Abstract
PI3Ks (phosphoinositide 3-kinases) are signalling molecules and drug targets with important biological functions, yet the regulation of PI3K gene expression is poorly understood. Key PI3Ks are the class IA PI3Ks that consist of a catalytic subunit (p110α, p110β and p110δ) in complex with a p85 regulatory subunit. Whereas p110α and p110β are ubiquitously expressed, high levels of p110δ are mainly found in white blood cells, with most non-leucocytes expressing low levels of p110δ. In the present paper we report that TNFα (tumour necrosis factor α) stimulation induces p110δ expression in human ECs (endothelial cells) and synovial fibroblasts, but not in leucocytes, through transcription start sites located in a novel promoter region in the p110δ gene (PIK3CD). This promoter is used in all cell types, including solid tumour cell lines that express p110δ, and is activated by TNFα in ECs and synovial fibroblasts. We further present a detailed biochemical and bioinformatic characterization of p110δ gene regulation, demonstrating that PIK3CD has distinct promoters, some of which can be dynamically activated by pro-inflammatory mediators. This is the first molecular identification of a PI3K promoter under the control of acute extracellular stimulation.
Keywords: bioinformatics, cytokine, inflammation, promoter, PIK3CD, tumour necrosis factor α (TNFα)
Abbreviations: ActD, actinomycin D; ChIP, chromatin immunoprecipitation; ChIP-seq, ChIP sequencing; DMEM, Dulbecco's modified Eagle's medium; EC, endothelial cell; ENCODE, Encyclopedia of DNA Elements; EST, expressed sequence tag; FBS, fetal bovine serum; H3K27Ac, histone 3 Lys27 acetylation; H3K4Me1, monomethylation of histone 3 Lys4; H3K4Me3, trimethylation of histone 3 Lys4; HUVEC, human umbilical vein EC; IκB, inhibitory κB; IKK, IκB kinase; IL, interleukin; NF-κB, nuclear factor κB; PI3K, phosphoinositide 3-kinase; qPCR, quantitative PCR; 5′RACE, rapid amplification of 5′ cDNA ends; SV40, simian virus 40; TF, transcription factor; TNFα, tumour necrosis factor α; TSS, transcription start site; UCSC, University of California Santa Cruz; UTR, untranslated region
INTRODUCTION
PI3Ks (phosphoinositide 3-kinases) phosphorylate inositol lipids in cellular membranes in response to a variety of stimuli. The involvement of PI3Ks in immunity, inflammation and cancer has made these enzymes important new targets for drug development [1–3]. This also applies to p110δ, a PI3K isoform mainly expressed in leucocytes [4,5], and Phase I/II trials with p110δ inhibitors are currently in progress for allergy and haematological malignancies [6,7].
Previous studies have started to uncover the molecular mechanism of the selective enrichment of p110δ in the haematopoietic lineage. p110δ expression is mainly regulated at the transcriptional level with the protein being produced from transcripts with different 5′-UTRs (untranslated regions) as a consequence of the presence of multiple TSSs (transcription start sites) in the p110δ gene (PIK3CD in humans and Pik3cd in mice) [8]. The 5′-UTR of most p110δ transcript types contains two untranslated exons, referred to as exons −1 and −2. Of the latter, two distinct species have been found in humans (−2a and −2b) and four in mice (−2a, −2b, −2c and −2d), with only exon −1 and a region of exon−2a being conserved between humans and mice [8]. In both human and mouse leucocytes, the p110δ transcript containing exons −2a and −1 is the most abundant, in line with the presence of a conserved region of predicted binding sites for leucocyte-specific TFs (transcription factors) in the proximal promoter region of the TSS of exon −2a. Upon transient transfection this region of the murine genome has a higher promoter activity in leucocytes than in non–leucocytes, and therefore probably contributes to the high expression of p110δ in haematopoietic cells [8].
In addition to leucocytes, some non-leucocytes such as melanocytes, breast cells and their transformed equivalents [8,10], neurons [11], ECs (endothelial cells) [12] and lung fibroblasts [13] also express p110δ, albeit at lower levels than in leucocytes. It is unclear how the expression of p110δ is controlled in these cells. In addition, p110δ expression can be increased in some non-leucocytes such as in rat aortic tissue upon long-term treatment (2–4 weeks) with hypertension-inducing agents [DOCA (deoxycorticosterone acetate) or Nω-nitro-L-arginine] [14], in rat and mouse cardiac fibroblasts upon a more acute stimulation (30–60 min) with aldosterone [15] and in the aortas of diabetic mice [16]. These studies, however, did not address the regulatory mechanism underlying the observed increase in p110δ levels.
We report in the present paper that TNFα (tumour necrosis factor α) stimulation induces the expression of p110δ in human ECs and synovial fibroblasts. We describe a novel and inducible human PIK3CD promoter region that gives rise to p110δ transcripts with previously unidentified 5′-UTRs. We further analyse and discuss these observations in the broader context of the distinct PIK3CD promoters that direct p110δ expression in different cell types.
EXPERIMENTAL
Antibodies and reagents
Antibodies were as follows: anti-p110α (C73F8) (catalogue number 4249), anti-p110β (C33D4) (catalogue number 3011), anti-[p38 MAPK (mitogen-activated protein kinase) (phospho-Thr180/Tyr182)] (3D7) (catalogue number 9215) and anti-[NF-κB (nuclear factor κB) p65 (phospho-Ser536)] (93H1) (catalogue number 3033) (Cell Signaling Technology); anti-p110δ (H-219) (catalogue number sc-7176) and anti-IκB (inhibitory κB)-α (catalogue number sc-371) (Santa Cruz Biotechnology); anti-p85 (catalogue number 06-195; Upstate Biotechnology); anti-α-tubulin (B-5-1-2) and anti-vinculin (clone hVIN-1) (Sigma). Carrier-free recombinant human TNFα was from R&D Systems and recombinant human IL (interleukin)-1β was from Peprotech. ActD (actinomycin D) was from Sigma and IKK (IκB kinase) inhibitor VII from Calbiochem.
Cell culture and cell stimulation
HUVECs (human umbilical vein ECs) were purchased from Lonza and cultured in EGM-2 medium (Lonza). HUVECs were grown on plastic coated with human fibronectin (10 μg/ml; Sigma) and used for experiments between passages 3 and 5. Culture media for cell lines were as follows: EA.hy926 (provided by Professor Anne Ridley, King's College London, University of London, London, U.K.), U-87 MG and MDA-MB-468 cells, DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% FBS (fetal bovine serum); SK-OV-3 cells, McCoy's 5A medium supplemented with 10% FBS; and THP-1, Jurkat and MCF-7 cells, RPMI 1640 supplemented with 10% FBS. Synovial tissues were obtained from patients undergoing total knee/hip replacement after informed consent (local research ethics committee reference number 05/Q0703/198) and used for isolation of synovial fibroblasts as described previously [17]. Synovial fibroblasts were cultured in DMEM/Ham's F12 supplemented with 10% FBS and 10 mM Hepes and used for experiments between passages 6 and 9 when the culture is devoid of contaminating lymphocytes and macrophages [18]. All cells were maintained at 37°C and 5% CO2. All cytokine stimulations were performed in complete culture medium, defined as medium containing FBS and antibiotics.
Western blotting
Cells were collected and lysed in lysis buffer [50 mM Tris/HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA and 1% (v/v) Triton X-100] supplemented with protease inhibitors. Equal amounts of protein were separated by SDS/PAGE (8% gels), immunoblotted with primary antibodies and HRP (horseradish peroxidase)-conjugated species-specific secondary antibodies and exposed to X-ray film. A Bio-Rad Laboratories GS-800 calibrated densitometer was used to quantify Western blot signals.
qPCR (quantitative PCR)
Total RNA was isolated using RNeasy Mini Kit (Qiagen). Equal amounts of RNA were transcribed to cDNA with SuperScript II reverse transcriptase using random primers (Invitrogen). TaqMan Gene Expression Assays were from Applied Biosystems: Hs00180679 (p110α), Hs00178872_m1 (p110β), Hs00192399_m1 (p110δ), Hs00933163 (p85α), Hs00178181_m1 (p85β), Hs99999034_m1 (IL-8), eukaryotic 18S rRNA (4310893E) and β-actin (4326315E). Custom-designed assays were used for specific human p110δ transcripts (sequences provided in Supplementary Table S1 at http://www.BiochemJ.org/bj/443/bj4430857add.htm). Samples were measured in triplicate using TaqMan Universal PCR Mastermix (4440044). The ΔΔCT method [19] was used for relative quantification. For absolute quantification of transcript copy numbers, a standard curve of linearized plasmid (molecules/μl) was generated for each amplicon. A dilution series of cDNA prepared from THP-1 cells was used as standard curve for β-actin.
5′RACE (rapid amplification of 5′ cDNA ends)
5′RACE was performed on 10 μg of total cellular RNA with the FirstChoice RLM-RACE kit (Ambion). Nested PCR products (primer sequences listed in Supplementary Table S1) were ligated into pGEM-T Easy (Promega) and transformed into Escherichia coli OneShot TOP10 competent cells (Invitrogen). Clones of transformed bacteria were isolated and purified DNA analysed by sequencing.
Cloning of pGL3 reporter constructs
pGL3-Basic (E1751), pGL3-SV40 (E1761), pRL-SV40 (Renilla; E2231) were from Promega. Regions surrounding human PIK3CD exons −2a and −2e (−139 to +92 and −385 to +197 relative to the start sites of exons −2a and −2e respectively) were PCR-amplified and inserted into pGL3-Basic using the MluI/BglII sites (primer sequences shown in Supplementary Table S1). The sequence surrounding exon −2c [−640 to +310 relative to start site of exon −2c according to EST (expressed sequence tag) DB216922] with flanking 5′ MluI and 3′ BglII sites was synthesized by OriGene Technologies and subcloned into pGL3-Basic using the MluI/BglII sites. The QuikChange Site-Directed Mutagenesis Kit (Stratagene) was used to mutate the NF-κB site in pGL3-exon-2e (primer sequences shown in Supplementary Table S1).
Transfection and reporter gene assays
Cells in 12-well plates were co-transfected in triplicate with 0.5 μg/well pGL3-luc plasmid and 0.25 μg/well pRL-SV40 using FuGENE. Cells received complete medium with TNFα (10 ng/ml) or complete medium only (unstimulated control) 24 h after transfection. Lysates were prepared 48 h after transfection using the Dual-Luciferase Reporter Assay System (Promega) and luciferase activity was analysed using a Wallac 1420 VICTOR3 Multilabel Counter (PerkinElmer). Renilla luciferase activity was used to normalize transfection efficiency. Promoter activity was expressed as the percentage of the activity of the pGL3-SV40 reporter. The promoter of the Vav gene [20] cloned into the pGL2 luciferase reporter was used as control for leucocyte-specific promoter activity [8].
Bioinformatic analysis of the human PIK3CD locus
The sequences of the p110δ exons −2c, −2d and −2e identified in 5′RACE clones were queried against the EST database using NCBI Blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The Ensembl genome database (GRCh37 assembly, release 64) was used to inspect annotated human p110δ transcripts. The TFSEARCH programme v.1.3 was used to predict NF-κB-binding sites using the vertebrate matrix with a threshold of 90 points (http://www.cbrc.jp/research/db/TFSEARCH.html).
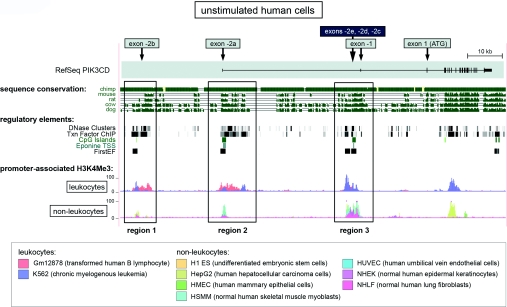
The PIK3CD locus (chr1:9,605,000-9,715,721) was analysed in the Human March 2006 (NCBI36/hg18) Assembly of the UCSC (University of California Santa Cruz) genome browser (http://genome.ucsc.edu/) for the presence of transcriptional regulatory elements using the genome-wide data generated by the ENCODE (Encyclopedia of DNA Elements) consortium [21]. Sequence conservation is depicted in the form of a multiple sequence alignment of the corresponding genomic sequences of chimp, mouse, rat, cow and dog generated using the MultiZ track. The DNase Clusters track shows DNase I-hypersensitivity sites known to be associated with gene-regulatory regions. The Txn Factor ChIP track shows TF binding to DNA as assessed by ChIP-seq [ChIP (chromatin immunoprecipitation)-sequencing]. The data for the DNase Clusters and Txn Factor ChIP tracks was from Gm12878 and K562 cells. The CpG islands track shows predictions of CpG islands using the following criteria: GC content>50%; >200 bp length; observed>expected CG dinucleotides ratio≥0.6. The Eponine track shows TSS predictions and the FirstEF track predictions of 5′-terminal exons and associated promoters. The track containing ChIP-seq data of promoter and enhancer-associated histone modifications [H3K4Me1 (monomethylation of histone 3 Lys4) and H3K4Me3 (trimethylation of histone 3 Lys4) and H3K27Ac (histone 3 Lys27 acetylation)] was divided into leucocytes [Gm12878 (transformed human B-lymphocyte) and K562 (chronic myelogenous leukaemia)] and non-leucocytes [H1 ES (undifferentiated embryonic stem cells), HMECs (human mammary epithelial cells), HSMMs (normal human skeletal muscle myoblasts), HUVECs, NHEKs (normal human epidermal keratinocytes), NHLFs (normal human lung fibroblasts) and HepG2 cells (human hepatocellular carcinoma cells; not included in the H3K4Me1 track)]. The data tracks were exported from the UCSC genome browser and compiled using Adobe Illustrator.
Bioinformatic analysis of the mouse Pik3cd locus
The Ensembl database (NCBIM37 assembly, release 64) was inspected for annotated mouse p110δ transcripts. The mouse Pik3cd locus was inspected in the UCSC genome browser (NCBI37/mm9 assembly). ChIP-seq data from the histone modification track [ENCODE/LICR (Ludwig Institute for Cancer Research)] for the promoter-associated H3K4Me3 modification in different tissue and cell types are shown in Figure 4, and Supplementary Figures S3 and S4 at http://www.BiochemJ.org/bj/443/bj4430857add.htm. The data tracks were exported from the UCSC genome browser and compiled using Adobe Illustrator.
Figure 4. Three human PIK3CD promoter regions revealed by bioinformatic analysis of functional regulatory elements.
The human PIK3CD locus (chromosome 1, bp 9605000–9715721) in the UCSC genome browser is shown. The RefSeq PIK3CD transcript (on top) contains the untranslated exons −2a and −1 and all of the protein coding exons. The approximate genomic locations of exons −2b, −2c, −2d and −2e are indicated with arrows on top of the RefSeq transcript. Sequence conservation shows a Multiz sequence alignment for the indicated species. For DNase clusters and TF ChIP data, a grey box indicates signal strength so that the darkness of the box is proportional to the maximum signal strength observed. ChIP-seq data for H3K4Me3 are shown separately for non-leucocytes and leucocytes, with data from individual cell types shown in different colours (lower panel). The promoter regions 1–3 identified are shown as boxed regions.
RESULTS
TNFα stimulation increases p110δ expression in human ECs and synoviocytes
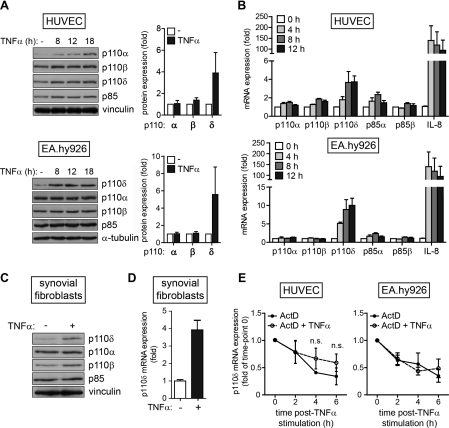
Given the involvement of p110δ in immune cell signalling and inflammation [22], we explored whether p110δ expression could be modulated by inflammatory cytokines such as TNFα and IL-1β, particularly in cell types that normally express low levels of this protein. The endothelium plays an important role in inflammation and we therefore analysed whether these inflammatory mediators could induce PIK3CD gene expression in ECs. Indeed, TNFα stimulation of HUVECs and the human endothelium-derived cell line EA.hy926 (a cell hybrid of HUVECs and A549 human pulmonary adenocarcinoma cells [23]) increased the levels of p110δ protein (Figure 1A) and mRNA (Figure 1B). A similar enhancement in the expression of p110δ protein (Figure 1C) and mRNA (Figure 1D) upon TNFα stimulation was also detected in human synovial fibroblasts isolated from patients with rheumatoid arthritis. In contrast with the consistent and highly reproducible increase in p110δ expression upon TNFα stimulation, only slight increases in the expression of p110α and p110β were noted in some, but not all, experiments. However, p110δ expression in TNFα-stimulated ECs remained lower compared with the basal levels of this PI3K isoform in the leucocyte cell line THP-1 (Supplementary Figure S1 at http://www.BiochemJ.org/bj/443/bj4430857add.htm).
Figure 1. TNFα stimulation induces p110δ expression in human ECs.
(A) PI3K isoform expression in HUVECs (upper panels) and EA.hy926 cells (lower panels) stimulated with TNFα (20 ng/ml for HUVECs and 10 ng/ml in EA.hy926 cells) for the indicated times. A representative immunoblot of three independent experiments is shown. Fold change in p110 isoform expression relative to unstimulated cells upon 18 h stimulation with TNFα (right-hand panels) as quantified by densitometry. Results are means±S.D. for three independent experiments. (B) PI3K mRNA expression in HUVECs (upper panels) and EA.hy926 cells (lower panels) stimulated with TNFα (10 ng/ml) for the indicated times and analysed by qPCR. Transcript expression was normalized to 18S rRNA and shown as fold increase over unstimulated levels for each transcript. Results are means±S.E.M. for three independent experiments. (C) PI3K isoform expression in synovial fibroblasts stimulated with TNFα (10 ng/ml) for 18 h. (D) PIK3CD mRNA expression in synovial fibroblasts stimulated with TNFα (10 ng/ml) for 18 h and analysed by qPCR. Transcript expression was normalized to 18S rRNA and shown as fold increase over unstimulated levels for each transcript. Results are means±S.E.M. for three independent experiments. (E) HUVECs (left-hand panel) and EA.hy926 cells (right-hand panel) were pre-treated with ActD (4 μg/μl) for 1 h and either exposed to 10 ng/ml TNFα or left unstimulated for the indicated times. p110δ mRNA levels were analysed by qPCR and normalized to 18S rRNA. The results are presented relative to the p110δ levels after 1 h of ActD treatment (t=0). Results are means±S.E.M. for three independent experiments. n.s., not significant.
The expression of PIK3CD mRNA was also increased upon IL-1β stimulation of HUVECs and synovial fibroblasts (Supplementary Figure S2 at http://www.BiochemJ.org/bj/443/bj4430857add.htm), but not EA.hy926 (results not shown), in line with the notion that EA.hy926 cells are not responsive to IL-1β [24,25]. To determine whether TNFα-induced p110δ expression was a common phenomenon or limited to ECs and synovial fibroblasts, we extended our analysis to include different human and murine cell types. To our surprise, TNFα stimulation had only a minor or no effect on p110δ mRNA or protein levels in the other cell types that we tested (Supplementary Table S2 at http://www.BiochemJ.org/bj/443/bj4430857add.htm). Interestingly, p110δ expression was not induced in murine ECs (primary lung or cardiac ECs) upon stimulation with TNFα (results not shown).
TNFα regulates cellular mRNA levels by inducing de novo transcription and/or by stabilizing existing transcripts. To determine whether TNFα increased cellular p110δ levels through a transcriptional mechanism, we pre-treated ECs with the transcriptional inhibitor ActD prior to stimulation with TNFα. The only way in which TNFα stimulation could increase PIK3CD mRNA levels in ActD-treated cells would be through enhancing the stability of existing p110δ transcripts. However, TNFα stimulation did not significantly alter the stability of p110δ transcripts in ActD-treated HUVECs or EA.hy926 cells (Figure 1E), demonstrating that TNFα induces p110δ expression by stimulating the transcription of PIK3CD.
TNFα stimulation induces expression of PIK3CD mRNA transcripts that contain novel 5′ untranslated exons
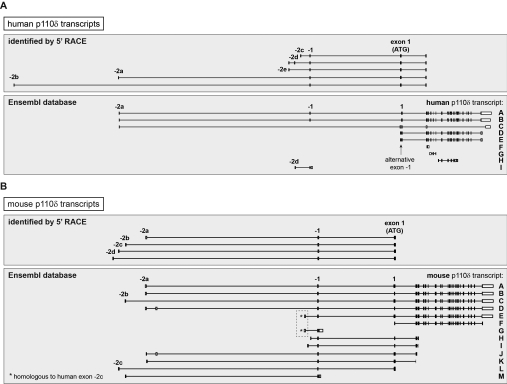
Baseline p110δ transcripts in ECs contained exon −1, without −2 exons (results not shown). We next used 5′RACE to identify the structure of the 5′-UTR of PIK3CD mRNA transcripts induced by TNFα stimulation of HUVECs and EA.hy926 cells (Figure 2A). A single PCR product was present in unstimulated cells, whereas TNFα stimulation led to the appearance of additional PCR products (Figures 2B and 2C). Cloning and sequencing of these PCR products revealed previously unidentified PIK3CD 5′-UTRs containing three new untranslated exons, located in close proximity to each other in the human genome, approximately 3 kb upstream of exon −1 (Figures 2D and 3A). In line with the nomenclature of currently known first exons of human PIK3CD (exon −2a and −2b), the new exons were named exons −2c, −2d and −2e (Figure 2D). All of the −2 exons identified contain a canonical splice donor site, with exon −2d also containing a splice acceptor site (Figure 2D and Table 1). Accordingly, each −2 exon was spliced to the 5′-end of exon −1 that is common to all PIK3CD transcripts (shown schematically in Figure 2D). In addition, a further 5′RACE product with exon −2d spliced between exons −2e and −1 was found in TNFα-stimulated EA.hy926 cells (Figures 2D and 3A).
Figure 2. Novel TNFα-inducible PIK3CD TSSs uncovered by 5′RACE in human ECs.
(A) Schematic representation of the 5′RACE strategy used to analyse the PIK3CD 5′-UTR in human ECs. The 5′RACE adapter is shown in grey, the 5′-UTR of the human PIK3CD mRNA transcript is shown in white and the protein-coding exons are shown in black. The approximate binding sites of the outer and inner primers used in the nested PCR are depicted with arrows. (B and C) 5′RACE was performed in unstimulated and TNFα-stimulated (10 ng/ml for 18 h) HUVECs (B) and EA.hy926 cells (C). Nested PCR was performed on 5′RACE cDNA using the outer and inner primer pairs shown in (A) and products of both reactions were analysed on a 1% agarose gel. Arrows indicate the cloned and sequenced RACE products. In unstimulated HUVECs, the 5′-UTR of p110δ transcripts contains a partial exon −1 spliced on to exon 1 without a −2 exon and in TNFα-stimulated HUVECs, exon −2c spliced on to the full-length exon−1. In both unstimulated and TNFα-stimulated EA.hy926 cells, the 5′-UTR of p110δ transcripts is formed of exon −2e spliced on to full-length exon −1 or exon −2d spliced between exons −2e and −1. No 5′RACE products with exon−2d as their first 5′ exon were found although a band with an estimated molecular mass corresponding to such a 5′RACE product is visible on the agarose gel in TNFα-stimulated EA.hy926 cells. The additional visible bands are degradation products containing partial sequences of exons −2c or −1. (D) Schematic representation of human PIK3CD 5′ untranslated exons and their splicing pattern as identified by 5′RACE or by PCR amplification. M, molecular mass marker.
Figure 3. Human and mouse PIK3CD/Pik3cd transcripts identified to date.
(A) Human PIK3CD transcripts identified by 5′RACE in ECs or different human cell types [8] (upper panel) or found in the Ensembl database (lower panel). The PIK3CD transcripts in the Ensembl database are labelled as follows: A, PIK3CD-202; B, PIK3CD -001; C, PIK3CD-002; D, PIK3CD-201; E, PIK3CD-003; F, PIK3CD-005; G, PIK3CD-006; H, PIK3CD-203; and I, PIK3CD-004). Transcripts C, D and E lack the canonical exon −1 and instead contain an alternative exon −1 (indicated with an arrow). Evidence for a PIK3CD transcript with an additional upstream exon that is spliced between exons −2a and −1 and that is located 23 kb upstream of exon −2e in the genome was found in an EST isolated from pre-B-cell acute lymphoblastic leukaemia cells (GenBank® accession number BE246970; not shown). (B) Mouse Pik3cd transcripts identified by 5′RACE [8] (upper panel) or found in the Ensembl database (lower panel). The Pik3cd transcripts in the Ensembl database are labelled as follows: A, Pik3cd-001; B, Pik3cd-003; C, Pik3cd-012; D, Pik3cd-201; E, Pik3cd-007; F, Pik3cd-002; G, Pik3cd-008; H, Pik3cd-005; I, Pik3cd-004; J, Pik3cd-009; K, Pik3cd-010; L, Pik3cd-011; and M, Pik3cd-006. The first 5′ exon in transcripts E and G is homologous with human exon −2c (indicated with an asterisk). Exons are shown as boxes and introns are shown as connecting lines.
Table 1. Summary of 5′ untranslated exons in human PIK3CD.
Details of all 5′ untranslated exons of human PIK3CD identified to date. Exonic sequences are shown in upper case and intronic in lower case with splice donor and acceptor sequences indicated in bold. A BLAST search for human ESTs found two containing exon −2c (GenBank® accession numbers DB216922 and DA318970 isolated from trachea and hippocampus respectively) with the TSS of exon −2c in DB216922 located 53 bp upstream of the 5′-end of the exon −2c present in the 5′RACE. One EST with a partial exon −2d sequence 2d was found (GenBank® accession number AA188126; transcript I in Figure 3A). No significant hits were obtained when the sequence of exon −2e was queried against the EST database.
| Exon | bp | TSS distance from ATG (kb) | Splice acceptor | 5′-end of exon | 3′-end of exon | Splice donor |
|---|---|---|---|---|---|---|
| −2e | 41 | 23 | ggctccgcc | CTCTCCCGGG | GTGCGGGCGG | gtgagtgcc |
| −2d | 183 | 22 | tcttctcag | GCACGAGGAA | TGTGGCAAAG | gtttgtctt |
| −2c (5′RACE) | 124 | 21 | ccctccggc | CCCCGCCGGC | GCGACACCCG | gtacggagc |
| −2c (EST) | 177 | 21 | ggccgcgcc | CCTCCGCCGA | GCGACACCCG | gtacggagc |
| −2b | 251 | 81 | cgggggtca | GAGGCGCCCA | ACTCTGACAG | gtgagtcta |
| −2a | 59 | 58 | gcgcccagc | GCAGTCGCTC | CGCCGGGACG | gtaagcgat |
| −1 | 105 | 19 | ccccaacag | ATAAGGAGTC | TTCCAGAGAG | gtaggttgg |
Mouse Pik3cd transcripts with 5′ untranslated exons with a genomic location corresponding to that of human exons −2c, −2d and −2e were present in the Ensembl database (Figure 3B, transcripts E, G, H and I), with the first exon of transcripts E and G (Figure 3B, boxed) being homologous with human exon −2c (results not shown). It is important to note that the previously adopted nomenclature for the murine p110δ exons −2c and −2d [8] (Figure 3B) does not correlate with the nomenclature and genomic location of the human p110δ exons −2c and −2d. Figure 3 shows a graphical representation of the different human and mouse PIK3CD/Pik3cd transcripts identified to date (full details of the 5′ untranslated exons in human PIK3CD are shown in Table 1).
Bioinformatic analysis of the PIK3CD/Pik3cd locus supports the existence of a novel PIK3CD/Pik3cd promoter, active in both leucocytes and non-leucocytes
We previously showed that a conserved genomic region of mouse Pik3cd exon −2a has higher promoter activity in leucocytes than in non-leucocytes, which is probably due to a cluster of binding sites for leucocyte-specific TFs in this region [8]. The promoters associated with the other known mouse TSSs and with the human PIK3CD TSSs have not been identified to date.
To gain more insight into the regulation of p110δ expression, we analysed the nature of potential regulatory elements associated with the different TSSs in both the human and mouse PIK3CD/Pik3cd genes. To this end, we used genome-wide data on regulatory elements compiled by the ENCODE project [21], available in the UCSC genome browser. Figure 4 shows the human PIK3CD locus in the UCSC genome browser and a selection of regulatory elements. The 5′-UTR in the PIK3CD reference mRNA transcript (RefSeq) is formed of exons −2a and −1. We extended the region of analysis approximately 25 kb upstream of exon −2a in order to include the TSS of exon −2b. A multiple species alignment showed that in addition to the coding region of PIK3CD, selected areas in the upstream genomic region showed conservation across mammals (Figure 4, sequence conservation panel). Interestingly, we found that cis- and trans-acting regulatory elements commonly associated with transcriptional regulation, including chromatin accessibility (DNase clusters), occupied TF-binding sites (Txn Factor ChIP) and Eponine and FirstEF promoter predictions, clustered in three regions (Figure 4, regions 1–3). As expected, two of these regions were associated with the previously identified exons −2b and −2a (Figure 4, regions 1 and 2). Interestingly, the location of the third cluster of regulatory elements (Figure 4, region 3) coincided with that of the newly identified exons −2c, −2d and −2e. This genomic region upstream of exon −1 was not known previously to contain a promoter.
Specific covalent modifications of histones have been shown to be associated with gene regulatory regions such as promoters and enhancers [21,26]. The ENCODE data on histone modifications (H3K4Me1, H3K27Ac and H3K4Me3) have been compiled from different human cell types (Figure 4 and Supplementary Figure S3 at http://www.BiochemJ.org/bj/443/bj4430857add.htm, represented by a different colours). As the expression of p110δ is highly polarized depending on the cell type, we analysed histone modifications at promoter regions 1–3 separately in leucocytes and non-leucocytes (Figure 4 and Supplementary Figure S3). A broad enrichment of the H3K4Me3 modification associated with active promoters was found in all three PIK3CD promoter regions in leucocytes (Figure 4). In contrast, in non-leucocytes, a strong H3K4Me3 signal was observed only in promoter region 3, with a much weaker H3K4Me3 signal seen in regions 1 and 2 (Figure 4). The pattern of H3K4Me3 enrichment suggests that all three human PIK3CD promoters are poised for activation in leucocytes, whereas in non-leucocytes, the promoter region 3 is likely to act as the preferred start site of PIK3CD transcription. The enhancer-associated histone modifications H3K4Me1 and H3K27Ac showed a similar pattern being highly enriched at all three promoter regions in leucocytes, but only at region 3 in non-leucocytes (Supplementary Figure S3).
We extended our analysis to the mouse Pik3cd locus, using ENCODE data in the UCSC genome browser. As expected, an enrichment of the H3K4Me3 modification was found at the genomic locations of the known mouse −2 exons in different mouse tissues and cell types (Supplementary Figure S4 at http://www.BiochemJ.org/bj/443/bj4430857add.htm). In addition, similar to the human PIK3CD locus, a strong H3K4Me3 signal was also observed upstream of exon −1 in the genomic region that corresponds to that of the human promoter region 3 in Figure 4 (Supplementary Figure S4).
Taken together, these results suggest that both the human and mouse PIK3CD/Pik3cd genes have an alternative, previously unidentified, promoter in the genomic region upstream of exon −1.
TNFα-stimulated expression of distinct PIK3CD transcripts
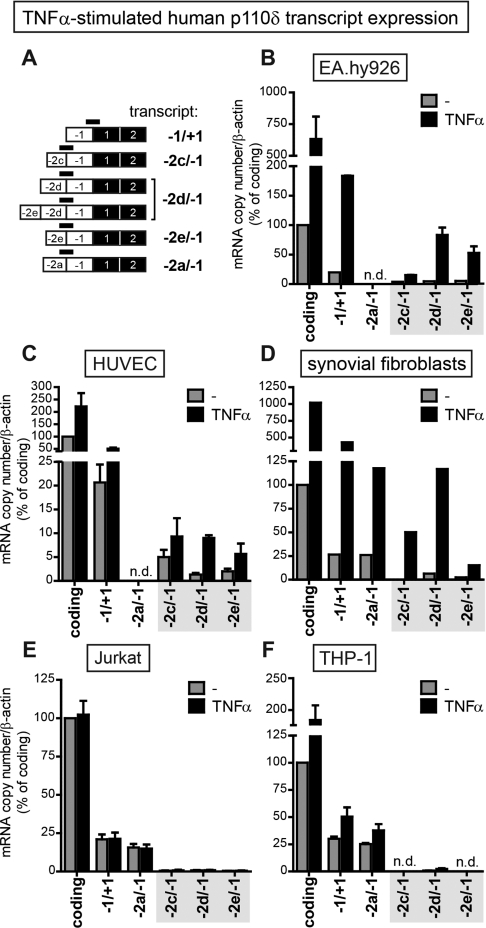
Given that our bioinformatic analysis strongly suggested that a functional promoter was associated with the TSSs of exons −2c, −2d and −2e, we analysed PIK3CD transcript expression by qPCR with primer/probe sets specific for the distinct untranslated exon–exon boundaries (Figure 5A), under basal conditions and after TNFα stimulation.
Figure 5. qPCR analysis of human PIK3CD transcript expression upon TNFα stimulation.
(A) Schematic representation of the distinct human p110δ 5′-UTR structures. The binding sites of the primer/probe sets used to amplify each transcript are depicted with black bars. (B–F) Quantitative analysis of PIK3CD transcript expression measured by qPCR in EA.hy926 cells (B), HUVECs (C), synovial fibroblasts (D), Jurkat cells (E) and THP-1 cells (F) before and after 8 h of stimulation with 10 ng/ml TNFα. Transcript expression was normalized to β-actin and expressed as the percentage of p110δ-coding transcripts in unstimulated cells. Results are means±S.E.M. for three independent experiments for HUVECs, EA.hy926 cells, Jurkat cells and THP-1 cells and the mean for one experiment for synovial fibroblasts; experiments were conducted in trplicate. n.d., not detected.
In ECs, basal levels of PIK3CD transcripts with exons −2c, −2d and −2e were low but increased upon TNFα stimulation (Figures 5B and 5C), in line with our 5′RACE results. Exon −2a transcripts were not detected in these cells (Figures 5B and 5C). In synovial fibroblasts, TNFα treatment also stimulated the expression of exon −2c, −2d and −2e transcripts, as well as that of exon −2a transcripts (Figure 5D).
In leucocytes (Jurkat and THP-1 cells), exon −2a transcripts made up the majority of the PIK3CD transcripts (Figures 5E and 5F), in accordance with a previous report [8]. In these cells, exon −2c-, −2d- or −2e-containing transcripts were only a minor or undetectable fraction of the total PIK3CD transcript pool, under both basal and TNFα-stimulated conditions (Figures 5E and 5F). However, TNFα stimulation induced exon −2a transcripts in THP-1 cells (Figure 5F), without detectably altering p110δ protein levels (results not shown), suggesting that p110δ expression may also be controlled by post-transcriptional mechanisms in these cells. In summary, PIK3CD transcripts with 5′-UTRs containing exons −2c, −2d or −2e are detectable at low levels in unstimulated cells and are further induced by TNFα stimulation in ECs and synovial fibroblasts, but not in leucocytes.
TNFα stimulation induces the activity of an exon −2e-associated NF-κB-dependent PIK3CD promoter in ECs
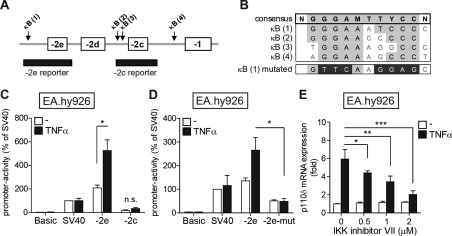
TNFα mediates most of its effects on the transcriptome via the NF-κB family of TFs. An in silico analysis of TF-binding sites in the PIK3CD locus identified four putative NF-κB-binding sites [κB(1)–(4)] in the genomic region surrounding exons −2c, −2d and −2e (Figure 6A) with sequences similar to that of the NF-κB consensus sequence [27] (Figure 6B).
Figure 6. TNFα-induced exon −2e promoter activity in ECs requires an intact NF-κB site.
(A) A schematic representation of the genomic region upstream of human PIK3CD exon −1 (not drawn to scale). PIK3CD exons are depicted as boxes and the genomic regions surrounding exons −2e and −2c that were inserted into pGL3-Basic are depicted as black bars. The arrows indicate the locations of predicted NF-κB sites [labelled κB(1)–(4)]. (B) The consensus NF-κB DNA-binding sequence (top row, in bold) and the sequences of κB(1)–(4) sites are shown. The bottom row shows the nucleotides that were mutated to disrupt NF-κB binding to the predicted κB(1) in the exon −2e-mut construct. (C and D) EA.hy926 cells were transfected with the indicated PIK3CD promoter reporters and stimulated for 24 h with 10 ng/ml TNFα. The promoter activity of each reporter is expressed relative to the activity of the SV40 promoter in unstimulated cells (set as 100%). Results are means±S.E.M. for three independent experiments. (E) EA.hy926 cells were pre-treated for 1 h with the indicated concentrations of IKK inhibitor VII or DMSO and stimulated with 10 ng/ml TNFα for 6 h. Expression of PIK3CD mRNA was analysed by qPCR and normalized to 18S rRNA. Results are the mean±S.E.M. fold increases over expression in unstimulated DMSO-treated cells of three independent experiments. *P<0.05, **P<0.01, ***P<0.001 by two-tailed unpaired Student's t test.
We next assessed the TNFα-responsiveness of promoters of the newly identified PIK3CD TSSs. The genomic regions surrounding exons −2e and −2c (Figure 6A) were inserted into the promoter-less pGL3-Basic luciferase reporter plasmid and their activities were analysed after transient transfection in unstimulated or TNFα-stimulated EA.hy926 cells. The basal activity of the exon −2e promoter was comparable with that of the SV40 (simian virus 40)-driven positive control promoter (Figure 6C, white bars) and, importantly, was further enhanced upon TNFα stimulation (Figure 6C, black bars). In contrast, the basal activity of the exon −2c promoter was much weaker and unresponsive to TNFα stimulation in these cells (Figure 6C).
To examine whether the predicted NF-κB site upstream of exon −2e was involved in the TNFα-induced promoter activity, the key residues in the κB(1) site required for NF-κB binding were mutated (Figure 6B, bottom row). The mutant exon −2e promoter (−2e-mut) was no longer able to induce luciferase expression upon TNFα stimulation in EA.hy926 cells (Figure 6D) demonstrating that the predicted κB(1) site is functional and required for the TNFα-responsiveness of the exon −2e promoter region.
To further evaluate the role of NF-κB in the TNFα-induced expression of endogenous p110δ, we used IKK inhibitor VII to block NF-κB activation and analysed the expression of PIK3CD mRNA upon TNFα stimulation in EA.hy926 cells. IKK inhibitor VII inhibited the TNFα-induced expression of PIK3CD mRNA in a dose-dependent manner (Figure 6E) at similar concentrations to those that also inhibited the expression of IL-8 (Supplementary Figure S5A at http://www.BiochemJ.org/bj/443/bj4430857add.htm) and the degradation of IκB-α (Supplementary Figure S5B). These results suggest that NF-κB is involved in the regulation of TNFα-induced p110δ expression also in the context of the endogenous p110δ promoter in EA.hy926 cells.
These data demonstrate that the promoter associated with human PIK3CD exon −2e is functional and further inducible in ECs upon stimulation with TNFα. An intact NF-κB-binding site upstream of exon −2e is required for the TNFα-induced exon −2e promoter activity. In contrast, the exon −2c promoter region has weaker basal activity, which cannot be further enhanced by TNFα stimulation.
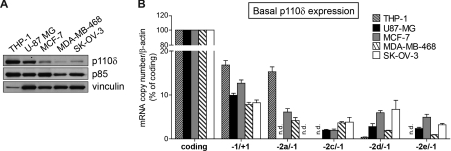
Expression of PIK3CD transcripts in human cancer cell lines
Most non-leucocytic cell types express low levels of p110δ under basal conditions. We chose four non-leucocytic human cancer cell lines that express intermediate (U-87 MG glioblastoma) or low (the breast cancer cell lines MCF-7 and MDA-MB-468 and the SK-OV-3 ovarian carcinoma) levels of p110δ (Figure 7A), to assess which PIK3CD promoters were used to express p110δ under unstimulated conditions in these cells.
Figure 7. p110δ expression under basal conditions in human cancer cell lines.
(A) p110δ protein expression in human cancer cell lines. Shown is a representative immunoblot of two independent experiments. (B) qPCR analysis of PIK3CD transcript expression in human cancer cell lines. Binding sites of the primer/probe sets used to amplify each transcript are shown schematically in Figure 5(A). The expression of each transcript was normalized to β-actin and expressed as the percentage of p110δ-coding transcripts. Results are means±S.E.M. for three independent experiments. n.d., not detected.
As expected, exon −2c, −2d and −2e transcripts were expressed in all non-leucocytic cells with exon −2a transcripts also found in the breast cancer cell lines MCF-7 and MDA-MB-468 (Figure 7B). Exon −2b transcripts were only detectable in THP-1 cells (results not shown). Interestingly, no exon−2a transcripts were detected in the U-87 MG cell line (Figure 7B) that expresses p110δ at levels comparable with that in leucocytes (Figure 7A), suggesting that transcription starting from the promoter(s) of exons −2c, −2d and −2e can lead to high cellular p110δ expression.
In conclusion, the novel PIK3CD promoter region identified in the present study not only is inducible by TNFα in human ECs and synovial fibroblasts, but also can allow transcriptional initiation in non-leucocytic cancer cell lines that express high levels of p110δ.
DISCUSSION
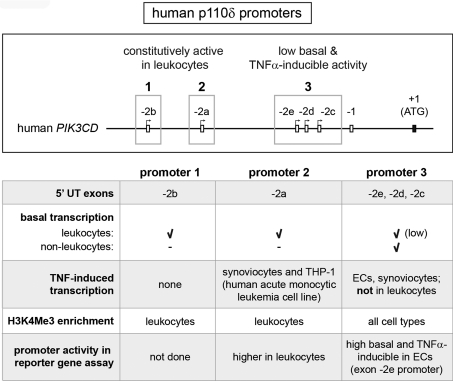
The functional role of the p110δ PI3K isoform has been widely studied in cells of the haematopoietic lineage in which p110δ expression is highly enriched. However, some non-leucocytes also express p110δ at moderate levels. Our results show that pro-inflammatory cytokines induce the expression of p110δ in human ECs and synovial fibroblasts, both important stromal cell types not often associated with p110δ expression or activity. In addition, we have identified a novel promoter region for human PIK3CD that gives rise to transcripts with previously unidentified 5′-UTRs. Figure 8 summarizes how this new promoter region compares with the other p110δ transcriptional elements known to date.
Figure 8. Overview of the three human PIK3CD promoters.
An overview of human PIK3CD promoters identified to date. The top panel shows a schematic representation of the three promoter regions (highlighted with boxes) and their associated exons with 5′ untranslated exons depicted as white boxes and the first protein-coding exon as a black box (not drawn to scale). The bottom panel summarizes the characteristics of each promoter. Promoters 1 and 2 are highly active in leucocytes, whereas promoter region 3 has low basal activity in all cell types which can be further enhanced by TNFα in some cell types.
We previously showed that the exon −2a promoter allows high basal expression of p110δ in mouse leucocytes [8]. Our results in human cells on exon −2a transcript expression, reporter gene assay (Supplementary Figure S6 at http://www.BiochemJ.org/bj/443/bj4430857add.htm) and analysis of promoter-associated histone modifications demonstrate that the promoter of exon −2a (Figure 8, promoter 2) is used to generate high basal expression of p110δ also in human leucocytes. In addition, expression of human exon −2b transcripts was highly restricted to leucocytes and the exon −2b promoter region (Figure 8, promoter 1) showed similar leucocyte-specific enrichment of H3K4Me3 as seen in the exon −2a promoter, suggesting that transcription from the exon −2b TSS also contributes to high p110δ expression in leucocytes. It is likely that a combination of regulatory mechanisms together allow enrichment of p110δ in leucocytes while keeping its expression low in other cell types. First, there is evidence to suggest that the exon −2a promoter region might be subject to active silencing, in a lineage-dependent manner. Indeed, a recent study demonstrated an inverse correlation between methylation of the genomic region upstream of PIK3CD exon −2a and p110δ expression in haematopoietic cells [28], suggesting that leucocyte-dependent hypomethylation contributes to the enrichment of p110δ in these cells while keeping p110δ expression low during early development and in non-haematological cells [28]. It is not known whether the exon −2b promoter is also regulated by cell/lineage-dependent methylation. Secondly, the expression of leucocyte-specific trans-acting factors have been shown to be involved in driving high p110δ expression from the exon −2a promoter [8,29]. In addition, the proteinase MT1-MMP (membrane-type 1 matrix metalloproteinase) was recently found to act as a TF that increases p110δ expression in macrophages through association with the genomic region upstream exon −1 [9]. Thirdly, recent genome-wide studies have highlighted the importance of enhancers in driving cell-type-specific gene expression [30]. The role of enhancers in regulating lineage-dependent PIK3CD transcription has not been investigated to date, but is likely to be of importance. However, there is evidence to suggest that the usage of the −2a and −2b promoters might not be completely restricted to leucocytes, as a modest enrichment of H3K4Me3 at these promoters was also observed in human and mouse non-leucocytes (Figure 4 and Supplementary Figure S4 respectively) and expression of exon −2a transcripts was detected in synovial fibroblasts and human breast cancer cells.
In addition to the exon −2a and −2b human PIK3CD promoters, we now report an alternative third PIK3CD promoter region (associated with exons −2c, −2d and −2e; Figure 8, promoter 3) that contributes to basal p110δ expression in all human cell types. Importantly, transcription starting from this promoter region can give rise to high p110δ expression as was seen in the glioblastoma cell line U-87 MG that does not express PIK3CD transcripts from the exon −2a and −2b promoters that are active in leucocytes (Figure 7). A corresponding mouse Pik3cd promoter has not been identified to date, but is likely to exist as mouse Pik3cd transcripts with 5′ untranslated exons in the corresponding genomic region are present in the Ensembl database (Figure 3B) and a clear enrichment of H3K4Me3 can be seen in the equivalent genomic region in different mouse cell types (Supplementary Figure S4).
As opposed to basal expression, our 5′RACE results showed that the PIK3CD promoter region 3 mediated the TNFα-induced expression of p110δ in human ECs. To our surprise, no TNFα-responsive inducibility of the promoter region 3 was seen in human leucocytes (Jurkat and THP-1 cells). In our reporter assays performed in ECs, an NF-κB site upstream of exon −2e was found to be required for TNFα-responsiveness. However, the TNFα-inducible expression of p110δ is likely to be more complex in an in vivo cellular context, and not limited to promoter region 3, given that exposure of synovial fibroblasts and THP-1 cells to TNFα also induced the expression of p110δ transcripts with the exon −2a (Figures 5D and 5F).
Our attempts to uncover a function for TNFα-induced p110δ in ECs or synovial fibroblasts were not successful, as p110δ inhibition did not affect survival, migration or production of IL-6 or IL-8 in TNFα-stimulated HUVECs, or production of IL-6, IL-8 or CCL2 [chemokine (C-C motif) ligand 2] in TNFα-stimulated synovial fibroblasts (results not shown). Others have shown that endothelial p110δ activity is involved in selectin-mediated capture of rolling leucocytes [12,31], an important endothelial process during inflammation, and it is possible that induced p110δ is important in this biology. Interestingly, a recent study found that TNFα and IL-1β stimulate the expression of p110δ in synovial fibroblasts and further implicated p110δ as a key isoform in PDGF (platelet-derived growth factor)-mediated Akt activation, cell growth and protection from apoptosis [32].
TNFα target genes differ in the timing of their induction upon TNFα stimulation: some genes are induced within minutes of exposure to TNFα and others with much slower kinetics [33]. Our findings would place p110δ into the latter group of TNFα target genes. The activity of p110δ in the endothelium (or synovium) might thus be dispensable in the early phase of inflammation, but become more important in the resolution phase or in chronic inflammatory states. Despite the high expression of p110α and p110β relative to p110δ in ECs, we noted an increase in the contribution of p110δ to the basal p85-associated PI3K activity in HUVECs upon TNFα stimulation (results not shown). It is possible, however, that activity from the TNFα-induced p110δ might be required to synergize with other PI3K isoforms as is the case in HepG2 cells where insulin-stimulated PKB phosphorylation is dependent on both p110α and p110δ activity [34]. Alternatively, TNFα-induced p110δ activity could be functionally important in a subcellularly localized manner and thus would not require expression levels similar to p110α or p110β. Indeed, p110δ activity has been demonstrated to be important for membrane fission in the trans-Golgi network in mouse macrophages [35]. Unfortunately, the lack of specific and high-affinity antibodies against p110δ, together with the low expression of p110δ in ECs, prevented our attempts to examine the subcellular localization of TNFα-induced p110δ. It was also of interest to note that TNFα stimulation did not increase the expression of p110γ, the other class I PI3K isoform whose expression is low in non-leucocytes. In fact, p110γ expression was reduced upon exposure of HUVECs to TNFα (results not shown).
Taken together, we have identified a third functional human PIK3CD promoter associated with the upstream exons −2c, −2d and −2e that has basal activity in both leucocytes and non-leucocytes, but TNFα-inducible activity only in selected human non-leucocytes. The differential enrichment of promoter- and enhancer-associated histone modifications support cell-type-specific usage of PIK3CD TSSs in line with the polarized expression of p110δ in different cell types. The fact that p110δ is a non-essential PI3K isoform in the organism (as opposed to p110α and p110β) might explain the observed versatility in transcriptional and post-transcriptional regulation of this PI3K isoform. In addition to the different PIK3CD transcripts generated through alternative TSS usage and splicing at the 5′-end, the role of alternative splicing in the p110δ protein-coding region is underexplored. Interestingly, a novel splice variant of human PIK3CD was recently identified in human leucocytes [36]. This splice variant, generated by usage of an alternative splice site in intron 5, encodes a C-terminally truncated variant of p110δ (designated p37δ) that is functionally distinct from the full-length p110δ protein [36]. Flexible use of exon assembly allows cells to dynamically respond to their environment, including therapy, as was recently shown for drug-resistant B-Raf [37], highlighting the importance of a thorough understanding of the transcriptional regulation of drug targets.
Online data
AUTHOR CONTRIBUTION
Maria Whitehead and Bart Vanhaesebroeck designed and analysed the experiments and wrote the paper. Maria Whitehead performed the experiments. Michele Bombardieri and Costatino Pitzalis provided human synovial fibroblasts and the relevant expertise. Bart Vanhaesebroeck obtained the funding.
ACKNOWLEDGEMENTS
We thank members of the Cell Signalling group for feedback on the paper, and Yvonne Kam (William Harvey Research Institute, Queen Mary University of London, London, U.K.) for providing human synovial fibroblasts.
FUNDING
This work was supported by Cancer Research UK [grant number C23338/A10200], the Ludwig Institute for Cancer Research and Queen Mary University of London. Personal support to M.A.W. was provided by an EU Marie Curie Actions Early-Stage Training fellowship, International Graduate Program in Molecular Medicine [grant number 20386]. Part of this work was supported by Arthritis Research UK [project grant number 18399 (to M.B. and C.P.)].
References
- 1.Engelman J. A. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat. Rev. Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 2.Holmes D. PI3K pathway inhibitors approach junction. Nat. Rev. Drug Discovery. 2011;10:563–564. doi: 10.1038/nrd3527. [DOI] [PubMed] [Google Scholar]
- 3.Jarvis L. M. PI3K at the clinical crossroads. Chem. Eng. News. 2011;89:15–19. [Google Scholar]
- 4.Vanhaesebroeck B., Welham M. J., Kotani K., Stein R., Warne P. H., Zvelebil M. J., Higashi K., Volinia S., Downward J., Waterfield M. D. p110δ, a novel phosphoinositide 3-kinase in leukocytes. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4330–4335. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chantry D., Vojtek A., Kashishian A., Holtzman D. A., Wood C., Gray P. W., Cooper J. A., Hoekstra M. F. p110δ, a novel phosphatidylinositol 3-kinase catalytic subunit that associates with p85 and is expressed predominantly in leukocytes. J. Biol. Chem. 1997;272:19236–19241. doi: 10.1074/jbc.272.31.19236. [DOI] [PubMed] [Google Scholar]
- 6.Furman R. R., Byrd J. C., Brown J. R., Coutre S. E., Benson D. M., Jr, Wagner-Johnston N. D., Flinn I. W., Kahl B. S., Spurgeon S. E., Lannutti B., et al. CAL-101, an isoform-selective inhibitor of phosphatidylinositol 3-kinase p110δ, demonstrates clinical activity and pharmacodynamic effects in patients with relapsed or refractory chronic lymphocytic leukemia. 2010. http://ash.confex.com/ash/2010/webprogram/Paper32870.html (Abstract)
- 7.Fruman D. A., Rommel C. PI3Kδ inhibitors in cancer: rationale and serendipity merge in the clinic. Cancer Discovery. 2011;1:562–572. doi: 10.1158/2159-8290.CD-11-0249. [DOI] [PubMed] [Google Scholar]
- 8.Kok K., Nock G. E., Verrall E. A., Mitchell M. P., Hommes D. W., Peppelenbosch M. P., Vanhaesebroeck B. Regulation of p110δ PI 3-kinase gene expression. PLoS ONE. 2009;4:e5145. doi: 10.1371/journal.pone.0005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimizu-Hirota R., Xiong W., Baxter B. T., Kunkel S. L., Maillard I., Chen X. W., Sabeh F., Liu R., Li X. Y., Weiss S. J. MT1-MMP regulates the PI3Kδ·Mi-2/NuRD-dependent control of macrophage immune function. Genes Dev. 2012;26:395–413. doi: 10.1101/gad.178749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawyer C., Sturge J., Bennett D. C., O'Hare M. J., Allen W. E., Bain J., Jones G. E., Vanhaesebroeck B. Regulation of breast cancer cell chemotaxis by the phosphoinositide 3-kinase p110δ. Cancer Res. 2003;63:1667–1675. [PubMed] [Google Scholar]
- 11.Eickholt B. J., Ahmed A. I., Davies M., Papakonstanti E. A., Pearce W., Starkey M. L., Bilancio A., Need A. C., Smith A. J., Hall S. M., et al. Control of axonal growth and regeneration of sensory neurons by the p110δ PI 3-kinase. PLoS ONE. 2007;2:e869. doi: 10.1371/journal.pone.0000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puri K. D., Doggett T. A., Douangpanya J., Hou Y., Tino W. T., Wilson T., Graf T., Clayton E., Turner M., Hayflick J. S., Diacovo T. G. Mechanisms and implications of phosphoinositide 3-kinase delta in promoting neutrophil trafficking into inflamed tissue. Blood. 2004;103:3448–3456. doi: 10.1182/blood-2003-05-1667. [DOI] [PubMed] [Google Scholar]
- 13.Conte E., Fruciano M., Fagone E., Gili E., Caraci F., Iemmolo M., Crimi N., Vancheri C. Inhibition of PI3K prevents the proliferation and differentiation of human lung fibroblasts into myofibroblasts: the role of class I P110 isoforms. PLoS ONE. 2011;6:e24663. doi: 10.1371/journal.pone.0024663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Northcott C. A., Poy M. N., Najjar S. M., Watts S. W. Phosphoinositide 3-kinase mediates enhanced spontaneous and agonist-induced contraction in aorta of deoxycorticosterone acetate-salt hypertensive rats. Circ. Res. 2002;91:360–369. doi: 10.1161/01.res.0000030861.13850.f1. [DOI] [PubMed] [Google Scholar]
- 15.Tsybouleva N., Zhang L., Chen S., Patel R., Lutucuta S., Nemoto S., DeFreitas G., Entman M., Carabello B. A., Roberts R., Marian A. J. Aldosterone, through novel signaling proteins, is a fundamental molecular bridge between the genetic defect and the cardiac phenotype of hypertrophic cardiomyopathy. Circulation. 2004;109:1284–1291. doi: 10.1161/01.CIR.0000121426.43044.2B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinho J. F., Medeiros M. A., Capettini L. S., Rezende B. A., Campos P. P., Andrade S. P., Cortes S. F., Cruz J. S., Lemos V. S. Phosphatidylinositol 3-kinase-delta up-regulates L-type Ca2+ currents and increases vascular contractility in a mouse model of Type 1 diabetes. Br. J. Pharmacol. 2010;161:1458–1471. doi: 10.1111/j.1476-5381.2010.00955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bombardieri M., Kam N. W., Brentano F., Choi K., Filer A., Kyburz D., McInnes I. B., Gay S., Buckley C., Pitzalis C. A BAFF/APRIL-dependent TLR3-stimulated pathway enhances the capacity of rheumatoid synovial fibroblasts to induce AID expression and Ig class-switching in B cells. Ann. Rheum. Dis. 2011;70:1857–1865. doi: 10.1136/ard.2011.150219. [DOI] [PubMed] [Google Scholar]
- 18.Hirth A., Skapenko A., Kinne R. W., Emmrich F., Schulze-Koops H., Sack U. Cytokine mRNA and protein expression in primary-culture and repeated-passage synovial fibroblasts from patients with rheumatoid arthritis. Arthritis Res. 2002;4:117–125. doi: 10.1186/ar391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT] method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Ogilvy S., Elefanty A. G., Visvader J., Bath M. L., Harris A. W., Adams J. M. Transcriptional regulation of vav, a gene expressed throughout the hematopoietic compartment. Blood. 1998;91:419–430. [PubMed] [Google Scholar]
- 21.Birney E., Stamatoyannopoulos J. A., Dutta A., Guigo R., Gingeras T. R., Margulies E. H., Weng Z., Snyder M., Dermitzakis E. T., Thurman R. E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okkenhaug K., Vanhaesebroeck B. PI3K in lymphocyte development, differentiation and activation. Nat. Rev. Immunol. 2003;3:317–330. doi: 10.1038/nri1056. [DOI] [PubMed] [Google Scholar]
- 23.Edgell C. J., McDonald C. C., Graham J. B. Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc. Natl. Acad. Sci. U.S.A. 1983;80:3734–3737. doi: 10.1073/pnas.80.12.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unger R. E., Krump-Konvalinkova V., Peters K., Kirkpatrick C. J. In vitro expression of the endothelial phenotype: comparative study of primary isolated cells and cell lines, including the novel cell line HPMEC-ST1.6R. Microvasc. Res. 2002;64:384–397. doi: 10.1006/mvre.2002.2434. [DOI] [PubMed] [Google Scholar]
- 25.Olszanecki R., Gebska A., Korbut R. Production of prostacyclin and prostaglandin E2 in resting and IL-1β-stimulated A549, HUVEC and hybrid EA.HY 926 cells. J. Physiol. Pharmacol. 2006;57:649–660. [PubMed] [Google Scholar]
- 26.Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 27.Zabel U., Schreck R., Baeuerle P. A. DNA binding of purified transcription factor NF-κB. Affinity, specificity, Zn2+ dependence, and differential half-site recognition. J. Biol. Chem. 1991;266:252–260. [PubMed] [Google Scholar]
- 28.Calvanese V., Fernandez A. F., Urdinguio R. G., Suarez-Alvarez B., Mangas C., Perez-Garcia V., Bueno C., Montes R., Ramos-Mejia V., Martinez-Camblor P., et al. A promoter DNA demethylation landscape of human hematopoietic differentiation. Nucleic Acids Res. 2012;40:116–131. doi: 10.1093/nar/gkr685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards H., Xie C., LaFiura K. M., Dombkowski A. A., Buck S. A., Boerner J. L., Taub J. W., Matherly L. H., Ge Y. RUNX1 regulates phosphoinositide 3-kinase/AKT pathway: role in chemotherapy sensitivity in acute megakaryocytic leukemia. Blood. 2009;114:2744–2752. doi: 10.1182/blood-2008-09-179812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heintzman N. D., Hon G. C., Hawkins R. D., Kheradpour P., Stark A., Harp L. F., Ye Z., Lee L. K., Stuart R. K., Ching C. W., et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Randis T. M., Diacovo N., Calafat J., Diacovo T. G. Shedding light on class I phosphoinositide 3-kinase activity in endothelium. Blood. 2008;111:4827–4828. doi: 10.1182/blood-2008-02-138396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartok B., Boyle D. L., Liu Y., Ren P., Ball S. T., Bugbee W. D., Rommel C., Firestein G. S. PI3 kinase δ: a key regulator of synoviocyte function in rheumatoid arthritis. Am. J. Pathol. 2012. doi:10.1016/j.ajpath.2012.01.030. [DOI] [PubMed]
- 33.Hao S., Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat. Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaussade C., Rewcastle G. W., Kendall J. D., Denny W. A., Cho K., Gronning L. M., Chong M. L., Anagnostou S. H., Jackson S. P., Daniele N., Shepherd P. R. Evidence for functional redundancy of class IA PI3K isoforms in insulin signalling. Biochem. J. 2007;404:449–458. doi: 10.1042/BJ20070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Low P. C., Misaki R., Schroder K., Stanley A. C., Sweet M. J., Teasdale R. D., Vanhaesebroeck B., Meunier F. A., Taguchi T., Stow J. L. Phosphoinositide 3-kinase delta regulates membrane fission of Golgi carriers for selective cytokine secretion. J. Cell Biol. 2010;190:1053–1065. doi: 10.1083/jcb.201001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fransson S., Uv A., Eriksson H., Andersson M. K., Wettergren Y., Bergo M., Ejeskar K. p37δ is a new isoform of PI3K p110δ that increases cell proliferation and is overexpressed in tumors. Oncogene. 2011. doi:10.1038/onc.2011.492. [DOI] [PubMed]
- 37.Poulikakos P. I., Persaud Y., Janakiraman M., Kong X., Ng C., Moriceau G., Shi H., Atefi M., Titz B., Gabay M. T., et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.