Abstract
Lengthy developmental programs generate cell diversity within an organotypic framework, enabling the later physiological actions of each organ system. Cell identity, cell diversity and cell function are determined by cell type-specific transcriptional programs; consequently, transcriptional regulatory factors are useful markers of emerging cellular complexity, and their expression patterns provide insights into the regulatory mechanisms at play. We performed a comprehensive genome-scale in situ expression screen of 921 transcriptional regulators in the developing mammalian urogenital system. Focusing on the kidney, analysis of regional-specific expression patterns identified novel markers and cell types associated with development and patterning of the urinary system. Furthermore, promoter analysis of synexpressed genes predicts transcriptional control mechanisms that regulate cell differentiation. The annotated informational resource (www.gudmap.org) will facilitate functional analysis of the mammalian kidney and provides useful information for the generation of novel genetic tools to manipulate emerging cell populations.
Keywords: Genome-scale expression screen, Transcriptional regulator, Kidney, Mouse
INTRODUCTION
The mammalian metanephric kidney is essential for the maintenance of water and electrolyte homeostasis, the regulation of blood pressure and blood cell composition, and bone formation (Koeppen and Stanton, 2001). Development of a functional kidney requires a complex interplay among its principal cellular components (Costantini and Kopan, 2010; Dressler, 2009).
The nephric duct-derived ureteric epithelium forms the arborized network of the collecting duct system in response to branching signals derived from adjacent mesenchyme. This network is crucial for urine transport from the nephron to the ureter, and the maintenance of pH and osmolarity in tissue fluids. Signals from the nascent ureteric epithelium regulate survival, proliferation and differentiation of mesenchymal, stem cell-like, nephron progenitors that cap each branch tip (Carroll et al., 2005; Kobayashi et al., 2008; Park et al., 2007). At each round of branching, a subset of these progenitors undergoes an epithelial transition that establishes a renal vesicle (RV). RVs undergo elaborate patterning and morphogenesis along a glomerular-collecting duct axis that is crucial for organ function. A highly developed vascular network (Gomez et al., 1997; Woolf and Loughna, 1998), interstitial support cells and a variety of distinct subregions with specialized properties that include contractile and sensory functions all contribute to the physiological actions of the kidney.
Not surprisingly, given the central role of the kidney in human health, the experimental analysis of mammalian kidney development has received considerable attention. The kidney was the first mammalian organ shown to replicate a developmental program in culture (Grobstein, 1953; Grobstein, 1955; Saxen, 1987). Since those pioneering studies in the 1950s, a wealth of genetic, molecular and cellular information has informed on the control of kidney development (Costantini and Kopan, 2010; Dressler, 2009). Collectively, these studies have underscored the complexity of kidney development, and highlighted the need for a more systematic analysis of cell types, cell relationships and cell interactions in the assembly of the kidney. The increasing evidence that developmental deficiencies increase the risk of kidney disease in later life emphasizes the need to enhance our understanding of developmental events (Bertram et al., 2011; Song and Yosypiv, 2011).
One barrier to progress is a dearth of molecular markers that can distinguish cell types and facilitate analysis of developmental programs. In many systems, transcriptional regulators have served as important cell type-specific markers and drivers of cell specification and differentiation. For example, large-scale expression screens of transcriptional regulators have proven fruitful in identifying organizational and developmental features in the assembly of the mammalian nervous system (Gray et al., 2004). We surmised that systematic identification of the expression patterns of a large set of transcriptional regulators in the developing urogenital system would provide new insights into the emergence of cell heterogeneity central to kidney formation and function. Furthermore, the accrued data could potentially facilitate in silico prediction of gene networks, and enable the design of novel genetic approaches for analysis of key cell types.
Here, we report on a genome-scale in situ analysis of the expression of genes encoding mammalian transcriptional regulatory factors in the developing mouse urogenital system. Our analysis provides a wealth of new markers of kidney development and reveals novel molecular subdomains in developing renal structures. A meta-analysis of selected transcriptional regulators highlights the potential of this dataset for discovery of networks governing differentiation of terminal cell fates.
MATERIALS AND METHODS
Further details of the protocols described below can be found at http://www.gudmap.org/Research/Protocols/McMahon.html and http://www.gudmap.org/Research/Protocols/Little.html. All experimental procedures with mice were performed in accordance with institutional and national animal welfare laws, guidelines and policies, and were approved by relevant institutional animal care committees.
Plasmid preparation and riboprobe production
Plasmid glycerol stocks from the Brain Molecular Anatomy Project (BMAP) cDNA library or selected cDNAs from Open Biosystems (Huntsville, AL, USA) were streaked on LB (Amp) plates and grown at 37°C overnight. Three colonies per clone were restreaked onto ‘master plates’. Plasmid DNA was purified from one colony per clone and sequenced to confirm the expected sequence, and determine the orientation of the cDNA. cDNA inserts were amplified from their plasmid backbone with polymerase chain reaction (PCR) using T7, T3 or SP6 primers depending on the vector backbone.
A large number of transcriptional regulator-specific probes were generated from a previously described collection (Gray et al., 2004). For these, cDNA inserts were amplified by PCR from a 1:100 dilution of Qiagen minipreps of cDNA plasmid subclones as above. An additional source of cDNA templates were provided by Fantom3 cDNA clones punched from a RIKEN DNAbook 2 (Source BioScience imaGenes, Berlin, Germany). PCR primers were designed to amplify a 500-700 bp region of the open reading frame for a given clone; a T7 promoter sequence (TAA TAC GAC TCA CTA TAG GG) appended to the 3′ clone-specific primer enabled direct generation of antisense riboprobes from T7-mediated transcription of PCR products. Finally, for manual cloning of cDNA inserts for probe production, specific primers were designed in Vector NTI (Invitrogen, Carlsbad, CA, USA) that would amplify ∼750 bp of Refseq annotated cDNA sequence encoding the target regulatory factor of interest, using the T7 3′ primer strategy above for direct synthesis of antisense riboprobes. Riboprobes were purified with Microspin columns (BioRad, Hercules, CA, USA) and diluted with prehybridization buffer [50% formamide, 5×SSC (pH 4.5), 50 μg/ml yeast tRNA, 1% SDS, 50 μg/ml Heparin] to 10 μg/ml.
Preparation of urogenital system samples
For whole-mount in situ hybridization, the entire mouse urogenital system with associated adrenal glands was dissected free of other embryonic tissues at embryonic day 15.5 (E15.5, TS23) and fixed in 4% paraformaldehyde (PFA) in PBS for 24 hours at 4°C. The specimens were dehydrated through a graded series of methanol/NaCl and stored in 100% methanol at –20°C before in situ hybridization.
For section in situ hybridization, E15.5 kidneys and ureters were dissected free of all surrounding tissues, fixed in 4% PFA for 24 hours at 4°C, and cryopreserved in 30% sucrose overnight. Samples were embedded in OCT, flash frozen in an ethanol/dry-ice bath and stored at –80°C. Blocks were sectioned at 20 μm for section in situ hybridization.
In situ hybridization of whole-mount urogenital systems with digoxigenin-labeled riboprobes
For whole-mount in situ hybridization, E15.5 urogenital system samples were rehydrated through a graded series of methanol/NaCl. After proteinase K (10 μg/ml) treatment for 30 minutes, samples were fixed again and prehybridized at 70°C for 1 hour before being hybridized with 500 ng/ml digoxigenin-labeled riboprobes at 70°C overnight. All steps post-hybridization were performed with a BioLane HTI liquid handling system (Intavis Bioanalytical Instruments AG, Widdersdorferstrasse, Koeln, Germany). Briefly, samples were washed with hot solutions at 65°C and treated with 100 μg/ml RNase A for 1 hour. After additional stringent hot washes, samples were washed with 1×MBST [0.1 M maleic acid, 0.15 M NaCl, 0.1% Tween-20 (pH 7.5)], blocked in blocking solution (10% sheep serum in MBST plus 2% Boehringer Mannheim Blocking Reagent) for 3-4 hours at room temperature, and incubated subsequently with anti-digoxigenin antibody conjugated with alkaline phosphatase (AP) (Roche, Indianapolis, IN, USA, 1:4000) overnight at 4°C. Extensive washing in 1×MBST was followed by incubation in NTMT [100 mM NaCl, 100 mM Tris-HCl (pH 9.5), 50 mM MgCl2, 0.1% Tween-20, 2 mM levamisole). Samples were then removed from the BioLane HTI, and the hybridization signal visualized by adding BM purple. Color reactions were terminated at 3-hour, 6-hour, 9-hour, 12-hour, 24-hour, 36-hour and 48-hour time points, when signals were strong or a background developed. After development of the colorimetric assay, samples were post-fixed, cleared through a graded series of glycerol and stored in 80% glycerol/PBS. Images were then captured with a Nikon DXM1200 digital camera (Nikon, Melville, NY, USA), attached to a Nikon SMZ1500 stereoscope (Nikon, Melville, NY, USA) at two different magnifications to view the entire urogenital system and for a higher resolution view of the kidney.
In situ hybridization and double in situ hybridization on frozen sections with digoxigenin-labeled and fluorescein-labeled riboprobes
For frozen section in situ hybridization, kidney sections were post-fixed in 4% PFA. After treatment with 10 μg/ml proteinase K for 10 minutes, sections were fixed again with 4% PFA, acetylated and dehydrated. Sections were then incubated with 500 ng/ml digoxigenin-labeled riboprobes at 68°C overnight. For double in situ hybridization, sections were incubated with both digoxigenin-labeled and fluorescein-labeled riboprobes. Sections were washed after hybridization and treated with 2 μg/ml RNase A for 15 minutes at 37°C. After stringent hot washes, sections were blocked for 1 hour or longer and then incubated with anti-digoxigenin-AP overnight at 4°C. After washing in 1×MBST and equilibration in NTMT, sections were incubated with BM purple and the slides developed for a maximum of 7 days. Following color reaction, sections were fixed and slides mounted with Glycergel mounting medium (DAKO, Carpinteria, CA, USA). Images were captured with a Leica MZ16F stereoscope equipped with a DFC300 FX camera (Leica, Buffalo Grove, IL, USA). For double in situ hybridization, after the BM purple color reaction, sections were re-fixed with 4% PFA for 30 minutes, washed with 1×MBST and incubated overnight at 4°C with an anti-Fluorescein-AP conjugated antibody that had been preabsorbed with mouse embryo powder to eliminate non-specific activity. After washing in 1×MBST and equilibration in NTMT, sections were incubated with INT/BCIP+10% polyvinyl alcohol (PVA) to visualize hybridization of the second probe over a development period that extended for a maximum of 7 days. The slides were then fixed and mounted in Glycergel mounting medium (DAKO, Carpinteria, CA, USA).
In situ hybridization on paraffin sections
The paraffin section in situ hybridization protocol has been previously published (Georgas et al., 2008; Little et al., 2007; Rumballe et al., 2008). For each gene examined by section in situ hybridization, a DNA template of 500-1000 bp was generated from either cDNA or plasmid clones by PCR and transcribed to produce a digoxigenin (DIG)-labeled antisense riboprobe (www.gudmap.org). The primer and probe sequences are available on the GUDMAP website within the section in situ hybridization data submissions for each gene. Kidneys were harvested from E15.5 CD1 mice and fixed overnight in 4% PFA at 4°C (Ethics IMB/180/10/NHMRC/NIF NF). Kidneys were paraffin embedded and sectioned at 7 μm. Slides were dewaxed manually and the remaining section in situ hybridization performed using a Tecan Freedom Evo 150 platform. Sections were hybridized at 64°C for 10 hours with 0.5-1.0 μg/ml of riboprobe, washed and incubated with anti-DIG-alkaline phosphatase Fab fragments (1:1000, Roche, Indianapolis, IN, USA) for 120 minutes at 25°C. Detection of alkaline phosphatase activity using BM Purple (Roche, Indianapolis, IN, USA) was performed manually. The signal intensity was monitored for up to 120 hours and the slides rinsed and fixed in 4% PFA prior to mounting in aqueous mounting medium. Images were captured with either an Olympus dotSlide System (Olympus, Mt Waverley, VIC, Australia) or a standard light microscope (Olympus BX51 DP70 color 12 megapixel digital camera, Mt Waverley, VIC, Australia).
Annotation of gene expression
Expression patterns observed in all whole-mount in situ hybridization and section in situ hybridization samples were annotated against a previously described comprehensive ontology of the developing urogenital tract (Little et al., 2007). Images and annotations are housed at www.gudmap.org.
Single molecule fluorescence in situ hybridization (FISH)
For each mRNA examined, a set of 48, 20-mer DNA oligonucleotides complementary to coding and 3′ untranslated regions were designed using an online available program (http://www.singlemoleculefish.com/designer.html) and synthesized with 3′-amino modifications by Biosearch Technologies. Probe sets were pooled at a concentration of 1 nM for each oligonucleotide, dried (Speedvac, medium heat), resuspended in 0.1 M sodium bicarbonate (pH 8.5) containing an excess of succinimidyl ester derivatives of Alexa594 (Invitrogen, Carlsbad, CA, USA) or Cy5 (GE Healthcare, Piscataway, NJ, USA), and incubated overnight at room temperature with gentle agitation. Excess fluorophore was removed by ethanol precipitation, and coupled oligonucleotides were separated from the uncoupled fraction by reverse phase high-pressure liquid chromatography.
Kidneys obtained from intercrosses of Hoxb7Cre transgenic mice (Yu et al., 2002) to ROSA-mT/mG double fluorescent reporter allele [Gt(ROSA)26Sortm4(ACTB-tdTomato-EGFP)Luo/J] (Muzumdar et al., 2007) were dissected at E15.5, fixed in 4% PFA for 1 hour at 4°C, and incubated overnight in 30% sucrose at 4°C before embedding in OCT compound. Tissue blocks were sectioned at 6 μm, fixed in 4% PFA at room temperature for 15 minutes, rinsed in PBS and incubated overnight in 70% ethanol at 4°C.
The detailed hybridization procedure is documented elsewhere (Raj et al., 2008). Briefly, sections were rehydrated in wash buffer before hybridizing with ∼0.3 ng/μl of labeled probe sets for each mRNA overnight at 30°C in the dark. Sections were washed twice for 30 minutes; DAPI (Invitrogen, Carlsbad, CA, USA) was added in the second wash to enable nuclear visualization and sections were mounted in anti-bleach buffer. Z-stack images were taken at 0.3 μm intervals with a Nikon Ti Eclipse inverted fluorescence microscope (Nikon, Melville, NY, USA) equipped with a 100× oil-immersion objective and a Roper Scientific Pixis CCD camera using MetaMorph software (Molecular Devices, Sunnyvale, CA, USA). For semi-automated counting of particles, images were filtered and processed as previously described (Raj et al., 2008). Spearman correlation coefficients of pairs of genes were calculated in Matlab. To obtain P-values, these correlation coefficients were compared with the correlation coefficients of 104 randomized datasets in which the values for one of the genes were shuffled among cells. The Z-scores of the experimental correlations compared with those of the randomized datasets were transformed to P-values based on the normal distribution.
Selection of potential transcription factor targets based upon synexpression across the developing kidney expression atlas
Target genes with synexpression to the compartment-specific transcription factor of interest were identified through an expression profile similarity measure using the GUDMAP embryonic kidney subcompartment microarray atlas (Brunskill et al., 2008) (www.gudmap.org) (GEO: GSE6290). The microarray expression data was RMA normalized and analyzed in Genespring version 7.3 (Agilent, Santa Clara, CA, USA). One-way-ANOVA (P<0.01) with FDR correction (Benjamini-Hochberg) was performed to identify differentially expressed probe sets across the entire dataset that was then used for subsequent analyses. Probe sets representing candidate target genes were required to have synexpression to the transcription factor probe set of interest, based on a minimum Pearson correlation similarity measure of 0.7. Where a transcription factor probe set in microarray did not correlate with in situ hybridization annotated expression (Pou3f1), we used a directed analysis approach specifying the required compartment-specific expression for target genes (FC≥2).
Prediction of transcription factor targets based on promoter analysis
For each transcription factor analyzed, a defined binding motif was identified from one of the Uniprobe (Newburger and Bulyk, 2009), Jaspar (Portales-Casamar et al., 2010) and TRANSFAC (Matys et al., 2006) motif databases. The promoters of potential targets were defined as extending from –1500 bp to +500 bp relative to the genomic location of each given target site. To predict statistically the most likely potential targets regulated by the selected transcription factors, binding sites (TFBS) were predicted using Monkey (Moses et al., 2004), which uses species conservation to predict binding sites for a given motif. We have found Monkey to be a reliable TFBS predictor (Hawkins et al., 2009). Monkey calculates the probability that a given motif binds to specific locations in a given set of promoters, using conservation and comparing the match of the motif to each site in the promoter sequences against that of randomly shuffled motifs. Monkey was preformed within a previously described pipeline specialized to this kind of prediction (Piper et al., 2010). As well as using the transcription factor motif and promoter sequences, Monkey uses a sequence alignment of the promoters with orthologous species. In this study, we generated data for two distinct sets of ortholog comparisons. In the first, mouse (mm9) genome was aligned with rat (rn4), guinea pig (cavPor2) and rabbit (oryCun1), a phylogenetic tree containing the orthologous species, with a 0-order background Markov model for mouse sequence data, with a motif database to ensure that shuffled motifs generated to test significance of the motif of interest are not real motifs and with a number of shuffled motifs to compare with the predicted motif. A second comparison was performed between mouse (mm9), rat (rn4), human (hg18) and zebrafish (danRer5). Both resulted in a statistical prediction for each promoter analyzed, based upon the most conserved predicted TFBS between all orthologs (see supplementary material Table S3).
RESULTS
In situ hybridization of known genes identified eight readily discernible expression patterns correlating with distinct cellular compartments in the E15.5 kidney and ureter
By E15.5, the ureteric network has branched eight or nine times (Cebrian et al., 2004) and an elaborate ductal tree is established. Development of the renal pelvis is under way, and the renal medullary zone, which is crucial for concentrating urine, is starting to emerge (Cebrian et al., 2004; Yu et al., 2009). Nephrogenesis is advancing at this time: in addition to early stage renal vesicles and comma- and S-shaped bodies, late stage renal corpuscles, proximal and distal convoluted tubules, and early stage loops of Henle (LOHs) are present, which is indicative of maturing nephrons. Consequently, analysis at E15.5 enables the identification of early and late emerging cell types at a time-point technically suited for a sensitive three-dimensional, low-resolution perspective of in situ gene expression through whole-mount in situ hybridization. The high-throughput whole-mount in situ hybridization primary screen provides a broad overview of the expression of transcriptional regulatory components throughout the entire urogenital system.
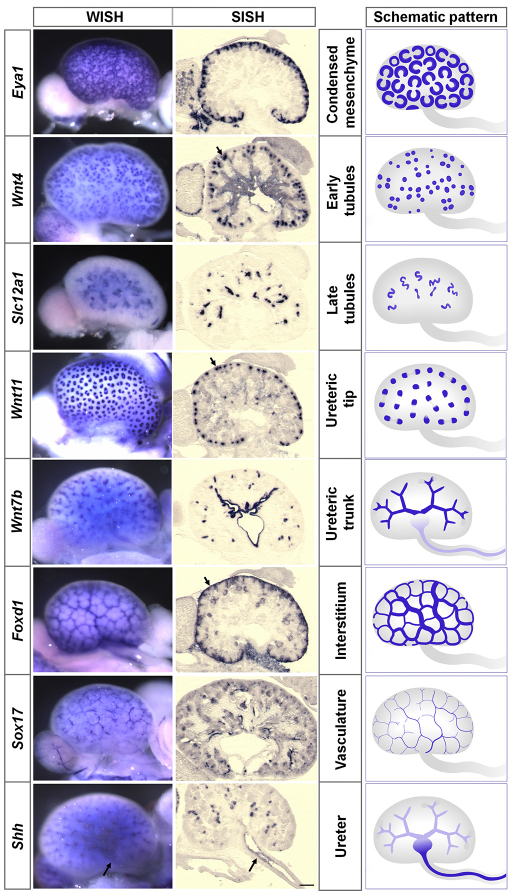
We first optimized whole-mount in situ hybridization around a series of genes that unambiguously score most major renal components at E15.5: Eya1, Wnt4, Slc12a1, Wnt11, Wnt7b, Foxd1, Sox17 and Shh (Fig. 1). As anticipated, each probe generated a distinctive and readily identifiable expression pattern, validated through high-resolution section in situ hybridization analysis (Fig. 1). Deep structures such as the ureteric tree (Wnt7b, Shh) and thick ascending limb of the LOH (Slc12a1) were uniformly labeled, and the technique was sufficiently sensitive to detect weak expression of Shh in the ureteric tree, deep within the kidney. The observed expression patterns could be readily grouped into eight broad categories: cap mesenchyme, early tubules (including pretubular aggregate, renal vesicle, comma- and S-shaped bodies), late tubules (nephrons beyond the S-shaped body stage), ureteric tip, ureteric trunk, renal interstitium, renal vasculature and ureter (including the ureter and the renal pelvis). These expression patterns, schematized in Fig. 1, form the basis of our subsequent comprehensive annotation of the expression of transcriptional regulatory factors.
Fig. 1.
Eight characteristic and readily discernible gene expression patterns are identified in the E15.5 kidney and ureter by in situ hybridization and associated with specific annotation terms (groups). Whole-mount in situ hybridization analysis (left panels) of E15.5 whole-mount kidneys and ureters reveals anatomically distinct expression profiles for eight kidney genes that are further resolved through high-resolution section in situ hybridization analysis (center panels). Together, these eight probes mark many of the major cell types/structures of the kidney and ureter (schematized in the right panels). When a gene is expressed in both superficial and internal structures, signals from superficial structures may mask internal signals in whole-mount in situ hybridization analysis. Arrows indicate expression associated with the respective annotation terms. Scale bar: 200 μm.
A comprehensive screen of the expression of genes encoding transcriptional regulatory factors identified molecularly distinct domains in the embryonic kidney and ureter
From gene ontology analysis of two independently generated genome-wide compilations of mammalian transcriptional regulators (Gray et al., 2004; Lee et al., 2007), we compiled a target list of 951 transcriptional regulatory factors for which there was a general consensus of this classification (see supplementary material Table S1). We performed whole-mount in situ hybridization on 921 of these genes (96.8% coverage) and annotated expression in regard to the annotation groupings documented in Fig. 1. Our emphasis has been to provide an accurate account of expression patterns that can be unambiguously discerned. The whole-mount in situ hybridization approach is subject to false-negative results; for example, where weak internal signals are masked by stronger superficial ones. And whole-mount in situ hybridization does not enable a high-resolution description of all expression domains – an outcome that can only be realized through serial-section, multi-probe based section in situ hybridization analysis. What the whole-mount in situ hybridization does generate is a broad framework of the expression of potential regulatory factors that greatly facilitates secondary analyses. All whole-mount in situ hybridization images and their annotations can be searched, viewed and mined at www.gudmap.org.
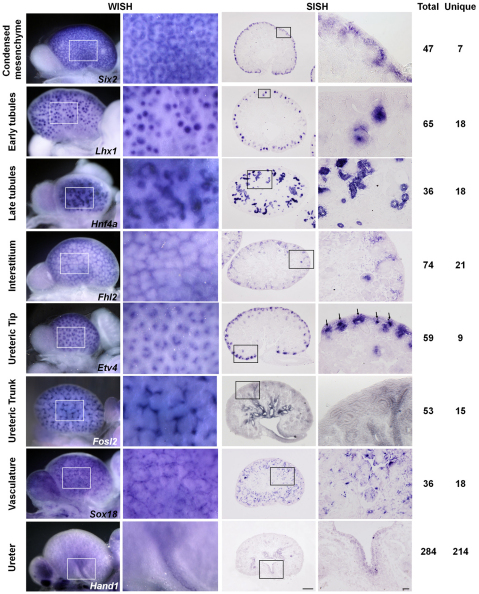
Of the large set of factors, 213 (23.1%) displayed regionally restricted expression in the kidney (bold genes in supplementary material Table S1) whereas 337 (36.6%) appeared to be ubiquitously expressed. In particular, we identified 106 (11.5%) genes expressed in only one renal category (excluding the ureter). Representative expression patterns of genes with localized expression are presented in Fig. 2, along with information on the number of genes displaying a given pattern, and the subset of these genes whose expression is unique to the domain of interest. Section in situ hybridization was performed on a subset of genes in each category to validate expression with cellular resolution, and to reveal expression domains potentially masked by whole-mount in situ hybridization (Figs 2, 3).
Fig. 2.
Expression of genes encoding transcriptional regulators within each annotation group. Low- and high-magnification whole-mount in situ hybridization images (left panels) display transcriptional regulators whose expression is restricted to one of the eight characteristic expression domains: the number of transcriptional regulators showing unique expression in each of these domains versus the total number of genes with a specific annotation to each term is shown (far right). Complementary low- and high-magnification section in situ hybridization images for each gene of interest provide cellular resolution of expression domains (right panels). ‘Unique’ genes may also be expressed in extrarenal compartments within the urogenital system. Most genes showing regionally restricted expression are expressed in more than one annotation group. Arrows indicate expression in ureteric tips. Scale bars: 200 μm (low magnification); 20 μm (high magnification).
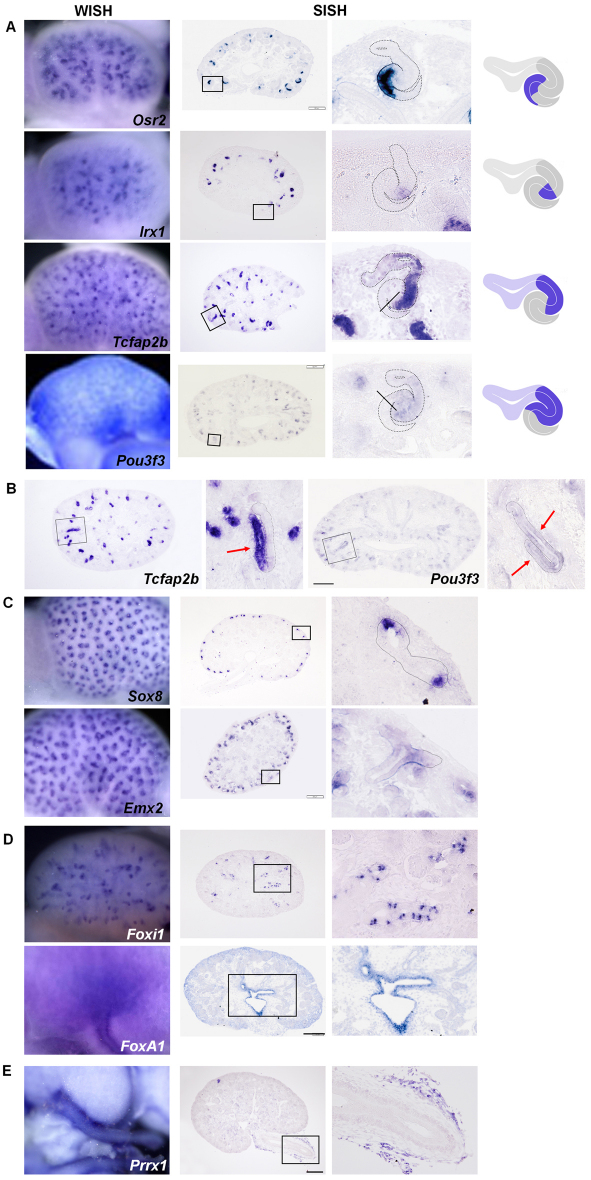
Fig. 3.
A selection of expression patterns potentially marking new compartment boundaries within the developing nephron, the ureteric epithelium and ureter. Primary whole-mount in situ hybridization analysis is shown in the left panels and high- and low-magnification section in situ hybridization in right panels. (A) Molecular subdivision of the S-shaped body from genes showing an early tubule annotation. Within the medial segment of the S-shaped body (SSB), Osr2 and Irx1 appear to display non-overlapping expression domains. The proximal boundary of Tcfap2b expression in the distal segment of the SSB (line) appears to lie slightly distal to that of Pou3f3 (line). The S-shaped bodies with their attaching ureteric epithelium are outlined in the high-magnification images and the expression domains of the genes are illustrated in the schematic drawings (far right). (B) Comparative expression of Tcfap2b and Pou3f3 in the loop of Henle (LOH) anlage suggests that Tcfap2b expression is restricted to a single arm of the LOH: the distal ascending limb (red arrows). (C) Differential expression of Sox8 and Emx2 in the bifurcating ureteric epithelium. (D) Ureteric trunk patterning and differentiation, as illustrated by the divergent expression domains of Foxi1 (intercalated cells of the collecting duct epithelium) and Foxa1 (urothelial lining of the pelvis and medullary collecting duct). (E) Prrx1 labels outer layers of loose ureteral mesenchyme. Scale bars: 200 μm.
Examination of complex transcription factor expression patterns identifies novel domains of regulatory activity and cellular subcompartments
The majority of documented expression patterns do not correspond to a specific anatomically defined compartment. For example, section in situ hybridization on a subset of 65 genes with whole-mount in situ hybridization expression patterns annotated to early tubules (supplementary material Table S1) revealed domains within the S-shaped body that do not readily match proximal, medial and distal segments defined in classical histological studies (Little et al., 2007). Rather, the data point to a finer patterning of the S-shaped body, with molecular subdivisions of each segment (Fig. 3). Both Osr2 and Irx1 are expressed in the medial segment of the S-shaped body; however, their expression domains do not span the entire medial segment, and may not be precisely congruent (Fig. 3A). Similarly, although both Tcfap2b and Pou3f3 (Brn1) are expressed in distal and medial segments of the S-shaped body, Pou3f3 expression extends slightly proximally to Tcfap2b (Fig. 3A). Pou3f3 is reportedly expressed in both limbs of the loop of Henle (LOH) anlage (Nakai et al., 2003), whereas data here suggest that Tcfap2b is expressed only in the ascending limb of the extending LOH (Fig. 3B). This raises the possibility that the proximal boundary of Tcfap2b marks the boundary between the ascending and descending limbs of the LOH, suggesting that the two limbs of the LOH may be differentially specified from the onset of LOH formation. Genetic fate mapping of these expression domains relative to the functional anatomy of the adult kidney will likely provide complementary insights into how nephron complexity is generated.
Within the ureteric epithelium, 25 transcriptional regulators were identified with a whole-mount in situ hybridization expression pattern restricted to the ureteric tips and not detected in ureteric trunks, regardless of their expression in other components of the kidney (supplementary material Table S1). Although all of these genes were excluded from the ureteric trunks, their whole-mount in situ hybridization expression patterns point to molecularly distinct cellular boundaries within the ureteric epithelium. Sox8 transcripts localized to each tip in a bifurcating branch (Fig. 3C), whereas Emx2 expression within the branches was almost complementary to Sox8 expression (Fig. 3C), providing evidence for the action of distinct transcriptional programs within the context of a highly dynamic branching process that may, for example, segregate cells between tip stem/progenitor and stalk domains.
Conversely, 19 transcriptional regulators were expressed specifically in the ureteric trunk but not the tip (supplementary material Table S1). Of these, only one gene, Foxi1, exhibited a variegated expression pattern in a subset of ureteric trunk cells, more readily discernible on section in situ hybridization analysis (Fig. 3D), concurring with a previous report of Foxi1 action in the mosaic differentiation of intercalated cells within the collecting duct epithelium (Blomqvist et al., 2004).
A large group of transcriptional regulators were annotated to the ureter and the renal pelvis (Fig. 2), most probably an overestimate that reflects background from probe and/or antibody trapping within these structures. However, a close examination of a select group of genes revealed previously unappreciated subdivisions in the pelvic/medullary region and the ureter. For example, whole-mount in situ hybridization expression of Foxa1 in the urothelium and renal pelvic epithelium was confirmed by section in situ hybridization analysis (Fig. 3D), in addition, section in situ hybridization examination showed an expression of Foxa1 in prospective medullary collecting ducts, the segment of the collecting ducts closest to the renal pelvic space (Fig. 3D). An independent study of microarray profiling results coupled with section in situ hybridization validation reported a similar molecular stratification of the ureteric epithelium (Thiagarajan et al., 2011). Whether the Foxa1 component prefigures a segmental patterning of the medullary collecting ducts or plays an active role in medullary cell specification remains to be determined, but collectively the data suggest an early molecular stratification of epithelial structures prefiguring the cortico-medullary axis of kidney organization and function.
The ureter consists of the urothelium and the ureteral mesenchyme, including the lamina propria, the ureteral smooth muscle and the adventitia. At E15.5, the ureteral mesenchyme comprises inner layers of condensed mesenchyme from which the smooth muscle is forming (Yu et al., 2002) and outer layers of loose mesenchyme whose fates and functions are poorly understood. Several genes are documented to show specific expression in inner condensed mesenchyme (Airik et al., 2006; Nie et al., 2010; Yu et al., 2002), but no specific markers have been reported for the outer loose mesenchyme. Interestingly, Prrx1, a paired-related homeobox gene, displayed a whole-mount in situ hybridization expression pattern indicative of superficial expression in the ureter (Fig. 3E). Section in situ hybridization analysis confirmed Prrx1 expression was restricted to the outermost layers of ureteral mesenchyme, providing a molecular inroad to this cell population (Fig. 3E).
Renal vasculature closely associates with nephron components and plays a particularly important role in renal physiology; for example, the capillary network within the Bowman’s capsule, where plasma filtration occurs, the peritubular capillaries at sites of tubular reabsorption and secretion, or pericyte-like mesangial cells and renin-secreting juxtaglomerular cells closely associate with blood vessels in or about the renal corpuscle, regulating blood pressure, glomerular blood flow and glomerular filtration. We identified a group of 36 transcriptional regulators that displayed a vascular-related whole-mount in situ hybridization expression pattern (Fig. 2, supplementary material Table S1). Remarkably, these genes exhibited diverse expression patterns, suggesting considerable spatial or temporal heterogeneity within renal vasculature-associated cell types.
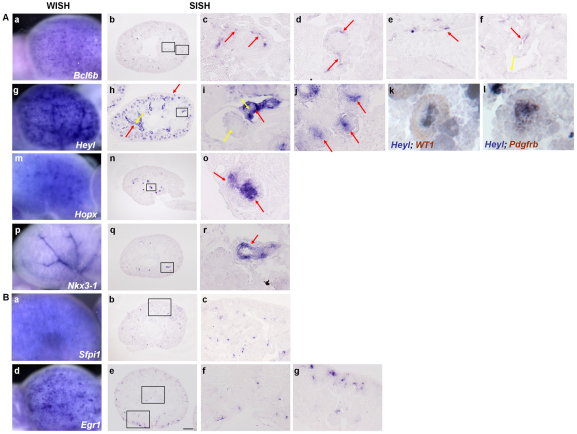
Bcl6b, which encodes a zinc-finger protein required for the enhanced level of the secondary response of memory CD8(+) T cells (Manders et al., 2005), marked isolated cells in the renal cortex and a chain of cells invading the lower cleft of the S-shaped body, probably of endothelial nature (Fig. 4Ac). Expression was also evident in the glomerular capillaries of the capillary loop stage renal corpuscle (stage III) with expression declining by the immature renal corpuscle stage (stage IV) (Fig. 4Ab,d). Bcl6b expression was also observed in the endothelium of the peritubular capillaries, renal arterioles and renal arteries, but not in renal veins (Fig. 4Ae,f). Expression of Bcl6b in endothelial cells was confirmed through colocalization analysis with the endothelial cell marker Pecam1 (data not shown).
Fig. 4.
Vasculature-associated gene expression and expression of genes with broad sporadic expression in the developing kidney. (A) Selected genes with vasculature-associated expression patterns. (a-f) Bcl6b is weakly expressed in the cortex and medulla. (c-f) High magnification images showing its expression in individual cells in the renal cortex (red arrow) and cells invading the lower cleft of the S-shaped body (red arrow) (c), the glomerular capillary of the capillary loop stage renal corpuscle (d, red arrow), and the peritubular capillaries (e, red arrow). Bcl6b is also expressed in the endothelial cells of the arterioles and renal arteries (red arrows) but not that of the renal veins (yellow arrow) (f). Panels c and f are from a different tissue section from that of panel b. (g-l) Heyl is expressed in the renal arterial (h, red arrow) and arteriolar (i, red arrows) smooth muscles, but not the arterial and arteriolar endothelium (h,i, yellow arrows). It is also expressed in the mesangium of the capillary loop stage renal corpuscle (j, red arrows) but decreased to almost undetectable levels in that of the immature renal corpuscle (i, yellow arrow). Double in situ hybridization studies shows that Heyl (blue) and Wt1 (brown, podocytes) do not overlap (k), whereas Heyl (blue) overlaps with Pdgfrb (brown) in glomerular mesangium (l). Heyl is also expressed in non-vascular locations, in early nephrons (g,h). Panels j, k and l are from different tissue sections from that of panel h. (m-o) Hopx shows very strong and specific expression in the glomerular and extraglomerular mesangium (o, red arrows), whereas Nkx3-1 expression (p-r) is restricted to smooth muscles of renal arteries and arterioles (r, red arrow). (Ba-g) Sfpi1 and Egr1 displayed punctate expression throughout the kidneys, the former reflecting interstitial macrophages and the latter potentially reflecting dynamic signaling responses. Scale bar: 200 μm.
In the non-endothelial components of the renal vasculature, Heyl was expressed both in smooth muscles surrounding the renal arteries and arterioles, and in the glomerular mesangium at the capillary loop stages (stage III) (Fig. 4Ah-l). Its expression in the glomerular mesangium at the immature renal corpuscle stages (stage IV) is greatly reduced to almost undetectable levels (Fig. 4Ai). Co-expression with pericytes/mesangium marker Pdgfrb confirmed Heyl expression in the glomerular mesangium (Fig. 4Al). Hopx, a homeodomain-containing transcriptional repressor required for cardiac development that displayed a late tubule expression pattern by whole-mount in situ hybridization, was shown by section in situ hybridization to be expressed in the mesangium, including both the glomerular mesangium and extraglomerular mesangium, but not in the smooth muscles surrounding the renal arteries or arterioles (Fig. 4An,o). Nkx3-1, which encodes a homeobox factor, was expressed in the smooth muscles surrounding the renal arteries and arterioles (Fig. 4Aq,r). However, in contrast to both Hopx and Heyl, it was not expressed in the mesangium (Fig. 4Ar).
Although the majority of expression patterns could be classified as including one or more of the eight reference expression patterns, we also identified two genes with punctate expression patterns throughout the kidney (Fig. 4Ba,d). Sfpi1 (PU.1), a macrophage lineage marker, displayed a punctate expression pattern concentrated in the outer cortex, the nascent renal medulla and the renal corpuscle (Fig. 4Bb,c). This expression pattern agrees with the previously described interstitial location of resident macrophages visualized using the Csf1r-EGFP transgenic mice (Rae et al., 2007). Egr1 (early growth response 1), a gene involved in cell proliferation, differentiation, stress response (Herschman, 1991; Liu et al., 1996), stem cell dormancy and stem cell localization (Min et al., 2008), is reported to be upregulated in specific segments of the kidney epithelia and in the glomerular tuft after ischemia-reperfusion (Bonventre et al., 1991). In the E15.5 kidney, Egr1 was expressed sporadically in diverse cell types but enriched in the nephrogenic zone, the nascent renal medulla and near or within the glomerulus (Fig. 4Bf,g). Given that Egr1 is a gene that is responsive to diverse growth factor signals, the pattern likely reflects active signaling in these regions. The identification of these spotted expression patterns representing single cells scattered throughout the organ demonstrates the resolution and sensitivity of whole-mount in situ hybridization.
Predicting target genes for specific transcriptional regulators defining specific developmental compartments or processes
Temporally or spatially restricted transcription factor activity presumably underpins a functional role in distinct developmental processes within disparate cell types in the developing kidney. Having identified novel spatially restricted patterns of transcription factor expression, we sought to identify potential transcription factor-target relationships. Five transcription factors displaying distinct patterns of spatial expression were selected for the analyses: Pou3f1 (late tubule), Tcfap2b (early tubule, and distal and medial segments of the S-shaped body), Sox8 (ureteric tip), Foxi1 (intercalated cells of collecting duct) and Irx1 (early tubule and the medial segment of the S-shaped body).
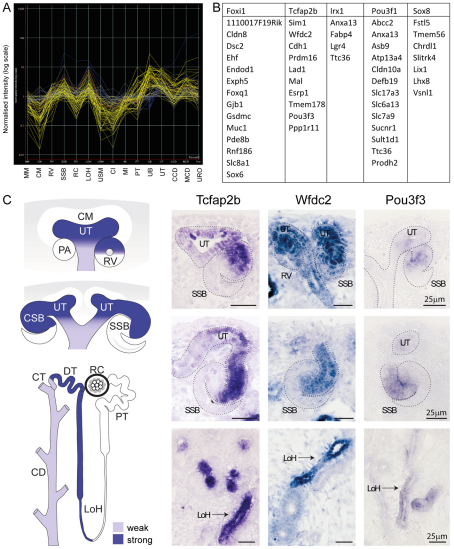
Initially, the expression of each transcription factor was compared across kidney development with previously published expression profiling of the developing kidney generated as another component of the GUDMAP initiative (Brunskill et al., 2008). Only genes displaying microarray-based synexpression matching expression of the canonical transcription factors across all 15 subcompartments of the developing kidney were selected as potential transcription factor targets (Fig. 5A, supplementary material Table S2). For each set of potential target genes, minimal promoters were defined and evidence sought for enriched transcription factor binding within these minimal promoters by the Monkey algorithm (Moses et al., 2004). Monkey analysis assesses evolutionary conservation of transcription factor-binding sites, and provides a statistical assessment of motif enrichment over chance. Monkey analysis was performed comparing mouse with two distinct orthologous groups: rat, guinea pig and rabbit, and rat, human and zebrafish. Fig. 5B lists all predicted targets (with a P-value <10–5) for each transcription factor examined in both ortholog groups. Table S3 in the supplementary material lists the full analysis including the number of predicted binding sites in each analysis and the statistical significance of top site predictions. Table S4 in the supplementary material lists the sequence and statistical significance of those predicted target sites in the putative target promoters for both ortholog comparisons. Section in situ hybridization analysis of the complex synexpression of two predicted target genes of Tcfap2b, Wfdc2 and Pou3f3, with Tcfap2b (Fig. 5C) provides evidence in support of this approach. The subtle distinctions in LOH and S-shaped body expression between Tcfap2b and Pou3f3 would argue that other transcription factors are also involved in Pou3f3 regulation.
Fig. 5.
Prediction and validation of transcriptional targets using synexpression and bioinformatics. (A) Genespring histogram representing all genes predicted to show a pattern of synexpression in comparison with the transcription factor Tcfap2b when compared with the 15 developing kidney subcompartments previously profiled by Brunskill et al. (Brunskill et al., 2008). (B) List of the most statistically robust putative targets for each of five transcription factors as assessed using Monkey. (C) An example of validation of such predicted targets. The transcription factor Tcfap2b displays a complex pattern of expression during kidney development, initiating at the ureteric tip (UT) and in the distal renal vesicle (RV) (top panels), distal comma-shaped body, and distal and medial S-shaped body (SSB, middle panels), then later in connecting ducts, distal tubules and the loop of Henle (LoH, lower panels). The same patterns of expression are observed for two predicted targets, Wfdc2 and Pou3f3. CD, collecting duct; CM, cap mesenchyme; CSB, comma-shaped body; CT, connecting tubule; PA, pretubular aggregate; PT, proximal tubule; RC, renal corpuscle; Scale bars: 25 μm.
Identification of a potential inhibitory role for Foxi1 in the suppression of the principal cell phenotype
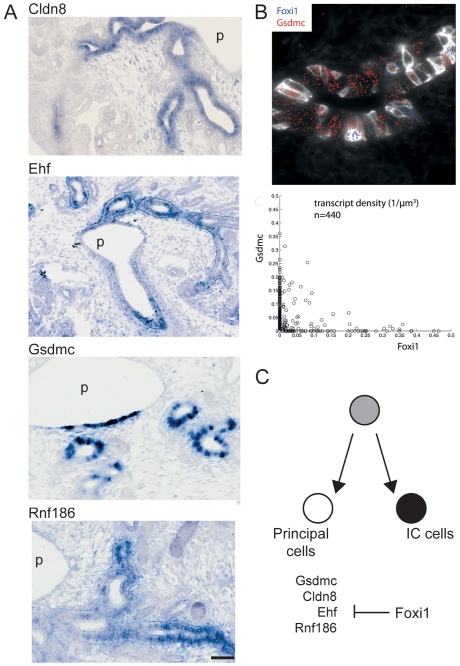
Of the chosen regulatory factors for synexpression-target prediction, the best understood is Foxi1, a determinant of intercalated cells within the collecting duct epithelium (Blomqvist et al., 2004). Intercalated cells comprise a distinct, dispersed differentiated cell type that is crucial for maintenance of acid-base homeostasis. A total of 52 potential targets were bioinformatically identified through synexpression; Monkey identified putative targets in 14 of these (P-value of <10–5 in both ortholog group analyses). On the assumption that a genuine target is more likely to be regulated by Foxi1 in tissues outside the kidney (ear, cartilage, skeleton), section in situ hybridization expression of each putative target was compared with data for whole embryo expression at E14.5 available through Eurexpress (www.eurexpress.org) (Diez-Roux et al., 2011). Four targets showed synexpression with Foxi1 beyond kidney development (Clnd8, Ehf, Dsc2 and Pde8). Section in situ hybridization was performed on a subset of putative targets (Cldn8, Ehf, Gsdmc, Rnf128). Importantly, all showed renal expression restricted to a subset of cells in the collecting duct epithelium (Fig. 6A; data not shown), but in a larger subset of cells than Foxi1, reminiscent of the water-salt regulating principal cell component of the collecting duct epithelium (Fig. 6A).
Fig. 6.
Foxi1 is negatively correlated with its predicted target gene Gsdmc. (A) Section in situ hybridization analysis of four predicted Foxi1 targets, Cldn8, Ehf, Gsdmc and Rnf186, showing expression in a region of the cells within the ureteric trunk epithelium. p, pelvis. Scale bar: 50 μm. (B) Two-color single molecule fluorescent in situ hybridization showing the negative correlation of expression patterns between Foxi1 and Gsdmc. Fixed E15.5 kidney sections were simultaneously hybridized with two differentially labeled probe libraries (Cy5 and Alexa594). Single mRNA molecules appear as diffraction-limited spots in an epifluorescence microscope. Transcripts were automatically detected and assigned to individual cells based on membrane staining via myristoylated GFP. Images show detected dots for Foxi1/Gsdmc in seven z-sections at 0.3 μm intervals and a correlation plot for Foxi1/Gsdmc. Each dot in the correlation plot denotes the absolute transcript density of a single cell for both genes. (C) Model of transcriptional role for Foxi1 in the switch between multi-potential progenitor and principal cell versus intercalated cell (IC) fate.
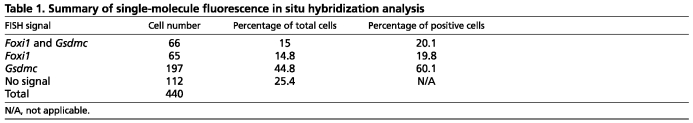
To examine this possible inverse relationship further, we used single molecule RNA fluorescence in situ hybridization on E15.5 kidney sections to label Foxi1 and one of these putative target genes, Gsdmc; this approach allows quantitative measurements of up to three different target RNAs in the same sections with single-cell resolution (Raj et al., 2008). Analysis of Foxi1 and Gsdmc transcripts in prospective medullary ureteric trunk epithelial cells (number of cells=440) (processed data, Fig. 6B; raw data, see supplementary material Fig. S1) indicated a significant negative correlation in their expression domains (Spearman correlation coefficient R=–0.59, P<10–4). Of the 328 transcript-positive cells expressing either gene, 80% showed a mutually exclusive expression pattern, i.e. the vast majority of cells either expressed Foxi1 or Gsdmc alone (Fig. 6B and Table 1). These data support a model in which Foxi1 directly silences Gsdmc to elicit intercalated cell fate determination. This is likely to also be the case for the other Foxi1 targets identified in this way.
Table 1.
Summary of single-molecule fluorescence in situ hybridization analysis
DISCUSSION
We have performed a comprehensive, stage-specific analysis of the expression of mouse transcriptional regulators by in situ hybridization in the developing urogenital system. Our goal – to use this regulatory subset of the mammalian genome to identify new cell markers for known cell types, identify novel cellular heterogeneity and develop predictive insights into regulatory interactions at play in the developing kidney – was realized in this study. Our analysis focused on a single stage of development identifying a subgroup of specifically expressed transcriptional regulators. Extending the analysis of this group to later stages and intersection of this data with other large-scale expression atlases [e.g. Eurexpress (www.eurexpress.org) and the Allen Brain Atlas (www.brain-map.org)] is likely to provide further information on cell diversity within the kidney and target relationships in other organ systems. Importantly, whereas we have focused our characterization on the kidney, all structures within the urogenital dataset have been annotated and these supply a wealth of new information for secondary analysis of gonads, reproductive ducts and lower urinary tract anatomy. The annotated resource is publicly available incorporated within the GUDMAP initiative database (www.gudmap.org). Together, these data will provide valuable information and a hypothesis-generating resource for the biomedical community.
Our screen identified a large set of transcriptional regulatory genes that are expressed in spatially or temporally restricted patterns in the developing kidney. The examination of the function of these genes, the development of genetic tools enabled by these genes and the analysis of synexpression groups overlapping transcription factor-defined cellular compartments will provide further insights into the molecular networks and cell-cell interactions underpinning kidney organogenesis. Initial analysis indicates novel tissue boundaries and cell groups marked by the expression of transcriptional regulators. The relationship of these domains to structures in the adult kidney is unclear, but it is reasonable to anticipate that investigating these issues will provide new insights into how regulatory programs in developing organs generate distinct physiological outputs in the functional organ system.
Supplementary Material
Acknowledgments
The authors thank Dave Tang and Han Chiu for their technical assistance, Jane Brennan for updating the bioinformatic information of the transcriptional regulator list, Dr Ariel Gomez for help with annotation of vascular genes, and Alison Lee for the mouse transcription factor list.
Footnotes
Funding
M.H.L. is a Principal Research Fellow of the National Health and Medical Research Council. The work was supported by a National Institutes of Health (NIH)/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant [5U01 DK070181 to A.P.M. and DK070136 to M.H.L.]. T.L.B. and P.M. were supported by an NIH R01 grant [RR021692]. Q.R. and J.Y. were supported by a NIH/NIDDK grant [1R01DK085080]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.74005/-/DC1
References
- Airik R., Bussen M., Singh M. K., Petry M., Kispert A. (2006). Tbx18 regulates the development of the ureteral mesenchyme. J. Clin. Invest. 116, 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram J. F., Douglas-Denton R. N., Diouf B., Hughson M. D., Hoy W. E. (2011). Human nephron number: implications for health and disease. Pediatr. Nephrol. 26, 1529–1533 [DOI] [PubMed] [Google Scholar]
- Blomqvist S. R., Vidarsson H., Fitzgerald S., Johansson B. R., Ollerstam A., Brown R., Persson A. E., Bergstrom G. G., Enerback S. (2004). Distal renal tubular acidosis in mice that lack the forkhead transcription factor Foxi1. J. Clin. Invest. 113, 1560–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonventre J. V., Sukhatme V. P., Bamberger M., Ouellette A. J., Brown D. (1991). Localization of the protein product of the immediate early growth response gene, Egr-1, in the kidney after ischemia and reperfusion. Cell Regul. 2, 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill E. W., Aronow B. J., Georgas K., Rumballe B., Valerius M. T., Aronow J., Kaimal V., Jegga A. G., Yu J., Grimmond S., et al. (2008). Atlas of gene expression in the developing kidney at microanatomic resolution. Dev. Cell 15, 781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll T. J., Park J. S., Hayashi S., Majumdar A., McMahon A. P. (2005). Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev. Cell 9, 283–292 [DOI] [PubMed] [Google Scholar]
- Cebrian C., Borodo K., Charles N., Herzlinger D. A. (2004). Morphometric index of the developing murine kidney. Dev. Dyn. 231, 601–608 [DOI] [PubMed] [Google Scholar]
- Costantini F., Kopan R. (2010). Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev. Cell 18, 698–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G., Banfi S., Sultan M., Geffers L., Anand S., Rozado D., Magen A., Canidio E., Pagani M., Peluso I., et al. (2011). A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 9, e1000582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler G. R. (2009). Advances in early kidney specification, development and patterning. Development 136, 3863–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgas K., Rumballe B., Wilkinson L., Chiu H. S., Lesieur E., Gilbert T., Little M. H. (2008). Use of dual section mRNA in situ hybridisation/immunohistochemistry to clarify gene expression patterns during the early stages of nephron development in the embryo and in the mature nephron of the adult mouse kidney. Histochem. Cell Biol. 130, 927–942 [DOI] [PubMed] [Google Scholar]
- Gomez R. A., Norwood V. F., Tufro-McReddie A. (1997). Development of the kidney vasculature. Microsc. Res. Tech. 39, 254–260 [DOI] [PubMed] [Google Scholar]
- Gray P. A., Fu H., Luo P., Zhao Q., Yu J., Ferrari A., Tenzen T., Yuk D. I., Tsung E. F., Cai Z., et al. (2004). Mouse brain organization revealed through direct genome-scale TF expression analysis. Science 306, 2255–2257 [DOI] [PubMed] [Google Scholar]
- Grobstein C. (1953). Inductive epithelio-mesenchymal interaction in cultured organ rudiments of the mouse. Science 118, 52–55 [DOI] [PubMed] [Google Scholar]
- Grobstein C. (1955). Inductive interaction in the development of the mouse metanephros. J. Exp. Zool. 130, 319–339 [Google Scholar]
- Hawkins J., Grant C., Noble W. S., Bailey T. L. (2009). Assessing phylogenetic motif models for predicting transcription factor binding sites. Bioinformatics 25, i339–i347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschman H. R. (1991). Primary response genes induced by growth factors and tumor promoters. Annu. Rev. Biochem. 60, 281–319 [DOI] [PubMed] [Google Scholar]
- Kobayashi A., Valerius M. T., Mugford J. W., Carroll T. J., Self M., Oliver G., McMahon A. P. (2008). Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3, 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppen B. M., Stanton B. A. (2001). Renal Physiology. St Louis, MO: Mosby; [Google Scholar]
- Lee A. P., Yang Y., Brenner S., Venkatesh B. (2007). TFCONES: a database of vertebrate transcription factor-encoding genes and their associated conserved noncoding elements. BMC Genomics 8, 441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little M. H., Brennan J., Georgas K., Davies J. A., Davidson D. R., Baldock R. A., Beverdam A., Bertram J. F., Capel B., Chiu H. S., et al. (2007). A high-resolution anatomical ontology of the developing murine genitourinary tract. Gene Expr. Patterns 7, 680–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Calogero A., Ragona G., Adamson E., Mercola D. (1996). EGR-1, the reluctant suppression factor: EGR-1 is known to function in the regulation of growth, differentiation, and also has significant tumor suppressor activity and a mechanism involving the induction of TGF-beta1 is postulated to account for this suppressor activity. Crit. Rev. Oncog. 7, 101–125 [PubMed] [Google Scholar]
- Manders P. M., Hunter P. J., Telaranta A. I., Carr J. M., Marshall J. L., Carrasco M., Murakami Y., Palmowski M. J., Cerundolo V., Kaech S. M., et al. (2005). BCL6b mediates the enhanced magnitude of the secondary response of memory CD8+ T lymphocytes. Proc. Natl. Acad. Sci. USA 102, 7418–7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V., Kel-Margoulis O. V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., et al. (2006). TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min I. M., Pietramaggiori G., Kim F. S., Passegue E., Stevenson K. E., Wagers A. J. (2008). The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell 2, 380–391 [DOI] [PubMed] [Google Scholar]
- Moses A. M., Chiang D. Y., Pollard D. A., Iyer V. N., Eisen M. B. (2004). MONKEY: identifying conserved transcription-factor binding sites in multiple alignments using a binding site-specific evolutionary model. Genome Biol. 5, R98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 [DOI] [PubMed] [Google Scholar]
- Nakai S., Sugitani Y., Sato H., Ito S., Miura Y., Ogawa M., Nishi M., Jishage K., Minowa O., Noda T. (2003). Crucial roles of Brn1 in distal tubule formation and function in mouse kidney. Development 130, 4751–4759 [DOI] [PubMed] [Google Scholar]
- Newburger D. E., Bulyk M. L. (2009). UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 37, D77–D82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X., Sun J., Gordon R. E., Cai C. L., Xu P. X. (2010). SIX1 acts synergistically with TBX18 in mediating ureteral smooth muscle formation. Development 137, 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. S., Valerius M. T., McMahon A. P. (2007). Wnt/beta-catenin signaling regulates nephron induction during mouse kidney development. Development 134, 2533–2539 [DOI] [PubMed] [Google Scholar]
- Piper M., Barry G., Hawkins J., Mason S., Lindwall C., Little E., Sarkar A., Smith A. G., Moldrich R. X., Boyle G. M., et al. (2010). NFIA controls telencephalic progenitor cell differentiation through repression of the Notch effector Hes1. J. Neurosci. 30, 9127–9139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E., Thongjuea S., Kwon A. T., Arenillas D., Zhao X., Valen E., Yusuf D., Lenhard B., Wasserman W. W., Sandelin A. (2010). JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 38, D105–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae F., Woods K., Sasmono T., Campanale N., Taylor D., Ovchinnikov D. A., Grimmond S. M., Hume D. A., Ricardo S. D., Little M. H. (2007). Characterisation and trophic functions of murine embryonic macrophages based upon the use of a Csf1r-EGFP transgene reporter. Dev. Biol. 308, 232–246 [DOI] [PubMed] [Google Scholar]
- Raj A., van den Bogaard P., Rifkin S. A., van Oudenaarden A., Tyagi S. (2008). Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumballe B., Georgas K., Little M. H. (2008). High-throughput paraffin section in situ hybridization and dual immunohistochemistry on mouse tissues. Cold Spring Harb. Protoc. 5030, 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxen L. (1987). Organogenesis of the Kidney. New York: Cambridge University Press; [Google Scholar]
- Song R., Yosypiv I. V. (2011). Genetics of congenital anomalies of the kidney and urinary tract. Pediatr. Nephrol. 26, 353–364 [DOI] [PubMed] [Google Scholar]
- Thiagarajan R. D., Georgas K. M., Rumballe B. A., Lesieur E., Chiu H. S., Taylor D., Tang D. T., Grimmond S. M., Little M. H. (2011). Identification of anchor genes during kidney development defines ontological relationships, molecular subcompartments and regulatory pathways. PLoS ONE 6, e17286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf A. S., Loughna S. (1998). Origin of glomerular capillaries: is the verdict in? Exp. Nephrol. 6, 17–21 [DOI] [PubMed] [Google Scholar]
- Yu J., Carroll T. J., McMahon A. P. (2002). Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development 129, 5301–5312 [DOI] [PubMed] [Google Scholar]
- Yu J., Carroll T. J., Rajagopal J., Kobayashi A., Ren Q., McMahon A. P. (2009). A Wnt7b-dependent pathway regulates the orientation of epithelial cell division and establishes the cortico-medullary axis of the mammalian kidney. Development 136, 161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.