Abstract
Targeted gene disruption in the murine TOP3β gene-encoding DNA topoisomerase IIIβ was carried out. In contrast to the embryonic lethality of mutant mice lacking DNA topoisomerase IIIα, top3β−/− nulls are viable and grow to maturity with no apparent defects. Mice lacking DNA topoisomerase IIIβ have a shorter life expectancy than their wild-type littermates, however. The mean lifespan of the top3β−/− mice is about 15 months, whereas that of their wild-type littermates is longer than 2 years. Mortality of the top3β−/− nulls appears to correlate with lesions in multiple organs, including hypertrophy of the spleen and submandibular lymph nodes, glomerulonephritis, and perivascular infiltrates in various organs. Because the DNA topoisomerase III isozymes are likely to interact with helicases of the RecQ family, enzymes that include the determinants of human Bloom, Werner, and Rothmund–Thomson syndromes, the shortened lifespan of top3β−/− mice points to the possibility that the DNA topoisomerase III isozymes might be involved in the pathogenesis of progeroid syndromes caused by defective RecQ helicases.
Keywords: RecQ helicases, human progeroid syndromes
In human as well as in mouse, DNA topoisomerase IIIβ is the newest member of the DNA topoisomerase family (1, 2). It belongs to the type IA subfamily of DNA topoisomerases that are characterized by their transient breakage of a DNA strand by transesterification between an enzyme tyrosyl group and a DNA 5′ phosphoryl group (3, 4). The type IA enzymes also display a strong specificity for negatively supercoiled DNA and DNA containing single-stranded regions (3, 4). Biochemically, mammalian DNA topoisomerase IIIβ resembles mammalian DNA topoisomerase IIIα as well as bacterial and yeast DNA topoisomerase III (2).
Information on the cellular functions of the type IA enzymes mostly is derived from studies of bacteria and yeasts. In bacteria, there are two type IA enzymes, DNA topoisomerases I and III. The major role of bacterial DNA topoisomerase I, encoded by the topA gene, appears to be the prevention of excessive negative supercoiling of intracellular DNA (3–5). The lethality resulting from inactivating Escherichia coli DNA topoisomerase I, for example, can be alleviated by mutations that reduce the cellular level of DNA gyrase (6–8), an enzyme that catalyzes DNA-negative supercoiling, or by the expression of eukaryotic or vaccinia virus DNA topoisomerase I (9, 10), a type IB enzyme that relaxes positively or negatively supercoiled DNA but is structurally and mechanistically very different from the type IA enzymes (3). The prominent role of E. coli DNA topoisomerase I in the removal of negative supercoils indicates that the other type IA enzyme in E. coli, the topB-encoded DNA topoisomerase III, is ineffective in this role. Purified E. coli DNA topoisomerase III is proficient in linking or unlinking DNA single strands, but is inefficient in the relaxation of negatively supercoiled DNAs (11). The two type IA E. coli enzymes may share, however, an essential cellular function unrelated to the regulation of DNA supercoiling; attempts of obtaining mutants lacking both DNA topoisomerases I and III, in cells with a compensatory mutation for topA, were unsuccessful (12).
Similar to E. coli DNA topoisomerase III, the product of the TOP3 gene of the budding yeast Saccharomyces cerevisiae is also inefficient in the removal of DNA negative supercoils (13). In yeasts, DNA topoisomerase III is the sole type IA enzyme. Nevertheless, despite the presence of DNA topoisomerases I and II, both of which are efficient in relaxing supercoiled DNA (3), S. cerevisiae top3 nulls display a range of phenotypes including slow growth, hyperrecombination between repetitive sequences, defective sporulation, and sensitivity to DNA-damaging agents (14–16). In the case of the fission yeast Schizosaccharomyces pombe, viability of cells lacking DNA topoisomerase III is severely compromised (17, 18). These results suggest that in yeasts, DNA topoisomerase III plays an important role or roles other than the removal of DNA-negative supercoils.
For S. cerevisiae top3 nulls, a number of phenotypes are suppressed by deleting the SGS1 gene encoding a DNA helicase (19). Yeast Sgs1 helicase is a member of the RecQ helicase family that includes E. coli RecQ, S. pombe Rqh1, and human RECQ1, BLM, WRN, RECQ4, and RECQ5 (20–22). Partial suppression of S. pombe top3 mutations by mutations in rqh1 also has been shown (17, 18). A large number of studies of the RecQ helicases suggest that these enzymes are involved in processing DNA intermediates that are formed in various cellular processes. It has been suggested, for example, that E. coli RecQ protein is involved in recombination and repair of double-stranded DNA breaks (23, 24), suppressing illegitimate recombination (25), and processing nascent DNA at a blocked replication fork (26). In S. cerevisiae, inactivation of Sgs1 leads to elevated mitotic recombination, aberrant chromosome segregation, and sensitivity to hydroxyurea and methylmethanesulfonate (19, 27–31). Among the human RECQ helicases, BLM and WRN were shown to be the determinants of the Bloom and Werner syndrome, respectively (32, 33), and two kindreds with Rothmund–Thomson syndrome were shown to segregate with mutations in RECQ4 (34). These RecQ helicase syndromes have been reported to exhibit genetic instability, and one of the clinical manifestations of Werner and Rothmund–Thomson syndromes patients is premature aging (35–37). Interestingly, sgs1 nulls also appear to exhibit a reduced lifespan, defined for the budding yeast as the total number of buds a mother cell can produce (38).
The identification of SGS1 as a suppressor of TOP3 suggests that Sgs1 helicase and DNA topoisomerase III are involved in the same cellular pathway and that Sgs1 helicase may create a deleterious intermediate in the absence of Top3 (19). Two-hybrid assays and biochemical measurements indicate that the two yeast proteins physically interact and that the amino-terminal region of the helicase is important in this interaction (19, 29, 38–41). Physical association between BLM and DNA topoisomerase IIIα (42, 43) and between the β-isoform of RECQ5 and DNA topoisomerase IIIα or IIIβ (44) also was reported. These results are reminiscent of the finding that the enzyme “reverse gyrase” of the archeon Sulfolobus acidocaldarius is composed of a type IA DNA topoisomerase fused to a polypeptide with DNA helicase motifs (45). It is uncertain, however, whether the archaeal enzyme provides an adequate model for a RecQ helicase–DNA topoisomerase pair. Reverse gyrase, which catalyzes ATP-dependent positive supercoiling of DNA, does not appear to exhibit a DNA helicase activity (46); conversely, to date, it has yet to be demonstrated that the combination of a eukaryotic RecQ helicase and a DNA topoisomerase III gives a DNA-supercoiling activity. Extensive biochemical studies indicate that all type IA DNA topoisomerases, including E. coli DNA topoisomerases I and III and yeast DNA topoisomerase III, are effective in the transport of one DNA single strand through another (3, 4). Thus, the most likely function of DNA topoisomerase III in the yeasts is the formation or resolution of DNA structures with intertwined single strands. The molecular details of such structures are yet to be identified, however.
There have been few studies of the physiological roles of mammalian DNA topoisomerases IIIα and IIIβ. We have reported previously that mouse embryos with a deletion in both copies of the TOP3α gene die shortly after implantation (47). Thus, mammalian DNA topoisomerase IIIβ apparently cannot supplant DNA topoisomerase IIIα, despite their very similar enzymatic characteristics. To gain insight into the function of mammalian DNA topoisomerase IIIβ, targeted deletion in the mouse TOP3β gene was carried out. We report here that top3β−/− mice lacking DNA topoisomerase IIIβ are viable and develop normally to maturity, but their mean lifespan is much shortened relative to their TOP3β+/+ or top3β+/− littermates. As they age, the top3β−/− nullizygotes show a high propensity of ulcerative dermatitis and lesions in multiple internal organs.
Experimental Procedures
Cloning of a Mouse TOP3β Segment for Targeted Disruption of the Gene.
A search of GenBank for mouse expression sequence tags homologous to the coding sequences of human DNA topoisomerase IIIβ (GenBank accession number AB015799) identified two entries, W50106 and W82793, and these clones were acquired and partially sequenced (the sequence of a full-length mouse TOP3β cDNA had been reported since; see ref. 2). From the sequencing results, several pairs of oligonucleotide primers were synthesized and used to amplify segments of mouse DNA by PCR. These amplified DNA samples then were analyzed by nested PCR, using primer pairs based on the known TOP3α and TOP3β cDNA sequences, to select a DNA segment derived from the TOP3β locus. A 3-kb product amplified from genomic DNA was chosen and used in the preparation of a 32P-labeled probe for screening of a mouse genomic DNA library in phage λ-FIX II (Stratagene). Several clones were picked and purified for further characterization.
To map the restriction sites flanking the active-site tyrosine codon Tyr-336 of DNA topoisomerase IIIβ, DNA from one of the genomic TOP3β clones obtained from the above screen was used in separate PCRs by using a reagent kit for obtaining long DNA products (Boehringer Mannheim). In the λ-FIX II library, mouse DNA inserts were placed between phage T3 and T7 promoter sequences. Two different pairs of primers, therefore, were used in restriction mapping: 5′-GGGCTATATCAGCTACCCACGGAC-3′ and a “T3 primer” (5′-AATTAACCCTCACTAAAGGG-3′), and 5′-TTCTGTCCGTGGGTAGCTGATATAGC-3′ and a “T7 primer” (5′-AATACGACTCACTATAGGG-3′). In each pair, the selection of the primer other than the T3 or T7 primer is based on the sequence of the exon containing Tyr-336. For each primer pair, two uniquely end-labeled PCR products were obtained by 32P-labeling of one of the primers at its 5′ end. Partial or complete digestion of the set of four uniquely end-labeled PCR products then was carried out, and the positions of the restriction sites were deduced from the sizes of the radiolabeled fragments.
Targeted Disruption of Mouse TOP3β.
From the map of the restriction sites, a 3.5-kb SacI-EcoRI fragment and a 4.5-kb SacI-XbaI fragment were chosen as the homologous arms for the targeted deletion of the TOP3β region that contains the active-site tyrosine codon. These fragments were inserted into pPGKNeo/TK (kindly provided by A. McMahon, Harvard University, Cambridge, MA) to give the targeting vector depicted in Results.
Electroporation of murine TC1 embryonic stem cells from strain 129SvEv (kindly provided by P. Leder and C. Deng, Harvard Medical School, Cambridge, MA) and selection of clones by plating the transfected cells in media containing 200 μg/ml G418 and 2 μM ganciclovir were carried out according to standard protocols (48). A total of 282 neomycin- and ganciclovir-resistant colonies were selected, and DNA samples prepared from these were screened by Southern blot hybridization, using radiolabeled probes that lie outside the homologous regions between the targeting vector and chromosomal TOP3β (see Results). Thirteen clones were found to carry the mutated allele in TOP3β.
Generation of top3β Mice.
Two of the 13 top3β clones from the screen described above were injected into C57BL/6 or BALB/c blastocysts to generate chimeric animals (carried out by the Center for Animal Resource and Comparative Medicine, Brigham and Women's Hospital, Boston). Male chimeras capable of transmitting the altered top3β allele were selected and crossed with female C57BL/6 mice (purchased from Taconic Farms), and the F1s from these crosses were genotyped (see below). The top3β+/− heterozygotes then were intercrossed to produce homozygous top3β−/− mice. During the early phase of this work, several F1 top3β+/− animals also were mated first with female C57BL/6 mice to produce a larger number of top3β+/−, which then were intercrossed.
For genotyping the TOP3β loci, DNA samples were prepared from tail snippets of mice by the use of a commercial reagent kit (Qiagen) or by phenol/chloroform extraction (48). These samples were examined by Southern blot hybridization, as described for screening of top3β+/− embryonic stem cells, or by a PCR assay using three primers: the first lies within the neo-cassette (5′-GCAGGCATGCTGGGGATGCGG-3′), the second lies within the TOP3β exon containing the active-site tyrosine (5′- GTGAAGATGCTAGAGAAGCAGACG-3′), and the third (5′-GAGGCCTGGGTCCATGGGTTATAC-3′) lies downstream of both. The third oligonucleotide served as the common primer, which combined with the first primer to give a 226-bp product derived from the disrupted allele or the second primer to give a 281-bp product derived from the unaltered allele.
Distribution of TOP3β mRNA.
Relative abundance of murine TOP3β message in various tissues and in embryos at different developmental stages was examined by probing an array of immobilized dots of poly(A)+ RNA samples (CLONTECH) with a 32P-labeled probe prepared by random copying of a gel-purified PCR product spanning base pairs 1858–2589 of the TOP3β coding sequence (2). According to the supplier, the amounts of poly(A)+ RNA samples dotted on the membrane had been normalized to the expression levels of a set of housekeeping genes, and, thus, the amount of radiolabel in a particular dot should reflect the relative amount of TOP3β message in that sample. Hybridization was done according to the protocol provided by the supplier. After hybridization, the amounts of radiolabel in the dots were quantitated by the use of a phosphoimager (Fuji).
Examination of TOP3β mRNA in Wild-Type and Mutant Mice.
Samples of liver RNA were prepared from mice of different genotypes by fusing reagents supplied by Life Technologies (Gaithersburg, MD) and following the manufacturer's instructions. A total of 2 μg of each RNA sample was used in 20 μl of a reverse transcriptase (RT) reaction containing 0.5 mM of each dNTP, 200 ng of dT16, 50 mM Tris⋅HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, and 200 units of Moloney murine leukemia virus RT. The reaction mixture was incubated at 37°C for 1 h to synthesize the cDNA strand complementary to mRNA. About 1/20 of the RT product then was used in a PCR, carried out in the Perkin–Elmer PCR buffer containing 2.5 mM MgCl2, 0.2 μM of each dNTP, 2 units of Taq polymerase, and 50 pmol of each of four primers: the first two (5′-GGAGATTGCACAGATGTTTTTAAAC-3′ and 5′-TTCTGTCCGTGGGTAGCTGATATAGC-3′) for the amplification of a 215-nt stretch of TOP3β coding sequence, and the other two (5′- GGCCCAGAGCAAGAGAGGTATCC-3′ and 5′-ACGCACGATTTCCCTCTCAGC-3′) for the amplification of a 460-nt stretch of β-actin, which served as an internal control. The TOP3β primers are located in two exons separated by an intron, and, therefore, the 215-bp product could originate only from the reverse-transcribed mRNA and not from genomic DNA. Each PCR mixture initially was heated to 94°C for 5 min and then subjected to 35 cycles of PCR (30 s at each of the temperature steps of 94°, 60°, and 72°C). After a final incubation at 72°C for 5 min, the products of each reaction were analyzed by agarose gel electrophoresis.
Results
Generation of top3β−/− Mice.
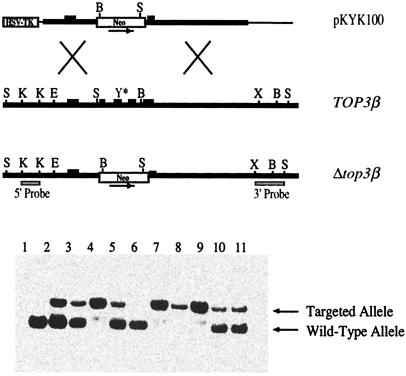
Fig. 1 Upper depicts sketches of the targeting vector, the region of TOP3β for targeted gene disruption, and the expected top3β allele from appropriate homologous recombination events between the two. The targeting construct was designed to replace a segment of TOP3β between nucleotides 777 and 1490 of the coding sequences (2) by a neomycin-resistance cassette. Several exons, including one containing the active-site tyrosine codon (Tyr-336) of murine DNA topoisomerase IIIβ, lie within this TOP3β segment; thus, replacing this segment by the neo-marker is expected to inactivate the gene.
Figure 1.
Targeted disruption of mouse TOP3β gene. (Upper) Sketches of the relevant regions of the targeting vector pKYK100, the endogenous TOP3β locus, and the expected Δtop3β allele. B, E, K, S, and X denote BglI, EcoRI, KpnI, SacI, and XbaI restriction sites, respectively. The 5′ and 3′ probes used in genotyping were derived from regions depicted under the Δtop3β allele. (Lower) Representative results of genotyping by Southern blot hybridization; the 5′ probe was used in this experiment.
Transfection of strain 129SvEv embryonic stem cells with the targeting vector, identification of colonies carrying a mutated copy of top3β, injection of the selected cells to obtain male germ-line chimeras, and crossing these chimeras with strain C57BL/6 females to obtain top3β+/− F1s were carried out as described in Experimental Procedures. The heterozygous top3β+/− F1s developed normally to adulthood and were phenotypically indistinguishable from their TOP3β+/+ littermates.
Mice Lacking DNA Topoisomerase IIIβ Are Viable and Develop Normally to Maturity.
In contrast to the embryonic lethality of mice deficient in DNA topoisomerase IIIα (47), mice lacking DNA topoisomerase IIIβ are viable. Intercrossing the top3β+/− F1s produced all three expected genotypes: TOP3β+/+, top3β+/−, and top3β−/− (Fig. 1 Lower). The ratio of wild type to heterozygotes to homozygotes for a total of 223 newborns was 1:1.68:0.80, which is not significantly different from the expected Mendelian ratio of 1:2:1.
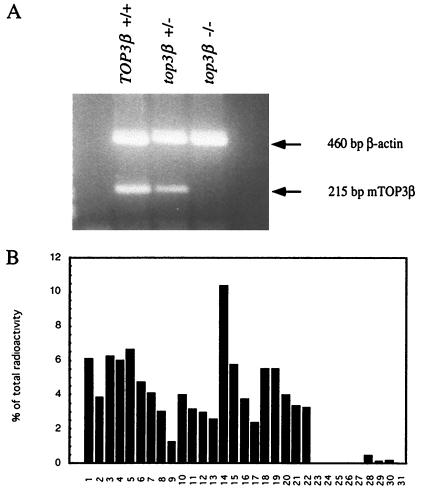
Homozygous top3β−/− mice, similar to the top3β+/− animals, developed to adulthood with apparent normalcy and showed no significant differences in size, weight, and agility relative to the wild-type TOP3β+/+ cohort at comparable stages of development. Examination of samples of liver RNA by a RT-PCR assay showed that TOP3β mRNA was undetectable in samples from top3β−/− mice but readily detectable in samples from TOP3β+/+ and top3β+/− mice (Fig. 2A). This result confirmed the expected absence of the TOP3β message and, thus, a functional DNA topoisomerase IIIβ in the nullizygotes. Clearly, DNA topoisomerase IIIβ is dispensable in embryonic development as well as during growth to maturity.
Figure 2.
(A) Analysis of message by RT and PCR (23). RNA samples prepared from the livers of TOP3β+/+, top3β+/−, and top3β−/− mice were reverse-transcribed and then amplified by PCR to give a 215-bp product. As an internal control, a 460-bp segment derived from β-actin message also was amplified in the PCR. (B) Relative amounts of TOP3β mRNA in embryos and various tissues of TOP3β+/+ mice. Lanes: 1, brain; 2, eye; 3, liver; 4, lung; 5, kidney; 6, heart; 7, skeletal muscle; 8, smooth muscle; 9, pancreas; 10, thyroid; 11, thymus; 12, submaxillary gland; 13, spleen; 14, testis; 15, ovary; 16, prostate; 17, epididymus; 18, uterus; 19–22, embryos 7, 11, 15, and 17 days old, respectively; 23, yeast total RNA; 24, yeast tRNA; 25, E. coli rRNA; 26, E. coli DNA; 27, poly(A)+ RNA; 28, Cot-1 DNA; 29, 100 ng mouse genomic DNA; 30, 500 ng mouse genomic DNA; 31, none.
The distribution of TOP3β message in different tissues of wild-type mouse also was examined. An array of membrane-attached poly(A)+ RNA samples was probed with a 32P-labeled TOP3β cDNA probe, and the relative amounts of TOP3β mRNA were estimated (Fig. 2B). The radiolabeled probe used in this experiment was prepared by random-primer labeling of a gel-purified PCR product spanning base pairs 1858–2589 of the TOP3β coding sequence (2). Within this region, the TOP3α and TOP3β coding sequences share little sequence homology, and no hybridization of the probe to mouse TOP3α cDNA was detected in a control experiment; thus, the presence of the TOP3α message in the membrane-bound RNA samples was not expected to contribute significantly to the 32P counts detected in the dot-blot. The results shown in Fig. 2B indicate that TOP3β mRNA is ubiquitously expressed in all tissues examined, with a higher level in the testis. This finding is in agreement with the earlier work of Seki et al. (2).
Shortened Lifespan of top3β−/− Mice.
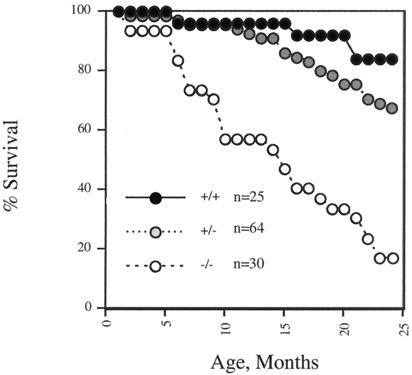
Whereas top3β−/− nulls developed to adulthood without any visible sign of abnormality, as they aged they showed a significantly higher mortality relative to their TOP3β+/+ and top3β+/− siblings of the same chronological age. Results from a total of 119 animals over a 2-year period are shown in Fig. 3. As described in Experimental Procedures, the top3β+/− heterozygotes used in these experiments were obtained from crossing founder male chimeras with female C57BL/6 or, in several cases, from a second round of breeding the top3β+/− mice produced with female C57 BL/6 to obtain a larger number of the heterozygotes; thus, all mice used in this study were in a 129SvEv-C57BL/6 mixed genetic background, with a roughly 50–75% contribution from the C57BL/6 strain.
Figure 3.
Mortality data for 25 TOP3β+/+ (●), 64 top3β+/− (shaded circles), and 30 top3β−/− (○) mice.
More than 80% of a group of TOP3β+/+ animals survived at the end of the 2-year period, whereas the medium lifespan of 30 top3β−/− nulls was about 15 months. The data shown in Fig. 3 are also suggestive of a reduction in the life expectancy of the group of 64 top3β+/− heterozygotes relative to their wild-type siblings, but the difference is small and its statistical significance is uncertain.
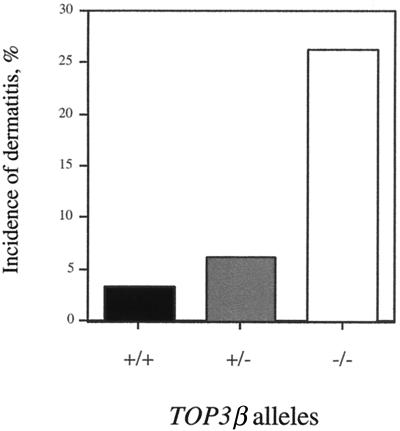
Dissection of two deceased animals, 8 and 10 months old, revealed lesions in multiple internal organs. The kidneys showed severe glomerulonephritis, the spleen was enlarged, with an increase in lymphocytic cells in the white pulp, and pancreatitis was noted in one of the animals. Hypertrophy of submandibular lymph nodes was also evident, and lymphocytic perivascular infiltrates were present in the salivary gland as well as in the lungs of one of the animals. To determine whether these lesions were already developing in top3β−/− mice that had yet to show apparent signs of physical deterioration, four 10-month-old nullizygotes were killed and dissected. Among these, one showed no significant lesion. Lesions similar to those described for the deceased mice, however, were seen in one or more of the others, although usually of a less severe nature. These include enlargement of the spleen and submandibular and axillary lymph nodes, hyperplasia of the pancreatic islet tissue, and perivascular infiltrates in the salivary glands and kidneys; in one case, gastric inflammation also was observed. These observations suggest that, as they age, the top3β−/− nullizygotes develop inflammatory responses of increasing severity in multiple organs. The underlying causes of these responses are unclear at present. Relative to their TOP3β+/+ and top3β+/− littermates, the top3β−/− nulls also showed a high propensity of ulcerative dermatitis (Fig. 4).
Figure 4.
Incidences of ulcerative dermatitis in wild-type and top3β mice 10–12 months of age. The cohort included 30 TOP3β+/+, 65 top3β+/−, and 38 top3β−/− animals.
Discussion
The findings that the type IA DNA topoisomerases are indispensable in bacteria and yeasts foretold their physiological importance in multicellular organisms. The major finding of the present study is that top3β−/− mice lacking DNA topoisomerase IIIβ can attain maturity without noticeable impairment, but their average lifespan is much shortened relative to that of their TOP3β+/+ littermates. Together with our earlier finding that mouse top3α−/− nullizygotes die during early embryogenesis (47), it is clear that in mice both DNA topoisomerases IIIα and IIIβ play important roles and that the two biochemically similar type IA DNA topoisomerases cannot fully substitute for each other.
The physical and functional interaction between RecQ helicase and DNA topoisomerase III in the yeasts (17–19, 29, 38–41), as well as evidence in favor of complex formation between mammalian RecQ helicases and type IA DNA topoisomerases (42–44), suggest that mammalian type IA DNA topoisomerases and RecQ helicases, similar to their yeast counterparts, interact functionally as well as physically. Thus, we suspect that the shortened life expectancy of the top3β−/− mice may reflect a key factor in the pathogenesis of the progeroid diseases resulting from defective RecQ helicases. Werner and Rothmund–Thomson patients are known to display signs of premature aging (35–37, 49). In the budding yeast, there is substantial evidence that a spectrum of phenotypes of sgs1 mutants probably reflect the effects of Sgs1 alterations on Top3 activity. Mutations in SGS1 lead to sensitivity to methylmethanesulfonate and hydroxyurea, hyperrecombination, aberrant chromosome segregation, and reduced sporulation efficiency and spore viability; these phenotypes are also seen in top3 mutants, sometimes in a more severe form (19, 27–30). Furthermore, deletion of the N-terminal region of Sgs1 that interacts with Top3 appears to lead to phenotypes similar to top3 nulls (29, 39, 40). Thus, some of the mutations in RecQ helicases may strongly affect the proper action of DNA topoisomerase III isozymes.
It remains unclear why the two rather similar type IA mammalian DNA topoisomerases cannot substitute for each other. The expression of the two could be regulated differently. Both DNA topoisomerase IIIα and IIIβ are expressed more prominently in testis than other tissues (2), but their expression patterns in different tissues at different developmental stages have not been examined. A second possibility is that the functions of the two enzymes are modulated by their specific interactions with other cellular proteins, especially the DNA helicases. The BLM protein, for example, is reported to bind DNA topoisomerase IIIα but not IIIβ (42). Given the functional importance of the physical interaction between yeast Sgs1 helicase and DNA topoisomerase III, the physiological roles of the mammalian type IA enzymes may be strongly influenced by their interaction with the particular RecQ helicases.
Acknowledgments
We thank Zhiping Liu and Wei Li for their help in the initial cloning of mouse TOP3β genomic DNA, Andy McMahon for materials and advice, Henry Warren for help in pathological examinations of mice, A. Sharpe and L. Du for performing blastocyst injections, and Nathan Ellis and Larry Loeb for their careful reading of this manuscript and suggestions. The generous gift of TC1 embryonic stem cells from P. Leder and C. Deng is also gratefully acknowledged. This work was supported by National Institutes of Health Research Grants CA47958 and GM24544.
Abbreviation
- RT
reverse transcriptase/transcription
References
- 1.Kawasaki K, Minoshima S, Nakato E, Shibuya K, Shintani A, Schmeits J L, Wang J, Shimizu N. Genome Res. 1997;7:250–261. doi: 10.1101/gr.7.3.250. [DOI] [PubMed] [Google Scholar]
- 2.Seki T, Seki M, Onodera R, Katada T, Enomoto T. J Biol Chem. 1998;273:28553–28556. doi: 10.1074/jbc.273.44.28553. [DOI] [PubMed] [Google Scholar]
- 3.Wang J C. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 4.Tse-Dinh Y C. Biochim Biophys Acta. 1998;1400:19–27. doi: 10.1016/s0167-4781(98)00125-0. [DOI] [PubMed] [Google Scholar]
- 5.Pruss G J, Manes S H, Drlica K. Cell. 1982;31:35–42. doi: 10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- 6.DiNardo S, Voelkel K A, Sternglanz R, Reynolds A E, Wright A. Cell. 1982;31:43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 7.Drlica K. Mol Microbiol. 1992;6:425–433. doi: 10.1111/j.1365-2958.1992.tb01486.x. [DOI] [PubMed] [Google Scholar]
- 8.Raji A, Zabel D J, Laufer C S, Depew R E. J Bacteriol. 1985;162:1173–1179. doi: 10.1128/jb.162.3.1173-1179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjornsti M-A, Wang J C. Proc Natl Acad Sci USA. 1987;84:8971–8975. doi: 10.1073/pnas.84.24.8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-Beros M E, Tse-Dinh Y C. J Bacteriol. 1992;174:7059–7062. doi: 10.1128/jb.174.21.7059-7062.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiGate R J, Marians K J. J Biol Chem. 1988;263:13366–13373. [PubMed] [Google Scholar]
- 12.Li Z, Hiasa H, Kumar U, DiGate R J. J Biol Chem. 1997;272:19582–19587. doi: 10.1074/jbc.272.31.19582. [DOI] [PubMed] [Google Scholar]
- 13.Kim R A, Wang J C. J Biol Chem. 1992;267:17178–17185. [PubMed] [Google Scholar]
- 14.Wallis J W, Chrebet G, Brodsky G, Rolfe M, Rothstein R. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 15.Gangloff S, de Massy B, Arthur L, Rothstein R, Fabre F. EMBO J. 1999;18:1701–1711. doi: 10.1093/emboj/18.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakraverty R K, Hickson I D. BioEssays. 1999;21:286–294. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 17.Goodwin A, Wang S W, Toda T, Norbury C, Hickson I D. Nucleic Acids Res. 1999;27:4050–4058. doi: 10.1093/nar/27.20.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maftahi M, Han C S, Langston L D, Hope J C, Zigouras N, Freyer G A. Nucleic Acids Res. 1999;27:4715–4724. doi: 10.1093/nar/27.24.4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gangloff S, McDonald J P, Bendixen C, Arthur L, Rothstein R. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis N A, German J. Hum Mol Genet. 1996;5:1457–1463. doi: 10.1093/hmg/5.supplement_1.1457. [DOI] [PubMed] [Google Scholar]
- 21.Shen J C, Loeb L A. Trends Genet. 2000;16:213–220. doi: 10.1016/s0168-9525(99)01970-8. [DOI] [PubMed] [Google Scholar]
- 22.Frei C, Gasser S M. J Cell Sci. 2000;113:2641–2646. doi: 10.1242/jcs.113.15.2641. [DOI] [PubMed] [Google Scholar]
- 23.Nakayama K, Irino N, Nakayama H. Mol Gen Genet. 1985;200:266–271. doi: 10.1007/BF00425434. [DOI] [PubMed] [Google Scholar]
- 24.Kusano K, Sunohara Y, Takahashi N, Yoshikura H, Kobayashi I. Proc Natl Acad Sci USA. 1994;91:1173–1177. doi: 10.1073/pnas.91.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H. Proc Natl Acad Sci USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Courcelle J, Hanawalt P C. Mol Gen Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- 27.Watt P M, Hickson I D, Borts R H, Louis E J. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamagata K, Kato J, Shimamoto A, Goto M, Furuichi Y, Ikeda H. Proc Natl Acad Sci USA. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullen J R, Kaliraman V, Brill S J. Genetics. 2000;154:1101–1114. doi: 10.1093/genetics/154.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saffi J, Pereira V R, Henriques J A. Curr Genet. 2000;37:75–78. doi: 10.1007/s002940050012. [DOI] [PubMed] [Google Scholar]
- 31.Frei C, Gasser S M. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 32.Ellis N A, Groden J, Ye T Z, Straughen J, Lennon D J, Ciocci S, Proytcheva M, German J. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 33.Yu C E, Oshima J, Fu Y H, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, et al. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 34.Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A. Genomics. 1998;54:443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- 35.Hilhorst-Hofstee Y, Shah N, Atherton D, Harper J I, Milla P, Winter R M. Clin Dysmorphol. 2000;9:79–85. doi: 10.1097/00019605-200009020-00001. [DOI] [PubMed] [Google Scholar]
- 36.Martin G M, Oshima J. Nature (London) 2000;408:263–266. doi: 10.1038/35041705. [DOI] [PubMed] [Google Scholar]
- 37.Nakura J, Ye L, Morishima A, Kohara K, Miki T. Cell Mol Life Sci. 2000;57:716–730. doi: 10.1007/s000180050036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinclair D A, Mills K, Guarente L. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 39.Bennett R J, Noirot-Gros M F, Wang J C. J Biol Chem. 2000;275:26898–26905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- 40.Duno M, Thomsen B, Westergaard O, Krejci L, Benedixen C. Mol Gen Genet. 2000;264:89–97. doi: 10.1007/s004380000286. [DOI] [PubMed] [Google Scholar]
- 41.Fricke W M, Kaliraman V, Brill S J. J Biol Chem. 2001;276:8848–8855. doi: 10.1074/jbc.M009719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu L, Davies S L, North P S, Goulaouic H, Riou J F, Turley H, Gatter K C, Hickson I D. J Biol Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 43.Johnson F B, Lombard D B, Neff N F, Mastrangelo M A, Dewolf W, Ellis N A, Marciniak R A, Yin Y, Jaenisch R, Guarente L. Cancer Res. 2000;60:1162–1167. [PubMed] [Google Scholar]
- 44.Shimamoto A, Nishikawa K, Kitao S, Furuichi Y. Nucleic Acids Res. 2000;28:1647–1655. doi: 10.1093/nar/28.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Confalonieri F, Elie C, Nadal M, de La Tour C, Forterre P, Duguet M. Proc Natl Acad Sci USA. 1993;90:4753–4757. doi: 10.1073/pnas.90.10.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Declais A C, Marsault J, Confalonieri F, de La Tour C B, Duguet M. J Biol Chem. 2000;275:19498–19504. doi: 10.1074/jbc.M910091199. [DOI] [PubMed] [Google Scholar]
- 47.Li W, Wang J C. Proc Natl Acad Sci USA. 1998;95:1010–1013. doi: 10.1073/pnas.95.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 49.Lindor N M, Furuichi Y, Kitao S, Shimamoto A, Arndt C, Jalal S. Am J Med Genet. 2000;90:223–228. doi: 10.1002/(sici)1096-8628(20000131)90:3<223::aid-ajmg7>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]