Summary
The early transcribed membrane proteins (ETRAMPs) are a family of small, highly charged transmembrane proteins unique to malaria parasites. Some members of the ETRAMP family have been localized to the parasitophorous vacuole membrane that separates the intracellular parasite from the host cell and thus presumably have a role in host-parasite interactions. Although it was previously shown that two ETRAMPs are critical for rodent parasite liver stage development, the importance of most ETRAMPs during the parasite life cycle remains unknown. Here, we comprehensively identify nine new etramps in the genome of the rodent malaria parasite P. yoelii, and elucidate their conservation in other malaria parasites. etramp expression profiles are diverse throughout the parasite life cycle as measured by RT-PCR. Epitope tagging of two ETRAMPs demonstrates protein expression in blood and liver stages, and reveals differences in both their timing of expression and their subcellular localization. Gene targeting studies of each of the nine uncharacterized etramps show that two are refractory to deletion and thus likely essential for blood stage replication. Seven etramps are not essential for any life cycle stage. Systematic characterization of the members of the ETRAMP family reveals the diversity in importance of each family member at the interface between host and parasite throughout the developmental cycle of the malaria parasite.
Introduction
Malaria is caused by Plasmodium parasites, which possess several multigene families important for growth and virulence in mammalian hosts (Templeton, 2009). These genes play roles in cytoadherence and host cell invasion, but many remain hypothetical, with no known function (Gardner et al., 2002). One multigene family that is conserved in, and specific to, Plasmodium parasites is the early transcribed membrane protein (ETRAMP) family (Spielmann et al., 2000). Members of this family were first recognized in the human malaria parasite Plasmodium falciparum, the genome of which contains fourteen single-exon etramp genes (Spielmann et al., 2003). etramps are typified by a well-conserved domain structure of the encoded proteins, including a signal peptide, followed by a short cationic N-terminal domain, a transmembrane domain, and a more variable and highly charged C-terminal domain (22-279 amino acids in length) (Spielmann et al., 2003). Most P. falciparum etramps are expressed during asexual intraerythrocytic parasite development, and a few are found in gametocytes and preerythrocytic stages (Spielmann et al., 2003, Silvestrini et al., 2005, MacKellar et al., 2010). Furthermore, etramps are highly expressed; several members are among the most abundant transcripts found in microarray and EST sequencing studies in blood stages of P. falciparum and P. vivax (Le Roch et al., 2003, Daily et al., 2004, Cui et al., 2005).
During asexual blood stage development, ETRAMPs localize to the parasitophorous vacuole membrane (PVM), which surrounds the intraerythrocytic parasite and acts as its interface with the host erythrocyte (Spielmann et al., 2003). The ETRAMPs are oriented in the membrane so that the polymorphic C-terminal domain is exposed to the host cell cytoplasm (Spielmann et al., 2003). Within the PVM, the ETRAMPs associate to form sub-domains that can exclude other ETRAMPs as well as other resident PVM proteins such as exported protein 1 (EXP-1, the homolog of the P. yoelii Hep17 protein) (Spielmann et al., 2006). Some ETRAMPs are exported beyond the PVM, where they localize to the Maurer's clefts, a group of parasite-derived vesicular compartments located beneath the erythrocyte plasma membrane (Vincensini et al., 2005, Birago et al., 2003, MacKellar et al., 2010). Two members of the ETRAMP family were previously identified as the pre-erythrocytic stage-specific genes UIS3 and UIS4 in the rodent malaria parasites P. yoelii and P. berghei (Matuschewski et al., 2002, Kaiser et al., 2004). UIS3 in P. berghei shares 34% amino acid identity with ETRAMP13 in P. falciparum, and UIS4 is syntenous with etramp10.3 (Mueller et al., 2005a, Mueller et al., 2005b), although the two syntenous proteins show little amino acid sequence conservation. Both UIS3 and UIS4 localize to the secretory organelles of sporozoites and the PVM of the liver stages, and each is essential for liver stage development (Mueller et al., 2005a, Mueller et al., 2005b, Mikolajczak et al., 2007). Furthermore, UIS3 binds to mouse liver-fatty acid binding protein (L-FABP), and this interaction is important for liver stage growth (Mikolajczak et al., 2007). This interaction appears conserved because P. falciparum UIS3/ETRAMP13 binds to human L-FABP, and co-crystallizes with the phospholipid phosphatidylethanolamine (Sharma et al., 2008). The data suggest that the UIS3-L-FABP interaction might mediate the uptake of lipids from the host cytoplasm. Two other blood stage-expressed P. falciparum ETRAMPs have been found to bind human proteins when expressed in yeast or as peptides, although the biological relevance of these interactions have not been studied (Vignali et al., 2008, Garcia et al., 2009).
Analyzing multiple members of a protein family throughout the complete P. falciparum life cycle is daunting (Moreno et al., 2007). However, tools for characterizing multiple genes throughout the entire life cycle exist for rodent malaria parasites, making these parasites attractive models to study multigene families. Conservation of the ETRAMP family in P. yoelii (Spielmann et al., 2003, Birago et al., 2003), and the relative ease of its genetic manipulation make this parasite suitable for the comprehensive characterization of this gene family.
Here, we conduct a systematic analysis of the etramp gene family in P. yoelii. We identified 11 etramps in the P. yoelii genome, and assayed for their expression throughout the parasite life cycle by RT-PCR. We added epitope tags to a subset of the ETRAMPs to analyze their expression and localization. Finally, we used targeted gene deletion to identify essential members of the family. We find that, of the nine previously uncharacterized genes, two appear essential for blood stage replication. For the remaining seven uncharacterized etramps that were successfully deleted, we analyzed the entire life cycle of mutants and determined that none are essential for any life cycle stage. Our results provide a broad view of this gene family in the rodent parasite model, and will be useful in directing future research on P. falciparum etramps.
Results
etramps in the genomes of rodent and human malaria parasites
We identified ETRAMP family members from the Pfam database (www.pfam.org), which uses alignments of representative members of a protein family to build a Hidden Markov Model (HMM) that permits the automated identification of additional family members from a protein database (Finn et al., 2010). This approach yielded 85 sequences across 11 species. We eliminated redundant entries and candidates that showed improper domain organization or poor alignment to known ETRAMPs. This resulted in a list of 66 putative proteins conserved in seven different species of Plasmodium (Supplementary Table 1). Not surprisingly, the number of etramps found in each species correlated positively with the extent of sequence coverage and annotation available in the draft genome sequences of these parasites. Gene identifiers for etramps found in two rodent and two human malaria parasites are presented in Table 1. These include one previously unpublished P. falciparum etramp, which we named etramp9, following the convention of naming these genes by the chromosome on which they are found. Eleven P. yoelii etramps were identified, two of which have been published previously as UIS3 and UIS4 (Matuschewski et al., 2002).
Table 1.
etramps genes identified in two human and two rodent malaria parasites
| Group | P. falciparum | P. vivax | P. yoelii | P. berghei | Syntenic |
|---|---|---|---|---|---|
| 1 | PFB0120w (ETRAMP2), PF11_0040 (ETRAMP11.2) | PVX_003565 | PY00205 | PbSEP3* | Pf-Pv, Py-Pb |
| 2 | PFD1120c (ETRAMP4) | PVX_090230 | PY03869 | PB106367 | Pv-Py |
| 3 | PFE1590w (ETRAMP5) | PVX_096070 | - | - | - |
| 4 | MAL8P1.6 (ETRAMP8) | PVX_088870 | - | - | Pf-Pv |
| 5 | PFI1745c (“ETRAMP9”) | PVX_086915 | - | - | - |
| 6 | PF10_0019 (ETRAMP10.1), PF14_0016 (ETRAMP14.1) | - | - | - | - |
| 7 | PF10_0323 (ETRAMP10.2) | PVX_111065 | PY07506 | PB106570, PB402471 | Pf-Pv-Pb-Py |
| 8 | PF10_0164 (ETRAMP10.3) | PVX_001715 | PY00204 (PyUIS4) | PB100551 (PbUIS4) | Pf-Pv-Py-Pb |
| 9 | PF11_0039 (ETRAMP11.1) | - | PY04799 | PB100384 | Py-Pb |
| 10 | PFL1945c (ETRAMP12) | - | - | - | - |
| 11 | PF13_0012 (ETRAMP13) | PVX_121950 | PY03011 (PyUIS3) | PB000892 (PbUIS3) | Pf-Pv-Pb |
| 12 | PF14_0729 (ETRAMP14.2) | PVX_118680 | - | - | - |
| 13 | - | PVX_090230 | - | - | - |
| 14 | - | - | PY02667 | PB105376 | - |
| 15 | - | - | PY03365, PY05433, PY06488 | - | - |
| 16 | - | - | PY03652 | - | - |
Not annotated in the P. berghei genome
We also sought to combine the ETRAMPs into groups of orthologs and paralogs, and consulted the OrthoMCL database (www.orthomcl.org) to accomplish this (Chen et al., 2006). This tool identifies orthologs via reciprocal best BLAST hits for proteins between two genomes, and paralogs by intraspecies reciprocal better BLAST hits. The resulting groups of etramps were further consolidated on the basis of synteny. This approach yielded 13 groups, arranged as rows in Table 1, and three individual genes for which no ortholog or paralog could be identified. The groups are representative of the possible states of homology, with some genes possessing exactly one ortholog in all species, other genes possessing only paralogs within a species, or combinations of orthologs and paralogs. Importantly, the polymorphic ETRAMP C-terminal domains, which likely impart specificity for parasite protein-host protein interactions, show conservation within several of these groups in terms of size, isoelectric point, or both (data not shown).
Expression of P. yoelii etramps throughout the parasite life cycle
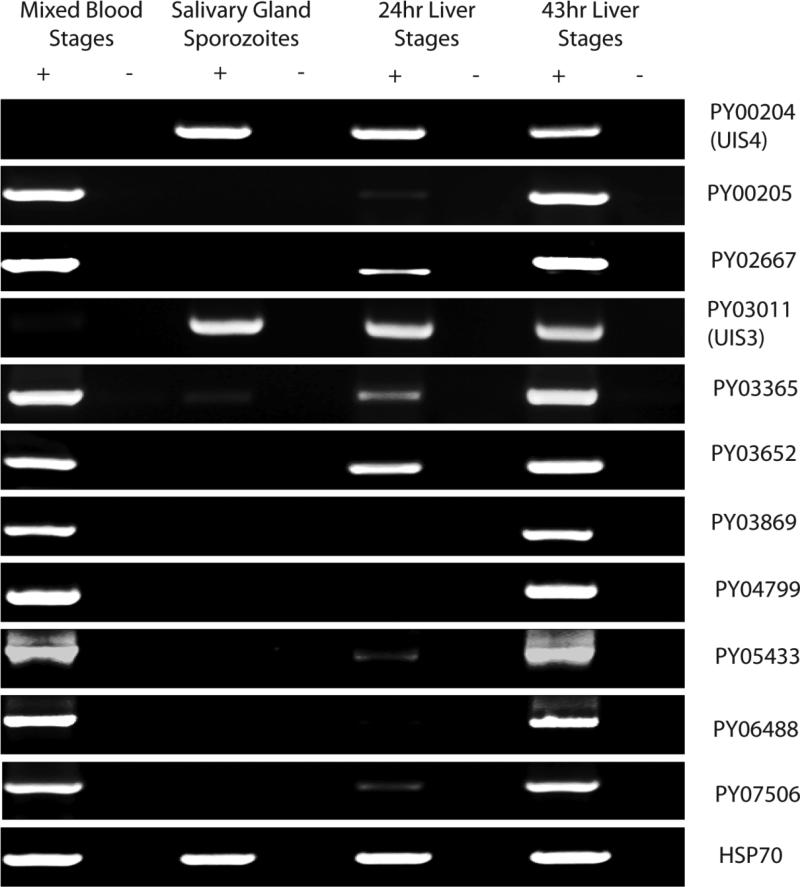
We assayed for expression of all P. yoelii etramps using RT-PCR (Fig. 1). Transcripts of all etramps except UIS3 and UIS4 were detected in mixed blood stages of P. yoelii 17XNL parasites. Conversely, only these two UISs/etramps were transcribed strongly in sporozoites isolated from mosquito salivary glands. Weaker evidence for expression of a third gene, PY03365, was also observed in sporozoites. Transcripts for eight of the etramps were detected in the livers of mice isolated 24hrs (early schizont) after infection with sporozoites. All transcripts were detected in late (43hr) liver stages derived from mouse infections, a time point when many blood stage proteins are also expressed in liver stages, likely due to the similarity of exoerythrocytic and intraerythrocytic merozoites (Tarun et al., 2008).
Figure 1.
RT-PCR analysis of etramps gene expression throughout the life cycle of P. yoelii. Each row represents a gene-specific amplicon for individual etramps, listed by their gene identifiers. Columns indicate the life cycle stage at which RNA was isolated, and then processed with (+) or without (-) reverse transcriptase. All etramps except UIS3 and UIS4 are expressed in blood stages. In addition to UIS3 and UIS4, only PY03365 is transcribed in salivary gland sporozoites. Transcripts are also detectable for eight of the 11 etramps in 24 hour liver stages. All 11 etramps are expressed in late liver stages. HSP70 is used as a positive control to verify RNA integrity.
Expression and localization of epitope-tagged ETRAMPs
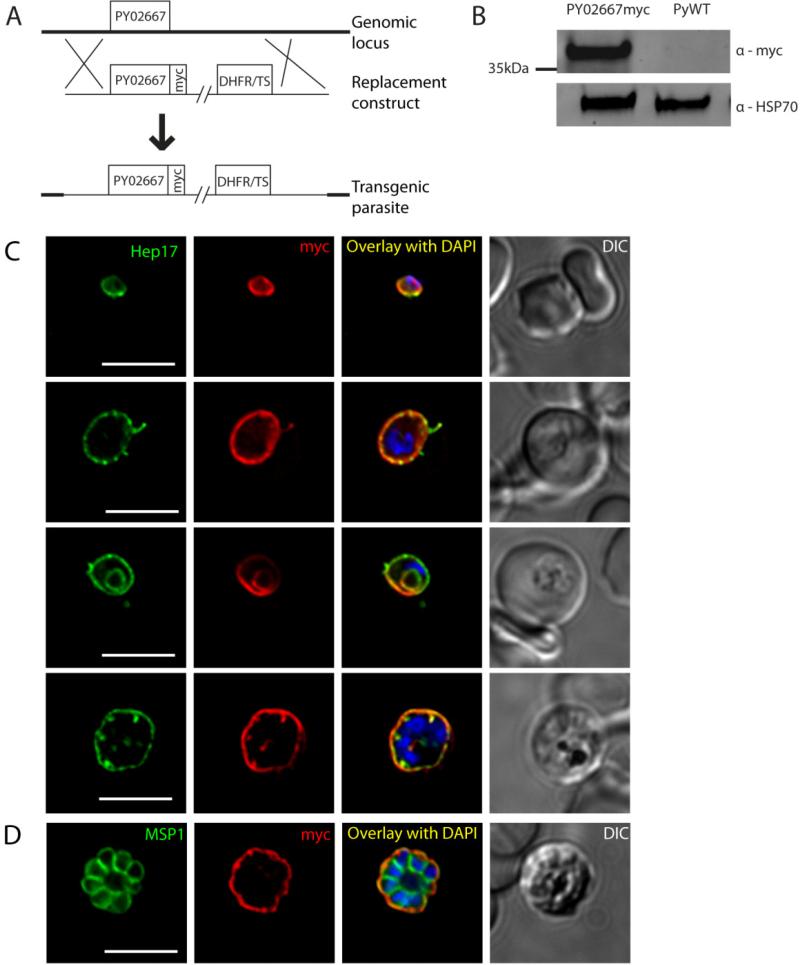
We selected two of the ETRAMPs for epitope-tagging in order to visualize protein localization throughout the life cycle. We developed constructs encoding the entire protein sequence of PY02667 and PY03652, with four tandem copies of the c-myc epitope fused to their C-termini, each expressed under control of their endogenous 5’ non-coding regions (Vaughan et al., 2009). These constructs were transfected into P. yoelii parasites and selected for integration of the construct into the genome at the endogenous locus of the respective gene. These parasites are hereafter referred to as PY02667myc and PY03652myc.
For PY02667myc, the construct was targeted to replace the endogenous ORF by double-crossover homologous recombination (Fig.2a). A Western blot of PY02667myc blood stage parasite lysates probed with anti-myc antibodies, shows the tagged protein as a single species of approximately 37kDa molecular mass (Fig.2b), which was absent in the wild-type (WT) control. This mobility is considerably slower than that predicted by the size of the protein after cleavage of the signal peptide (21kDa). Other ETRAMPs have been reported to migrate slowly in electrophoresis, presumably because of their high density of positive charges (Spielmann et al., 2003). Interestingly, however, PY02667 is one of the few ETRAMPs that carries a net negative charge, with a predicted isoelectric point of 4.4. Immunofluorescence analysis of blood stages shows that PY02667myc is expressed throughout the asexual developmental cycle, where it appears to localize to the PVM, as indicated by its proximity to the PVM resident protein Hep17 (Fig.2c). While PY02667myc is in the same compartment as Hep17, its distribution in the PVM is more asymmetrical. Specifically, whereas Hep17 aggregates into foci, PY02667myc shows a smoother pattern locally, but the fluorescent signal often shows intensity differences across the PVM. This agrees with previous evidence that ETRAMP proteins associate in sub-domains of the membrane, which can exclude other ETRAMPs and other PVM proteins (Spielmann et al., 2006). Expression of PY02667myc continues through late schizogony, when it remains within the PVM surrounding the new daughter merozoites, as delineated by staining with the parasite plasma membrane (PPM) marker MSP1 (Fig.2d). The protein is not exported beyond the PVM to the host erythrocyte. In trophozoites, PY02667myc and Hep17 both also localized in a concentric ring pattern within the parasite (Fig.2c). It has recently been shown that ring stages adopt a cup-shaped conformation within the erythrocyte, and the PVM could conceivably be distorted in three dimensions such that a single image plane represents this continuous structure as concentric rings (Abu Bakar et al., 2010). Observation of a deconvoluted stack of images, however, was inconclusive in confirming or excluding this possibility. Another possibility is that the structure observed here is part of the food vacuole. Proteins within the PVM could hypothetically be taken up during bulk feeding via the cytostome, and processed EXP-1, the P. falciparum homolog of Hep17, has previously been observed in the food vacuole of mature blood stage parasites (Adisa et al., 2003), as has ETRAMP5 (Lamarque et al., 2008).
Figure 2.
Protein expression and localization of PY02667myc in blood stages. (A) Schematic representation of the strategy used to replace PY02667 in P. yoelii with a construct encoding a c-terminal fusion of the 4x myc epitope to PY02667. (B) Western blot of lysates from mouse blood infected with asynchronous PY02667myc parasites or WT control. (C) Immunofluorescence (IFA) microscopy images of PY02667myc blood stages labeled with anti-myc or anti-Hep17 antibodies, and DAPI (to label DNA). A differential interference contrast (DIC) image of the liver section is shown to the right. Overlap of myc signal with Hep17 indicates localization of PY02667myc to the PVM. PY02667myc is expressed in all stages of intraerythrocytic development. Stages are distinguishable on the basis of morphology and DAPI staining; small uninucleate ring stages (top) grow into uninucleate trophozoites (middle rows). Myc staining persists throughout karyokinesis with the onset of schizogony (bottom). Infected erythrocytes are shown with differential interference contrast (DIC). Scale bars = 5 micrometers. (D) IFA of blood stage PY02667myc schizont, labeled with anti-myc or anti-MSP1 antibodies, and DAPI. Myc signal remains in the PVM surrounding the newly formed daughter merozoites, indicated with the PPM marker MSP1. Scale bar = 5 micrometers.
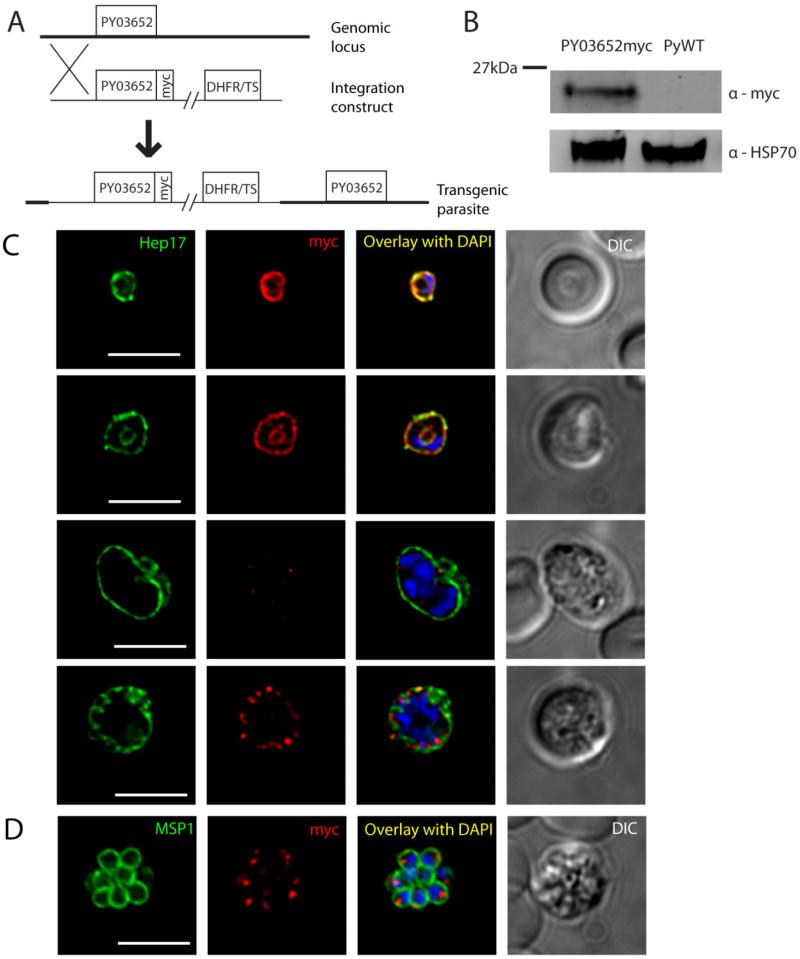
For PY03652myc, the construct encoding the epitope-tagged protein was integrated into the genome upstream of the endogenous ORF, resulting in two copies of the gene (Fig.3a). Western blots with PY03652myc transgenic mixed blood stage lysates and probed with anti-myc antibodies revealed a band approximately 20kDa in size that was not detected in WT control lysates, which is larger than the predicted mass of 10kDa (Fig.3b). The apparent mass of either PY02667myc or PY03652myc did not change under stronger reducing conditions including 1mM dithiothreitol, suggesting that the discrepancy between the predicted and observed masses is not due to residual disulfide bonds, and is better explained by the high charge density of these proteins (data not shown). Immunofluorescence analysis of blood stages shows that PY03652myc is expressed in ring stages and early trophozoites, but then expression is not observed again until late schizogony (Fig.3c). This is evident from lack of PY03652myc expression in parasites with 2-4 nuclei, which is in contrast with clear PY02667myc expression at a similar developmental time point (compare Fig.2c, third row and Fig.3c, third row). In early stages of the intraerythrocytic developmental cycle, PY03652myc localizes in a circumferential manner that overlapped with Hep17 and thus indicates PVM localization. Similar to PY02667myc, an inner ring containing PY03652myc that is separate from the PVM is visible in trophozoites, and may indicate additional localization to the food vacuole. PY03652myc is not observed to be exported beyond the PVM. PY03652myc in late schizonts, however, localizes to punctate structures near the parasite periphery but distinct from the PVM. When late schizonts are co-stained with MSP1 individual daughter merozoites are each seen to possess distinct foci of PY03652myc, suggesting packaging of the protein within the new invasive stages, possibly into secretory organelles (Fig.3d). In order to ensure that these foci are not part of a more convoluted structure in three dimensions, we recombined multiple deconvolved cross sectional images into a three dimensional stack, and find that the foci were indeed separate and distinct, found in roughly a 1:1 ratio with each daughter merozoite (see Video S1). We hypothesize that the 03652myc expressed in secretory organelles of blood stage schizonts is released and incorporates into the PVM of the early ring stage after merozoite invasion.
Figure 3.
Protein expression and localization of PY03652myc in blood stages. (A) Schematic representation of the strategy used to insert a construct encoding a c-terminal fusion of the 4x myc epitope to PY03652 upstream of the endogenous PY03652 ORF. (B) Western blot of lysates from mouse blood infected with asynchronous PY03652myc parasites or WT control. (C) IFA images of PY03652myc blood stages labeled with anti-myc or anti-Hep17 antibodies, and DAPI (to label DNA). A differential interference contrast (DIC) image of the liver section is shown to the right. Overlap of myc signal with Hep17 indicates localization of PY03652myc to the PVM in ring and trophozoite stages. Myc staining was weak or absent in early schizonts, but appears again in late schizogony, when it localizes to vesicular structures in the parasite periphery, not in the PVM. Scale bars = 5 micrometers. (D) IFA of blood stage PY03652myc schizont, labeled with anti-myc or anti-MSP1 antibodies, and DAPI. The myc signal appears in small foci within the daughter merozoites, likely within secretory organelles. Scale bar = 5 micrometers.
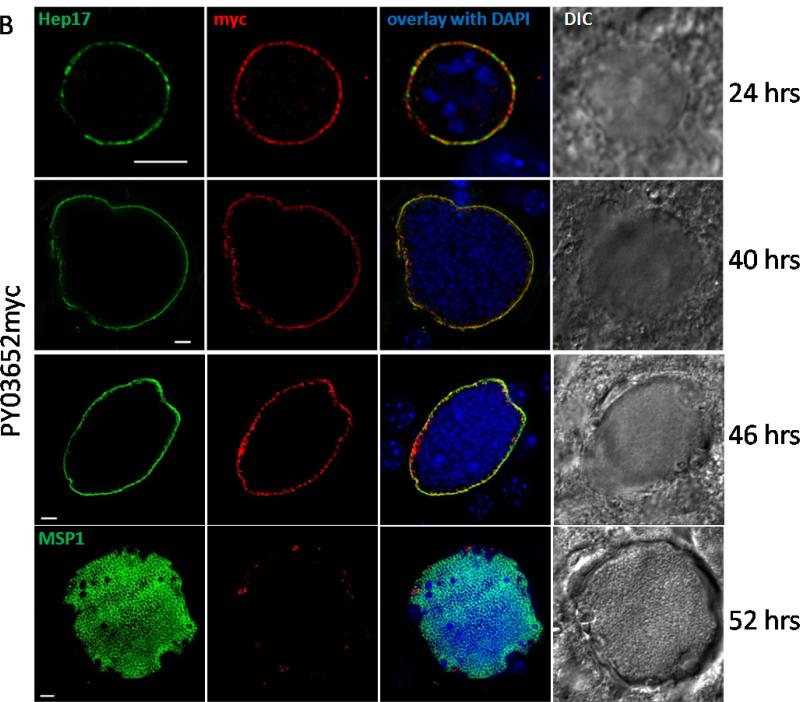
We next infected mosquitoes with PY02667myc or PY03652myc parasites to visualize protein expression in mosquito stages. We do not detect expression of either protein by IFA in salivary gland sporozoites produced in these mosquitoes (data not shown), in agreement with our RT-PCR analysis. To look at expression of the myc-tagged proteins early in liver stage development, we infected the hepatoma cell line HepG2:CD81 with PY02667myc and PY03652myc sporozoites. However, no discernible expression of either myc-tagged protein was seen up to 12 hours after sporozoite infection of cells. When PY02667myc or PY03652myc sporozoites are injected into mice, however, the developing liver stages express PY02667myc and PY03652myc in a circumferential manner that overlaps with Hep17 at 24, 40 and 46 hours post-infection, indicating PVM localization in liver stage schizonts (Fig.4a, b). PY02667myc expression was also seen at 18 hours after infection (Fig. 4a, upper panel) and at 52 hours post infection (Fig. 4a, lower panel), where expression appeared to be localized to the PVM as well as internal sites of the late liver stage schizont that showed a degree of co-localization with MSP1. In contrast, PY03652myc expression was absent at 18 hours after infection (data not shown) and is almost entirely absent from very late liver stage schizonts when exoerythrocytic merozoites have formed (52 hours post infection, Fig. 4b, lower panel). Some punctate structures that appeared to co-localize to the PVM were seen. In contrast to PY03652myc labeling of structures within erythrocytic merozoites, no localization of PY03652myc was seen within exo-erythrocytic merozoites of the late liver stage parasite (compare Fig. 3d to Fig. 4b, lower panel). The liver stage protein expression we see is in agreement with the RT-PCR data showing transcripts for PY02667 and PY03652 at 24 and 43 hours of liver stage development (Fig.1).
Figure 4.
PY02667myc (A) and PY03652myc (B) are expressed in liver stages. IFA of liver stage PY02667myc or PY03652myc parasites 18, 24, 40, 46 and 52 hours after infecting BALB/c mice with 1 × 106 sporozoites. Liver sections were labeled with anti-Hep17, MSP1 or anti-myc antibodies, and DAPI (to label DNA). A differential interference contrast (DIC) image of the liver section is shown to the right. Overlap of myc signal with Hep17 indicates localization of PY02667myc and PY03652myc to the PVM. Scale bar = 5 micrometers.
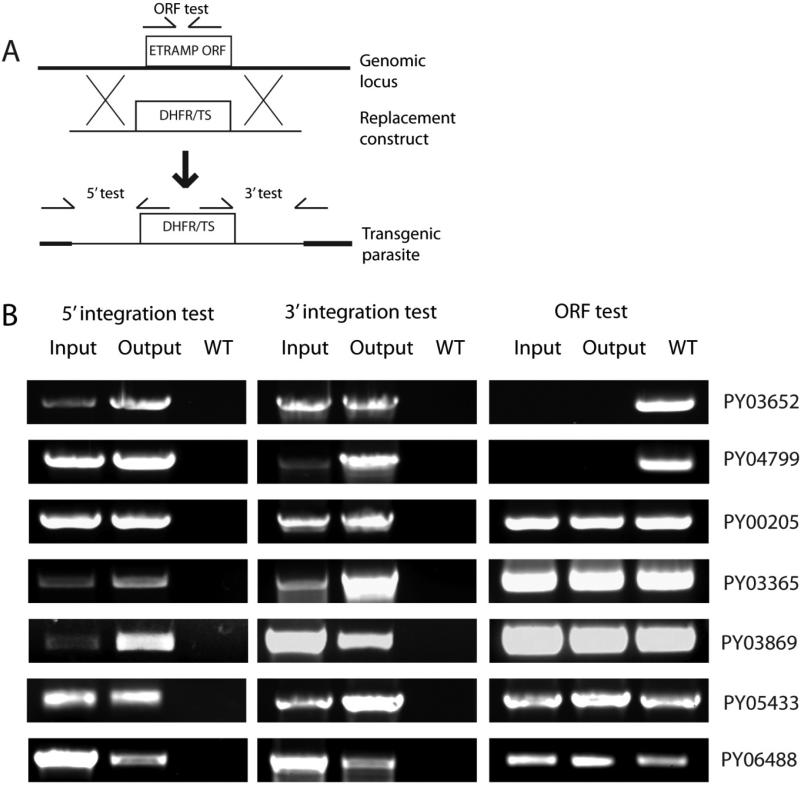
Deletion of P. yoelii etramps and phenotypic characterization of mutants
Finally, we attempted to delete each of the nine uncharacterized etramps in P. yoelii as single-gene knockouts (KO). Each gene was targeted by double-homologous recombination for replacement of the endogenous ORF with a cassette containing a mutant copy of the dihydrofolate reductase/thymidylate synthase gene from Toxoplasma gondii (TgDHFR/TS), which confers resistance to the antimalarial drug pyrimethamine (Fig.5a) (Ménard et al., 1997). Replacement was confirmed in each case by PCRs that use oligonucleotide primers annealing within the construct or within the flanking genomic sequence. These result in products specific for integration of the construct into the genomic locus as detected from independent 5’ and 3’ PCR genotyping tests. Two of the etramps, PY02667 and PY07506, could not be deleted in any of three independent transfections, each performed in duplicate, strongly suggesting that these genes perform functions that are essential for blood stage replication. Of the remaining seven etramps, two knockout parasites, PY03652- and PY04799, were cloned and analyzed for the entire life cycle. To ascertain if the blood stage replication was affected by the knockout of PY03562 and PY04799, one million parasitized red blood cells from wildtype, PY03652- or PY04799- parasite lines were injected into groups of five Swiss Webster mice and subsequently the increase in parasitemia was followed for 11 days. No significant differences were seen during this time period during on the increase in blood stage parasitemia (Fig. S1), suggesting that the deletion of PY03652 and PY04799 has no effect on blood stage replication and growth. Similarly, the development of the mosquito stage of the life cycle of the knockout parasite lines was similar to wildtype. This was determined by feeding parasitized blood containing gametocytes from mice harboring either wildtype, PY03652- or PY04799- parasite lines to female Anopheles stephensi mosquitoes and determining the sporozoite load in mosquito salivary glands 14 days post infectious blood meal. This revealed no differences in the number of sporozoites produced per mosquito (Table S3). To ascertain whether the gene knockouts affected liver stage development, intravenous injections of sporozoites into BALB/c mice were carried out and the day to blood stage patency determined. This revealed no change in liver stage development when comparing the gene knockouts with the wildtype as measured by the time elapsed before onset of blood stage parasitemia (Table 2). Furthermore, the size of liver stage schizonts at 40 hours post infection was similar when comparing wildtype with PY03562- and the PY04799- (Fig. S2).
Figure 5.
Individual deletion of seven etramps in blood stages and passage through mosquito and liver stages reveals no critical function in any stage. (A) Generalized schematic representation of the strategy used to replace P. yoelii etramp ORFs with TgDHFR/TS. (B) Genotyping of etramp KO blood stage parasites before (“Input”) and after (“Output”) passage through mosquito and liver stages. The presence of knockout parasites in the output blood stages indicates that each individual etramp is not essential for completion of the parasite life cycle. Specific oligonucleotide primer amplicons indicated in (A) are used to test for integration of the KO construct from the 5’ end or 3’ end, and also to test for the presence of the endogenous etramp ORF. PY03652- and PY04799- parasites were analyzed as clones; the remaining strains were analyzed as mixed parental populations of gene deletion parasites and WT parasites, and therefore show evidence for both 5’ and 3’ integration, as well as the unmodified ORF, in different parasites.
Table 2.
Patency in mice infected with P. yoelii etramps knockout clones, or P. yoelii WT
| Parasite strain | Sporozoites injected | Mice patent/Mice injected | Avg prepatent period (to within 0.5 days) |
|---|---|---|---|
| PY03562- | 10,000 | 5/5 | 3.0 |
| 1,000 | 5/5 | 3.5 | |
| PY04799- | 10,000 | 5/5 | 3.0 |
| 1,000 | 5/5 | 3.5 | |
| wildtype | 10,000 | 5/5 | 3.0 |
| 1,000 | 5/5 | 3.5 |
For expediency, the remaining etramp transfection-positive parasite populations – PY00205, PY03365, PY03869, PY05433, and PY06488 – were analyzed as enriched populations of transgenic parasites containing residual P. yoelii WT. These five knockout strains show no defect in blood stage growth or salivary gland sporozoite production (data not shown). When knockout sporozoites were injected into susceptible mice, all knockout parasites completed liver stage development as detected by genotyping of the emerging blood stage parasites (Fig.5b). While these data show that none of these five etramps are essential for any stage of the parasite life cycle, we cannot rule out the possibility that the knockouts might have minor defects in blood stage and/or liver stage development that we cannot detect by analysis of parental populations.
Discussion
Malaria parasites are separated from their host cell by the PVM throughout intracellular development. The PVM serves as the interface between parasite and host, and any host factors required for parasite growth and parasite factors required for interaction with the host cell must either localize to the PVM or cross this barrier. Despite its critical role in host-parasite interactions, few PVM resident proteins are known. The ETRAMPs constitute the largest family of known PVM proteins, are unique to Plasmodium parasites, and are frequently expressed at high levels and in a stage-specific manner (Spielmann et al., 2003). Therefore, we have undertaken the characterization of this family in the rodent malaria parasite P. yoelii, in which the importance of these proteins can be assessed in vivo throughout the life cycle.
P. yoelii possesses 11 currently identifiable etramps, a number similar to P. falciparum. Six of these genes have putative orthologs in P. falciparum, four of which have exactly one ortholog in each of the four Plasmodium species considered in this study. The remaining five etramps in P. yoelii appear to be specific to rodent malaria parasites, and may even have undergone lineage-specific amplification since divergence from the sibling species P. berghei. Such assignments of orthology are predictions, however, and should be tested empirically before being accepted as correct. For example, a recent study from our laboratory assessed the functional orthology of P. yoelii UIS4 and syntenous P. falciparum ETRAMP10.3 (MacKellar et al., 2010). We observed differences in the expression profiles and localization patterns of these proteins, although both showed expression within the PVM. Additionally, when etramp10.3 was used to replace UIS4 in the genome of P. yoelii parasites, it localized to the PVM of liver stages but failed to complement the function(s) of UIS4, which is essential for liver stage development (MacKellar et al., 2010). These results underscore the importance of experimental testing when assigning potential functional orthology, and further demonstrate that rodent malaria models are valuable tools for clarifying relationships between members of this gene family.
Comparisons between the etramps are also complicated by the fact that not all of the genomes of malaria parasites have been sequenced and annotated to the same depth, and additional members of the family are likely yet to be detected. Still, the available data support the conclusion that this gene family, specific to Plasmodium, contains some members that are conserved in all species and others that are specific to a subset of the malaria parasites. It is interesting that our analysis registered five of the 16 groups of homologs as specific to either human or murine parasites. While parasite phylogeny often correlates with host phylogeny, it is thought that P. falciparum and P. vivax are not more closely related to each other than either species is with the murine parasites (Escalante et al., 1994, Escalante et al., 1998, Perkins et al., 2002). If the divergence of the parasite proteins was not constrained by host-specific factors, we would predict no greater sequence similarity between ETRAMPs of P. falciparum and P. vivax than of either species with the rodent malaria parasites. Thus, the pattern of homology seen is consistent with the hypothesis that these proteins mediate host-parasite interactions.
Similar to P. falciparum, the etramps of P. yoelii show varied transcription profiles. Nine of the etramps were transcribed in blood stages, but only three could be detected in sporozoites, and eight in liver stages 24 hours after infection. More than half of the etramps were detected in the blood stages and the liver stages, raising the question of whether they might perform the same functions in both stages. If so, this assumption would limit the number of candidate functions that could be performed by proteins at the host-parasite boundary, since the requirements for parasite survival differ between the two host cells. For instance, interactions with host cell organelles, which has been observed in liver stages (Bano et al., 2007), would be irrelevant in blood stages. Similarly, any role in cytostome organization or hemoglobin uptake into the food vacuole is excluded in the hepatocyte, since liver stages lack these structures (Meis et al., 1988). ETRAMPs expressed in both intracellular stages would have to perform distinct functions in either stage, or else not participate in such processes.
Differences in expression were also seen within the intraerythrocytic developmental cycle for the two proteins to which we added an epitope tag. PY02667myc was expressed throughout, whereas PY03652myc appeared to play a role in invasion or early development, then to disappear with the onset of schizogony, since multinucleate parasites lacked the protein until new merozoites had formed. This differential regulation within the blood stages was previously observed at the transcript level for the P. falciparum etramps, and at the protein level for epitope-tagged ETRAMP2, which disappeared from the PVM partway through blood stage development (Spielmann et al., 2003, Spielmann et al., 2006). Clearly, malaria parasites modulate the constituents of the PVM throughout development, even within a single host cell. Interestingly, we did not see the localization of PY03652myc in internal structures of exo-erythrocytic merozoites and its expression appeared to be localized to the PVM throughout liver stage development, although it was absent late in schizogony when merozoites had already formed. The data suggest that PY03652myc may be playing alternative roles in the blood stage and liver stage parasite and this could be associated with the different host cells the parasite finds itself within.
ETRAMPs have previously been shown to oligomerize in membranes and form subdomains within the PVM (Spielmann et al., 2006). Accordingly, we observed blood stage parasites in which the epitope-tagged ETRAMPs were distributed in an asymmetrical or punctate circumferential pattern. The functional significance of such aggregation is unclear. The authors of another study observed a similar distribution of the Maurer's cleft protein Membrane Associated Histidine Rich Protein 1 (MAHRP-1) during its transient occupation of the PVM, and suggest this may represent preliminary steps in the formation of the Maurer's clefts from budded segments of the PVM (Spycher et al., 2006). It is possible, therefore, that the formation of subdomains of the PVM facilitates the reorganization of the host erythrocyte by establishing regions of the membrane that will be trafficked to the host cell. If true, however, this hypothesis would not explain the asymmetrical distribution of ETRAMPs such as PY02667 that appear to remain in the PVM throughout blood stage development.
The phenotypes of knockout parasites analyzed herein suggest that the majority of the etramps are not essential for any life cycle stage in P. yoelii. We identified two genes (PY02667 and PY07506) that appear essential to blood stages, since we were unable to delete them despite several attempts. Importantly, our replacement of PY02667 with an epitope-tagged copy suggests this failure was not due to inaccessibility of the genomic locus. Two other P. yoelii etramps, UIS3 and UIS4, have previously been shown to be expressed exclusively in preerythrocytic stages and are essential for liver stage development (Mueller et al., 2005a, Mueller et al., 2005b). Knockouts of the remaining seven members of this family, however, showed no strong phenotype throughout the parasite life cycle, including liver stages. Five of the seven gene knockouts were analyzed as mixed parental populations that still included WT parasites, and such analysis might fail to uncover a subtle negative phenotype during liver stage development that would manifest as a delay in patency relative to WT. It seems likely, however, that these ETRAMPs may have at least partially overlapping functions; three of the five nonessential genes that were analyzed as mixed parental populations (PY03365, PY05433, and PY06488) share over 80% sequence identity. Such close paralogs might compensate for the loss of one gene, and therefore a strong phenotype would not be expected. It will be of interest to determine whether combinatorial deletions of etramps within a single parasite would reveal such functional redundancy. While the tools for achieving sequential genetic manipulations are limited in rodent malaria parasites, our use of TgDHFR/TS as the selectable marker would potentially permit one additional deletion in any of these strains by using the human DHFR in conjunction with the drug WR99210(Sultan et al., 2001).
The ETRAMPs have no domains of known homology to any characterized protein, and their functions remain unknown. Here we have elucidated the expression and essentiality of the etramp genes in P. yoelii, and the localization of two of their products. The next important experiments to characterize this family will be the identification of host factors that they bind directly at the PVM. To date, three studies have described binding between ETRAMPs and host proteins (Mikolajczak et al., 2007, Vignali et al., 2008, Garcia et al., 2009). Vignali et al. observed an interaction between P. falciparum ETRAMP5 and human apolipoproteins in a yeast two-hybrid system, and Garcia et al. found specific binding between synthetic peptides representing parts of ETRAMP10.2 and unidentified molecules on the surface of human erythrocytes. In the latter study, pre-incubation of erythrocytes with an excess of synthetic peptide caused partial inhibition of invasion by merozoites (Garcia et al., 2009). While this was the only study so far to suggest a role for ETRAMPs in invasion, the localization that we observe here for PY03652myc does suggest that some members of this family may be packaged into secretory organelles and could function in invasion or early intraerythrocytic development. Mikolajczak et al. identified an interaction between UIS3 and L-FABP. The importance of L-FABP for liver stage growth was corroborated by the use of RNAi against, or overexpression of, L-FABP in hepatoma cells. When L-FABP was knocked down, parasite development was retarded relative to parasites in untreated host cells, mimicking the liver stage arrest seen with uis3- parasites in mice (Mikolajczak et al., 2007). Interestingly, when L-FABP was over-expressed, parasite growth improved over unmodified cells, suggesting that L-FABP concentration may be limiting for growth, at least in vitro (Mikolajczak et al., 2007). These studies present a cross-section of the tools available for identifying host-parasite interactions. Applying them in a systematic manner to the remaining ETRAMPs, which represent the largest known family of PVM proteins, will greatly advance our understanding of the malaria parasite's complex interactions with its host cells.
Experimental procedures
Bioinformatics tools
ETRAMPs were identified from their entry (PF09716) in the Pfam database (Finn et al., 2010). Members that showed poor alignment with other ETRAMPs or more than one exon were excluded from Table S1. These included sexual stage antigens s16 (Pfs16 and Pvs16), as well as a sequence from Clostridium botulinum. Potentially homologous ETRAMPs were identified from their entry in the OrthoMCL database (Chen et al., 2006). Some groups of homologs were then combined on the basis of synteny.
Experimental animals, parasites, and cell lines
Six to eight week-old female BALB/c and Swiss Webster (SW) mice were purchased from Harlan Laboratories (Indianapolis, IN). SW mice were used for routine parasite life cycle maintenance; all experimental results were obtained in BALB/c mice. All experiments were conducted in accordance with protocols approved by the Seattle Biomed Institutional Animal Care and Use Committee (IACUC). P. yoelii strain 17XNL non-lethal parasites were used for genetic manipulations and as WT controls. Anopheles stephensi mosquitoes were used for sporogonic cycles.
RT-PCR
Liver-stage cDNA was prepared from total RNA from whole infected mouse livers. BALB/c mice were injected with one million P. yoelii sporozoites per mouse, and livers were collected 24 or 43 hours post-injection. Livers were homogenized in 3mL of Trizol, and RNA collected according to manufacturer's instructions (Invitrogen). Collection of total RNA from other life cycle stages, as well as cDNA synthesis and PCR cycling conditions, are as previously described (Aly et al., 2008). Transcripts were detected with oligonucleotide primers that anneal to the ORF of the specified etramps. All primers used in this study are listed in Supplementary Table 2. Primers used for detecting the ORFs are listed as XXXX orfF and XXXX orfR, where ‘XXXX’ are the final four digits in the PlasmoDB identifier for each gene.
Generation of transgenic P. yoelii parasites
etramp knockout constructs were made by cloning the 5’ and 3’ sequences flanking each etramp ORF into the b3D.DTH.D vector (catalog# MRA-80 in MR4-Malaria Research and Reference Reagent Resource Center; http://www.malaria.mr4.org/) (Waters et al., 1997). The 5’ flank for each gene was amplified with the respective Rep1F and Rep2R primers, and the 3’ flank was amplified with the Rep3F and Rep4R primers. These constructs were then digested with SacII and SpeI and transfected into P. yoelii 17XNL schizonts to target the genomic locus for replacement of the ORF by homologous recombination. Transfection, selection, and cloning of parasites were performed as described previously (Tarun et al., 2006). Epitope-tagged etramp constructs were made by cloning the endogenous promoter sequence and ORF of specified etramps into the b3D vector that has been modified to encode four tandem copies of the c-myc epitope fused in frame to the 3’ end of the ORF (Vaughan et al., 2009). These constructs were transfected into P. yoelii schizonts to target the genomic locus for integration as a second copy of the gene, or for replacement of the endogenous ORF.
Parasite genotyping
Transgenic parasites were genotyped with oligonucleotide primers to detect integration of the relevant construct at the genomic locus that was targeted. Integration from the 5’ end was detected by PCR amplification using the primers TgR and a specific forward primer that anneals in the genomic sequence 5’ to the integration site (listed as XXXX TestF in Table S2, where ‘XXXX’ are the final four digits of the PlasmoDB identifier for each gene). Integration from the 3’ end was detected by PCR using the primers TgF and a specific reverse primer that anneals in the sequence 3’ to the integration site (listed as XXXX TestR in Table S2). The presence or absence of the ORF was confirmed by PCR using the same oligonucleotide primers used for detecting transcripts by RT-PCR.
Western blots
Asynchronous blood stage PY02667myc, PY03652myc, or PyWT parasites were collected from mouse blood and lysed in Laemmli sample buffer containing 5% beta-mercaptoethanol or 1mM dithiothreitol. Lysates were separated by electrophoresis in a 4-20% SDS-PAGE gel (Thermo Scientific, Rockford, IL). Proteins were transferred to a nitrocellulose membrane via semi-dry electrophoresis (Bio-Rad, Hercules, CA). Blots were blocked with Tris-buffered saline and 5% bovine serum albumin prior to incubation with antibodies against PyHSP70 or the c-myc epitope, and imaged on the Odyssey imaging platform with far-red secondary antibodies (Li-Cor Biosciences, Lincoln, NE).
Patency assay
Mosquitoes were fed on mice infected with transgenic or WT blood stage parasites to generate sporozoites. Sporozoites were obtained on day 14 post-feed from salivary glands of dissected mosquitoes, and injected into the tail vein of BALB/c mice. These mice were monitored twice daily for parasitemia by microscopy of Giemsa-stained thin blood smears.
Phenotypic analysis of blood stage P. yoelii PY03652- and PY04799- parasites
To assay growth of non-lethal P. yoelii 17XNL wildtype (wt), PY03652- and PY04799- blood stages, blood was removed from infected SW mice when parasitemia was between 0.5% and 1.5%. The blood was diluted in RPMI-1640 media (HyClone, Logan, UT) so that 100 μl contained 106 parasites. SW mice (5 in each group) were then injected intravenously with the parasites. Percentage parasitemia was followed as often as daily for eleven days by assay of Giemsa-stained blood smear.
Immunofluorescence assays
Infected erythrocytes were fixed in 4% paraformaldehyde, permeabilized, and incubated with the indicated primary antibodies, then with fluorophore-conjugated goat anti-rabbit or goat anti-mouse secondary antibodies, as well as 4’, 6-diamidino-2-phenylindole (DAPI). Liver stage in vitro assays were conducted using the human hepatoma cell line HepG2 expressing the tetraspanin CD81 (HepG2 CD81) cultured in Dulbecco's modified Eagle's medium with 10% fetal calf serum at 37°C and 5% CO2 as previously described (Vaughan et al., 2009). Liver stage in vivo assays were carried out in BALB/c mice. Livers were harvested from mice infected 24, 40, 46 or 52 hours earlier with 106 sporozoites. Livers were fixed in 4% paraformaldehyde and 50μm sections were made with a Vibratome (Ted Pella, Inc., Redding, CA). Sections were permeabilized and incubated with the indicated primary antibodies, then the appropriate secondary antibodies and DAPI. Images were captured on a Deltavision Spectris RT microscope (Applied Precision, Issaquah WA) using a 40x or 100x oil objective, and subjected to deconvolution with the softWoRx software package. Three-dimensional reconstruction was made with the Volume Viewer tool in the softWoRx package.
Supplementary Material
Acknowledgements
This study was funded by grants from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health Initiative and the National Institutes of Health (RO1 AI053709-07).
References
- Abu Bakar N, Klonis N, Hanssen E, Chan C, Tilley L. Digestive-vacuole genesis and endocytic processes in the early intraerythrocytic stages of Plasmodium falciparum. J Cell Sci. 2010;123:441–450. doi: 10.1242/jcs.061499. [DOI] [PubMed] [Google Scholar]
- Adisa A, Rug M, Klonis N, Foley M, Cowman AF, Tilley L. The signal sequence of exported protein-1 directs the green fluorescent protein to the parasitophorous vacuole of transfected malaria parasites. J Biol Chem. 2003;278:6532–6542. doi: 10.1074/jbc.M207039200. [DOI] [PubMed] [Google Scholar]
- Aly AS, Mikolajczak SA, Rivera HS, Camargo N, Jacobs-Lorena V, Labaied M, et al. Targeted deletion of SAP1 abolishes the expression of infectivity factors necessary for successful malaria parasite liver infection. Mol Microbiol. 2008;69:152–163. doi: 10.1111/j.1365-2958.2008.06271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bano N, Romano JD, Jayabalasingham B, Coppens I. Cellular interactions of Plasmodium liver stage with its host mammalian cell. Int J Parasitol. 2007;37:1329–1341. doi: 10.1016/j.ijpara.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Birago C, Albanesi V, Silvestrini F, Picci L, Pizzi E, Alano P, et al. A gene-family encoding small exported proteins is conserved across Plasmodium genus. Mol Biochem Parasitol. 2003;126:209–218. doi: 10.1016/s0166-6851(02)00275-x. [DOI] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Stoeckert CJ, Jr., Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui LW, Fan Q, Hu Y, Karamycheva SA, Quackenbush J, Khuntirat B, et al. Gene discovery in Plasmodium vivax through sequencing of ESTs from mixed blood stages. Mol. Biochem. Parasitol. 2005;144:1–9. doi: 10.1016/j.molbiopara.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Daily JP, Le Roch KG, Sarr O, Fang XM, Zhou YY, Ndir O, et al. In vivo transcriptional profiling of Plasmodium falciparum. Malar. J. 2004;3 doi: 10.1186/1475-2875-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Ayala FJ. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proceedings of the National Academy of Sciences USA. 1994;91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc Natl Acad Sci U S A. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J, Curtidor H, Obando-Martinez AZ, Vizcaino C, Pinto M, Martinez NL, et al. Synthetic peptides from conserved regions of the Plasmodium falciparum early transcribed membrane and ring exported proteins bind specifically to red blood cell proteins. Vaccine. 2009;27:6877–6886. doi: 10.1016/j.vaccine.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser K, Matuschewski K, Camargo N, Ross J, Kappe SH. Differential transcriptome profiling identifies Plasmodium genes encoding pre-erythrocytic stage-specific proteins. Mol Microbiol. 2004;51:1221–1232. doi: 10.1046/j.1365-2958.2003.03909.x. [DOI] [PubMed] [Google Scholar]
- Lamarque M, Tastet C, Poncet J, Demettre E, Jouin P, Vial H, Dubremetz JF. Food vacuole proteome of the malarial parasite Plasmodium falciparum. Proteom. Clin. Appl. 2008;2:1361–1374. doi: 10.1002/prca.200700112. [DOI] [PubMed] [Google Scholar]
- Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- MacKellar DC, O'Neill MT, Aly ASI, Sacci JB, Cowman AF, Kappe SHI. Plasmodium falciparum PF10_0164 (ETRAMP10.3) Is an Essential Parasitophorous Vacuole and Exported Protein in Blood Stages. Eukaryot. Cell. 2010;9:784–794. doi: 10.1128/EC.00336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuschewski K, Ross J, Brown SM, Kaiser K, Nussenzweig V, Kappe SH. Infectivity-associated changes in the transcriptional repertoire of the malaria parasite sporozoite stage. J Biol Chem. 2002;277:41948–41953. doi: 10.1074/jbc.M207315200. [DOI] [PubMed] [Google Scholar]
- Meis JF, Verhave JP. Exoerythrocytic development of malarial parasites. Adv Parasitol. 1988;27:1–61. doi: 10.1016/s0065-308x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- Ménard R, Janse C. Methods: a Companion to Methods in Enzymology- Analysis of Apicomplexan Parasites. Academic Press, Inc.; Orlando, FL: 1997. Gene targeting in malaria parasites. pp. 148–157. [DOI] [PubMed] [Google Scholar]
- Mikolajczak SA, Jacobs-Lorena V, MacKellar DC, Camargo N, Kappe SH. L-FABP is a critical host factor for successful malaria liver stage development. Int J Parasitol. 2007;37:483–489. doi: 10.1016/j.ijpara.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Moreno A, Perignon JL, Morosan S, Mazier D, Benito A. Plasmodium falciparum-infected mice: more than a tour de force. Trends Parasitol. 2007;23:254–259. doi: 10.1016/j.pt.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Mueller AK, Camargo N, Kaiser K, Andorfer C, Frevert U, Matuschewski K, Kappe SH. Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc Natl Acad Sci U S A. 2005a;102:3022–3027. doi: 10.1073/pnas.0408442102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller AK, Labaied M, Kappe SH, Matuschewski K. Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature. 2005b;433:164–167. doi: 10.1038/nature03188. [DOI] [PubMed] [Google Scholar]
- Perkins SL, Schall JJ. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol. 2002;88:972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Sharma A, Yogavel M, Akhouri RR, Gill J. Crystal structure of soluble domain of malaria sporozoite protein UIS3 in complex with lipid. J Biol Chem. 2008;283:24077–24088. doi: 10.1074/jbc.M801946200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestrini F, Bozdech Z, Lanfrancotti A, Di Giulio E, Bultrini E, Picci L, et al. Genome-wide identification of genes upregulated at the onset of gametocytogenesis in Plasmodium falciparum. Mol Biochem Parasitol. 2005;143:100–110. doi: 10.1016/j.molbiopara.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Spielmann T, Beck HP. Analysis of stage-specific transcription in plasmodium falciparum reveals a set of genes exclusively transcribed in ring stage parasites. Mol Biochem Parasitol. 2000;111:453–458. doi: 10.1016/s0166-6851(00)00333-9. [DOI] [PubMed] [Google Scholar]
- Spielmann T, Fergusen DJ, Beck HP. etramps, a New Plasmodium falciparum Gene Family Coding for Developmentally Regulated and Highly Charged Membrane Proteins Located at the Parasite-Host Cell Interface. Mol Biol Cell. 2003;14:1529–1544. doi: 10.1091/mbc.E02-04-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann T, Gardiner DL, Beck HP, Trenholme KR, Kemp DJ. Organization of ETRAMPs and EXP-1 at the parasite-host cell interface of malaria parasites. Mol. Microbiol. 2006;59:779–794. doi: 10.1111/j.1365-2958.2005.04983.x. [DOI] [PubMed] [Google Scholar]
- Spycher C, Rug M, Klonis N, Ferguson DJP, Cowman AF, Beck HP, Tilley L. Genesis of and trafficking to the Maurer's clefts of Plasmodium falciparum-infected erythrocytes. Mol. Cell. Biol. 2006;26:4074–4085. doi: 10.1128/MCB.00095-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan AA, Thathy V, de Koning-Ward TF, Nussenzweig V. Complementation of Plasmodium berghei TRAP knockout parasites using human dihydrofolate reductase gene as a selectable marker. Mol Biochem Parasitol. 2001;113:151–156. doi: 10.1016/s0166-6851(01)00209-2. [DOI] [PubMed] [Google Scholar]
- Tarun AS, Baer K, Dumpit RF, Gray S, Lejarcegui N, Frevert U, Kappe SH. Quantitative isolation and in vivo imaging of malaria parasite liver stages. Int J Parasitol. 2006;36:1283–1293. doi: 10.1016/j.ijpara.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Tarun AS, Peng X, Dumpit RF, Ogata Y, Silva-Rivera H, Camargo N, et al. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc Natl Acad Sci U S A. 2008;105:305–310. doi: 10.1073/pnas.0710780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton TJ. The varieties of gene amplification, diversification and hypervariability in the human malaria parasite, Plasmodium falciparum. Mol Biochem Parasitol. 2009;166:109–116. doi: 10.1016/j.molbiopara.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Vaughan AM, O'Neill MT, Tarun AS, Camargo N, Phuong TM, Aly AS, et al. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell Microbiol. 2009;11:506–520. doi: 10.1111/j.1462-5822.2008.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali M, McKinlay A, LaCount DJ, Chettier R, Bell R, Sahasrabudhe S, et al. Interaction of an atypical Plasmodium falciparum ETRAMP with human apolipoproteins. Malar J. 2008;7:211. doi: 10.1186/1475-2875-7-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincensini L, Richert S, Blisnick T, Van Dorsselaer A, Leize-Wagner E, Rabilloud T, Braun Breton C. Proteomic analysis identifies novel proteins of the Maurer's clefts, a secretory compartment delivering Plasmodium falciparum proteins to the surface of its host cell. Mol Cell Proteomics. 2005;4:582–593. doi: 10.1074/mcp.M400176-MCP200. [DOI] [PubMed] [Google Scholar]
- Waters AP, Thomas AW, van Dijk MR, Janse CJ. Methods: a Companion to Methods in Enzymology- Analysis of Apicomplexan Parasites. Academic Press, Inc.; Orlando, FL: 1997. Transfection of malaria parasites. pp. 134–147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.