Abstract
It has been suggested that delayed DNA replication underlies fragility at common human fragile sites, but specific sequences responsible for expression of these inducible fragile sites have not been identified. One approach to identify such cis-acting sequences within the large nonexonic regions of fragile sites would be to identify conserved functional elements within orthologous fragile sites by interspecies sequence comparison. This study describes a comparison of orthologous fragile regions, the human FRA3B/FHIT and the murine Fra14A2/Fhit locus. We sequenced over 600 kbp of the mouse Fra14A2, covering the region orthologous to the fragile epicenter of FRA3B, and determined the Fhit deletion break points in a mouse kidney cancer cell line (RENCA). The murine Fra14A2 locus, like the human FRA3B, was characterized by a high AT content. Alignment of the two sequences showed that this fragile region was stable in evolution despite its susceptibility to mitotic recombination on inhibition of DNA replication. There were also several unusual highly conserved regions (HCRs). The positions of predicted matrix attachment regions (MARs), possibly related to replication origins, were not conserved. Of known fragile region landmarks, five cancer cell break points, one viral integration site, and one aphidicolin break cluster were located within or near HCRs. Thus, comparison of orthologous fragile regions has identified highly conserved sequences with possible functional roles in maintenance of fragility.
Characterization and sequencing of inherited fragile sites have determined the specific cause of fragility at a number of rare fragile sites; for example, FRA11B is caused by expansion of CGG triplets and FRA16B by AT rich minisatellite repeats (1, 2). These rare fragile sites were isolated by positional cloning of the relevant genomic loci from DNA of family members segregating these rare fragile sites. However, the common non-familial fragile sites (n < 100) are considered to be normal chromosome structures. To date, common fragile sites FRA3B (3), FRA7H (4), FRA7G (5), and FRA16D (6, 7) have been identified, cloned, and sequenced. Analyses and comparisons of these sequences have not revealed the mechanism of their fragility, although they revealed that common fragile sites are actually large fragile regions (≈150 to 1,000 kbp). Expanded repeats were not found in these fragile regions (4, 8, 9).
The human FRA3B locus at chromosome region 3p14.2 is the most inducible common fragile site, exhibiting apparent breaks in up to 50% of metaphases after exposure to aphidicolin (10). Deletions and structural rearrangements in FRA3B have been observed in a large fraction of tumor types. The tumor suppresser gene FHIT encompasses the FRA3B fragile region and is altered by deletion or translocation in many types of cancer, including those of lung, cervix, esophagus, bladder, and kidney carcinomas (11–16). We have been interested in the mechanism of fragility in this region and its contribution to cancer susceptibility. Previously, we have reported the sequence of a large portion of the human FHIT gene, including the epicenter of fragility surrounding exon 5 (8, 9). The sequence of the region enabled us to find the exact end points of deletions within FHIT/FRA3B in many cancer cell lines. These studies indicated that (i) the FRA3B locus is AT rich with numerous short and long repeats throughout the region, (ii) many cancer cell deletion end points are located near or in LINE elements, suggesting that homologous end joining was important in repair of fragile breaks (8, 9), (iii) triplet or minisatellites repeats are not the cause of fragility, and (iv) cancer deletion end points are not coincident with aphidicolin-induced breaks (8, 9, 17).
Recently, there have been several reports of comparisons of orthologous regions between two different species, e.g., human vs. mouse and human vs. dog (18–21). Generally, the sequences of introns and non-coding regions are quite different, presumably because there was no selective pressure for conservation. Thus, investigation of conserved sequences could be useful in the study of gene regulatory elements or other conserved functions. For example, Oeltjen et al. (22) have reported that conserved non-coding sequence of the Bruton's tyrosine kinase (BTK) loci regulates specific expression of BTK, and Loots et al. (23) have identified the regulator of gene expression of interleukins 4, 5, and 13 in the sequence conserved between human and mouse.
The murine Fhit locus has also been cloned and characterized (24, 25). It exhibits 10 exons with a start codon in exon 5, as in the human FHIT gene. The murine Fhit exon-intron structure, large size, and position near the Ptprg gene are also similar to the human FHIT gene. Interestingly, the mouse Fhit locus, near the centromere of mouse chromosome 14, is an aphidicolin inducible common fragile site (Fra14A2; ref. 24). In addition, several tumorigenic mouse cell lines exhibit homozygous deletion of Fhit exons (25). So, the mouse Fhit/Fra14A2 locus is an ideal model for the study of mechanisms of fragility of common fragile regions and their contribution to tumorigenesis. In this study, we sequenced over 600 kbp of the mouse Fra14A2 locus and compared it with the corresponding human FRA3B sequence to determine what the comparison would reveal about conservation of fragility and evolution of the Fhit gene.
Materials and Methods
DNA Sequencing Templates.
Bacterial artificial chromosome (BAC) clones including mouse Fhit exon 4 and 5 were identified by screening of a mouse BAC library by PCR amplification with primer pairs specific for exons 4 and 5. Overlapping BACs in intron 4 were identified by using the BAC end library database (http://www.tigr.org/tdb/bac_ends/mouse/bac_end_intro.html). Positions of BACs within the mouse Fhit gene are shown in Fig. 1. These BACs were obtained from Research Genetics (RPCI-145C4, RPCI-331I7, CITB-228L5, RPCI-255P3, RPCI-258D17, CITB-225H3, RPCI-409H24, CITB-167E2, RPCI-197H24, RPCI-19O13 and RPCI-223O23; mouse BAC library, Research Genetics, Huntsville, AL).
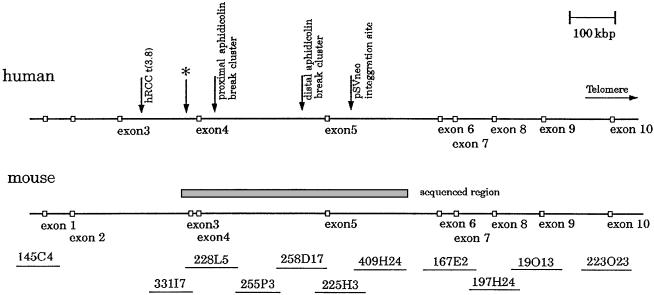
Figure 1.
The genomic structure of human and mouse Fhit loci. The top line indicates the position of known landmarks and exons. The gray box is the region sequenced in this study, which is covered by six BAC clones. The asterisk indicates the location of sequence corresponding to mouse Fhit exon 3.
BAC DNA was prepared by PSAI BAC DNA isolation kit (Princeton Separations, Adelphia, NJ) according to the manufacturer's protocol. Shotgun libraries of these BACs were constructed, sequenced, and assembled as previously described (8).
Computer Analysis of Sequence.
To search for other genes in this region, the final sequence was analyzed by using the genefinder (26) and genescan (27) programs. In addition, sequence homology searches were carried out against the GenBank and expressed sequence tag (EST) databases by the blastn and tblastn programs (28).
For the large-scale comparison between human FRA3B and mouse Fra14A2, we used the pipmaker computer program (http://bio.cse.psu.edu/pipmaker/; ref. 29). To identify highly conserved regions, we used strong hit computer program (http://bio.cse.psu.edu/pipmaker/tools.html). Repetitive elements were analyzed by the repeatmasker computer program (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker). We predicted the matrix attachment regions (MAR) by mar finder (http://www.ncgr.org/MarFinder/; ref. 30). The human and mouse sequences used in our analysis, along with annotations and detailed results, can be obtained at the website http://bio.cse.psu.edu/.
Inverse PCR.
We performed inverse PCR, as previously described (9, 31), to determine the deletion end points within the Fhit gene of RENCA cells, a cell line established from a primary carcinogen-induced renal cell carcinoma isolated from a BALB/c mouse.
Results and Discussion
Sequence of the Fra14A2/Fhit Region from Exon 3 to Intron 5.
The organization of this locus and the location of the BACs containing sequences from the FHIT gene are diagrammed in Fig. 1. Shotgun sequencing was performed for the region encompassing mouse Fhit exons 3, 4, and 5. Sequence fidelity was confirmed by amplification of identical sequence fragments from mouse DNA templates. The complete finished sequence of 612 kbp was submitted to GenBank (accession nos. AF332859, AF332860, AF332861, and AF332862). Several studies reported that human FRA3B is characterized by high AT content and repetitive elements distributed throughout (8, 9). GC content of the mouse and human Fhit regions thus far sequenced were 35.1% and 38.9%, respectively. Although there have been suggestions of other genes in this region (32), homology searches and gene prediction program analyses did not reveal putative genes aside from Fhit exons 3, 4, and 5.
Comparison of Human FRA3B and Mouse Fra14A2 Sequences.
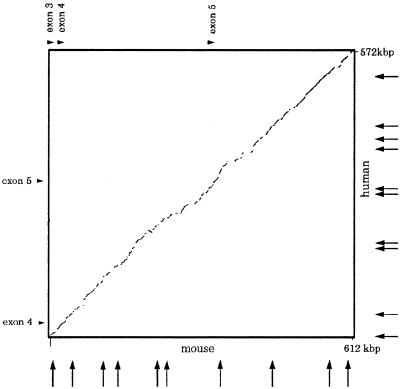
We Compared 600 kbp of the murine Fra14A2 and human FRA3B regions to analyze the sequence homology and distribution of repetitive elements. A dot plot (Fig. 2) shows the nearly linear pattern obtained by plotting human and mouse sequence. The weight percent identity was 72.6%, calculated by ΣCiFi/N, where Ci is the length in base pairs of each individual aligned sequence, Fi is percent identity of the aligned sequence, and N is total number in base pairs of compared sequence, as described previously (22).
Figure 2.
Dotplot comparison of human and mouse Fhit sequence. This plot was made by the advanced pipmaker program, which also eliminated the repetitive elements. The arrows on horizontal and vertical axes indicate the positions of matrix attachment regions, predicted by mar finder in human and mouse sequences. Note that the almost linear pattern of dotplot means that there were many conserved regions in conserved positions and that the MAR positions were different in mouse and human sequences.
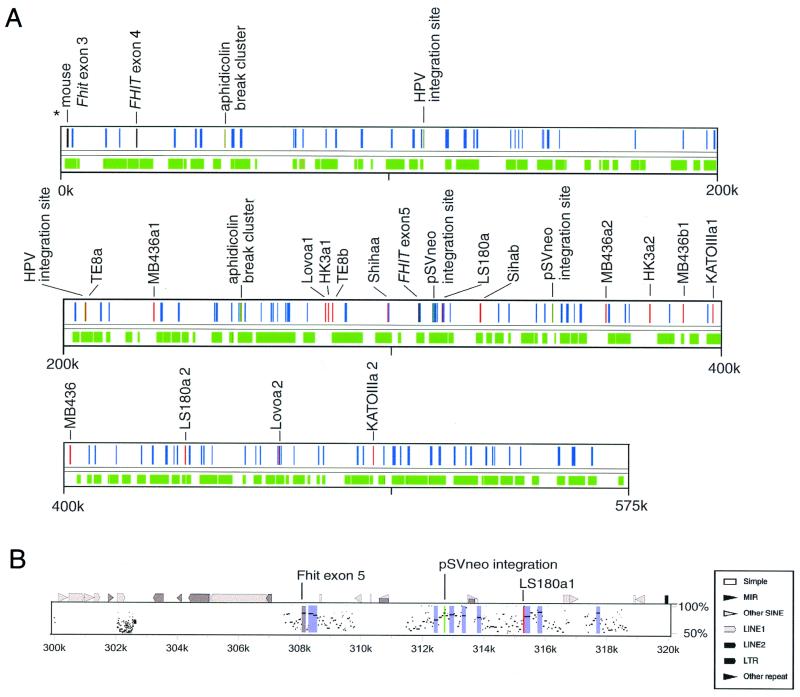
To visualize the small conserved regions in more detail, we analyzed the sequence by the advanced pipmaker program. This program transformed the alignment into percent positional identity plots (Pips) in the two sequences. Fig. 3A shows the positions of aligned regions over the entire sequence. The portion of region aligned by this program was 55.8%. Fig. 3B shows the pipmaker plots around exon 5. Human FHIT exon 5 (coding exon) was 85% homologous to mouse exon 5, and there were many homologous regions around exon 5.
Figure 3.
Alignment and landmarks. (A) The relation between landmarks of this region and conserved regions. Red, gray, and dark green bars indicate the positions of break points of cancer cell lines, FHIT gene exons, and aphidicolin break cluster or integration sites, respectively. Green zones are the regions aligned with mouse sequence, and blue zones are HCRs. (B) Percent identity plots in 20 kbp of the human sequence around exon 5. The top line shows the position of repeats (by repeatmasker) and several land marks. The gray zone is the region of FHIT exon 5, and the blue zones are HCRs. Note that FHIT exon 5 (coding exon) was 85% homologous to mouse sequence and that two landmarks, pSVneo integration sites and LS180a break point, are located within or near HCRs.
To assess the relative degree of conservation of this region, we analyzed the CFTR (cystic fibrosis gene) region. This region has low GC content, contains one gene, and is not fragile. The degree of conservation in these two regions is remarkably similar, except that the human sequence is shorter than the mouse around FHIT, and longer than the mouse around CFTR (Table 1). This result may suggest the low GC content of the two regions, CFTR and FHIT, as the basis for possibly spurious matches throughout the region, a possibility that will require further study.
Table 1.
Comparison between FHIT region and CFTR region
| FHIT | CFTR | |
|---|---|---|
| Region of human, bp | 571992 | 422439 |
| Region of mouse, bp | 610791 | 357088 |
| GC contents | 38.4% | 37.4% |
| Aligned portions in human | 55.8% | 48.7% |
| Aligned portions in mouse | 50.5% | 55.2% |
| Putative insertions in human | 25.0% | 31.9% |
| Putative insertions in mouse | 31.5% | 21.9% |
We hypothesized that there might be well-conserved sequences underlying fragility, specificity of splicing mechanisms, or other functions in the large FHIT introns. Such sequences should exhibit higher conservation than non-functional sequences. We extracted regions in the human sequence that we considered to be highly conserved (HCR) by a strong hit computer program. Specifically, we chose regions that align without a gap for at least 100 bp and with at least 70% nucleotide identity. These criteria gave 171 HCR segments of total length 26,851 bp. We searched for sequence homologies of HCRs against the GenBank and htgs (high throughput genomic sequence) databases by blastn programs, but found no homologous sequences in the human genome, including at other common fragile sites, FRA7G, FRA7H, and FRA16D.
We also extracted a set of regions that was not highly conserved. Specifically, we removed from the human sequence all exons, regions masked by repeatmasker, and HCRs, then retained each remaining interval of at least 100 bp, leaving 575 sequences of total length 300,935 bp (non-HCR). We looked for differences in composition between HCRs and non-HCRs. In terms of nucleotide content, they were essentially indistinguishable. In the conserved regions, the fraction of A, C, G, and T nucleotides were 30.4%, 17.6%, 18.8%, and 33.2%, respectively, whereas in the non-conserved regions they were 30.0%, 17.9%, 19.4%, and 32.8%. Among hexamers, we noted that strings with all A, or As and one G, were common in the non-conserved regions but much less common in the conserved regions. Overall, the difference in hexamer content between conserved and non-conserved regions was not statistically significant.
There have been several reports about the relation between fragile regions and replication elements. Wang et al. (17) reported that there were MARs and topoisomerase II consensus sequences around aphidicolin-induced break points. Palin et al. (33) reported that there were many replication-related elements, such as replication origin consensus, autonomous replicating consensus, and scaffold attachment consensus sequences in an aphidicolin-sensitive hamster sequence. We looked for MARs by computer prediction software. The mar finder software allows prediction of matrix attachment regions by analysis for several motifs: origin of replication, TG-rich sequence, and curved sequence (30). The software recognized 10 MAR candidates in the human and mouse Fhit sequences (Fig. 2), but the locations of MARs were not conserved. For a more detailed analysis, further information about the structure of the DNA replication signals for mammalian cells is required.
To investigate whether the fragility of the human FRA3B locus is reflected in a higher rate of genomic deletions over evolutionary time, we determined the positions of potential deletions in each species. From the human and mouse sequences lying between successive aligned regions, we removed all segments masked by repeatmasker, with the exception of MIR and LINE2 elements, which are believed to have inserted before human–mouse divergence. Our working assumption was that segments that have inserted since human–mouse divergence would be detected by repeatmasker, and hence any other large difference between the sequences could be ascribed to deletion. According to this objective approach, there were 17 deletions of at least 1,000 bp in the lineage leading to humans, and 22 in the lineage leading to mice. The largest 2 of these were in humans, indicating a deletion of 4,062 bp corresponding to contemporary human positions 110,363–117,298 and a deletion of 3,973 bp at 318,669–324,367. By comparison, in the shorter CFTR sequence (spanning 422,439 human bp, compared with 571,992 bp for the FHIT alignments) there were apparently 26 deletions of at least 1,000 bp in the human lineage, but only 10 in the mouse lineage. Thus, we concluded that fragility of FRA3B is not reflected by an unusual tendency to suffer deletions over evolutionary time.
Comparison of Repetitive Elements.
In the murine Fra14A2 locus, long interspersed nuclear elements (LINE, L1, and L2), short interspersed nuclear elements (B1 and MIR), elements with long terminal repeats (HERVs and MalRVs), and DNA transposons (mariner and MER) were spread throughout the region, as in the FRA3B region. Total interspersed repeats represented 32.6% of the sequenced region. The repeat contents of the mouse and human loci are summarized in Table 2.
Table 2.
Occurrence of individual repetitive elements within the FRA3B and FRA14A2 sequences
| Fra14A2 | Length, bp (percent of sequence) | FRA3B | Length, bp (percent of sequence) |
|---|---|---|---|
| SINEs | 32,995 (3.9%) | SINEs | 50,364 (8.8%) |
| B1s | 7,947 (0.9%) | ALUs | 34,004 (5.9%) |
| B2–B4 | 21,009 (2.5%) | MIRs | 16,360 (2.9%) |
| Ids | 454 (0.1%) | ||
| MIRs | 3,585 (0.4%) | ||
| LINEs | 178,340 (20.9%) | LINEs | 104,847 (18.3%) |
| LINE1 | 175,677 (20.6%) | LINE1 | 84,392 (14.8%) |
| LINE2 | 2,633 (0.3%) | LINE2 | 18,314 (3.2%) |
| LTR elements | 57,121 (6.7%) | LTR elements | 46,145 (8.2%) |
| MaLRs | 30,313 (3.6%) | MaLRs | 17,198 (3.0%) |
| Retroviral | 16,336 (1.9%) | Retroviral | 11,374 (2.0%) |
| MER4_group | 370 (0.0%) | MER4_group | 12,195 (2.1%) |
| DNA elements | 7,312 (0.9%) | DNA elements | 26,882 (4.7%) |
| MER1_type | 5,325 (0.6%) | MER1_type | 13,182 (2.3%) |
| MER2_type | 1,830 (0.2%) | MER2_type | 10,885 (1.9%) |
| Mariners | 0 (0.0%) | Mariners | 1,515 (0.3%) |
| Unclassified | 1,802 (0.2%) | Unclassified | 0 (0.0%) |
| Total repeats | 277,570 (32.6%) | Total repeats | 228,238 (39.9) |
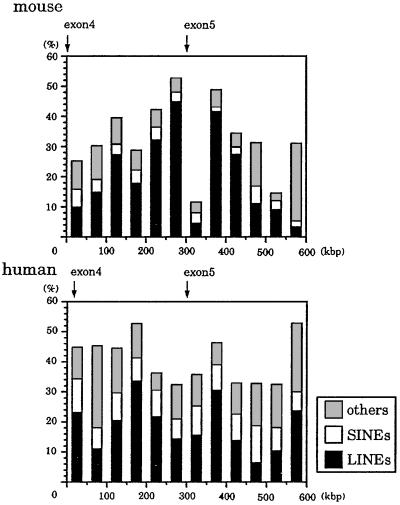
Fig. 4 shows the distribution of the LINE and SINE repeats in 50-kbp intervals of mouse and human sequence. In the entire orthologous region, there are 86 and 155 LINE1 elements in the murine and human loci, respectively. Almost all of the LINE1 elements were inserted after divergence of the human and mouse ancestor, because these elements disrupt the alignment. It is surprising that the length of intron 4 was conserved, despite insertion of numerous repetitive elements after divergence of human and mouse.
Figure 4.
The distribution of the repetitive elements in mouse and human sequences. LINE included LINE1 and LINE2 elements. Mouse SINEs included B1s, B2-B4, and ID repeats, and human SINEs included Alu and MIR repeats.
The Exon Structure of the Fhit Gene.
Batzoglou et al. (34) have reported that intron length was not conserved in 1,196 orthologous human and mouse gene pairs. In this study, mouse Fhit has a large intron 4 (308 kbp) with a size similar to human intron 4 (285 kbp).
Pekarsky et al. (25) reported that the mouse Fhit gene had a unique exon 3 that contained a start codon in addition to the exon 5 start codon; nevertheless, the major transcript of the mouse Fhit gene was an alternatively spliced isoform without exon 3. Thus, the exon 5 start codon was generally used, as observed for human FHIT. We found a sequence corresponding to mouse Fhit exon 3 in the human genome by comparing the two sequences. Fig. 1 shows the exon-intron structure of Fhit genes of human and mouse. The mouse Fhit exon 3 has no homology to the human exon 3, but has 70% homology to the human sequence at the corresponding position (asterisk of Fig. 1). These results suggested that the human FHIT gene changed exon structure during evolution. We designed specific primers for this ancient human exon 3 and performed reverse transcription (RT)-PCR by using RNA from human cell lines, but were unable to detect transcription of this fossil (or ancient) human exon 3.
Fra14A2/Fhit Landmarks.
The RENCA cell line had been demonstrated to exhibit a homozygous deletion of Fhit exon 5 (25). To localize more precisely the end points of the deletion, we used the sequence around exon 5 to design sets of primer pairs spanning the region for use in PCR analysis of RENCA DNA. We were able to assign the two break point regions to a region small enough (≈1 kbp) to allow end point sequencing by inverse PCR. Unfortunately, the RENCA break in intron 5 is within or adjacent to LINE sequence, so we were unable to do inverse PCR to obtain the sequence adjoining the LINE element. The RENCA break point located in intron 4 was successfully characterized by inverse PCR. The RENCA DNA was cut with AluI and circularized. Nested PCR amplification was performed, and a single PCR product was generated and sequenced. Primers were designed from the sequence from the other side of the deletion, and we were able to map it, by PCR, to BAC clone 167E2. Thus, one allele of RENCA exhibits a deletion from 20.6 kbp 5′ of exon 5 to a region near exon 6 (Fig. 1).
Cancer cell deletion or translocation break points, aphidicolin break clusters, and several integration sites have been precisely mapped in human FRA3B (8, 9). These landmark sequences have been examined for specific features, but specific DNA sequences were not associated with cancer cell break points, aphidicolin break points, or several integration sites.
Fig. 3A shows the relation between landmarks of this region and HCRs. The sequence at the distal aphidicolin break cluster had 58% homology with corresponding mouse sequence, a level of homology that hovers just above background level, and was located near an HCR. There was no mouse sequence corresponding to the aphidicolin proximal break cluster. There were also no sequences corresponding to HPV integration sites in the mouse genome. One pSVneo integration site had 70% homology to mouse sequence and was near an HCR. Among previously identified cancer cell break points (8, 9), seven had corresponding sequence in the mouse genome. The Lovoa2 break point was located in an HCR. The homologies of six other break points were at background level. But the break points of LS180a1, Sihaa, and MB436a2 were located near HCRs (<1,000 bp away). The break points HK3a1, HK3a2, MB436a2, and TE8b were located in repetitive elements that had no corresponding repeats in the mouse genome, and the break points KATOIIIa2, Lovo1a1, MB436a1, and TE8a were located in human unique sequence (8, 9). As for murine fragile site landmarks, one break point of RENCA was located in a LINE1 repeat and the other was located in mouse unique sequence <500 bp from an HCR. Thus, five cancer cell break points, an integration site, and an aphidicolin break cluster were located within or near HCRs. Previous studies had shown that many break points of cancer cell lines were located within or near LINE 1 repetitive elements (8, 9); this study has shown that positions of such repetitive elements were not conserved.
Conclusions
Fragile sites have been reported in mouse, rat, hamster, cow, cat, and dog (35–40). They appear to be inherent and universal structures of the mammalian genome. However, the understanding of fragility is still incomplete. This study, an attempt to further our understanding of common fragile sites by use of orthologous sequence comparison, revealed interesting features of the FRA3B/Fra14A2 orthologous region. Both region were characterized by low GC content, and the frequency of insertion/deletion change within these fragile regions was apparently not elevated during evolution. Thus, these regions are stable over evolutionary time, even though they are highly recombinogenic when on inhibition of DNA replication. The region exhibited unusual HCRs: five cancer cell break points, one pSVneo integration site, and one aphidicolin break cluster were located within or near HCRs, suggesting that the HCRs could have important roles in fragility-associated chromosome alterations. Comparison of other fragile region orthologs will be needed to confirm and extend our knowledge of the importance of conserved features of common fragile regions.
Acknowledgments
This research was supported by National Cancer Institute Grants PO1-CA77738 and HG02238 and Cancer Center Support Grant CCSG-CA56036. L.B. was supported by U.S. Public Health Service Training Grant T32-CA09678 from the National Cancer Institute.
Abbreviations
- BAC
bacterial artificial chromosome
- HCR
highly conserved region
- MAR
matrix attachment region
Footnotes
References
- 1.Jones C, Penny L, Mattina T, Yu S, Baker E, Voullaire L, Langdon W Y, Sutherland G R, Richards R I, Tunnacliffe A. Nature (London) 1995;376:145–149. doi: 10.1038/376145a0. [DOI] [PubMed] [Google Scholar]
- 2.Yu S, Mangelsdorf M, Hewett D, Hobson L, Baker E, Eyre H J, Lapsys N, Le Paslier D, Doggett N A, Sutherland G R, Richards R I. Cell. 1997;88:367–374. doi: 10.1016/s0092-8674(00)81875-9. [DOI] [PubMed] [Google Scholar]
- 3.Wilke C M, Hall B K, Hoge A, Paradee W, Smith D I, Glover T W. Hum Mol Genet. 1996;5:187–195. doi: 10.1093/hmg/5.2.187. [DOI] [PubMed] [Google Scholar]
- 4.Mishmar D, Rahat A, Scherer S W, Nyakatura G, Hinzmann B, Kohwi Y, Mandel-Gutfroind Y, Lee J R, Drescher B, Sas D E, Margalit H, Platzer M, Weiss A, Tsui L C, Rosenthal A, Kerem B. Proc Natl Acad Sci USA. 1998;95:8141–8146. doi: 10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang H, Qian C, Jenkins R B, Smith D I. Genes Chromosomes Cancer. 1998;21:152–159. doi: 10.1002/(sici)1098-2264(199802)21:2<152::aid-gcc11>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 6.Mangelsdorf M, Ried K, Woollatt E, Dayan S, Eyre H, Finnis M, Hobson L, Nancarrow J, Venter D, Baker E, Richards R I. Cancer Res. 2000;60:1683–1689. [PubMed] [Google Scholar]
- 7.Paige A J, Taylor K J, Stewart A, Sgouros J G, Gabra H, Sellar G C, Smyth J F, Porteous D J, Watson J E. Cancer Res. 2000;60:1690–1697. [PubMed] [Google Scholar]
- 8.Inoue H, Ishii H, Alder H, Snyder E, Druck T, Huebner K, Croce C M. Proc Natl Acad Sci USA. 1997;94:14584–14589. doi: 10.1073/pnas.94.26.14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mimori K, Druck T, Inoue H, Alder H, Berk L, Mori M, Huebner K, Croce C M. Proc Natl Acad Sci USA. 1999;96:7456–7461. doi: 10.1073/pnas.96.13.7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glover T W, Stein C K. Am J Hum Genet. 1988;43:265–273. [PMC free article] [PubMed] [Google Scholar]
- 11.Ohta M, Inoue H, Cotticelli M G, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, et al. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 12.Virgilio L, Shuster M, Gollin S M, Veronese M L, Ohta M, Huebner K, Croce C M. Proc Natl Acad Sci USA. 1996;93:9770–9775. doi: 10.1073/pnas.93.18.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sozzi G, Veronese M L, Negrini M, Baffa R, Cotticelli M G, Inoue H, Tornielli S, Pilotti S, De Gregorio L, Pastorino U, Pierotti M A, Ohta M, Huebner K, Croce C M. Cell. 1996;85:17–26. doi: 10.1016/s0092-8674(00)81078-8. [DOI] [PubMed] [Google Scholar]
- 14.Mori M, Mimori K, Shiraishi T, Alder H, Inoue H, Tanaka Y, Sugimachi K, Huebner K, Croce C M. Cancer Res. 2000;60:1177–1182. [PubMed] [Google Scholar]
- 15.Baffa R, Gomella L G, Vecchione A, Bassi P, Mimori K, Sedor J, Calviello C M, Gardiman M, Minimo C, Strup S E, McCue P A, Kovatich A J, Pagano F, Huebner K, Croce C M. Am J Pathol. 2000;156:419–424. doi: 10.1016/S0002-9440(10)64745-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Druck T, Hadaczek P, Fu T B, Ohta M, Siprashvili Z, Baffa R, Negrini M, Kastury K, Veronese M L, Rosen D, et al. Cancer Res. 1997;57:504–512. [PubMed] [Google Scholar]
- 17.Wang L, Paradee W, Mullins C, Shridhar R, Rosati R, Wilke C M, Glover T W, Smith D I. Genomics. 1997;41:485–488. doi: 10.1006/geno.1997.4690. [DOI] [PubMed] [Google Scholar]
- 18.Koop B F, Hood L. Nat Genet. 1994;7:48–53. doi: 10.1038/ng0594-48. [DOI] [PubMed] [Google Scholar]
- 19.Kuo C L, Chen M L, Wang K, Chou C K, Vernooij B, Seto D, Koop B F, Hood L. Proc Natl Acad Sci USA. 1998;95:3839–3844. doi: 10.1073/pnas.95.7.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubchak I, Brudno M, Loots G G, Pachter L, Mayor C, Rubin E M, Frazer K A. Genome Res. 2000;10:1304–1306. doi: 10.1101/gr.142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koop B F. Trends Genet. 1995;11:367–371. doi: 10.1016/s0168-9525(00)89108-8. [DOI] [PubMed] [Google Scholar]
- 22.Oeltjen J C, Malley T M, Muzny D M, Miller W, Gibbs R A, Belmont J W. Genome Res. 1997;7:315–329. doi: 10.1101/gr.7.4.315. [DOI] [PubMed] [Google Scholar]
- 23.Loots G G, Locksley R M, Blankespoor C M, Wang Z E, Miller W, Rubin E M, Frazer K A. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 24.Glover T W, Hoge A W, Miller D E, Ascara-Wilke J E, Adam A N, Dagenais S L, Wilke C M, Dierick H A, Beer D G. Cancer Res. 1998;58:3409–3414. [PubMed] [Google Scholar]
- 25.Pekarsky Y, Druck T, Cotticelli M G, Ohta M, Shou J, Mendrola J, Montgomery J C, Buchberg A M, Siracusa L D, Manenti G, et al. Cancer Res. 1998;58:3401–3408. [PubMed] [Google Scholar]
- 26.Solovyev V V, Salamov A A, Lawrence C B. Nucleic Acids Res. 1994;22:5156–5163. doi: 10.1093/nar/22.24.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burge C, Karlin S. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 28.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz S, Zhang Z, Frazer K A, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh G B, Kramer J A, Krawetz S A. Nucleic Acids Res. 1997;25:1419–1425. doi: 10.1093/nar/25.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue H, Sawada M, Ryo A, Tanahashi H, Wakatsuki T, Hada A, Kondoh N, Nakagaki K, Takahashi K, Suzumura A, et al. Glia. 1999;28:265–271. doi: 10.1002/(sici)1098-1136(199912)28:3<265::aid-glia10>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 32.Julicher K, Marquitan G, Werner N, Bardenheuer W, Vieten L, Brocker F, Topal H, Seeber S, Opalka B, Schutte J. J Natl Cancer Inst. 1999;91:1563–1568. doi: 10.1093/jnci/91.18.1563. [DOI] [PubMed] [Google Scholar]
- 33.Palin A H, Critcher R, Fitzgerald D J, Anderson J N, Farr C J. J Cell Sci. 1998;111:1623–1634. doi: 10.1242/jcs.111.12.1623. [DOI] [PubMed] [Google Scholar]
- 34.Batzoglou S, Pachter L, Mesirov J P, Berger B, Lander E S. Genome Res. 2000;10:950–958. doi: 10.1101/gr.10.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Djalali M, Adolph S, Steinbach P, Winking H, Hameister H. Hum Genet. 1987;77:157–162. doi: 10.1007/BF00272384. [DOI] [PubMed] [Google Scholar]
- 36.Lin M S, Takabayashi T, Wilson M G, Marchese C A. Cytogenet Cell Genet. 1984;38:211–215. doi: 10.1159/000132062. [DOI] [PubMed] [Google Scholar]
- 37.Smeets D F, van de Klundert F A. Cytogenet Cell Genet. 1990;53:8–14. doi: 10.1159/000132885. [DOI] [PubMed] [Google Scholar]
- 38.Stone D M, Jacky P B, Prieur D J. Am J Med Genet. 1991;40:223–229. doi: 10.1002/ajmg.1320400220. [DOI] [PubMed] [Google Scholar]
- 39.Vitetta L, Sali A, Little P, Nayman J, Elzarka A. HPB Surg. 1991;4:209–220. doi: 10.1155/1991/76160. ; discussion 221–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson T J, Elder F F. Chromosoma. 1987;96:45–49. doi: 10.1007/BF00285882. [DOI] [PubMed] [Google Scholar]