Abstract
Background
Decreased expression of cardiac myosin binding protein C (cMyBPC) as a result of genetic mutations may contribute to the development of hypertrophic cardiomyopathy (HCM); however, the mechanisms that link cMyBPC expression and HCM development, especially contractile dysfunction, remain unclear.
Methods and Results
We evaluated cardiac mechanical function in vitro and in vivo in young mice (8–10 weeks of age) carrying no functional cMyBPC alleles (cMyBPC−/−) or 1 functional cMyBPC allele (cMyBPC±). Skinned myocardium isolated from cMyBPC−/− hearts displayed significant accelerations in stretch activation cross-bridge kinetics. Cardiac MRI studies revealed severely depressed in vivo left ventricular (LV) magnitude and rates of LV wall strain and torsion compared with wild-type (WT) mice. Heterozygous cMyBPC± hearts expressed 23±5% less cMyBPC than WT hearts but did not display overt hypertrophy. Skinned myocardium isolated from cMyBPC± hearts displayed small accelerations in the rate of stretch induced cross-bridge recruitment. MRI measurements revealed reductions in LV torsion and circumferential strain, as well reduced circumferential strain rates in early systole and diastole.
Conclusions
Modest decreases in cMyBPC expression in the mouse heart result in early-onset subtle changes in cross-bridge kinetics and in vivo LV mechanical function, which could contribute to the development of HCM later in life.
Keywords: cardiomyopathy, MRI, mechanics, myocardial contraction
Hypertrophic cardiomyopathy (HCM) is one of the most commonly occurring genetic myocardial disorders, affecting approximately 1 in 500 people.1 Cardiac myosin binding protein C (cMyBPC) is a thick filament protein that modulates actin-myosin interactions and thereby the rate of muscle contraction.2,3 It is well established that the gene encoding cMyBPC is one of the most common causes of inherited HCM, with nearly 200 known mutations identified4 since the first reported mutation in this gene.5,6 However, many younger individuals who carry disease-causing mutations in the cMyBPC gene do not exhibit overt LVH, because increases in LV wall thickness are often only detectable with advanced age.7,8 Because these seemingly asymptomatic carriers are at risk for the development of HCM and cardiac disease later in life, the diagnosis and treatment of these patients is a major clinical challenge.
The majority of mutations in the gene that encode cMyBPC are heterozygous and are predicted to result in expression of truncated cMyBPC lacking the C-terminal regions of the protein that binds to myosin and titin.9 However, analysis of myocardial biopsy samples from patients with cMyBPC mutations have not detected truncated cMyBPC, but rather a reduction in the amount of full-length cMyBPC protein has been noted.10–14 It is therefore likely that mutant cMyBPC mRNA or proteins are rapidly degraded by nonsense mediated mRNA decay or the ubiquitin-proteosome system, thereby preventing mutant proteins from incorporating into the sarcomere.15 Therefore, the allele generating mutant cMyBPC effectively functions as a null allele, causing cMyBPC haploinsufficiency. However, the mechanisms that link reduced cMyBPC levels in the heart with the development and progression of HCM have remained elusive.
Considering the functional importance of cMyBPC in regulating myofilament contractile properties, it is reasonable to suppose that decreased cMyBPC expression could affect in vivo mechanical function. In this regard, it has been shown that homozygous cMyBPC knockout mice (cMyBPC−/−) with 2 null cMyBPC alleles (ie, a model of pure insufficiency) display early-onset impairments in systolic and diastolic contractile function and severe LVH.16–18 In contrast, heterozygous cMyBPC knockout mice (cMyBPC±) with 1 null cMyBPC allele develop a phenotype later in life, displaying modest hypertrophy despite preserved systolic and diastolic contractile function.19 Interestingly, cMyBPC± hearts expressed ≈ 25% less total cMyBPC than aged-matched wild-type (WT) mice,19 which is similar to the amount of full length cMyBPC in patients with heterozygous cMyBPC mutations.12,13 These results suggest that modest decreases in cMyBPC expression in the heart may be sufficient to produce cardiac dysfunction and/or LVH; however, it is has not been established if these changes are related to altered cardiac mechanical performance.
Therefore, in the present study we examined the effects of variable cMyBPC expression on in vitro and in vivo mechanical function in young (8–10 weeks of age) cMyBPC−/− and cMyBPC± mice to determine if cMyBPC insufficiency can cause mechanical dysfunction early in life. We used MRI to quantify both global and regional mechanical indices such as LV twist, torsion, and principal strains, over the whole cardiac cycle to observe subtle changes in mechanical function. Our goal was to link cMyBPC expression and cross-bridge function with LV strain and torsion, which are direct measures of myocardial wall deformation, to characterize the functional consequences of cMyBPC insufficiency.
Methods
Animal Models
Adult male cMyBPC null (cMyBPC−/−), heterozygous cMyBPC mice (cMyBPC±), and WT mice of the SV/129 strain (8–10 weeks of age) were used in this study.16 All procedures involving animal care and handling were performed according to institutional guidelines set forth by the Animal Care and Use Committee at Case Western Reserve University.
Skinned Fiber Experiments
Force-pCa relationships and stretch activation kinetics were measured in skinned ventricular myocardium isolated from WT, cMyBPC−/−, and cMyBPC± hearts as previously described.20,21 Myofibrillar protein content and phosphorylation were determined in LV homogenates and skinned myocardium as previously described.20,22
In Vivo MRI Measurements of Cardiac Function
Two-dimensional LV myocardial motion was quantified at the apex, mid, and basal levels using multiphase displacement encoding with stimulated-echo (DENSE) MRI in a 9.4-T Bruker Biospec (Billerica, MA) horizontal scanner.23 LV twist, torsion, circumferential and radial strain, as well as torsion and strain rates were quantified, using custom software as previously described.24
Statistical Analysis
Cross-sectional areas of skinned preparations were calculated by measuring the width of the mounted preparation and assuming a cylindrical cross section. Submaximal Ca2+-activated force (P) was expressed as a fraction of the force (Po) generated at pCa 4.5, that is, P/Po. Rate constants of stretch-induced force decay (krel) and development (kdf) were obtained by fitting the time course trace with a single exponential. Data are reported as mean±SD or mean±SEM. Means for fiber data were generated by averaging data for each variable for each mouse and then averaging the means for all mice within a group. Approximately 3 fibers were studied from each mouse. Comparisons of force-pCa relationships and stretch activation variables between groups were done using a 1-way ANOVA and a Tukey-Kramer post hoc test. Comparisons of strain and twist angles were independently analyzed at each ventricular level by 1-way ANOVA and Tukey-Kramer test. Strain and torsion time course data were evaluated by 1-way ANOVA with a Bonferroni adjustment for multiple comparisons. Additionally, a separate analysis examining only peak rates of torsion and strain in systole and diastole was performed using 1-way ANOVA and Tukey-Kramer post hoc test. Statistical significance was established at a level of P<0.05.
See the expanded Methods section in the online Data Supplement Material for detailed descriptions of methods.
Results
Myofilament Protein Expression and Phosphorylation in WT and cMyBPC Mutant Myocardium
We examined the expression and phosphorylation status of myofilament proteins in age-matched male WT, cMyBPC−/−, and cMyBPC± LV tissue homogenates. As expected, cMyBPC was not detected in cMyBPC−/− hearts in either Coomassie-stained gels or Western blots probed with a cMyBPC specific antibody (Santa Cruz, CA); however, cMyBPC± hearts expressed 23±5% less cMyBPC than WT hearts (P<0.05, online Data Supplement Figure 1). Consistent with previous findings,16 expression of β-myosin heavy chain (MHC) was slightly elevated in cMyBPC−/− hearts (16±3%, P<0.05) compared with WT hearts, but there was no significant difference in MHC isoform expression between cMyBPC± and WT hearts (online Data Supplement Figure 2). No significant differences in the relative abundance or phosphorylation status of other myofilament proteins in WT, cMyBPC−/−, and cMyBPC± myocardium were detected (online Data Supplement Figure 3). The cMyBPC content of skinned myocardium isolated from cMyBPC± LV used for mechanical experiments was also quantified (online Data Supplement Table) and was nearly identical to the cMyBPC content measured in LV tissue homogenates prepared from cMyBPC± hearts (ie, reduced 24±6% compared with WT).
Mechanical Properties of Skinned Myocardium Isolated From WT and cMyBPC Mutant Hearts
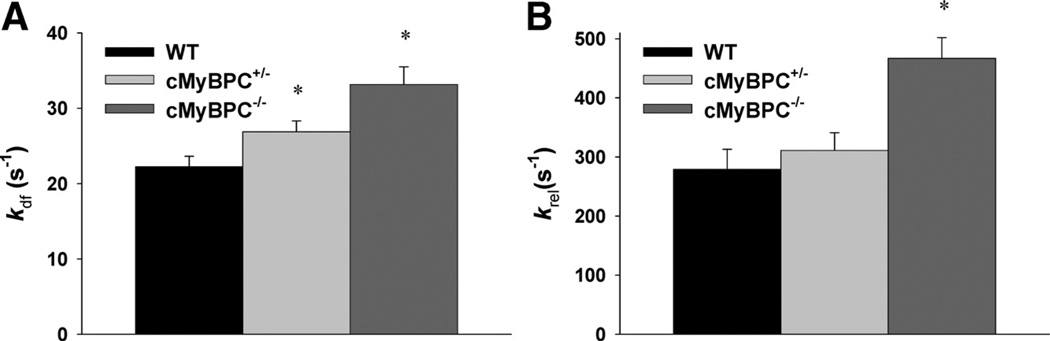
The steady-state mechanical properties of skinned myocardium isolated from WT, cMyBPC−/−, and cMyBPC± hearts are summarized in the online Data Supplement Table. Skinned preparations from the 3 groups of mice exhibited similar force-pCa relationships. There were no differences in force generation at maximal and submaximal [Ca2+] or in the steepness of the force-pCa relationship (Hill coefficient, nH). Consistent with previous studies,22 cMyBPC−/− myocardium displayed dramatically accelerated rates of stretch-induced force decay (krel) and delayed force development (kdf) at submaximal Ca2+ activations as well as greater stretch-induced force decay (P2 amplitude) and stretch activation amplitude (Pdf), compared with WT myocardium (Figure 1). Skinned myocardium isolated from cMyBPC± hearts displayed small but significant (21%) accelerated stretch-induced delayed force development (kdf) at submaximal Ca2+ activations, but P2 and Pdf amplitude were not significantly different compared with WT (Table 1).
Figure 1.
Stretch activation kinetics in skinned myocardium. The rates of A, delayed force development (kdf), and B, force relaxation (krel) after a stretch of 1% of muscle length, were recorded from wild-type (WT), cardiac myosin binding protein C (cMyBPC)±, and cMyBPC−/− skinned myocardium activated with Ca2+ to a prestretch isometric force of ≈50% maximal. Data are mean±SD. *Significantly different from WT.
Table 1.
Stretch Activation Parameters of Skinned Fibers Isolated From WT, cMyBPC+/−, and cMyBPC−/− Myocardium
| kdf (S−1) | krel (S−1) | P2 | P3 | ||
|---|---|---|---|---|---|
| WT | 22.24±1.39 | 279±34 | 0.04±0.01 | 0.10±0.01 | 0.06±0.01 |
| cMyBPC+/− | 26.88±1.45* | 311±30 | 0.02±0.01 | 0.10±0.01 | 0.08±0.01 |
| cMyBPC−/− | 33.15±2.35* | 467±35* | −0.03±0.01* | 0.10±0.01 | 0.13±0.01* |
Data are mean±SD. Skinned ventricular myocardium was isolated from 5–6 mice per group (16–19 fibers total/group). Rate constants were calculated from force transients in response to stretches of 1% of muscle length at approximately half-maximal levels of activation. Stretch activation parameters are described in the online-only Data Supplement.
WT indicates wild-type; cMyBPC, cardiac myosin binding protein C.
Significantly different from WT.
In Vivo Assessment of Cardiac Morphology and Mechanical Function
LV Morphology and Contractile Function
Cardiac morphology and global contractile function analysis of WT, cMyBPC±, and cMyBPC−/− hearts are presented in Table 2. The cMyBPC−/− hearts displayed significant LV hypertrophy as demonstrated by increased end-diastolic wall thickness and decreased ejection fraction compared with WT controls. In contrast, cMyBPC± hearts displayed a small increase in apical thickening at end-diastole compared with WT hearts (Table 2, P<0.05), but systolic function was preserved, as indicated by near-normal ejection fraction values. Global hypertrophy was seen in cMyBPC−/− hearts, as demonstrated by greater heart weight–to–body-weight (HTWT/BWT) ratios compared with WT hearts. There was no difference in HTWT/BWT ratio between cMyBPC± and WT hearts (online Data Supplement Figure 4). Molecular markers of hypertrophy, including transcript expression of ANF, BNP, and α-skeletal actin were also significantly upregulated in cMyBPC−/− hearts but not in cMyBPC± hearts (online Data Supplement Figure 4).
Table 2.
Cardiac Morphology and Cardiovascular Function of WT, cMyBPC+/−, and cMyBPC−/− Hearts
| WT | cMyBPC+/− | cMyBPC−/− | |
|---|---|---|---|
| Body weight, g | 25.6±4.0 | 24.1±2.2 | 22.2±2.5 |
| Ejection fraction, % | 68.2±8 | 62.7±6 | 32.5±8* |
| End-diastolic volume, mm3 | 0.31±0.05 | 0.34±0.07 | 0.60±0.08* |
| End-systolic volume, mm3 | 0.10±0.03 | 0.12±0.04 | 0.41±0.09* |
| End-diastolic wall thickness, mm | |||
| Base | 0.71±0.08 | 0.71±0.06 | 0.88±0.09* |
| Mid-ventricle | 0.68±0.07 | 0.65±0.04 | 0.80±0.20 |
| Apex | 0.58±0.07 | 0.67±0.06* | 0.74±0.10* |
| End-systolic wall thickness, mm | |||
| Base | 1.19±0.2 | 1.16±0.1 | 1.12±0.1 |
| Mid-ventricle | 1.20±0.2 | 1.07±0.1 | 1.01±0.2* |
| Apex | 0.95±0.1 | 0.99±0.2 | 0.90±0.2 |
| Heart rate, bpm | 562±27 | 573±31 | 571±34 |
| Body temperature, °C | 35.3±0.4 | 35.5±0.3 | 35.5±0.4 |
| Respiratory rate, per minute | 80±12 | 84±13 | 85±9 |
Data are mean±SD.
WT indicates wild-type; cMyBPC, cardiac myosin binding protein C.
Significantly different from WT.
Myocardial Strain
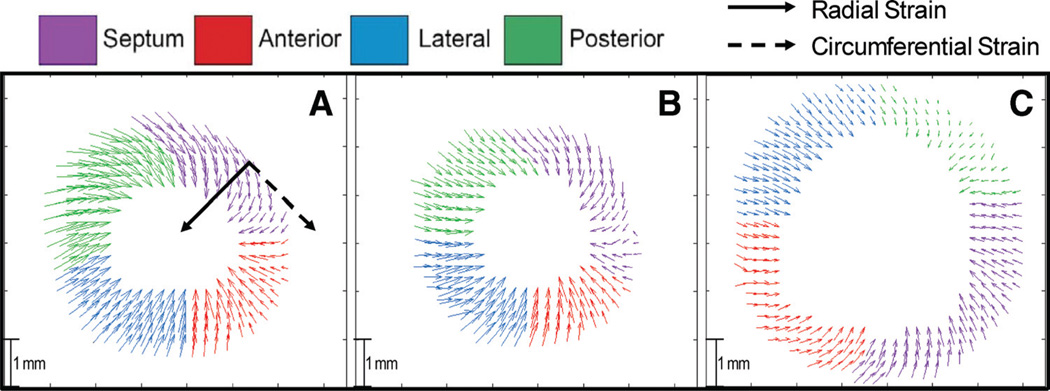
Representative displacement maps for the in-plane motion of the WT, cMyBPC±, and cMyBPC−/− groups in the midventricle at peak systole are presented in Figure 2. The overall reduction in displacement magnitude (Figure 2C) for strain vectors displayed by cMyBPC−/− hearts compared with WT hearts is consistent with significant contractile dysfunction.
Figure 2.
Representative MRI principal strain vector displacement maps. Representative DENSE displacement maps at the mid-ventricle during peak systole for A, wild-type (WT); B, cardiac myosin binding protein C (cMyBPC)±; and C, cMyBPC−/−. The direction of radial (solid arrow) and circumferential strain (dashed arrow) is labeled in A).
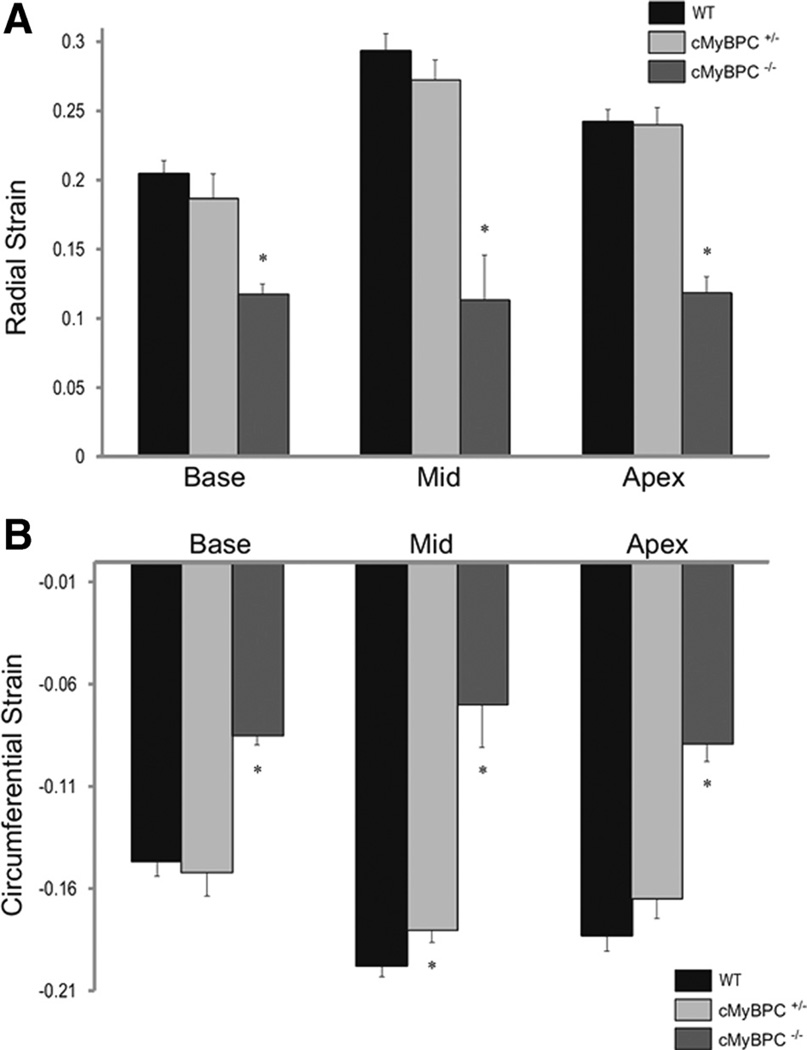
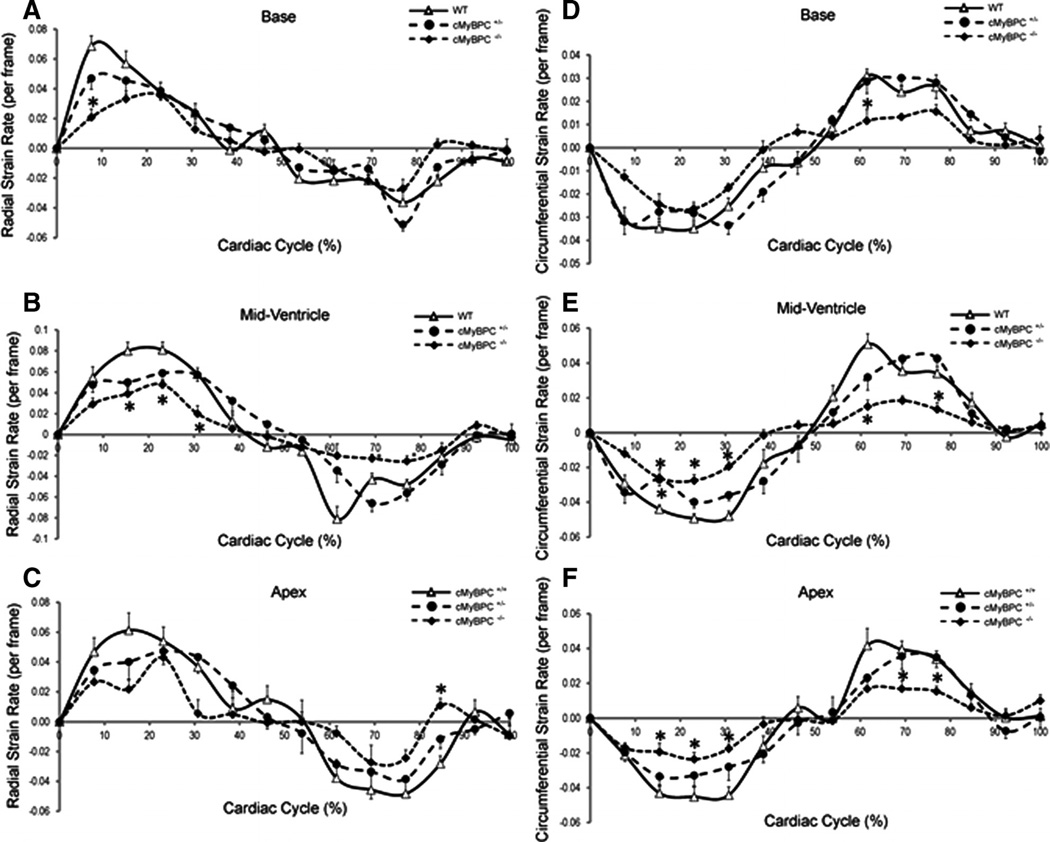
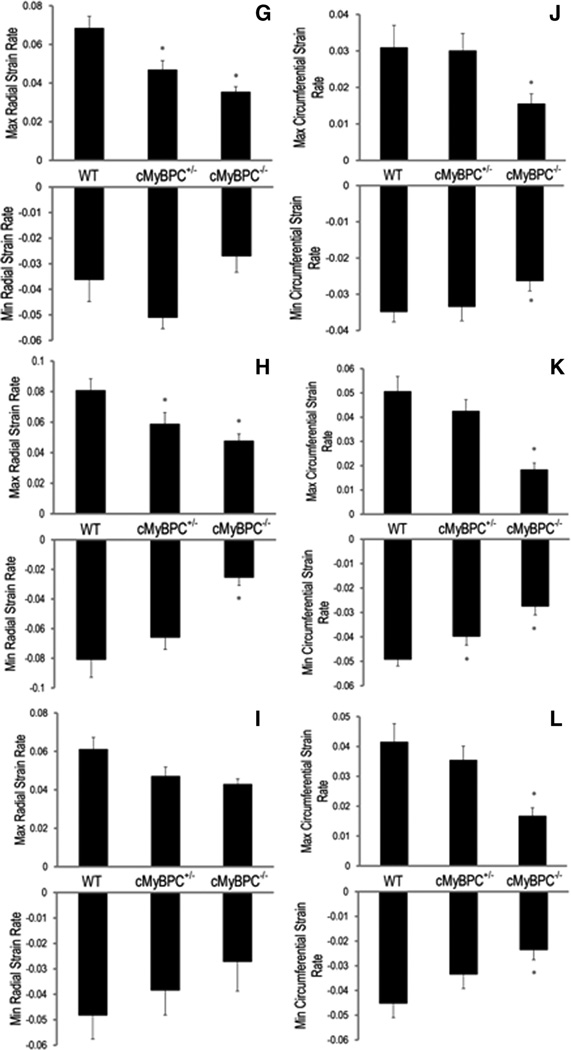
Mean LV strain was calculated at the apex, base, and mid-ventricle for each group. There was a significant reduction in maximal LV radial and circumferential strains in the cMyBPC−/− mice for all 3 ventricular levels (Figure 3), and there was a significant reduction in maximal LV circumferential strain at the mid-LV of cMyBPC± mice compared with WT mice (Figure 3B, P<0.05). Measurements of average radial (Figure 4A through 4C) and circumferential strain rates (Figure 4D through 4F) were significantly reduced in cMyBPC−/− mice throughout the cardiac cycle. Reduced early systolic strain rates were observed in cMyBPC± mice at the mid-LV (Figure 4). Analysis of peak strain rates revealed that cMyBPC± mice displayed lower peak radial systolic strain rates in the base and mid-LV, and lower peak circumferential diastolic strain rates in the mid-LV (Figure 4G through 4L).
Figure 3.
Measurements of left ventricular (LV) global principal strains. Global radial (A) and circumferential (B) strain at the base, mid, and apex in LV of wild-type (WT), cardiac myosin binding protein C (cMyBPC)±, and cMyBPC−/− mice. Data are mean±SE. *Significantly different from WT.
Figure 4.
Analysis of principal strain rates. Time course of radial (A through C) and circumferential (D through F) strain rates were quantified throughout the cardiac cycle, and peak radial (G through I) and circumferential (J–L) strain rates were quantified (from top to bottom) at the base, mid-ventricle, and apical segments in wild-type (WT), cardiac myosin binding protein C (cMyBPC)±, and cMyBPC−/− mice. Data are mean±SE. *Significantly different from WT by 1-way ANOVA with Bonferroni (A through F) or Tukey-Kramer (G through L).
Ventricular Twist and Torsion
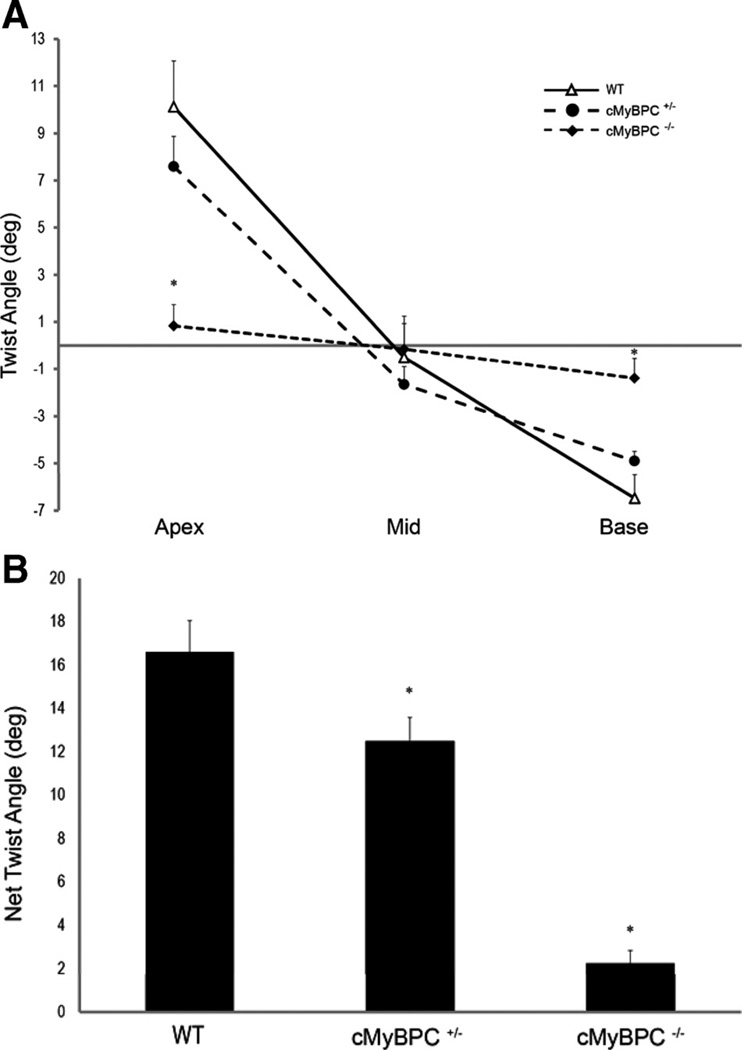
Figure 5A illustrates the difference in twist (rotation) angles between WT, cMyBPC±, and cMyBPC−/− mice at the apical, mid-LV, and basal levels during peak systole. There was a clear decrease in overall rotation in cMyBPC−/− hearts. LV twisting occurred in the counterclockwise direction at the apex in the WT (10.12±1.9°) and cMyBPC± hearts (7.59±1.3°, P=NS), but minimal apical twisting was observed in cMyBPC−/− hearts (0.83±0.9°, P<0.05). Clockwise twisting was observed at the base in WT (−6.48±1.0°) and cMyBPC± hearts (−4.90±0.40°, P=NS), but minimal rotation occurred in cMyBPC−/− hearts (−1.39±0.84°, P<0.05).
Figure 5.
Left ventricular twist relationships during systole. A, Maximal twist angles (degrees) from the apex, mid-ventricle, and base during systole, and B, net twist angles (apex-base) during systole of the wild-type (WT), cardiac myosin binding protein C (cMyBPC)±, and cMyBPC−/− mice. Data are mean±SE. *Significantly different from WT.
The net twist, defined as the difference between the twist angles at the base and apex, is plotted in Figure 5B. There was a significant reduction in net twist in cMyBPC−/− (2.23±0.61°) and cMyBPC± (12.49±1.08°) hearts compared with WT hearts (16.60±1.46°). In particular, net twist between the mid and basal LV segments was reduced in cMyBPC± hearts compared with WT (Figure 5A, 3.24±2.0° and 5.98±4.5°, respectively), although this difference fell short of statistical significance (P=0.08).
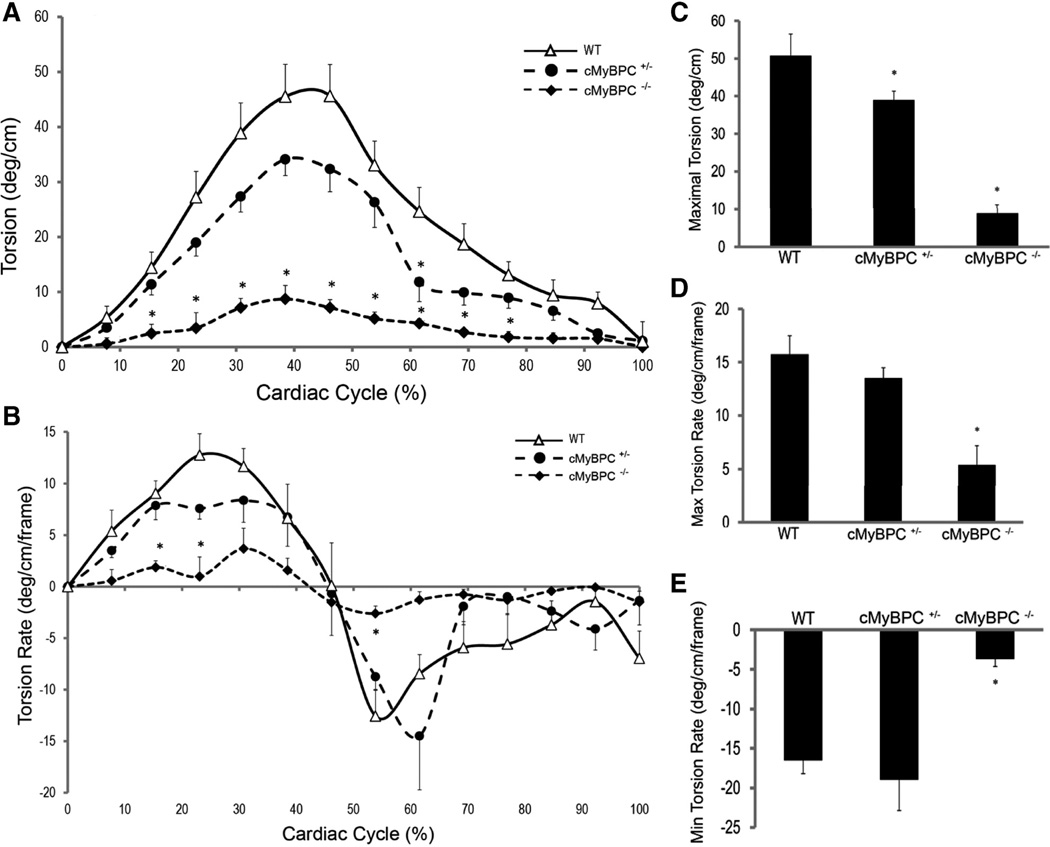
There was a significant reduction in the magnitude of peak torsion generated by cMyBPC−/− and cMyBPC± hearts compared with WT (50.8°/cm ±5.7): 8.9°/cm ±2.2 in cMyBPC−/− and 39.0°/cm ±2.4 in cMyBPC± (Figure 6A and 6C), and peak systolic torsion occurred earlier in cMyBPC−/− hearts. Maximal systolic and diastolic torsion rates were lower in cMyBPC−/− hearts (Figure 6B, 6D, 6E).
Figure 6.
Analysis of left ventricular torsion. Time course of ventricular torsion (A) and torsion rates (B) in 1 cardiac cycle for wild-type (WT), cardiac myosin binding protein C (cMyBPC)±, and cMyBPC−/− mice. Peak systolic torsion amplitude (C), peak rates of systolic torsion (D), and diastolic torsion (E) in WT, cMyBPC±, and cMyBPC−/− mice. Data are mean±SE. *Significantly different from WT by 1-way ANOVA with Bonferroni (A and B) or Tukey-Kramer (C through E).
Discussion
There is growing evidence that decreased cMyBPC expression may be a common feature of HCM related to mutations in the gene encoding cMyBPC12,13; however, the link between cMyBPC expression, cardiac mechanical function, and the development of HCM is not well understood. We show that decreased cMyBPC expression in young cMyBPC± mice leads to altered myofilament function and in vivo torsion and principal strains in the absence of overt hypertrophy. Our data suggest that mechanical dysfunction exists in preclinical cMyBPC related HCM.
Myofilament Function in cMyBPC Mutant Myocardium
Levels of full-length cMyBPC have been shown to be decreased (24–33%) in patients with mutations in cMyBPC.12,13 Similarly, levels of cMyBPC were decreased in heterozygous mouse models expressing 1 functional cMyBPC allele (the present study and Carrier et al19). Mutations in cMyBPC predicted to produce C-terminal truncated proteins are not found in patients.10–14 The mutant mRNA and/or proteins are thought to be unstable and are degraded before incorporation in the sarcomere,15 such that the mutant allele effectively acts as a null allele, leading to cMyBPC haploinsufficiency.
Consistent with previous studies, we found that skinned myocardium isolated from cMyBPC+/−16 or cMyBPC−/− hearts22 does not display changes in Ca2+-sensitivity of force generation. This suggests that cMyBPC content in the sarcomere does not directly determine myofilament Ca2+ sensitivity. A recent study12 showed that skinned myocytes isolated from myectomy samples obtained from patients with HCM-causing cMyBPC mutations displayed increases in the Ca2+ sensitivity of force generation. However, changes in force generation did not correlate with cMyBPC expression and were likely a result of the large decrease in troponin I (TnI) phosphorylation (84%) in myectomy samples compared with control donor samples.
It is well established that skinned myocardium isolated from cMyBPC−/− hearts displays significantly accelerated rates of cross-bridge kinetics22,25,26 because myosin heads are in closer juxtaposition to actin molecules, thereby enhancing the probability of acto-myosin interactions.27 In the present study, we found that a relatively small decrease in cMyBPC expression (24%) in cMyBPC± skinned myocardium produced acceleration in the rate of delayed cross-bridge recruitment (kdf) after rapid stretch at submaximal Ca2+ activations compared with WT skinned myocardium (Table 1). Decreased cMyBPC content may result in nonuniform incorporation of cMyBPC into the sarcomere and regional inhomogeneities in rates of force development. Thus, sarcomeres with reduced levels of cMyBPC could develop force at a faster rate compared with sarcomeres with normal cMyBPC content.28 This would result in simultaneous shortening and stretch of different regions of the sarcomere, which would disrupt the timing and synchronization of fiber shortening during systole. Additionally, there may be a delay in force relaxation of regions that are being actively stretched (stretch-activated) 22 by regions of the sarcomere that are shortening, thereby prolonging relaxation in vivo.16–18
Effects of Decreased cMyBPC Expression on In Vivo Mechanical Function
Studies of in vivo cardiac contractile function in cMyBPC−/− mice16,17,19,29 have demonstrated global systolic and diastolic dysfunction and severe LVH. However, in vivo cardiac function and morphology of cMyBPC± hearts has been shown to be normal at a young age,16,19,29 with unexplained asymmetrical hypertrophy developing at 10–11 months of age despite preserved systolic and diastolic function.19 Cardiac MRI can detect subtle changes in LV morphology and mechanical function, so we examined LV motion indices of torsion and strain throughout the cardiac cycle in cMyBPC−/− and cMyBPC± hearts. In this study, we used DENSE MRI to quantify myocardial mechanics in the mouse heart, a technique that has been validated against rotating phantoms or traditional MRI tagging protocols in mice and humans.30,31 We show that the lack of cMyBPC nearly abolished LV torsion and twist in young mice. Furthermore, the magnitude of LV torsion and strain are reduced and the pattern of LV mechanical function is altered in cMyBPC± hearts in the absence of overt hypertrophy. The latter may be the early mechanical manifestation of cMyBPC haploinsufficiency, which could contribute to the progression and development of LVH with advanced age.
The counterclockwise rotation of the apex and the clockwise rotation of the base32 produces the wringing motion that maximizes pumping of blood into the circulation. We observed that the magnitude of LV torsion (Figure 6) and net twist (Figure 5) were severely impaired in cMyBPC−/− hearts such that LV systolic rotation was minimal. These results are consistent with overt hypertrophy and fibrosis,16 which renders the cMyBPC−/− LV extremely stiff. Additionally, myofiber architecture abnormalities disrupt the transmural progression of the helical arrangement of subendocardial and subepicardial fibers.33 This disruption may decrease the generation of mechanical torque by subepicardial fibers, which propels the rotation of the LV during systole.34 Interestingly, despite a significantly reduced magnitude of LV torsion in cMyBPC−/− hearts, peak systolic torsion occurred earlier in systole compared with WT hearts. The resulting shortening of the ejection phase in cMyBPC−/− hearts is consistent with findings that cMyBPC is crucial for modulating the rate of pressure development during isovolumic contraction and the period of systolic ejection.18
In contrast to severe mechanical dysfunction and hypertrophy in cMyBPC−/− hearts, a modest 23% decrease in cMyBPC content in cMyBPC± hearts resulted in a small but significant reduction in the magnitude of peak systolic torsion (Figure 6). The decrease in the magnitude and duration of systolic torsion in cMyBPC± hearts was due to a reduction in the overall net twist angle between the apex and base (Figure 5). Specifically, there was a more prominent reduction in twist between the mid-LV and the basal LV segments (Figure 5), although this difference did not reach statistical significance (P=0.08). Because systolic rotation and fiber shortening in the LV proceeds in an apex-to-base sequence,35 a reduction in net twist could be due to global or regional abnormalities in circumferential strain. Thus, the small reduction in circumferential strain at the mid-LV in cMyBPC± hearts could impair the progression of mechanical rotation from the apex to the base during systole resulting in diminished twist between the mid-LV and basal-LV. These data are also consistent with the idea that cMyBPC is important for prolongation fiber shortening during late systole,18 perhaps through continued rotation of the later-activated LV base. Accelerated rates of cross-bridge kinetics and fiber shortening in the early isovolumic contraction phase18,22 in cMyBPC± hearts may disrupt the timing of fiber shortening in later-activated regions such as the base, thereby reducing LV rotation in late systole.
Slower systolic and diastolic circumferential strain rates36,37 are a common feature in HCM carriers. In the present study, cMyBPC−/− hearts displayed significant reductions in radial and circumferential strain rates during systole and diastole compared with WT hearts (Figure 4), and cMyBPC± hearts displayed lower early systolic and peak diastolic circumferential strain rates at the mid-LV, and lower peak radial systolic strain rates in the base and mid-LV (Figure 4). Diastolic dysfunction is thought to be an early manifestation of impaired cardiac function in preclinical HCM.38 The early phase of LV reverse rotation after systole is an important determinant of diastolic filling39,40, and decreases in diastolic mid-LV circumferential strain rates in cMyBPC± hearts may result in a slowing of the rate of LV relaxation and decreased diastolic filling, as has been noted previously in cMyBPC−/− hearts.16,17
Potential Functional Consequences of cMyBPC Haploinsufficiency
Our data show that small reductions in cMyBPC content in the mouse heart, similar to decreases reported in myocardial samples obtained from patients expressing cMyBPC mutations, can significantly accelerate rates of cross-bridge kinetics and alter myocardial mechanical function in vivo. Non-uniform incorporation of cMyBPC in sarcomeres may result in regional inhomogeneities in rates of force development within sarcomeres along with uncoordinated sarcomere shortening and stretching. This can result in abnormal patterns of LV mechanical torsion in vivo that decrease the efficiency of contraction and relaxation and thereby, chamber ejection and filling. Furthermore, both increased ATP turnover due to accelerated cross-bridge cycling and reduced LV mechanical efficiency would be expected to increase energy expenditure by myocytes,41,42 which promotes apoptotic pathways43 that lead to decreased myocyte survival and replacement of myocytes with fibrotic tissue.44 The induction of LV fibrosis progressively stiffens the myocardium and impairs active fiber shortening and stretch. Fibrotic myocardium has a reduced capacity to generate sufficient strain and torsion to meet circulatory demands, which leads to compensatory LV hypertrophy.
Clinical Implications
It is possible that phenotypic unpredictability in contractile function and LVH, as well as the age at which disease symptoms are apparent in patients expressing cMyBPC mutations, is related to variability in overall levels of cMyBPC expression.44 In addition, the regional distribution of cMyBPC expression in the myocardium, such that patients with more significant cMyBPC loss present with more severe dysfunction and LVH at an earlier age. Although mutations in the gene encoding cMyBPC have typically been associated with late-onset cardiac dysfunction and LVH, there is growing evidence that clinical symptoms of the disease are apparent in a significant number of cMyBPC mutation carriers at a young age, including infancy.45,46 It is noteworthy that decreased levels of cMyBPC have been reported in myocardial samples from young HCM patients (20–30 years of age)12,13 carrying heterozygous cMyBPC mutations, which parallel our findings in young cMyBPC± mice. Collectively, these data imply that clinical symptoms of cardiac dysfunction related to mutations in cMyBPC, and perhaps mechanical dysfunction specifically, manifest earlier than previously thought and should be screened for in young relatives of HCM patients.45 With the emergence of improved cardiac MRI technology,47 it may now be possible to identify subtle changes in mechanical function and hypertrophy in cMyBPC carriers at an earlier age, thus significantly reducing risks for development of cardiac disease later in life.
Limitations
The mechanical manifestation and degree of cardiac hypertrophy in human HCM is often diverse due to the underlying cause of the disease, which often involves multiple contributing factors and may or may not involve mutations in sarcomeric genes. The mechanical data presented here were collected from a homogeneous population of mice in the absence of environmental modifiers that could impact disease progression in humans. Therefore, the animal models of cardiac disease used in this study may not fully reproduce all aspects of human disease. Furthermore, the mice used in this study were deficient in cMyBPC, and thus our data may not be representative of a general population of HCM in which cardiac dysfunction and hypertrophy is not related to mutations in cMyBPC. For example, echocardiography studies in a general cohort of HCM patients of unknown genotype revealed slightly increased systolic circumferential strain compared with controls,48 whereas we observed lower circumferential strains in cMyBPC-deficient mice. However, consistent with our data, lower systolic circumferential strains have been shown in HCM patients with cMyBPC mutations.36 Thus, it can be speculated that divergent cardiac mechanical function in different HCM populations may reflect the underlying cause of the disease.
CLINICAL PERSPECTIVE.
Hypertrophic cardiomyopathy (HCM) is a disease caused by mutations in genes that encode sarcomeric proteins. Mutations in cardiac myosin binding protein C (cMyBPC) are among the most common causes genetically explained HCM, accounting for more than 40% of the total known cases worldwide. A large number of disease-causing mutations in cMyBPC in humans are predicted to produce C-terminal truncated proteins that are not incorporated into the sarcomere, resulting in cMyBPC haploinsufficiency, which reduces the total amount of cMyBPC in the sarcomere. However, the link between decreased cMyBPC expression in the sarcomere and the development of cardiac disease is unclear. Our data demonstrate that a mouse model of cMyBPC haploinsufficiency with only 1 functional cMyBPC allele (cMyBPC+/−) displays a 24% reduction in total cMyBPC expression and accelerated cross-bridge kinetics, which underlies in vivo impairments in systolic and diastolic cardiac mechanics measured by MRI. Importantly, reduced mechanical indices of torsion, twist, and strain were apparent in young cMyBPC+/− mice 8–10 weeks of age, suggesting that cardiac dysfunction in cMyBPC-related HCM is not necessarily late-onset, as is commonly accepted, but may be detected early in life using high-resolution cardiac MRI. Furthermore, in contrast to the severe mechanical dysfunction and cardiac hypertrophy displayed by cMyBPC-null mice, mechanical dysfunction in cMyBPC+/− mice was apparent before the development of overt cardiac hypertrophy, implicating altered mechanical function as the first link between reduced cMyBPC expression in the sarcomere and the development of pathological hypertrophy and cardiac disease.
Supplementary Material
Acknowledgments
Sources of Funding
This study was supported by the National Institutes of Health and American Heart Association grants HL-73315 and HL-86935 (Dr Yu) and 09SDG2050195 (Dr Stelzer).
Footnotes
Disclosures
None.
References
- 1.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in the general population of young adults: echocardiographic analysis of 4111 subjects in the CARDIA study: Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 2.Carrier L. Cardiac myosin-binding protein C in the heart. Arch Mal Coeur Vaiss. 2007;100:238–243. [PubMed] [Google Scholar]
- 3.Barefield D, Sadayappan S. Phosphorylation and function of cardiac myosin binding protein-C in health and disease. J Mol Cell Cardiol. 2010;48:866–875. doi: 10.1016/j.yjmcc.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris SP, Lyons RG, Bezold KL. In the thick of it: HCM-causing mutations in myosin binding proteins of the thick filament. Circ Res. 2011;108:751–764. doi: 10.1161/CIRCRESAHA.110.231670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watkins H, Conner D, Thierfelder L, Jarcho JA, MacRae C, McKenna WJ, Maron BJ, Seidman JG, Seidman CE. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 6.Bonne G, Carrier L, Bercovici J, Cruaud C, Richard P, Hainque B, Gautel M, Labeit S, James M, Beckmann J, Weissenbach J, Vosberg HP, Fiszman M, Komajda M, Schwartz K. Cardiac myosin binding protein-C gene splice acceptor site mutation is associated with familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:438–440. doi: 10.1038/ng1295-438. [DOI] [PubMed] [Google Scholar]
- 7.Maron BJ, Seidman JG, Seidman CE. Proposal for contemporary screening strategies in families with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:2125–2132. doi: 10.1016/j.jacc.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 8.Niimura H, Bachinski LL, Sangwatanaroj S, Watkins H, Chudley AE, McKenna W, Kristinsson A, Roberts R, Sole M, Maron BJ, Seidman JG, Seidman CE. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. N Engl J Med. 1998;338:1248–1257. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 9.Carrier L, Bonne G, Bährend E, Yu B, Richard P, Niel F, Hainque B, Cruaud C, Gary F, Labeit S, Bouhour JB, Dubourg O, Desnos M, Hagège AA, Trent RJ, Komajda M, Fiszman M, Schwartz K. Organization and sequence of human cardiac myosin binding protein C gene (MYBPC3) and identification of mutations predicted to produce truncated proteins in familial hypertrophic cardiomyopathy. Circ Res. 1997;80:427–434. [PubMed] [Google Scholar]
- 10.Rottbauer W, Gautel M, Zehelein J, Labeit S, Franz WM, Fischer C, Vollrath B, Mall G, Dietz R, Kübler W, Katus HA. Novel splice donor site mutation in the cardiac myosin binding protein-C gene in familial hypertrophic cardiomyopathy. Characterization Of cardiac transcript and protein. J Clin Invest. 1997;100:475–482. doi: 10.1172/JCI119555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moolman JA, Reith S, Uhl K, Bailey S, Gautel M, Jeschke B, Fischer C, Ochs J, McKenna WJ, Klues H, Vosberg HP. A newly created splice donor site in exon 25 of the MyBP-C gene is responsible for inherited hypertrophic cardiomyopathy with incomplete disease penetrance. Circulation. 2000;101:1396–1402. doi: 10.1161/01.cir.101.12.1396. [DOI] [PubMed] [Google Scholar]
- 12.van Dijk SJ, Dooijes D, dos Remedios C, Michels M, Lamers JM, Winegrad S, Schlossarek S, Carrier L, ten Cate FJ, Stienen GJ, van der Velden J. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation. 2009;119:1473–1483. doi: 10.1161/CIRCULATIONAHA.108.838672. [DOI] [PubMed] [Google Scholar]
- 13.Marston S, Copeland O, Jacques A, Livesey K, Tsang V, McKenna WJ, Jalilzadeh S, Carballo S, Redwood C, Watkins H. Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ Res. 2009;105:219–222. doi: 10.1161/CIRCRESAHA.109.202440. [DOI] [PubMed] [Google Scholar]
- 14.Jacques A, Hoskins AC, Kentish JC, Marston SB. From genotype to phenotype: a longitudinal study of a patient with hypertrophic cardiomyopathy due to a mutation in the MYBPC3 gene. J Muscle Res Cell Motil. 2008;29:239–246. doi: 10.1007/s10974-009-9174-0. [DOI] [PubMed] [Google Scholar]
- 15.Schlossarek S, Mearini G, Carrier L. Cardiac myosin-binding protein C in hypertrophic cardiomyopathy: mechanisms and therapeutic opportunities. J Mol Cell Cardiol. 2011;50:613–620. doi: 10.1016/j.yjmcc.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 16.Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, Powers PA, Moss RL. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res. 2002;90:594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 17.Palmer BM, Georgakopoulos D, Janssen PM, Wang Y, Alpert NR, Belardi DF, Harris SP, Moss RL, Burgon PG, Seidman CE, Seidman JG, Maughan DW, Kass DA. Role of cardiac myosin binding protein C in sustaining left ventricular systolic stiffening. Circ Res. 2004;94:1249–1255. doi: 10.1161/01.RES.0000126898.95550.31. [DOI] [PubMed] [Google Scholar]
- 18.Nagayama T, Takimoto E, Sadayappan S, Mudd JO, Seidman JG, Robbins J, Kass DA. Control of in vivo left ventricular contraction/relaxation kinetics by myosin binding protein C: protein kinase A phosphorylation dependent and independent regulation. Circulation. 2007;116:2399–2408. doi: 10.1161/CIRCULATIONAHA.107.706523. [DOI] [PubMed] [Google Scholar]
- 19.Carrier L, Knöll R, Vignier N, Keller DI, Bausero P, Prdhon B, Isnard R, Ambroisine ML, Fiszman M, Ross J, Jr, Schwartz K, Chien KR. Asymetric septal hypertrophy in heterozygous cMyBP-C null mice. Cardiovasc Res. 2004;63:293–304. doi: 10.1016/j.cardiores.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Somji A, Yu X, Stelzer JE. Altered in vivo left ventricular torsion and principal strains in hypothyroid rats. Am J Physiol Heart Circ Physiol. 2010;299:H1577–H1587. doi: 10.1152/ajpheart.00406.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stelzer JE, Larsson L, Fitzsimons DP, Moss RL. Activation dependence of stretch activation in mouse skinned myocardium: implications for ventricular function. J Gen Physiol. 2006;127:95–107. doi: 10.1085/jgp.200509432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stelzer JE, Patel JR, Walker JW, Moss RL. Differential roles of cardiac myosin-binding protein C and cardiac troponin I in the myofibrillar force responses to protein kinase A phosphorylation. Circ Res. 2007;101:503–511. doi: 10.1161/CIRCRESAHA.107.153650. [DOI] [PubMed] [Google Scholar]
- 23.Liu W, Ashford MW, Chen J, Watkins MP, Williams TA, Wickline SA, Yu X. MR tagging demonstrates quantitative differences in regional ventricular wall motion in mice, rats, and men. Am J Physiol Heart Circ Physiol. 2006;291:H2515–H2521. doi: 10.1152/ajpheart.01016.2005. [DOI] [PubMed] [Google Scholar]
- 24.Zhong J, Liu W, Yu X. Characterization of three-dimensional myocardial deformation in the mouse heart: an MR tagging study. J Magn Reson Imaging. 2008;27:1263–1270. doi: 10.1002/jmri.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stelzer JE, Fitzsimons DP, Moss RL. Ablation of myosin binding protein-C accelerates force development in mouse myocardium. Biophys J. 2006;90:4119–4127. doi: 10.1529/biophysj.105.078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer BM, Noguchi T, Wang Y, Heim JR, Alpert NA, Burgon PG, Seidman CE, Seidman JG, Maughan DW, LeWinter MM. Effect of cardiac myosin binding protein-C on mechanoenergetics in mouse myocardium. Circ Res. 2004;94:1615–1622. doi: 10.1161/01.RES.0000132744.08754.f2. [DOI] [PubMed] [Google Scholar]
- 27.Colson BA, Bekyarova T, Fitzsimons DP, Irving TC, Moss RL. Radial displacement of myosin cross-bridges in mouse myocardium due to ablation of myosin binding protein-C. J Mol Biol. 2007;367:36–41. doi: 10.1016/j.jmb.2006.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Theis JL, Bos JM, Theis JD, Miller DV, Dearani JA, Schaff HV, Gersh BJ, Ommen SR, Moss RL, Ackerman MJ. Expression patterns of cardiac myofilament proteins: genomic and protein analysis of surgical myectomy tissue from patients with obstructive hypertrophic cardiomyopathy. Circ Heart Fail. 2009;2:325–333. doi: 10.1161/CIRCHEARTFAILURE.108.789735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McConnell BK, Fatkin D, Semsarian C, Jones KA, Georgakopoulos D, Maguire CT, Healey MJ, Mudd JO, Moskowitz IP, Conner DA, Giewat M, Wakimoto H, Berul CI, Schoen FJ, Kass DA, Seidman CE, Seidman JG. Comparison of two murine models of familial hypertrophic cardiomyopathy. Circ Res. 2001;88:383–389. doi: 10.1161/01.res.88.4.383. [DOI] [PubMed] [Google Scholar]
- 30.Kim D, Gilson WD, Kramer CM, Epstein FH. Myocardial tissue tracking with two-dimensional cine displacement-encoded MR imaging: development and initial evaluation. Radiology. 2004;230:862–871. doi: 10.1148/radiol.2303021213. [DOI] [PubMed] [Google Scholar]
- 31.Gilson WD, Yang Z, French BA, Epstein FH. Complementary displacement-encoded MRI for contrast-enhanced infarct detection and quantification of myocardial function in mice. Magn Reson Med. 2004;51:744–752. doi: 10.1002/mrm.20003. [DOI] [PubMed] [Google Scholar]
- 32.Streeter DD, Jr, Spotnitz HM, Patel DP, Ross J, Jr, Sonnenblick EH. Fiber orientation in the canine left ventricle during diastole and systole. Circ Res. 1969;24:339–347. doi: 10.1161/01.res.24.3.339. [DOI] [PubMed] [Google Scholar]
- 33.Wang TT, Kwon HS, Dai G, Wang R, Mijailovich SM, Moss RL, So PTC, Wedeen VJ, Gilbert RJ. Resolving myoarchitectural disarray in the mouse ventricular wall with diffusion spectrum magnetic resonance imaging. Ann Biomed Eng. 2010;38:2841–2450. doi: 10.1007/s10439-010-0031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taber LA, Ming Y, Podszus WW. Mechanics of ventricular torsion. J Biomech. 1996;29:745–752. doi: 10.1016/0021-9290(95)00129-8. [DOI] [PubMed] [Google Scholar]
- 35.Sengupta PP, Tondato F, Khanderia BK, Belohavek M, Jahangir A. Electromechanical activation sequence in normal heart. Heart Failure Clin. 2008;4:303–314. doi: 10.1016/j.hfc.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Germans T, Rüssel IK, Götte MJ, Spreeuwenberg MD, Doevendans PA, Pinto YM, van der Geest RJ, van der Velden J, Wilde AA, van Rossum AC. How do hypertrophic cardiomyopathy mutations affect myocardial function in carriers with normal wall thickness? Assessment with cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2010;12:13. doi: 10.1186/1532-429X-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ennis DB, Epstein FH, Kellman P, Fananapazir L, McVeigh ER, Arai AE. Assessment of regional systolic and diastolic dysfunction in familial hypertrophic cardiomyopathy using MR tagging. Magn Reson Med. 2003;50:638–642. doi: 10.1002/mrm.10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho CY, Carlsen C, Thune JJ, Havndrup O, Bundgaard H, Farrohi F, Rivero J, Cirino AL, Andersen PS, Christiansen M, Maron BJ, Orav EJ, Køber L. Echocardiographic strain imaging to assess early and late consequences of sarcomere mutations in hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2009;2:314–321. doi: 10.1161/CIRCGENETICS.109.862128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doucende G, Schuster I, Rupp T, Startun A, Dauzat M, Obert P, Nottin S. Kinetics of left ventricular strains and torsion during incremental exercise in healthy subjects: the key role of torsional mechanics for systolic-diastolic coupling. Circ Cardiovasc Imaging. 2010;3:586–594. doi: 10.1161/CIRCIMAGING.110.943522. [DOI] [PubMed] [Google Scholar]
- 40.Carasso S, Yang H, Woo A, Jamorski M, Wigle ED, Rakowski H. Diastolic myocardial mechanics in hypertrophic cardiomyopathy. J Am Soc Echocardiogr. 2010;23:164–171. doi: 10.1016/j.echo.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 41.Crilley JG, Boehm EA, Blair E, Rajagopalan B, Blamire AM, Styles P, McKenna WJ, Ostman-Smith I, Clarke K, Watkins H. Hypertrophic cardiomyopathy due to sarcomeric gene mutations is characterized by impaired energy metabolism irrespective of the degree of hypertrophy. J Am Coll Cardiol. 2003;41:1776–1782. doi: 10.1016/s0735-1097(02)03009-7. [DOI] [PubMed] [Google Scholar]
- 42.Timmer SA, Germans T, Götte MJ, Rüssel IK, Dijkmans PA, Lubberink M, ten Berg JM, ten Cate FJ, Lammertsma AA, Knaapen P, van Rossum AC. Determinants of myocardial energetics and efficiency in symptomatic hypertrophic cardiomyopathy. Eur J Nucl Med Mol Imaging. 2010;37:779–788. doi: 10.1007/s00259-009-1350-3. [DOI] [PubMed] [Google Scholar]
- 43.Eijssen LM, van den Bosch BJ, Vignier N, Lindsey PJ, van den Burg CM, Carrier L, Doevendans PA, van der Vusse GJ, Smeets HJ. Altered myocardial gene expression reveals possible maladaptive processes in heterozygous and homozygous cardiac myosin-binding protein C knockout mice. Genomics. 2008;91:52–60. doi: 10.1016/j.ygeno.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 44.Morita H, Nagai R, Seidman JG, Seidman CE. Sarcomere gene mutations in hypertrophy and heart failure. J Cardiovasc Transl Res. 2010;3:297–303. doi: 10.1007/s12265-010-9188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaski JP, Syrris P, Esteban MT, Jenkins S, Pantazis A, Deanfield JE, McKenna WJ, Elliott PM. Prevalence of sarcomere protein gene mutations in preadolescent children with hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2009;2:436–441. doi: 10.1161/CIRCGENETICS.108.821314. [DOI] [PubMed] [Google Scholar]
- 46.Rodríguez-García MI, Monserrat L, Ortiz M, Fernández X, Cazón L, Núñez L, Barriales-Villa R, Maneiro E, Veira E, Castro-Beiras A, Hermida-Prieto M. Screening mutations in myosin binding protein C3 gene in a cohort of patients with hypertrophic cardiomyopathy. BMC Med Genet. 2010;11:67. doi: 10.1186/1471-2350-11-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maron MS. The current and emerging role of cardiovascular magnetic resonance imaging in hypertrophic cardiomyopathy. J Cardiovasc Trans Res. 2009;2:415–425. doi: 10.1007/s12265-009-9136-3. [DOI] [PubMed] [Google Scholar]
- 48.Carasso S, Yang H, Woo A, Vannan MA, Jamorski M, Wigle ED, Rakowski H. Systolic myocardial mechanics in hypertrophic cardiomyopathy: novel concepts and implications for clinical status. J Am Soc Echocardiogr. 2008;21:675–683. doi: 10.1016/j.echo.2007.10.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.