., Missal K., Tanzer A., Washietl S., Korbel J., Emanuelsson O., Pedersen J.S., Holroyd N., Taylor R., Swarbreck D., Matthews N., Dickson M.C., Thomas D.J., Weirauch M.T., Gilbert J., Drenkow J., Bell I., Zhao X., Srinivasan K.G., Sung W.K., Ooi H.S., Chiu K.P., Foissac S., Alioto T., Brent M., Pachter L., Tress M.L., Valencia A., Choo S.W., Choo C.Y., Ucla C., Manzano C., Wyss C., Cheung E., Clark T.G., Brown J.B., Ganesh M., Patel S., Tammana H., Chrast J., Henrichsen C.N., Kai C., Kawai J., Nagalakshmi U., Wu J., Lian Z., Lian J., Newburger P., Zhang X., Bickel P., Mattick J.S., Carninci P., Hayashizaki Y., Weissman S., Hubbard T., Myers R.M., Rogers J., Stadler P.F., Lowe T.M., Wei C.L., Ruan Y., Struhl K., Gerstein M., Antonarakis S.E., Fu Y., Green E.D., Karaoz U., Siepel A., Taylor J., Liefer L.A., Wetterstrand K.A., Good P.J., Feingold E.A., Guyer M.S., Cooper G.M., Asimenos G., Dewey C.N., Hou M., Nikolaev S., Montoya-Burgos J.I., Loytynoja A., Whelan S., Pardi F., Massingham T., Huang H., Zhang N.R., Holmes I., Mullikin J.C., Ureta-Vidal A., Paten B., Seringhaus M., Church D., Rosenbloom K., Kent W.J., Stone E.A., Batzoglou S., Goldman N., Hardison R.C., Haussler D., Miller W., Sidow A., Trinklein N.D., Zhang Z.D., Barrera L., Stuart R., King D.C., Ameur A., Enroth S., Bieda M.C., Kim J., Bhinge A.A., Jiang N., Liu J., Yao F., Vega V.B., Lee C.W., Ng P., Shahab A., Yang A., Moqtaderi Z., Zhu Z., Xu X., Squazzo S., Oberley M.J., Inman D., Singer M.A., Richmond T.A., Munn K.J., Rada-Iglesias A., Wallerman O., Komorowski J., Fowler J.C., Couttet P., Bruce A.W., Dovey O.M., Ellis P.D., Langford C.F., Nix D.A., Euskirchen G., Hartman S., Urban A.E., Kraus P., Van Calcar S., Heintzman N., Kim T.H., Wang K., Qu C., Hon G., Luna R., Glass C.K., Rosenfeld M.G., Aldred S.F., Cooper S.J., Halees A., Lin J.M., Shulha H.P., Zhang X., Xu M., Haidar J.N., Yu Y., Ruan Y., Iyer V.R., Green R.D., Wadelius C., Farnham P.J., Ren B., Harte R.A., Hinrichs A.S., Trumbower H., Clawson H., Hillman-Jackson J., Zweig A.S., Smith K., Thakkapallayil A., Barber G., Kuhn R.M., Karolchik D., Armengol L., Bird C.P., de Bakker P.I., Kern A.D., Lopez-Bigas N., Martin J.D., Stranger B.E., Woodroffe A., Davydov E., Dimas A., Eyras E., Hallgrimsdottir I.B., Huppert J., Zody M.C., Abecasis G.R., Estivill X., Bouffard G.G., Guan X., Hansen N.F., Idol J.R., Maduro V.V., Maskeri B., McDowell J.C., Park M., Thomas P.J., Young A.C., Blakesley R.W., Muzny D.M., Sodergren E., Wheeler D.A., Worley K.C., Jiang H., Weinstock G.M., Gibbs R.A., Graves T., Fulton R., Mardis E.R., Wilson R.K., Clamp M., Cuff J., Gnerre S., Jaffe D.B., Chang J.L., Lindblad-Toh K., Lander E.S., Koriabine M., Nefedov M., Osoegawa K., Yoshinaga Y., Zhu B., de Jong P.J. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J., Pierani A., Jessell T.M., Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Broihier H.T., Skeath J.B. Drosophila homeodomain protein dHb9 directs neuronal fate via crossrepressive and cell-nonautonomous mechanisms. Neuron. 2002;35:39–50. doi: 10.1016/s0896-6273(02)00743-2. [DOI] [PubMed] [Google Scholar]
- Cvekl A., Yang Y., Chauhan B.K., Cveklova K. Regulation of gene expression by Pax6 in ocular cells: a case of tissue-preferred expression of crystallins in lens. Int. J. Dev. Biol. 2004;48:829–844. doi: 10.1387/ijdb.041866ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund H. Pancreatic organogenesis—developmental mechanisms and implications for therapy. Nat. Rev. Genet. 2002;3:524–532. doi: 10.1038/nrg841. [DOI] [PubMed] [Google Scholar]
- Epstein J., Cai J., Glaser T., Jepeal L., Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J. Biol. Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- Ferrier D.E., Brooke N.M., Panopoulou G., Holland P.W. The Mnx homeobox gene class defined by HB9, MNR2 and amphioxus AmphiMnx. Dev. Genes Evol. 2001;211:103–107. doi: 10.1007/s004270000124. [DOI] [PubMed] [Google Scholar]
- Flanagan-Steet H., Fox M.A., Meyer D., Sanes J.R. Neuromuscular synapses can form in vivo by incorporation of initially aneural postsynaptic specializations. Development. 2005;132:4471–4481. doi: 10.1242/dev.02044. [DOI] [PubMed] [Google Scholar]
- Gilmour D.T., Maischein H.M., Nusslein-Volhard C. Migration and function of a glial subtype in the vertebrate peripheral nervous system. Neuron. 2002;34:577–588. doi: 10.1016/s0896-6273(02)00683-9. [DOI] [PubMed] [Google Scholar]
- Gradwohl G., Dierich A., LeMeur M., Guillemot F. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grapin-Botton A., Majithia A.R., Melton D.A. Key events of pancreas formation are triggered in gut endoderm by ectopic expression of pancreatic regulatory genes. Genes Dev. 2001;15:444–454. doi: 10.1101/gad.846001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habener J.F., Kemp D.M., Thomas M.K. Minireview: transcriptional regulation in pancreatic development. Endocrinology. 2005;146:1025–1034. doi: 10.1210/en.2004-1576. [DOI] [PubMed] [Google Scholar]
- Harrison K.A., Thaler J., Pfaff S.L., Gu H., Kehrl J.H. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat. Genet. 1999;23:71–75. doi: 10.1038/12674. [DOI] [PubMed] [Google Scholar]
- Hartl M., Karagiannidis A.I., Bister K. Cooperative cell transformation by Myc/Mil(Raf) involves induction of AP-1 and activation of genes implicated in cell motility and metastasis. Oncogene. 2006;25:4043–4055. doi: 10.1038/sj.onc.1209441. [DOI] [PubMed] [Google Scholar]
- Hauptmann G., Gerster T. Multicolor whole-mount in situ hybridization. Methods Mol. Biol. 2000;137:139–148. doi: 10.1385/1-59259-066-7:139. [DOI] [PubMed] [Google Scholar]
- Huang H.P., Chu K., Nemoz-Gaillard E., Elberg D., Tsai M.J. Neogenesis of beta-cells in adult BETA2/NeuroD-deficient mice. Mol. Endocrinol. 2002;16:541–551. doi: 10.1210/mend.16.3.0784. [DOI] [PubMed] [Google Scholar]
- Itkin-Ansari P., Marcora E., Geron I., Tyrberg B., Demeterco C., Hao E., Padilla C., Ratineau C., Leiter A., Lee J.E., Levine F. NeuroD1 in the endocrine pancreas: localization and dual function as an activator and repressor. Dev. Dyn. 2005;233:946–953. doi: 10.1002/dvdy.20443. [DOI] [PubMed] [Google Scholar]
- Jenny M., Uhl C., Roche C., Duluc I., Guillermin V., Guillemot F., Jensen J., Kedinger M., Gradwohl G. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 2002;21:6338–6347. doi: 10.1093/emboj/cdf649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y., Uchikawa M., Tanouchi A., Sekido R., Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 2001;15:1272–1286. doi: 10.1101/gad.887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 2004;77:201–222. doi: 10.1016/s0091-679x(04)77011-9. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., Agathon A., Alexander J., Thisse C., Waldron S., Yelon D., Thisse B., Stainier D.Y. casanova encodes a novel Sox-related protein necessary and sufficient for early endoderm formation in zebrafish. Genes Dev. 2001;15:1493–1505. doi: 10.1101/gad.892301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel R.A., Meyer D. Molecular regulation of pancreas development in zebrafish. Methods Cell Biol. 2010;100:261–280. doi: 10.1016/B978-0-12-384892-5.00010-4. [DOI] [PubMed] [Google Scholar]
- Kinkel M.D., Prince V.E. On the diabetic menu: zebrafish as a model for pancreas development and function. Bioessays. 2009;31:139–152. doi: 10.1002/bies.200800123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.K., Pfaff S.L. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38:731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Lee S.K., Jurata L.W., Funahashi J., Ruiz E.C., Pfaff S.L. Analysis of embryonic motoneuron gene regulation: derepression of general activators function in concert with enhancer factors. Development. 2004;131:3295–3306. doi: 10.1242/dev.01179. [DOI] [PubMed] [Google Scholar]
- Lee S., Lee B., Joshi K., Pfaff S.L., Lee J.W., Lee S.K. A regulatory network to segregate the identity of neuronal subtypes. Dev. Cell. 2008;14:877–889. doi: 10.1016/j.devcel.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Edlund H. Persistent expression of Hlxb9 in the pancreatic epithelium impairs pancreatic development. Dev. Biol. 2001;240:247–253. doi: 10.1006/dbio.2001.0440. [DOI] [PubMed] [Google Scholar]
- Li H., Arber S., Jessell T.M., Edlund H. Selective agenesis of the dorsal pancreas in mice lacking homeobox gene Hlxb9. Nat. Genet. 1999;23:67–70. doi: 10.1038/12669. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D.J., Evans R.M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- McGaughey D.M., Stine Z.E., Huynh J.L., Vinton R.M., McCallion A.S. Asymmetrical distribution of non-conserved regulatory sequences at PHOX2B is reflected at the ENCODE loci and illuminates a possible genome-wide trend. BMC Genomics. 2009;10:8. doi: 10.1186/1471-2164-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro E., Gnugge L., Braghetta P., Bortolussi M., Argenton F. Analysis of beta cell proliferation dynamics in zebrafish. Dev. Biol. 2009;332:299–308. doi: 10.1016/j.ydbio.2009.05.576. [DOI] [PubMed] [Google Scholar]
- Nakano T., Windrem M., Zappavigna V., Goldman S.A. Identification of a conserved 125 base-pair Hb9 enhancer that specifies gene expression to spinal motor neurons. Dev. Biol. 2005;283:474–485. doi: 10.1016/j.ydbio.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Naya F.J., Huang H.P., Qiu Y., Mutoh H., DeMayo F.J., Leiter A.B., Tsai M.J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls S., Zecchin E., Tiso N., Bortolussi M., Argenton F. Function and regulation of zebrafish nkx2.2a during development of pancreatic islet and ducts. Dev. Biol. 2007;304:875–890. doi: 10.1016/j.ydbio.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Roh T.Y., Wei G., Farrell C.M., Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17:74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A.J., Ruiz-Perez V., Wang Y., Hagan D.M., Scherer S., Lynch S.A., Lindsay S., Custard E., Belloni E., Wilson D.I., Wadey R., Goodman F., Orstavik K.H., Monclair T., Robson S., Reardon W., Burn J., Scambler P., Strachan T. A homeobox gene, HLXB9, is the major locus for dominantly inherited sacral agenesis. Nat. Genet. 1998;20:358–361. doi: 10.1038/3828. [DOI] [PubMed] [Google Scholar]
- Rubio-Cabezas O., Jensen J.N., Hodgson M.I., Codner E., Ellard S., Serup P., Hattersley A.T. Permanent neonatal diabetes and enteric anendocrinosis associated with biallelic mutations in NEUROG3. Diabetes. 2011;60:1349–1353. doi: 10.2337/db10-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rukstalis J.M., Habener J.F. Neurogenin3: a master regulator of pancreatic islet differentiation and regeneration. Islets. 2009;1:177–184. doi: 10.4161/isl.1.3.9877. [DOI] [PubMed] [Google Scholar]
- Saha M.S., Miles R.R., Grainger R.M. Dorsal-ventral patterning during neural induction in Xenopus: assessment of spinal cord regionalization with xHB9, a marker for the motor neuron region. Dev. Biol. 1997;187:209–223. doi: 10.1006/dbio.1997.8625. [DOI] [PubMed] [Google Scholar]
- Sarrazin A.F., Villablanca E.J., Nunez V.A., Sandoval P.C., Ghysen A., Allende M.L. Proneural gene requirement for hair cell differentiation in the zebrafish lateral line. Dev. Biol. 2006;295:534–545. doi: 10.1016/j.ydbio.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Schwitzgebel V.M. Programming of the pancreas. Mol. Cell. Endocrinol. 2001;185:99–108. doi: 10.1016/s0303-7207(01)00628-1. [DOI] [PubMed] [Google Scholar]
- Schwitzgebel V.M., Scheel D.W., Conners J.R., Kalamaras J., Lee J.E., Anderson D.J., Sussel L., Johnson J.D., German M.S. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- Shin C.H., Chung W.S., Hong S.K., Ober E.A., Verkade H., Field H.A., Huisken J., Stainier D.Y. Multiple roles for Med12 in vertebrate endoderm development. Dev. Biol. 2008;317:467–479. doi: 10.1016/j.ydbio.2008.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki R., Pfaff S.L. Transcriptional codes and the control of neuronal identity. Annu. Rev. Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Stafford D., White R.J., Kinkel M.D., Linville A., Schilling T.F., Prince V.E. Retinoids signal directly to zebrafish endoderm to specify insulin-expressing beta-cells. Development. 2006;133:949–956. doi: 10.1242/dev.02263. [DOI] [PubMed] [Google Scholar]
- Tanabe Y., William C., Jessell T.M. Specification of motor neuron identity by the MNR2 homeodomain protein. Cell. 1998;95:67–80. doi: 10.1016/s0092-8674(00)81783-3. [DOI] [PubMed] [Google Scholar]
- Thaler J., Harrison K., Sharma K., Lettieri K., Kehrl J., Pfaff S.L. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron. 1999;23:675–687. doi: 10.1016/s0896-6273(01)80027-1. [DOI] [PubMed] [Google Scholar]
- Verbruggen V., Ek O., Georlette D., Delporte F., Von Berg V., Detry N., Biemar F., Coutinho P., Martial J.A., Voz M.L., Manfroid I., Peers B. The Pax6b homeodomain is dispensable for pancreatic endocrine cell differentiation in zebrafish. J. Biol. Chem. 2010;285:13863–13873. doi: 10.1074/jbc.M110.108019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendik B., Maier E., Meyer D. Zebrafish mnx genes in endocrine and exocrine pancreas formation. Dev. Biol. 2004;268:372–383. doi: 10.1016/j.ydbio.2003.12.026. [DOI] [PubMed] [Google Scholar]
- Wichterle H., Lieberam I., Porter J.A., Jessell T.M. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Wilson M.E., Scheel D., German M.S. Gene expression cascades in pancreatic development. Mech. Dev. 2003;120:65–80. doi: 10.1016/s0925-4773(02)00333-7. [DOI] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Walter K., Abnizova I., Gilks W., Edwards Y.J., Cooke J.E., Elgar G. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Stopka T., Golestaneh N., Wang Y., Wu K., Li A., Chauhan B.K., Gao C.Y., Cveklova K., Duncan M.K., Pestell R.G., Chepelinsky A.B., Skoultchi A.I., Cvekl A. Regulation of alphaA-crystallin via Pax6, c-Maf, CREB and a broad domain of lens-specific chromatin. EMBO J. 2006;25:2107–2118. doi: 10.1038/sj.emboj.7601114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecchin E., Filippi A., Biemar F., Tiso N., Pauls S., Ellertsdottir E., Gnugge L., Bortolussi M., Driever W., Argenton F. Distinct delta and jagged genes control sequential segregation of pancreatic cell types from precursor pools in zebrafish. Dev. Biol. 2007;301:192–204. doi: 10.1016/j.ydbio.2006.09.041. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Anderson D.J. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zhou Y.H., Zheng J.B., Gu X., Saunders G.F., Yung W.K. Novel PAX6 binding sites in the human genome and the role of repetitive elements in the evolution of gene regulation. Genome Res. 2002;12:1716–1722. doi: 10.1101/gr.188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials.