Abstract
Ion channels play crucial roles in transport and regulatory functions of living cells. Understanding the gating mechanisms of these channels is important to understanding and treating diseases that have been linked to ion channels. One potential model peptide for studying the mechanism of ion channel gating is alamethicin, which adopts a split alpha/310 helix structure and responds to changes in electric potential. In this study, sum frequency generation vibrational spectroscopy (SFG-VS), supplemented by attenuated total reflectance Fourier transform infrared spectroscopy (ATR-FTIR), has been applied to characterize interactions between alamethicin (a model for larger channel proteins) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid bilayers in the presence of an electric potential across the membrane. The membrane potential difference was controlled by changing the pH of the solution in contact with the bilayer, and measured using fluorescence spectroscopy. The orientation angle of alamethicin in POPC lipid bilayers was then determined at different pH values using polarized SFG amide I spectra. Assuming that all molecules adopt the same orientation (a δ distribution), at pH=6.7 the α-helix at the N-terminus and the 310 helix at the C-terminus tilt at about 72° (θ1) and 50° (θ2) versus the surface normal, respectively. When pH increases to 11.9, θ1 and θ2 decrease to 56.5° and 45°, respectively. The δ distribution assumption was verified using a combination of SFG and ATR-FTIR measurements, which showed a quite narrow distribution in the angle of θ1 for both pH conditions. This indicates that all alamethicin molecules at the surface adopt a nearly identical orientation in POPC lipid bilayers. The localized pH change in proximity to the bilayer modulates the membrane potential and thus induces a decrease in both the tilt and bend angles of the two helices in alamethicin. This is the first reported application of SFG to the study of model ion channel gating mechanisms in model cell membranes.
1. Introduction
Ion channel proteins are transmembrane proteins that regulate ionic permeability in cell membranes. They are key elements in signaling and sensing pathways, and provide selective transport in and out of cells.1 Defective ion channels may be a factor in diseases such as cystic fibrosis, cardiac arrhythmias, and Parkinson’s disease.2–9 The study of ion channel structures is important to understanding disease mechanisms and developing effective treatments.
Voltage-gated ion channels are a subset of transmembrane ion channels, activated by changes in electrical potential across the cell membrane.10,11 Much research has been done in obtaining ion channel crystal structures and understanding how these channels sense and respond to membrane potential changes.12–24 Although these studies have substantially improved our understanding of ion channel voltage sensing, the detailed mechanism for this process is still an open question. Currently, three models are under debate: the transporter model, the helical screw model, and the paddle model.22,23 We believe that direct, in situ structural measurements of ion channels and related membrane proteins will elucidate the precise mechanism of ion channel gating.
Alamethicin, frequently used as a model for larger channel proteins,25–35 is a 20-residue peptide extracted from the fungus Trichoderma viride. Alamethicin molecules can form voltage-gated ion channels in membranes.25–38 The molecular structure and conformational features of alamethicin have been studied extensively.25–42 The X-ray crystal structure of alamethicin crystallized from methanol was determined to be predominantly helical with an N-terminal α-helix and a C-terminal domain containing a 310-helical element.39 The Pro14 residue separating the two domains induces a 20° to 35° bend in alamethicin.39
The mechanisms of alamethicin action within cell membranes have been widely studied.25–38 It is currently believed that alamethicin interacts with cell membranes through the barrel-stave mode, in which conducting pores in the membrane are formed by parallel bundles of three to twelve helical alamethicin monomers surrounding a central, water-filled pore.27,28,37–39,43–45 However, some molecular-level details of the mechanism of alamethicin channel formation remain unclear,46,47 with contradictory results regarding its orientation in cell membranes in the absence of membrane potential. It has been variously suggested that alamethicin adopts a transmembrane orientation,40–42, 48–52 lies down on the membrane surface,53–55 or both.56,57 A continuous distribution of orientations has also been proposed.58 Recently, we applied sum frequency generation vibrational spectroscopy (SFG-VS) to investigate the interactions between alamethicin and different lipid bilayers without the presence of a membrane potential.59 SFG-VS is a nonlinear optical spectroscopic technique which provides vibrational spectra of surfaces and interfaces.59–71 Our previous studies have indicated that alamethicin inserts into fluid-phase lipid bilayers, but lies down and/or aggregates on gel-phase bilayer surfaces.59
In order to understand the gating mechanism of ion channels, it is important to investigate these biological molecules in the presence of a membrane potential. In this study, for the first time, we investigate the interactions between alamethicin and model membranes in situ with the presence of a transmembrane potential.
Membrane potential (e.g., lipid membrane dipole potential) plays important roles in protein-membrane interactions.72–74 In addition to an external electric field, protons and hydroxides (or pH variations) can also effectively modulate lipid membrane potential by altering surface charge and dipole potential.75–79 Upon exposing a lipid membrane to aqueous solutions of varying pH, the phosphate and choline groups of lipid molecules may change charge distribution at the membrane interface, resulting in alterations of their Debye length, membrane surface charge density, and zeta potential.75–79 This can lead to a change in membrane potential and an increase in channel protein conductance.79 Because pH plays a crucial role in modulating the electric characteristics of zwitterionic-based lipid membranes, changes in pH have been used to modulate the transport properties of alamethicin oligomers inserted in zwitterionic-based model lipid membranes.75 In addition, earlier studies have indicated that high pH values increased the open-state probability of the channel of Entamoeba histolytica, which has a mechanism of action similar to alamethicin.80 With this in mind, we modulated the membrane potential by changing the pH of the subphase in contact with the lipid bilayer. We then investigated how changes in subphase pH affected the orientation of alamethicin in membranes using SFG-VS and attenuated total reflectance-Fourier transform infrared spectroscopy (ATR-FTIR). Results from this in situ study will help to understand ion channel proteins in action with the presence of a membrane potential.
2. Experimental Section
2.1 Materials and Sample Preparations
Alamethicin from Trichoderma viride was purchased from Sigma-Aldrich (St. Louis, MO) with a minimum purity of 90%. The lipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) ammonium salt (Rhod-DMPE) were purchased from Avanti Polar Lipids (Alabaster, AL). Potassium chloride (KCl), potassium phosphate (K3PO4), and deuterated water (D2O) were ordered from Aldrich (Milwaukee, WI) with a purity of >99.0% and were used as received. The stock salt solution (1.0 M) of pH 6.7 or 11.9 was prepared by dissolving KCl or K3PO4 into ultrapure water from a Millipore system (Millipore, Bedford, MA). Right-angle CaF2 prisms were purchased from Altos (Trabuco Canyon, CA). They were thoroughly cleaned as described elsewhere,59 and were verified using SFG to be free of contamination before lipid deposition.
Single lipid bilayers were prepared on CaF2 substrates using Langmuir-Blodgett and Langmuir-Schaefer (LB/LS) methods with a KSV2000 LB system.59,69,71 Lipid bilayers were immersed in water inside a 1.6 mL reservoir throughout the entire experiment. No SFG C-H stretching signal was detected after depositing a deuterated lipid monolayer or bilayer on a CaF2 surface, indicating that there was no contamination on the CaF2 surface when constructing the lipid monolayer or bilayer. For alamethicin-bilayer interaction experiments, ~15 μL alamethicin solution (in methanol with a concentration of 2.5 mg/mL) was injected into the reservoir, resulting in a final alamethicin concentration of 23 μg/mL. A magnetic micro stirrer was used to ensure a homogeneous concentration of peptide molecules in the subphase below the bilayer.
2.2 Polarized ATR-FTIR and SFG-VS Experiments
Details regarding SFG theories and instruments have been reported previously59–71. The SFG and ATR-FTIR experimental details and data analysis methods used in this research are presented in the supporting information.
2.3 Estimation of Membrane Potential Difference
It has been demonstrated that membrane potentials can be measured directly using fluorescence spectroscopy.81–83 In this study, we estimated the membrane potential change based on the fluorescent intensity change of (10%Rhod-DMPE+90%POPC (in mass ratio))/POPC and POPC/(10%Rhod-DMPE+90%POPC) bilayers using our SFG experimental geometry (shown in supporting information). The potential difference across the POPC/POPC bilayer was estimated to be −16.2 mV when the subphase pH was changed from 7.3 to 11.8. This value agrees with the results of phospholipid 1-stearoyl-2-oleoyl-phosphatidyl-choline reported by Zhou et al.76
3. Results and Discussion
3.1 SFG Results at pH= 6.7
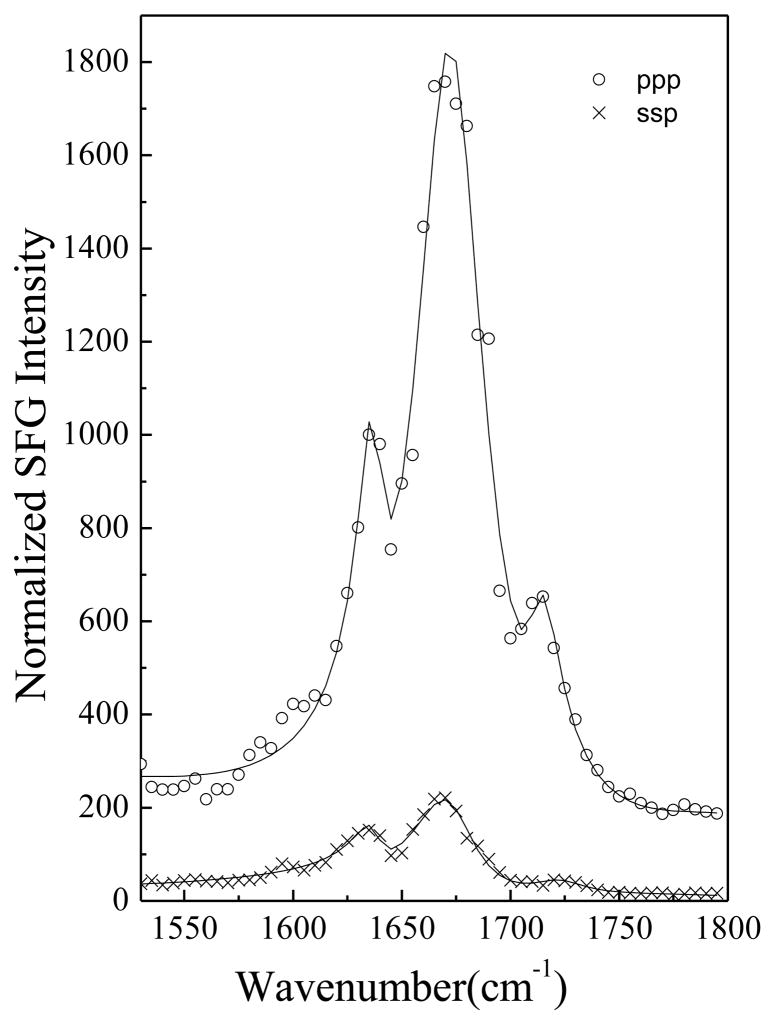
SFG ssp and ppp spectra of alamethicin in a POPC/POPC bilayer at pH=6.7 are shown in Fig. 1. Spectra were collected after an injection of a 15 μL alamethicin/methanol solution into the subphase (~1.6 mL) of a POPC/POPC bilayer at pH 6.7. The resulting concentration of alamethicin in the subphase is 23 μg/mL. We observed three peaks in these spectra: a 1670 cm−1 peak originating from a helical structure dominated by α-helix with minor contribution from a 310-helix,59 a 1635 cm−1 peak due to the 310-helical structure,59 and a 1720 cm−1 signal generated by the carbonyl groups of the lipid bilayer. More details on the amide I peak assignments can be found in ref.59 and the references therein.
Figure 1.
SFG spectra of alamethicin in a POPC/POPC bilayer at pH=6.7.
Alamethicin consists of two helical segments due to the presence of the helix-breaking Pro14 residue.39 Here, we define the tilt angles between the principal axis of the alpha helix (residues 1 to 13) or 310 helix (residues 14 to 20) and the POPC/POPC bilayer surface normal to be θ1 and θ2, respectively. Molecular orientation information can be obtained by relating SFG susceptibility tensor elements χijk (i, j, k = x, y, z) to the SFG molecular hyperpolarizability tensor elements βlmn(l, m, n= a, b, c).59–71 For α-helices and 310-helices, the details of orientation determination using polarized SFG spectra have been published previously.69 Using the relationship between the measured ppp and ssp spectral intensity ratios of the peaks at 1670 cm−1 and 1635 cm−1, it is possible to determine the orientation angles θ1 and θ2.59 Here the experimentally measured χppp/χssp ratios of the peaks at 1670 cm−1 and 1635 cm−1 for alamethicin in the POPC/POPC bilayer at pH =6.7 are 2.65 and 2.18. The resulting orientation angles θ1 and θ2 are calculated to be about 72 ° and 50 °, assuming a δ-orientation distribution.
Our previous SFG studies indicated that alamethicin molecules also adopt a tilted orientation in a DMPC/DMPC bilayer with θ1 and θ2 calculated to be 63° and 43°.59 In the POPC/POPC bilayer used in this work, the alamethicin molecules are tilted more towards the bilayer surface. “Hydrophobic matching” between the hydrophobic region of a peptide and the hydrocarbon region of a lipid bilayer is the major requirement for the stable transmembrane insertion of peptaibols.42,50–52 However, factors such as the hydrophobic moment of the peptide, the lipid saturated/unsaturated fatty acyl chain compositions, and the lipid bilayer phase transition temperature are also important.31,50–52 Although the hydrophobic thickness of a POPC bilayer is larger than that of a DMPC bilayer, the phase transition temperature of POPC is much lower than that of DMPC because of the unsaturated chain in POPC. This may lead to the difference in peptide tilt angle observed.
3.2 SFG Results at pH=11.9
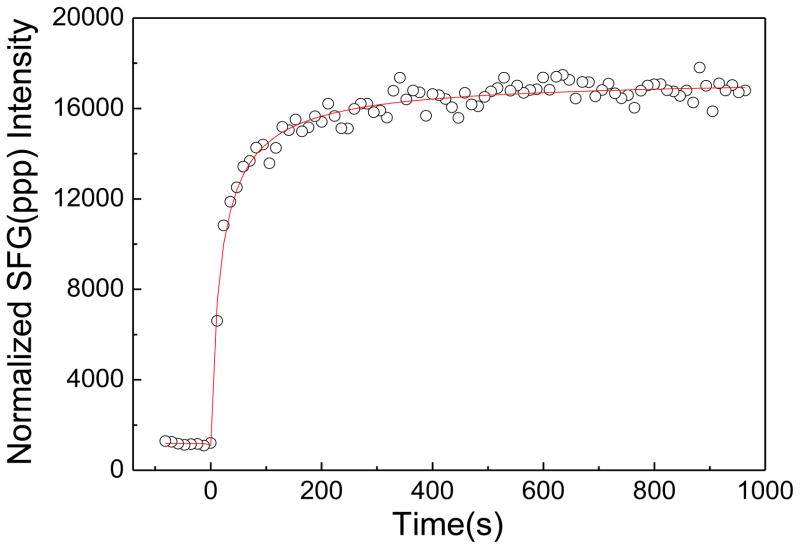
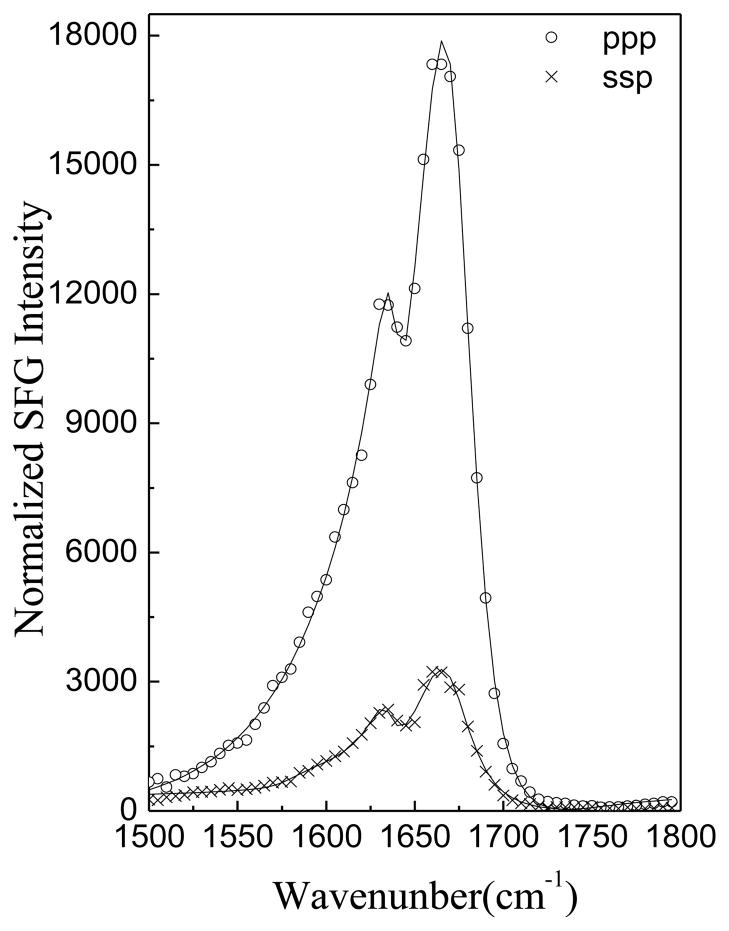
After all measurements were performed at pH=6.7, the subphase pH was raised to 11.9 by adding 100 μL K3PO4 (in 1M solution) into the subphase (~1.6 mL DI water), which resulted in a 0.06 M K3PO4 solution. ATR-FTIR experiments confirmed that the POPC/POPC bilayer is stable at pH 11.9 in the absence of peptide based on the ATR-FTIR signals observed in the C-H stretching frequency region (not shown). Figure 2 shows the time dependent intensity change of the SFG amide I peptide signals (1665 cm−1) in the ppp polarization after changing the subphase pH from 6.7 to 11.9. The intensity increased immediately after adjusting the pH, indicating that the pH change rapidly affected the alamethicin interaction with the bilayer. Figure 3 presents the SFG spectra of alamethicin in a POPC/POPC bilayer at pH =11.9, which shows that the SFG amide I intensity from alamethicin increased substantially in the POPC/POPC bilayer.
Figure 2.
The time-dependent intensity change of ppp SFG spectra at 1665 cm−1 in the alamethicin bound POPC/POPC bilayer after a change in pH from 6.7 to 11.9.
Figure 3.
SFG ssp and ppp spectra of alamethicin in a POPC/POPC bilayer at pH=11.9.
After adjusting the pH, the fitted χppp/χssp ratios for the peaks shown in Fig. 3 changed to 1.93 and 2.34. From these ratios, the orientation angles were determined to be θ1 = 56.5° and θ2 = 45°. We compared the fitted χppp and χssp of the 1670 cm−1 peak at different pH value and found that χppp(pH=11.9)/χppp(pH=6.7) = 3.63 and χssp(pH=11.9)/χssp(pH=6.7) = 4.11. It is well known that SFG signal intensity is related to the number of molecules detected and their orientations. Since the orientation was deduced above from the χppp/χssp ratio, the effect of a change in the number of molecules can be isolated. Thus by comparing spectral intensities, we quantified that the number of the molecules that inserted into the lipid bilayer increased about two times after adjusting the pH from 6.7 to 11.9.
To verify that the observed effect arose from the pH change alone instead of a change in the ionic strength of the subphase, we also investigated the effect of salt on the SFG signals from alamethicin in a POPC/POPC bilayer at pH=6.7. After adding 300 μL KCl solution (1.0 M) (pH=6.7) into the aqueous subphase (resulting in a 0.16 M KCl solution), alamethicin amide I intensity decreased (data shown in the supporting information). After the addition of KCl, the fitted χssp and χppp values for the alamethicin amide I signal decreased by a factor of 1.5 and 1.7, respectively. Overall, the ratio χppp/χssp decreased slightly in this case, which would indicate59,69 that alamethicin stood up relative to the bilayer. In that case, an increase in signal intensity would be expected if the number of peptide molecules in the lipid bilayer remained the same. Therefore, the observed decrease in SFG amide I intensity must come from the “salting out” effect, which results in fewer peptides in the bilayer. Such an effect has been observed in an earlier study of the effect of NaCl addition on the incorporation of alamethicin to a dioleoylphosphatidylcholine (DOPC) bilayer.84 The salt experiment presented here confirms that the pH change induced more alamethicin to insert into the lipid bilayer. Indeed, this change in intensity and the number of peptide molecules present was found to be fully reversible when the pH of the subphase was returned to 7. After the SFG spectra in Figure 3 were collected, about 40 μL H3PO4 (in 1.0 M stock solution) solution was injected into the subphase and SFG spectra were collected after equilibration (while the subphase pH is ~8.0). After that, an additional 20 μL H3PO4 (1.0 M) solution was injected into the subphase and SFG spectra were again collected after further equilibration (at a subphase pH ~7.0). The SFG spectra and the time-dependent SFG signal change are shown in the supporting information. With the addition of H3PO4, the alamethicin SFG amide I intensity decreased quickly. Based on this, we believe that the pH effect is not reversible. The final SFG amide I signal intensity of alamethicin after the pH returned to ~7 is lower that the initial SFG intensity, which may be due to the combination of pH and salt effects. Another possible reason may be due to the slightly altered lipid bilayer structure induced by the inserted alamethicin. As shown in Figure 1, a peak at 1710–1720 cm−1 generated by the carbonyl groups of the lipid bilayer was observed while this peak disappeared in Figure 3.
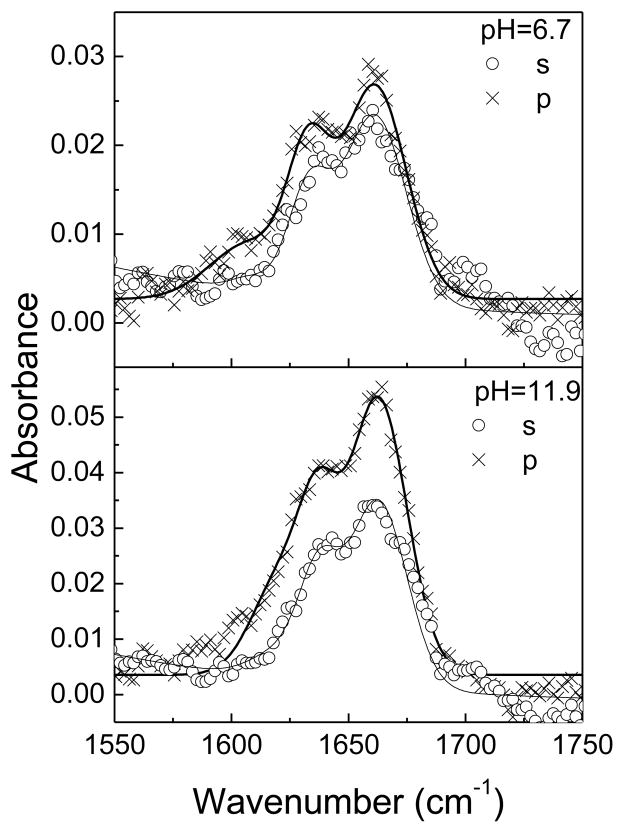
3.3 ATR-FTIR Results
ATR-FTIR was used as a supplemental technique to substantiate SFG results. Polarized ATR-FTIR spectra of alamethicin in a POPC/POPC bilayer are displayed in Fig 4. According to our previous results,85 spectra were fitted using three peaks centered at 1623 cm−1, 1635 cm−1, and 1660 cm−1. The intensity ratio (R) of the signal measured using the p- versus s- polarized beam for the 1660 cm−1 peak is 1.15 at pH=6.7 and 1.50 at pH=11.9. Assuming a δ orientation distribution, the orientation angle is determined to be θ1 = 74° at pH=6.7 and θ1 = 58° at pH=11.9. The results from these ATR-FTIR experiments are comparable to the SFG results of θ1 =72° at pH=6.7 and θ1 = 56.5° at pH=11.9 presented above. In addition, the fitted absorbance area of the 1660 cm−1 peak increased 1.6 and 2.1 times for s and p polarizations, respectively. Based on the changes in absorbance area and dichroic ratio, we can deduce that the number of molecules inserted into the lipid bilayer increased by ~2x after adjusting the pH from 6.7 to 11.9, in agreement with our SFG result.
Figure 4.
Polarized ATR-FTIR spectra of alamethicin in a POPC/POPC bilayer at pH=6.7 and 11.9.
3.4 Combined SFG and ATR-FTIR Studies
In sections 3.1 and 3.2, we assumed that all molecules adopted the same orientation (a δ distribution). This is supported by the good agreement between results from SFG and ATR-FTIR (which measure different orientational parameters). By combining these results, it is possible to characterize a Gaussian distribution directly and measure both the average orientation angle θ and the distribution width σ simultaneously. As discussed previously,86 SFG can measure <cosθ> and <cos3θ> (where “< >” means average). Here the χppp/χssp ratio measures <cosθ>/<cos3θ>. ATR-FTIR measures <cos2θ>. Table 1 lists the deduced combinations of the average angle and angle distribution from measured SFG and ATR-FTIR data when a Gaussian distribution is used. Combining SFG and ATR-FTIR measurements, at pH=6.7, θ1 =75°, σ =5°. At pH=11.9, θ1 =58.5°, σ =8°. Nevertheless, the average angles are similar to those deduced by the delta orientation distribution, and the angle distribution widths are quite narrow. We believe that the narrow orientation distribution width is due to the specific interactions between lipid bilayer and alamethicin. This is different from protein-polymer interactions, where many different interactions can be involved, leading to broad orientation distributions for proteins adsorbed onto polymers.87
Table 1.
The deduced orientation angle and angle distribution widths from SFG or ATR-FTIR measurements, when a Gaussian distribution is used.
| pH | Method | Gaussian width
|
|||
|---|---|---|---|---|---|
| σ =0 | σ =5 | σ =8 | σ =10 | ||
| 6.7 | SFG | 72 | 75 | 79 | 85 |
| 6.7 | ATR-FTIR | 74 | 75 | 77 | 78 |
| 11.9 | SFG | 56.5 | 57.5 | 58.5 | 60 |
| 11.9 | ATR-FTIR | 58 | 58.5 | 58.5 | 59 |
3.5 Further Discussion
Surface charge induced χ(3) contributions have been extensively discussed in second harmonic generation spectroscopy.88–90 We have considered the possibility that the surface charge (or surface electric field) could induce χ(3) contributions to SFG amide I signals.91 Such a method was also reported in a review article by Space and his colleagues.92 We found that for a static field of 2×108 V/m, the χ(3) contribution is at least one order of magnitude smaller than the χ(2) contribution. In the experiments presented in this work, our measured membrane potential is much smaller: only −16.2 mV surface potential difference for two different pH conditions. If we take the lipid bilayer thickness to be 5 nm, the calculated field is 3×106 V/m, which is almost two orders of magnitude smaller than the case discussed previously.91 Therefore we expect the contribution from the χ(3) term to be negligible compared to the SFG signals (χ(2) term).
Since orientation analysis depends on a known secondary structure, we next considered the possibility that the change in pH would affect protein conformation. In general, when alamethicin is denatured, a coiled-coil structure is formed,93 which shifts the peak centers of the SFG amide I signal and requires additional spectral fitting using new peak centers. Both ssp and ppp SFG spectra collected from alamethicin at pH=6.7 and pH=11.9 can be fitted using the same two amide I peaks between 1600 and 1700 cm−1. Previous studies have also indicated that alamethicin is stable above its pKa (5.2~5.8).94 Moreover, we performed unpolarized ATR-FTIR measurements and CD experiments on alamethicin in the POPC/POPC bilayer at pH 6.7 and pH 11.9 (supporting information), and no substantial secondary structural changes were observed. Together, these findings indicate that no denaturation occurred for alamethicin at pH=11.9.
Our SFG and ATR-FTIR studies both indicate that increasing the pH alters not only the orientation of alamethicin, but also the number of alamethicin molecules present in the lipid bilayer.
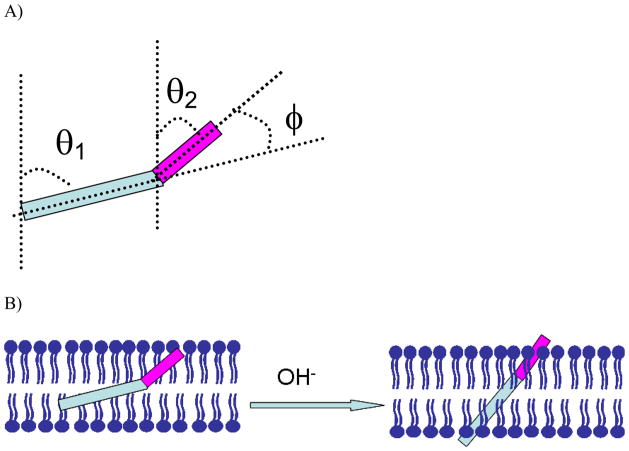
It is interesting to observe that the bend angle between the two helical components in alamethicin (θ1−θ2) is about 22° at pH=6.7 and 11.5° at pH=11.9 when we assume the plane containing both the helical components is perpendicular to the membrane surface. These results suggest that an increase in subphase pH changes not only the peptide orientation (θ1, θ2), but also the angle of the bend between helical segments (φ=θ1−θ2) (as shown in Figure 5). The change in pH can alter the membrane surface charge density and membrane dipole potential. In conjunction with possible lateral pressure effects within the lipid membrane,75–79 these changes in the membrane environment can induce alamethicin to stand up more and unbend as the pH is increased. The observed pH-dependent alamethicin action is quite similar to the models proposed by Stockner, et al.95 to explain the effect of lateral pressure upon conformational transition of an ion channel from the closed to the open state. They built these ion channel models based on bacterial homologues of the nAChR channel with different structural properties:96,97 (i) In the asymmetric tilted helix channel model, the orientation angle becomes more narrow in the open state. (ii) In the symmetric bent helix model, the bend angle between the two helical components becomes smaller in the active state. Stockner’s simulation results support both models and a change in lateral pressure led to a more pronounced effect for the bent-helix model.95 Our experimental results presented above also support both models.
Figure 5.
A). The definition of tilt angle and bend angle of alamethicin in POPC/POPC bilayer. B) A schematic to show the pH dependent channel gating action of alamethicin.
Generally, the ion channel will not completely open unless the potential difference is larger than 100 mV. The potential difference used in this study was only ~ −16.2 mV, which was not enough to open the channel completely. However, this potential difference was enough to lead to a detectable change in alamethicin orientation, which will provide molecular information on the voltage dependence of the alamethicin channels formed in membranes.
Although crystal structures are an excellent source of high-resolution structural information, proteins are far more dynamic and can interact quite differently in their native environments (membrane or otherwise) than in the crystal lattice. Although crystal structures reveal little direct information on how these proteins interact with membranes, they are certainly valuable in guiding interpretations. In this study, we successfully applied SFG-VS and ATR-FTIR to study alamethicin in lipid bilayer in situ, using the alamethicin crystal structure as a starting point to interpret our observations. We found that upon membrane potential change, alamethicin changes its membrane orientation. Even though we could not directly relate our observations to interpret gating mechanisms for larger ion channel proteins, in the future, the application of similar experimental approaches on more complex systems, e.g., larger ion channel proteins, with the help of newly developed data analysis software,98 will provide further insight into different gating mechanisms. Recently we developed a software package which can read crystal structures from the protein data bank and calculate SFG amide I signals of helical structures in the protein with different polarization combinations as a function of protein orientation (or orientations of various helical structures within the protein). In the literature, it is stated that the models for the modes of voltage sensing under debate are a transporter model, helical screw model, and paddle model. These models were proposed for complex ion channel protein systems, examples of which include the voltage-gated potassium channel protein KvAP, which contains six hydrophobic segments per subunit, S1–S6.15–17 The SFG spectra response due to these three models is different. In the transporter model, orientation of S4 helix changes; in the helical screw model, none of the helices changes orientation; while in the paddle model, substantial orientation changes of both S3 and S4 helices occur.99 When studying large ion channel proteins in the future, we will be able to use the methodology developed in this study, along with the software package mentioned above, to investigate orientation changes of helices in situ upon a change in membrane potential. This will provide direct evidence on which model best interprets the ion channel working mechanisms.
4. Conclusion
SFG has developed into a powerful tool to study peptide and protein structures at interfaces.100–115 We have applied SFG to characterize interactions between the model ion-channel peptide alamethicin and POPC lipid bilayers, in situ in real time with the presence of a membrane potential and without the need for exogenous labels. The membrane potential was controlled by altering the pH of the bilayer subphase. The orientation of alamethicin in POPC/POPC lipid bilayers was determined by deducing the orientations of the α- and 310- helical structural segments using polarized SFG amide I spectra, and substantiated by polarized ATR-FTIR studies. We found that at pH=6.7, the N-terminal α-helix tilts at about 72° versus the surface normal in POPC bilayer (θ1) and the C-terminal 310 helix (beyond the Pro14 residue) tilts at about 50° versus the surface normal (θ2). When pH increased to 11.9, the orientation angles θ1 and θ2 decreased to 56.5° and 45°, respectively, indicating a change in peptide orientation as well as in the bend between helical segments. The change in membrane potential caused by the change in pH also induced the immediate insertion of more alamethicin molecules into the membrane. Our results from SFG and ATR-FTIR were in good agreement, showing that the orientation distribution of alamethicin in the POPC bilayer is quite narrow. This work provides a basis for future elucidation of gating mechanisms of larger ion channel proteins using SFG in physiologically relevant environments in situ.
Supplementary Material
Acknowledgments
SY gratefully acknowledges the support from National Natural Science Foundation of China (21073175, 91127042), National Basic Research Program of China (2010CB923300) and the Fundamental Research Funds for the Central Universities. ZC thanks the support from USA National Institute of Health (1R01GM081655-01A2).
Supporting material: Details about ATR-FTIR, SFG, fluorescence, and CD experiments, as well as data analysis procedures, can be found in the supporting material. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kelkar DA, Chattopadhyay A. Biochim Biophys Acta. 2007;1768:2011–2025. doi: 10.1016/j.bbamem.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 2.Cooper EC, Jan LY. Proc Natl Acad Sci USA. 1999;96:4759–4766. doi: 10.1073/pnas.96.9.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jentsch TJ, Hübner CA, Fuhrmann JC. Nat Cell Biol. 2004;6:1039–1047. doi: 10.1038/ncb1104-1039. [DOI] [PubMed] [Google Scholar]
- 4.Verkman AS, Galietta LJV. Nat Rev Drug Discov. 2009;8:153–171. doi: 10.1038/nrd2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger AL, Ikuma M, Welsh MJ. Proc Natl Acad Sci USA. 2005;102:455–460. doi: 10.1073/pnas.0408575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gadsby DC, Vergani P, Csanady L. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan HL, Bink-Boelkens MTE, Bezzina CR, Viswanathan PC, Beaufort-Krol GCM, van Tintelen PJ, van den Berg MP, Wilde AAM, Balser JR. Nature. 2001;409:1043–1047. doi: 10.1038/35059090. [DOI] [PubMed] [Google Scholar]
- 8.Armstrong N, Sun Y, Chen GQ, Gouaux E. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 9.Stutts MJ, Canessa CM, Olsen JC, Hamrick M, Cohn JA, Rossier BC, Boucher RC. Science. 1995;269:847–850. doi: 10.1126/science.7543698. [DOI] [PubMed] [Google Scholar]
- 10.Mortimer JT, Bhadra N. Fundamentals of electrical stimulation. In: Krames E, Peckham PH, Rezai AR, editors. Neuromodulation. Vol. 2. 2009. p. p111. [Google Scholar]
- 11.Armstrong CM, Hille B. Neuron. 1998;20:371–380. doi: 10.1016/s0896-6273(00)80981-2. [DOI] [PubMed] [Google Scholar]
- 12.Gribkoff VK, Leonard KK, editors. Structure, Function and Modulation of Neuronal Voltage-Gated Ion Channels. John Wiley & Sons; 2009. [Google Scholar]
- 13.Lorincz A, Nusser Z. Science. 2010;328:906–909. doi: 10.1126/science.1187958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki M, Takagi M, Okamura Y. Science. 2006;312:589–592. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- 15.Doyle DA, Cabral JM, Pfuetzner RA, Kuo AL, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 16.Long SB, Campbell EB, MacKinnon R. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 17.Long SB, Tao X, Campbell EB, MacKinnon R. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 18.Wu YK, Yang Y, Ye S, Jiang YX. Nature. 2010;466:393-U148. doi: 10.1038/nature09252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swartz KJ. Nature. 2008;456:891–897. doi: 10.1038/nature07620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green DR, Reed JC. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 21.Giancotti FG, Ruoslahti E. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 22.MacKinnon R. Science. 2004;306:1304–1305. doi: 10.1126/science.1105528. [DOI] [PubMed] [Google Scholar]
- 23.Sands Z, Grottesi A, Sansom MS. Curr Biol. 2005;15:R44–47. doi: 10.1016/j.cub.2004.12.050. [DOI] [PubMed] [Google Scholar]
- 24.Vinothkumar KR, Henderson R. Quart Rev Biophys. 2010;43:65–158. doi: 10.1017/S0033583510000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sansom MSP. Prog Biophys Mol Biol. 1991;55:139–235. doi: 10.1016/0079-6107(91)90004-c. [DOI] [PubMed] [Google Scholar]
- 26.Woolley GA, Wallace BA. J Mem Biol. 1992;129:109–136. doi: 10.1007/BF00219508. [DOI] [PubMed] [Google Scholar]
- 27.Sansom MSP. Eur Biophys J. 1993;22:105–124. doi: 10.1007/BF00196915. [DOI] [PubMed] [Google Scholar]
- 28.Sansom MSP. Quart Rev Biophys. 1993;26:365–421. doi: 10.1017/s0033583500002833. [DOI] [PubMed] [Google Scholar]
- 29.Cafiso DS. Annu Rev Biophys Biomol Struct. 1994;23:141–165. doi: 10.1146/annurev.bb.23.060194.001041. [DOI] [PubMed] [Google Scholar]
- 30.Marsh D. Biochem J. 1994;23:345–361. doi: 10.1042/bj3150345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bechinger B. J Mem Biol. 1997;156:197–211. doi: 10.1007/s002329900201. [DOI] [PubMed] [Google Scholar]
- 32.Duclohier H, Wroblewski H. J Mem Biol. 2001;184:1–12. doi: 10.1007/s00232-001-0077-2. [DOI] [PubMed] [Google Scholar]
- 33.Nagaraj R, Balaram P. Acc Chem Res. 1981;14:356–362. [Google Scholar]
- 34.Kessel A, Cafiso DS, Ben-Tal N. Biophys J. 2000;78:571–583. doi: 10.1016/S0006-3495(00)76617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathew MK, Balaram P. Mol Cell Biochem. 1983;50:47–64. doi: 10.1007/BF00225279. [DOI] [PubMed] [Google Scholar]
- 36.Hall JE, Vodyanoy I, Balasubramanian TM, Marshall GR. Biophys J. 1984;45:233–247. doi: 10.1016/S0006-3495(84)84151-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathew MK, Balaram P. FEBS Lett. 1983;157:1–5. doi: 10.1016/0014-5793(80)80733-2. [DOI] [PubMed] [Google Scholar]
- 38.Leitgeb B, Szekeres A, Manczinger L, Vagvolgyi C, Kredics L. Chem BioDivers. 2007;4:1027–1051. doi: 10.1002/cbdv.200790095. [DOI] [PubMed] [Google Scholar]
- 39.Fox RO, Richards FM. Nature. 1982;300:325–330. doi: 10.1038/300325a0. [DOI] [PubMed] [Google Scholar]
- 40.North CL, Barranger-Mathys M, Cafiso DS. Biophys J. 1995;69:2392–2397. doi: 10.1016/S0006-3495(95)80108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bak M, Baywater RP, Hohwy M, Thomsen JK, Adelhorst K, Jakobsen HJ, Sorensen OW, Nielsen NC. Biophys J. 2001;81:1684–1698. doi: 10.1016/S0006-3495(01)75822-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salnikov ES, Friedrich H, Li X, Bertani P, Reissmann S, Hertweck C, O’Neil JDJ, Raap J, Bechinger B. Biophys J. 2009;96:86–100. doi: 10.1529/biophysj.108.136242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duclohier H. Eur Biophys J. 2004;33:169–174. doi: 10.1007/s00249-003-0383-y. [DOI] [PubMed] [Google Scholar]
- 44.Laver DR. Biophys J. 1994;66:355–359. doi: 10.1016/s0006-3495(94)80784-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vodyanoy I, Hall JE, Balasubramanian TM. Biophys J. 1983;42:71–82. doi: 10.1016/S0006-3495(83)84370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milov AD, Samoilova RI, Tsvetkov YD, Zotti MD, Toniolo C, Raap J. J Phys Chem B. 2008;112:13469–13472. doi: 10.1021/jp8046714. [DOI] [PubMed] [Google Scholar]
- 47.Stella L, Burattini M, Mazzuca C, Palleschi A, Venanzi M, Coin I, Peggion C, Toniolo C, Pispisa B. Chem Biodivers. 2007;4:1299–1312. doi: 10.1002/cbdv.200790111. [DOI] [PubMed] [Google Scholar]
- 48.Kessel A, Cafiso DS, Ben-Tal N. Biophys J. 2000;78:571–583. doi: 10.1016/S0006-3495(00)76617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marsh D, Jost M, Peggion C, Toniolo C. Biophys J. 2007;92:473–481. doi: 10.1529/biophysj.106.092775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marsh D, Jost M, Peggion C, Toniolo C. Biophys J. 2007;92:4002–4011. doi: 10.1529/biophysj.107.104026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marsh D. Biochemistry. 2009;48:729–737. doi: 10.1021/bi801279n. [DOI] [PubMed] [Google Scholar]
- 52.Salnikov ES, Zotti MD, Formaggio F, Li X, Toniolo C, O’Neil JDJ, Raap J, Dzuba SA, Bechinger B. J Phys Chem B. 2009;113:3034–3042. doi: 10.1021/jp8101805. [DOI] [PubMed] [Google Scholar]
- 53.Banerjee U, Zidovetzki R, Birge RR, Chan SI. Biochemistry. 1985;24:7621–7627. doi: 10.1021/bi00347a019. [DOI] [PubMed] [Google Scholar]
- 54.Ionov R, El-Abed A, Angelova A, Goldmann M, Peretti P. Biophys J. 2000;78:3026–3035. doi: 10.1016/S0006-3495(00)76841-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mottamal M, Lazaridis T. Biophys Chem. 2006;122:50–57. doi: 10.1016/j.bpc.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 56.Chen FY, Lee MT, Huang HW. Biophys J. 2002;82:908–914. doi: 10.1016/S0006-3495(02)75452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang HW, Wu Y. Biophys J. 1991;60:1079–1087. doi: 10.1016/S0006-3495(91)82144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spaar A, Munster C, Salditt T. Biophys J. 2004;87:396–407. doi: 10.1529/biophysj.104.040667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye SJ, Nguyen KT, Chen Z. J Phys Chem B. 2010;114:3334–3340. doi: 10.1021/jp911174d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miranda PB, Shen YR. J Phys Chem B. 1999;103:3292–3307. [Google Scholar]
- 61.Kim J, Somorjai GA. J Am Chem Soc. 2003;125:3150–3158. doi: 10.1021/ja028987n. [DOI] [PubMed] [Google Scholar]
- 62.Kim J, Cremer PS. ChemPhysChem. 2001;2:543–546. doi: 10.1002/1439-7641(20010917)2:8/9<543::AID-CPHC543>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 63.Li G, Ye S, Morita S, Nishida T, Osawa M. J Am Chem Soc. 2004;126:12198–12199. doi: 10.1021/ja046183x. [DOI] [PubMed] [Google Scholar]
- 64.Voges AB, Al-Abadleh HA, Musorrariti MJ, Bertin PA, Nguyen ST, Geiger FM. J Phys Chem B. 2004;108:18675–18682. [Google Scholar]
- 65.Li QF, Hua R, Chea IJ, Chou KC. J Phys Chem B. 2008;112:694–697. doi: 10.1021/jp072147j. [DOI] [PubMed] [Google Scholar]
- 66.Ye HK, Gu ZY, Gracias DH. Langmuir. 2006;22:1863–1868. doi: 10.1021/la052030r. [DOI] [PubMed] [Google Scholar]
- 67.Yatawara AK, Tiruchinapally G, Bordenyuk AN, Andreana PR, Benderskii AV. Langmuir. 2009;25:1901–1904. doi: 10.1021/la803113h. [DOI] [PubMed] [Google Scholar]
- 68.Liu J, Conboy JC. Biophys J. 2005;89:2522–2532. doi: 10.1529/biophysj.105.065672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nguyen KT, Le Clair S, Ye SJ, Chen Z. J Phys Chem B. 2009;113:12169–12180. doi: 10.1021/jp904153z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ye SJ, Nguyen KT, Le Clair S, Chen Z. J Struct Biol. 2009;168:61–77. doi: 10.1016/j.jsb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen X, Wang J, Boughton AP, Kristalyn CB, Chen Z. J Am Chem Soc. 2007;129:1420–1427. doi: 10.1021/ja067446l. [DOI] [PubMed] [Google Scholar]
- 72.Zheng C, Vanderkooi G. Biophys J. 1992;63:935–941. doi: 10.1016/S0006-3495(92)81673-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cladera J, O’Shea P. Biophys J. 1998;74:2434–2442. doi: 10.1016/S0006-3495(98)77951-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luchian T, Mereuta L. Langmuir. 2006;22:8452–8457. doi: 10.1021/la0613777. [DOI] [PubMed] [Google Scholar]
- 75.Chiriac R, Luchian T. Biophys Chem. 2007;130:139–147. doi: 10.1016/j.bpc.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 76.Zhou Y, Raphael RM. Biophys J. 2007;92:2451–2462. doi: 10.1529/biophysj.106.096362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Capone S, Walde P, Seebach D, Ishikawa T, Caputo R. Chem Bio Divers. 2008;5:16–30. doi: 10.1002/cbdv.200890006. [DOI] [PubMed] [Google Scholar]
- 78.Garcia-Manyes S, Oncins G, Sanz F. Electrochimica Acta. 2006;51:5029–5036. [Google Scholar]
- 79.Boggara MB, Faraone A, Krishnamoorti R. J Phys Chem B. 2010;114:8061–8066. doi: 10.1021/jp100494n. [DOI] [PubMed] [Google Scholar]
- 80.Keller F, Hanke W, Trissl D, Bakkergrunwald T. Biochim Biophys Acta. 1989;982:89–93. doi: 10.1016/0005-2736(89)90178-8. [DOI] [PubMed] [Google Scholar]
- 81.Shapiro HM. Methods. 2000;21:271–279. doi: 10.1006/meth.2000.1007. [DOI] [PubMed] [Google Scholar]
- 82.Cohen LB, Salzberg BM. Rev Physiol Biochem Pharmacol. 1978;83:35–88. doi: 10.1007/3-540-08907-1_2. [DOI] [PubMed] [Google Scholar]
- 83.Plfigek J, Sigler K. J Photochem Photobio B. 1996;33:101–124. doi: 10.1016/1011-1344(96)07283-1. [DOI] [PubMed] [Google Scholar]
- 84.Stankowski S, Schwarz UD, Schwarz G. Biochim Biophys Acta. 1988;941:11–18. doi: 10.1016/0005-2736(88)90208-8. [DOI] [PubMed] [Google Scholar]
- 85.Haris PI, Chapman D. Biochim Biophys Acta. 1988;943:375–380. doi: 10.1016/0005-2736(88)90571-8. [DOI] [PubMed] [Google Scholar]
- 86.Wang J, Paszti Z, Clarke ML, Chen X, Chen Z. J Phys Chem B. 2007;111:6088–6095. doi: 10.1021/jp070383o. [DOI] [PubMed] [Google Scholar]
- 87.Wang J, Lee S-H, Chen Z. J Phys Chem B. 2008;112:2281–2290. doi: 10.1021/jp077556u. [DOI] [PubMed] [Google Scholar]
- 88.Zhao XL, Ong S, Eisenthal KB. Chem Phys Lett. 1993;202:513–520. [Google Scholar]
- 89.Ong S, Zhao XL, Eisenthal KB. Chem Phys Lett. 1992;191:327–335. [Google Scholar]
- 90.Geiger FM. Annu Rev Phys Chem. 2009;60:61–83. doi: 10.1146/annurev.physchem.59.032607.093651. [DOI] [PubMed] [Google Scholar]
- 91.Chen X, Wang J, Paszti Z, Wang F, Schrauben JN, Tarabara VV, Schmaier AH, Chen Z. Anal Bioanal Chem. 2007;388:65–72. doi: 10.1007/s00216-006-0999-8. [DOI] [PubMed] [Google Scholar]
- 92.Perry A, Neipert C, Space B, Moore PB. Chem Rev. 2006;106:1234–1258. doi: 10.1021/cr040379y. [DOI] [PubMed] [Google Scholar]
- 93.Woolley GA, Wallace BA. Biochemistry. 1993;32:9819–9825. doi: 10.1021/bi00088a037. [DOI] [PubMed] [Google Scholar]
- 94.Payne JW, Jakes R, Hartley BS. Biochem J. 1970;117:757–766. doi: 10.1042/bj1170757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jerabek H, Pabst G, Rappolt M, Stockner T. J Am Chem Soc. 2010;132:7990–7997. doi: 10.1021/ja910843d. [DOI] [PubMed] [Google Scholar]
- 96.Hilf RJC, Dutzler R. Nature. 2009;457:115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 97.Hilf RJC, Dutzler R. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 98.Boughton AP, Yang P, Tesmer VM, Ding B, Tesmer JJ, Chen Z. Proc Natl Acad Sci USA. 2011;108:E667–E673. doi: 10.1073/pnas.1108236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tombola F, Pathak MM, Isacoff EY. Annu Rev Cell Dev Biol. 2006;22:23–52. doi: 10.1146/annurev.cellbio.21.020404.145837. [DOI] [PubMed] [Google Scholar]
- 100.Mermut O, Phillips DC, York RL, McCrea KR, Ward RS, Somorjai GA. J Am Chem Soc. 2006;128:3598–3607. doi: 10.1021/ja056031h. [DOI] [PubMed] [Google Scholar]
- 101.Phillips DC, York RL, Mermut O, McCrea KR, Ward RS, Somorjai GA. J Phys Chem C. 2007;111:255–261. [Google Scholar]
- 102.Jung S, Lim YSM, Albertorio F, Kim G, Gurau MC, Yang RD, Holden MA, Cremer PS. J Am Chem Soc. 2003;125:12782–12786. doi: 10.1021/ja037263o. [DOI] [PubMed] [Google Scholar]
- 103.Kim G, Gurau M, Kim J, Cremer PS. Langmuir. 2002;18:2807–2811. [Google Scholar]
- 104.Kim G, Gurau MC, Lim S-M, Cremer PS. J Phys Chem B. 2003;107:1403–1409. [Google Scholar]
- 105.Evans-Nguyen KM, Fuierer RR, Fitchett BD, Tolles LR, Conboy JC, Schoenfisch MH. Langmuir. 2006;22:5115–5121. doi: 10.1021/la053070y. [DOI] [PubMed] [Google Scholar]
- 106.Weidner T, Apte JS, Gamble LJ, Castner DG. Langmuir. 2009;26:3433–3440. doi: 10.1021/la903267x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weidner T, Samuel NT, McCrea K, Gamble LJ, Ward RS, Castner DG. Biointerphases. 2010;5:9–16. doi: 10.1116/1.3317116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Weidner T, Breen NF, Li K, Drohny GP, Castner DG. Proc Natl Acad Sci U S A. 2010;107:13288–13293. doi: 10.1073/pnas.1003832107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fu L, Ma G, Yan EC. J Am Chem Soc. 2010;132:5405–5412. doi: 10.1021/ja909546b. [DOI] [PubMed] [Google Scholar]
- 110.Chen X, Wang J, Sniadecki JJ, Even MA, Chen Z. Langmuir. 2005;21:2662. doi: 10.1021/la050048w. [DOI] [PubMed] [Google Scholar]
- 111.Clarke ML, Wang J, Chen Z. J Phys Chem B. 2005;109:22027–22035. doi: 10.1021/jp054456k. [DOI] [PubMed] [Google Scholar]
- 112.Chen X, Chen Z. Biochem Biophys Acta. 2006;1758:1257–1273. doi: 10.1016/j.bbamem.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 113.Doyle AW, Fick J, Himmelhaus M, Eck W, Graziani I, Prudovsky I, Grunze M, Maciag T, Neivandt DJ. Langmuir. 2004;20:8961–8965. doi: 10.1021/la0484220. [DOI] [PubMed] [Google Scholar]
- 114.Nguyen KT, Soong R, lm S, Waskell L, Ramamoorthy A, Chen Z. J Am Chem Soc. 2010;132:15112–15115. doi: 10.1021/ja106508f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nguyen KT, King JT, Chen Z. J Phys Chem B. 2010;114:8291–8300. doi: 10.1021/jp102343h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.