Abstract
Objectives
To investigate the expression of recently identified human mucin genes in an in vitro model of cultured mouse middle ear epithelial cells (MMEEC).
Methods
MMEEC were established, RNA was extracted and primers were designed for RT-PCR to assess for expression of mucin genes Muc1, Muc2, Muc3, Muc4, Muc5AC, Muc5B, Muc6, Muc7, Muc8, Muc9, Muc10, Muc11/12, Muc13, Muc15, Muc16, Muc17, Muc18, Muc19 and Muc20 expression.
Results
Mucin genes Muc1, Muc2, Muc3, Muc4, Muc5AC, Muc5B, Muc9, Muc10, Muc13, Muc15, Muc16, Muc18, Muc19 and Muc20 were identified and expressed in MMEEC. The genes Muc6, Muc7, Muc8, Muc11/12 and Muc17 were not identified.
Conclusion
Many of the mucin genes that have been recently identified in human MEE and chinchilla MEE are also expressed in MMEEC. There are differences in expression, however, which may have implications in utilizing various animal models for study of middle ear physiology and pathogenesis; specifically as it relates to mucin gene expression.
Keywords: mucin gene, mouse, middle ear epithelium, otitis media
Introduction
Recent efforts to understand the molecular underpinnings of otitis media (OM) have highlighted the importance of mucin gene expression and regulation in the middle ear epithelium (MEE) [1–4]. Composed primarily of respiratory-type epithelium, the ME contains numerous mucin secreting cells. As a family of glycoproteins, there are currently 19 uniquely described mucin genes that have been identified. In the epithelium throughout the respiratory tract, including the ME, mucins participate in a number of processes important for the protection and function of the underlying epithelium. These include mechanical protection of the epithelium, mucociliary clearance of pathogens and particulate matter, antigen presentation, and prevention of pathogen adherence and host invasion [1,5]. In the ME, mucins determine ME fluid viscosity and are one of the important components in determining how readily ME fluid is cleared from the ME through the eustachian tube. Although more viscous fluid may provide a greater barrier to pathogen destruction and invasion it also can result in increased difficulty in mucociliary clearance leading to mucostasis and associated hearing loss in children. Each mucin gene product has somewhat different characteristics and, as such, understanding the totality of mucin gene expression in any given experimental model is important for allowing conclusions utilizing that particular model.
The specific aim of this current investigation was to perform a comprehensive investigation of mucin gene expression in mouse ME epithelium from an in vitro murine source to characterize the mucin gene expression in this model of MEE and compare these findings to an in vivo mouse ME model. This characterization will provide the foundation for future investigations into the potential role of these various mucins in the physiology and pathophysiology of the MEE and specifically as these mucins relate to otitis media.
Methods
Mouse Middle Ear
Mouse middle ear materials were obtained from 8 adult 129Sv mice. Animal handlings were in accordance with IACUC approved protocol. Following euthanasia, temporal bones were harvested and the middle ear cavity was rinsed with TRIzol reagent (Invitrogen). Total RNA was extracted following the manufacturer’s instruction.
Mouse Middle Ear Epithelial Cell Cultures (MMEEC)
Cells used in mouse middle ear cultures were MMEEC whose primary characterization regarding transformation and growth properties have been published [6]. MMEEC were grown in full growth media comprised Ham’s F-12K media supplemented with 1.5g/L sodium bicarbonate, 2mM L-glutamine (ATCC), 10ng/ml epidermal growth factor (EGF, R&D Systems), 10mg/ml insulin-transferrin-sodium selenite (Sigma), 2.7g/L glucose, 500ng/ml hydrocortisone, 0.1mM non-essential amino acids (Invitrogen), 4% fetal bovine serum (Invitrogen) and 1% Penicillin/streptomycin (Cambrex). Culture was incubated at 33°C in humidified incubator containing 5% CO2. Media was changed every 3 days.
Mouse Mucin Primers
Each of the mucin genes studied has been primarily identified in a tissue outside of the middle ear [7]. Primer pairs utilized for measurement of mucin genes in the MMEEC are listed in Table 1. References to primer sequence are listed as PubMed ID number or the probe sequence ID for the published sequence, and GenBank accession number of the sequence used in primer design. Utilizing this approach, primers were readily obtained for mucin genes: Muc1, Muc2, Muc3, Muc4, Muc5AC, Muc5B, Muc6, Muc9, Muc10, Muc13, Muc15, Muc16, Muc18, Muc19 and Muc20. However, the mouse genome would suggest that, in the mouse, there do not exist corresponding genes for Muc 7, Muc 8, Muc 11/12 and Muc 17. To investigate the possibility of expression of genes corresponding to these previously identified human mucin genes primer pairs were developed utilizing human mucin gene sequences. As a designed set of primers failed to demonstrate an expression on RT-PCR additional steps to ensure the lack of expression were taken. This included designing and testing multiple sets of primer pairs with the potential to amplify non-identified mucin genes and literature search for possible related sequence.
Table 1.
Mouse mucin primer sequences.
| Gene | Fwd Primer | Amplicon (bp) |
RE | Expected size (bp) | PrimerReferences |
|---|---|---|---|---|---|
| Muc1 | TCTCC AGCCA CCAGC CCTCT AA TGGCC ATGGT AGGAG AAACA GG |
436 | KpnI SphI |
259+177 237+199 |
PMID: 16158528 |
| Muc2 | GGGAG GGTGG AAGTG GCATT GT TGCTG GGGTT TTTGT GAATC TC |
619 | SmaI PstI |
402+217 488+131 |
PMID: 16158528 |
| Muc3 | AACTG CAGCT ACGGC AAATG TC AGGTT TCGCC TACCA TCGTA AC |
656 | EcoRI | 529+127 | BC058768 |
| Muc4 | CATAT TCAAT ACCAC CGGTG TTC AAGGA TGGAA TTGGT GCTTT GTC |
466 | HinfI | 349+117 | NM_080457 |
| Muc5AC | CACCA TCTCT ACAAC CCAAA CT TGAGG TCCAG GTCTT TGTGT CT |
518 | PstI | 274+244 | PMID: 16158528 |
| Muc5B | GCCCT CACTG CCTCT GCTCC AC TTTTA CAGTG CCAGG GTTTA TT |
387 | BsmFI DdeI |
266+121 248+114+25 |
PMID: 16158528 |
| Muc6 | AGTCC TGCAG CCAGT CGTCA G GCACG CAGGC CTCAT AGTAG |
953 | BamHI PstI |
826+127 674+153+116+10 |
PMID: 12676567 |
| Muc9 | ACTTA TTATG GGTTT CCCCA CC TGGTG GTCTT AGAGA TCCCA GT |
745 | SmaI | 516+229 | Riboprobe ID: RP_050505_03_D08 |
| Muc10 | GGTTT CATTC CAAGC TCTCC TTAGG AGAAC GGCGA CTGAT |
101 | PMID: 16514118 | ||
| Muc13 | TCCCT GGGGA CATTA GCA GGCTA GGGAG GGTTC CAA |
919 | PvuII | 617+302 | Riboprobe ID: RP_050125_02_A11 |
| Muc15 | ATCCT TTACA GGTCT CCGAA CA CTGCA GCCAT CTTTC TCCTA AT |
639 | DdeI | 361+278 | Riboprobe ID:RP_060606_02_G12 |
| Muc16 | CCAAT CTACT GTACG GAGAA CATG CATAG AGACT GTCCT GATCC AG |
315 | PstI | 183+132 | XM_911929 |
| Muc18 | GGAAC CAACT ATTCA AGCCA ATG GGTTG AGGGT TGCCA TCTGT C |
429 | PstI | 337+92 | NM_023061 |
| Muc19 | GATTA TGCGA TTGGT TCATC CT GTGCA ATGTC CCTGA ACTCA TA |
349 | EcoRI PstI |
298+51 176+173 |
PMID: 12882755 |
| Muc20 | ACCCT TTGTA CCGAT GACAG CTCTG AAGAG CAAGC AGTGG ATGCA GATGT TGTAG GATG |
884 | HindIII | 508+376 | PMID: 14565953 |
Mucin gene expression
Mucin gene expression analysis was determined by using standard RT-PCR techniques. Total RNA of MMEEC was harvested using the RNeasy Mini Kit (Qiagen). Yield and purity was determined by spectrophotometry. Genomic DNA digestion was performed using RQ1 RNase-Free DNase set (Promega). cDNA was obtained using Superscript III RNase H− Reverse Transcriptase (Invitrogen), following the manufacturer instruction. Each reverse transcriptase reaction used 2 µg of the previously purified RNA. The cDNA was amplified in 50 µl reaction contained 1.0 unit Platinum Taq DNA polymerase (Invitrogen), 0.2mM each of dNTPs (Invitrogen), 0.2 µM of each primer, and 1 µl of cDNA template using GeneAmp 2400 thermocycler (Perkin-Elmer). A negative control contained every component except the reverse transcriptase was used for each reaction. The PCR reactions were run on 2% agarose gel at 100V. The PCR product was visualized with GelStar (Cambrex).
Reliability of the PCR product generated by the selected primer pairs and identified in the gels was further assessed by exposing the product to appropriate endonuclease digestion listed in Table 1. Ability to “cut” the PCR product into expected size bands was interpreted as confirmation that the generated cDNA sequence did, in fact, represent sequence from the mucin gene in question. In cases where enzymatic digestion was not possible the generated cDNA sequences were sequenced and confirmed by comparison to previously published sequence of the mucin gene in question.
Results
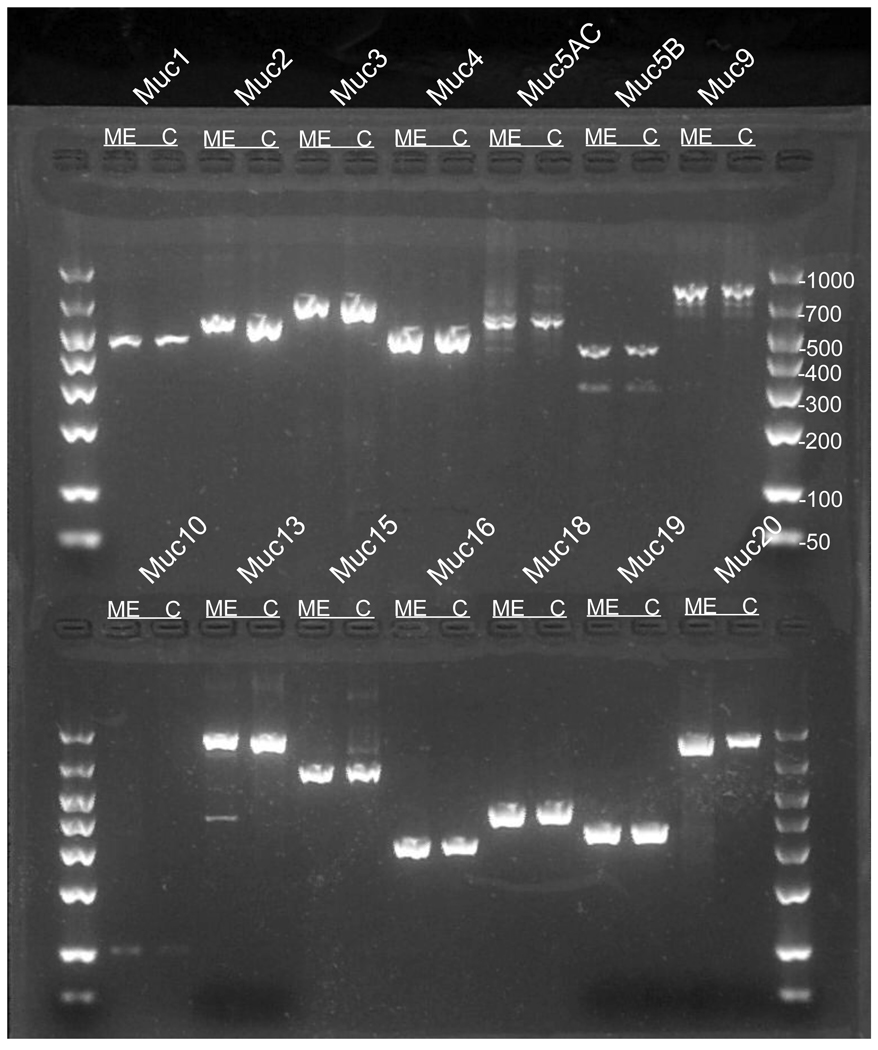
Primer pairs used for each of the mucin gene assessments in MMEEC are listed in Table 1. Mucin genes Muc1, Muc2, Muc3, Muc4, Muc5AC and Muc5B, which have been previously studied in mouse middle ear, were each assessed and found to be present in both in vivo and in vitro preparations (Figure 1).
Figure 1. Mucin gene expression in mouse middle ear epithelium.
Mucin amplicons generated from mouse middle ear epithelium (ME) and MMEEC (C) were analysis in agarose gel electrophoresis.
Of the more recently identified mucin genes Muc9, Muc10, Muc13, Muc15, Muc16, Muc18, Muc19 and Muc20 were identified.
Muc3 is not in the class of recently identified and characterized mucin genes. It has not been discussed previously as an important middle ear epithelial mucin and has not received identification as being present in most studies examining middle ear mucosal mucin gene expression. However, Muc3 was also identified in both in vitro and in vivo specimens in this investigation (Figure 1).
Use of restriction endonucleases (Table 1), to ensure the identified PCR product represented the mucin gene in question, demonstrated appropriate digested products to the corresponding gel bands for mucin genes Muc1, Muc2, Muc3, Muc4, Muc5AC, Muc5B, Muc9, Muc13, Muc15, Muc16, Muc18, Muc19, and Muc20. The Muc10 cDNA scarcely generated with the chosen primer pair, did not lend itself to enzymatic digestion and therefore was subjected to DNA sequencing. The sequence obtained demonstrated the complete homology to previously published sequence of mouse Muc10, GENBANK accession number NM_008644.
Muc6 was absent in the middle ear both in vivo and in vitro. Ability of the primer pair to amplify Muc6 was confirmed using mouse small intestine cDNA in which Muc6 was reported to be highly expressed. The obtained small intestine cDNA sequence demonstrated homology to the previously published mouse Muc6, GENBANK accession number NM_181729 (data not shown).
The result of mucin gene expression analysis in mouse middle ear epithelium both in vivo and in vitro specimens was summarized in Table 2.
Table 2.
Summary of mucin gene expression in study models: mouse middle ear epithelium; mME, mouse middle ear epithelial culture; MMEEC, human middle ear epithelium; HsME, and human middle ear epithelial culture; HMEEC. (presence; +, absence; −, not tested; nt).
| Mucin | mME | MMEEC | HsME | HMEEC |
|---|---|---|---|---|
| 1 | + | + | + | + |
| 2 | + | + | + | + |
| 3 | + | + | + | + |
| 4 | + | + | + | + |
| 5AC | + | + | + | + |
| 5B | + | + | + | + |
| 6 | − | − | − | − |
| 7 | n/t | n/t | + | + |
| 8 | n/t | n/t | + | + |
| 9 | + | + | + | + |
| 10 | + | + | n/t | n/t |
| 11/12 | n/t | n/t | + | + |
| 13 | + | + | + | + |
| 15 | + | + | + | + |
| 16 | + | + | + | + |
| 17 | n/t | n/t | − | − |
| 18 | + | + | + | + |
| 19 | + | + | + | + |
| 20 | + | + | + | + |
Discussion
Mouse models as an investigative paradigm for pathogenesis in many areas have become increasingly important with sequencing of the mouse genome. This scientific achievement has allowed for development of sophisticated molecular tools to assess genetic function, molecular interactions, protein properties and the ability to create specific in vivo constructs through utilization of knock-out, knock-in and knock-down techniques. Given these advantages, utilization of the mouse model in the study of the middle ear (ME) and otitis media (OM) pathogenesis has also received increased attention in recent years with many investigators utilizing mouse models as important research tools [7]. Advances in cell culture techniques have also enabled investigations of middle ear epithelium in culture providing a means to conduct in vitro investigations that are not limited by the variables and difficulties inherent in in vivo studies. Establishment of primary cell cultures, although technically feasible, is limited by the need for repeated harvest of tissue and inherent variability in underlying cultures obtained. Immortalization of middle ear tissue to allow for consistent and reliable cells for culture has developed as an alternative to repeated primary cell culture [6, 9]. Although the transformation of normal epithelial cells into immortalized cells provides challenges with respect to the potential disruption in normal phenotypic or genotypic expression, these techniques have proven effective in multiple laboratories. Importantly, these investigations have also demonstrated genetic, functional and mechanistic similarities between immortalized and normal in vivo epithelium rather than differences. [4, 10].
Having a reliable in vitro method to study MEE provides advantages in conducting focused molecular research into the physiology and pathophysiology of the ME. The results in this manuscript demonstrate complete homology between in vivo specimens and the in vitro cell culture model of mouse middle ear epithelium with regards to the mouse mucin genes Muc1, Muc2, Muc4, Muc5AC and Muc5B which have previously been demonstrated to be important in otitis media and otitis media with effusion [11]. In addition, this study characterized the expression of a number of new mucin genes that have been identified in other tissues and in human MEE [4] but have either not previously been reported or compared in these two model systems in the mouse. Of these recently identified mucin genes, Muc9, Muc13, Muc15, Muc16, Muc18, Muc19 and Muc20 were consistently identified in the in vitro model of MMEEC as well as in the ME of healthy mice without ME disease. In addition, the two models demonstrated complete homology with respect to expression of these mucin genes. The mucin gene Muc3, although less recently identified, was also found to be consistently present in both ME models. Further, in cases where mucin genes were found to not be expressed in vitro: Muc6, Muc7, Muc8, Muc11/12 and Muc17; these genes were also found to be absent in vivo. However, the mouse genome would suggest that, in the mouse, there do not exist corresponding genes for Muc 7, Muc 8, Muc 11/12 and Muc 17. [12, 13] These findings of complete homology of mucin gene expression and non-expression between in vitro and in vivo tissues would suggest that MMEEC models represent a reasonable surrogate for in vivo studies when investigating mouse ME mucin mechanisms.
However, the investigators acknowledge that certain limitations in these results should be considered with respect to glandular architecture and absence of complete cell-cell interactions, such as immunocytes, when considering ME pathophysiology and mucin interactions in this in vitro model system.
The importance of mucins in respiratory epithelium, in the ME in particular, and in the pathophysiology of OM has prompted numerous investigations into the function, regulation, and potential modulation of these large (500–600kDa) glycoproteins. Our laboratory and others have detailed the importance of mucin proteins with gel-forming qualities, MUC2 [3], MUC5AC [14–15], MUC5B [16–18] , and the more recently described MUC19 [19], given that these proteins are secreted. These secretory properties allow these mucins to function at the apical surface of the epithelium and beyond as they interact with pathogens and protect the underlying epithelium from invasion and mechanical damage. However, the ability to respond in this fashion also provides the opportunity for gel-forming mucin “over-production” leading to more highly viscous fluids with difficulty in ME clearance and subsequent hearing loss. This current investigation demonstrates that the MMEEC model is analogous to in vivo mouse models, as well as published human models, with respect to expression of these important gel-forming mucins (Table 2.) and provides a reasonable platform for further investigations.
It has also been recently recognized that in the human ME a much broader range of mucin genes are expressed than previously thought [4]. Given that individual mucin products have specific characteristics, size and interactions with the underlying epithelium and surrounding environment, including pathogens, it would be anticipated that the other mucin genes expressed also have an important role in the MEE. As such, our laboratory has investigated the ME for each of previously described mucins to assess the potential for expression and, ultimately, functionality of these other mucins in the ME. The work from this current investigation would suggest that there is significant uniformity between the mouse and human in not only the gel-forming mucins but also in other, membrane bound, mucins. Both human and mouse epithelia express MUC1, MUC4, MUC9, MUC10, MUC13, MUC15, MUC16, MUC18 and MUC20. And both epithelia do not express MUC6 or MUC17. However, there are differences in mucin gene expression between the mouse and the human. The lack of expression of Muc7, Muc8, Muc11/12 and Muc17 is notable in considering potential functions of these glycoproteins. Differential expression of these mucins has been reported in malignant and inflammatory diseases of epithelial cells [20–23].
Differences between human models and mouse models are certainly not unique to the ME. However, future investigations should be cognizant of these variations in designing and interpreting results, at least as they specifically relate to mucin function in the ME. However, the concordance between the MMEEC and mouse in vivo tissue samples regarding these genes also suggests that findings in either model system, as they relate to mouse ME physiology, may be translatable to the other mouse model.
Conclusion
This current investigation is the first to comprehensively analyze mouse MEE mucin gene expression. The results demonstrate that the in vitro MMEEC model has complete correlation to in vivo mouse tissue. There also exists complete homology between these mouse models and previously describe human models for most mucins which have been investigated to date. However, there do exist difference between the mouse and human models which warrant future examination.
Acknowledgements
This work was supported by NIH grants NIDCD: DC007903 (JEK), and was supported in part through funding provided by the Department of Otolaryngology and Communication Sciences, Medical College of Wisconsin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Portions of this manuscript presented at the Association for Research in Otolaryngology Annual Meeting, Baltimore, MD, February XY, 2009.
Conflict of interest statement
None of the authors of this manuscript have any financial or non-financial competing interests to disclose.
References
- 1.Kerschner JE, Meyer TK, Burrows A. Chinchilla middle ear epithelial mucin gene expression in response to inflammatory cytokines. Arch Otolaryngol Head Neck Surg. 2004 Oct;130(10):1163–1167. doi: 10.1001/archotol.130.10.1163. [DOI] [PubMed] [Google Scholar]
- 2.Samuel EA, Burrows A, Kerschner JE. Cytokine regulation of mucin secretion in a human middle ear epithelial model. Cytokine. 2008 Jan;41(1):38–43. doi: 10.1016/j.cyto.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ubell ML, Kerschner JE, Wackym PA, Burrows A. MUC2 expression in human middle ear epithelium of patients with otitis media. Arch Otolaryngol Head Neck Surg. 2008 Jan;134(1):39–44. doi: 10.1001/archoto.2007.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerschner JE. Mucin gene expression in human middle ear epithelium. American Laryngological, Rhinological and Otological Society Thesis. Laryngoscope. 2007 Sep;117(9):1666–1676. doi: 10.1097/MLG.0b013e31806db531. [DOI] [PubMed] [Google Scholar]
- 5.Diamond G, Legarda D, Ryan LK. The innate immune response of the respiratory epithelium. Immunol Rev. 2000 Feb;173:27–38. doi: 10.1034/j.1600-065x.2000.917304.x. [DOI] [PubMed] [Google Scholar]
- 6.Tsuchiya K, Kim Y, Ondrey FG, Lin J. Characterization of a temperature-sensitive mouse middle ear epithelial cell line. Acta Otolaryngol. 2005 Aug;125(8):823–829. doi: 10.1080/00016480510031533. [DOI] [PubMed] [Google Scholar]
- 7.Escande F, Porchet N, Bernigaud A, Petitprez D, Aubert JP, Buisine MP. The mouse secreted gel forming micin gene cluster. Biochem Biophys Acta. 2004 Feb 20;1676(3):240–250. doi: 10.1016/j.bbaexp.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Trune DR, Zheng QY. Mouse models for human otitis media. Brain Res. 2009 Jun 24;1277:90–103. doi: 10.1016/j.brainres.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun YM, Moon SK, Lee HY, Webster P, Brackmann DE, Rhim JS, Lim DJ. Immortalization of normal adult human middle ear epithelial cells using a retrovirus containing the E6/E7 genes of human papillomavirus type 16. Ann Otol Rhinol Laryngol. 2002 Jun;111(6):507–517. doi: 10.1177/000348940211100606. [DOI] [PubMed] [Google Scholar]
- 10.Shuto T, Imasato A, Jono H, Sakai A, Xu H, Watanabe T, Rixter DD, Kai H, Andalibi A, Linthicum F, Guan YL, Han J, Cato AC, Lim DJ, Akira S, Li JD. Glucocorticoids synergistically enhance nontypeable Haemophilus influenzae-induced Toll-like receptor 2 expression via a negative cross-talk with p38 MAP kinase. J Biol Chem. 2002 May 10;277(19):17263–17270. doi: 10.1074/jbc.M112190200. [DOI] [PubMed] [Google Scholar]
- 11.Kubba H, Pearson JP, Birchall JP. The aetiology of otitis media with effusion: a review. Clin Otolaryngol Allied Sci. 2000 Jun;25(3):181–194. doi: 10.1046/j.1365-2273.2000.00350.x. [DOI] [PubMed] [Google Scholar]
- 12.Lang T, Hansson GC, Samuelsson T. Gel-forming mucins appeared early in metazoan evolution. Proc Natl Acad Sci USA. 2007 Oct 9;104(41):16209–16214. doi: 10.1073/pnas.0705984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malmberg EK, Noaksson KA, Phillipson M, Johansson ME, Hinojosa-Kurtzberg M, Holm L, Gendler SJ, Hansson GC. Increased levels of mucins in the cystic fibrosis mouse small intestine, and modulator effects of the MUC1 mucin expression. Am J Physiol Gastrointest Liver Physiol. 2006 Aug;291(2):G203–G210. doi: 10.1152/ajpgi.00491.2005. [DOI] [PubMed] [Google Scholar]
- 14.Caramori G, Casolari P, Di Gregorio C, Saetta M, Baraldo S, Boschetto P, Ito K, Fabbri LM, Barnes PJ, Adcock IM, Cavallesco G, Chung KF, Papi A. MUC5AC expression is increased in bronchial submucosal glands of stable COPD patients. Histopathology. 2009 Sep;55(3):321–331. doi: 10.1111/j.1365-2559.2009.03377.x. [DOI] [PubMed] [Google Scholar]
- 15.Dogru M, Matsumoto Y, Okada N, Igarashi A, Fukagawa K, Shimazaki J, Tsubota K, Fujishima H. Alterations of the ocular surface epithelial MUC16 and globlet cell MUC5AC in patients with atopic keratoconjunctivitis. Allergy. 2008 Oct;63(10):1324–1334. doi: 10.1111/j.1398-9995.2008.01781.x. [DOI] [PubMed] [Google Scholar]
- 16.Kirkham S, Kolsum U, Rousseau K, Singh D, Vestbo J, Thornton DJ. MUC5B is the major mucin in the gel phase of sputum in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008 Nov 15;178(10):1033–1039. doi: 10.1164/rccm.200803-391OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elsheikh MN, Mahfouz ME. Up-regulation of MUC5AC and MUC5B mucin genes in nasopharyngeal respiratory mucosa and selective up-regulation of MUC5B in middle ear in padiatric otitis media with effusion. Laryngoscope. 2006 Mar;116(3):365–369. doi: 10.1097/01.MLG.0000195290.71090.A1. [DOI] [PubMed] [Google Scholar]
- 18.Lin J, Tsuboi Y, Rimell F, Liu G, Toyama K, Kawano H, Paparella MM, Ho SB. Expression of mucins in mucoid otitis media. J Assoc Res Otolaryngol. 2003 Sep;4(3):384–393. doi: 10.1007/s10162-002-3023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerschner JE, Khampang P, Erbe CB, Kolker A, Cioffi JA. Mucin gene 19 (MUC19) expression and response to inflammatory cytokines in middle ear epithelium. Glycoconj J. 2009 Dec;26(9):1275–1284. doi: 10.1007/s10719-009-9245-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samuels TL, Handler E, Syring ML, Pajewski NM, Blumin JH, Kerschner JE, Johnston N. Ann Otol Rhinol Laryngol. 2008 Sep;117(9):688–695. doi: 10.1177/000348940811700911. [DOI] [PubMed] [Google Scholar]
- 21.Packer LM, Williams SJ, Callaghan S, Gotley DC, McGuckin MA. Expression of the cell surface mucin gene family in adenocarcinomas. Int J Oncol. 2004 Oct;25(4):1119–1126. [PubMed] [Google Scholar]
- 22.Lee HM, Kim DH, Kim JM, Lee SH, Hwang SJ. MUC8 mucin gene up-regulation in chronic rhinosinusitis. Ann Otol Rhinol Laryngol. 2004 Aug;113(8):662–666. doi: 10.1177/000348940411300812. [DOI] [PubMed] [Google Scholar]
- 23.Choi JY, Kim JY, Kim CW, Ho JS, Lee KD, Yoo JB, Ahn YE, Yoon JH. IL-1beta promotes the ciliogenesis of human middle ear epithelial cells: possible linkage with the expression of mucin gene 8. Acta Otolaryngol. 2005 Mar;125(3):260–265. doi: 10.1080/00016480410022985. [DOI] [PubMed] [Google Scholar]