Abstract
Hypoxia-inducible factor (HIF) is a transcription factor that regulates fundamental cellular processes in response to changes in oxygen concentration. HIFα protein levels are increased in most solid tumours and correlate with patient prognosis. The link between HIF and apoptosis, a major determinant of cancer progression and treatment outcome, is poorly understood. Here we show that Caenorhabditis elegans HIF-1 protects against DNA-damage-induced germ cell apoptosis by antagonizing the function of CEP-1, the homologue of the tumour suppressor p53. The antiapoptotic property of HIF-1 is mediated by means of transcriptional upregulation of the tyrosinase family member TYR-2 in the ASJ sensory neurons. TYR-2 is secreted by ASJ sensory neurons to antagonize CEP-1-dependent germline apoptosis. Knock down of the TYR-2 homologue TRP2 (also called DCT) in human melanoma cells similarly increases apoptosis, indicating an evolutionarily conserved function. Our findings identify a novel link between hypoxia and programmed cell death, and provide a paradigm for HIF-1 dictating apoptotic cell fate at a distance.
Hypoxia-inducible factor (HIF) mediates cellular and systemic adaptive responses to maintain oxygen homeostasis in all metazoan species. HIF heterodimer consists of an oxygen-dependent α-subunit and a constitutively expressed β-subunit. Under normal oxygen levels, prolyl hydroxylases (PHD1–PHD3) use oxygen as a substrate to modify proline residues of HIFα1–3. Hydroxylated HIFα is recognized by the von Hippel–Lindau tumour suppressor (VHL), a subunit of the VCB–Cul2 ubiquitin-ligase, and targeted for proteasomal degradation4. Inactivation of VHL results in accumulation of HIFα protein and is the underlying basis of von Hippel–Lindau disease5, a multisystem cancer syndrome. HIFα levels are also increased as a consequence of intratumoral hypoxia as well as genetic alterations in a variety of cancer types6,7. Increased HIFα levels have been shown to be associated with increased mortality and treatment failure in many solid tumours8. However, the molecular mechanisms underlying this correlation are still poorly understood.
A major determinant of tumour progression and cancer therapy is the ability of cancer cells to activate apoptotic cell death9. Understanding how aberrant signalling within tumours can interfere with apoptosis is therefore of particular importance.
The HIF and the apoptotic pathway are both evolutionarily highly conserved and are well characterized in the nematode C. elegans. The C. elegans hif-1, aha-1 and vhl-1 genes encode the single worm homologues of HIFα, HIFβ and VHL, whereas EGL-9 is the single HIFα prolyl hydroxylase1,10. Furthermore, germline apoptosis induced by DNA damage involves a conserved machinery of checkpoint proteins, the p53 homologue CEP-1, and the core apoptotic machinery consisting of CED-9 (Bcl-2), CED-4 (Apaf1) and CED-3 (caspase)11–13. Here we use C. elegans to analyse the link between HIF-1 and apoptosis.
HIF-1 inhibits ionizing-radiation-induced apoptosis
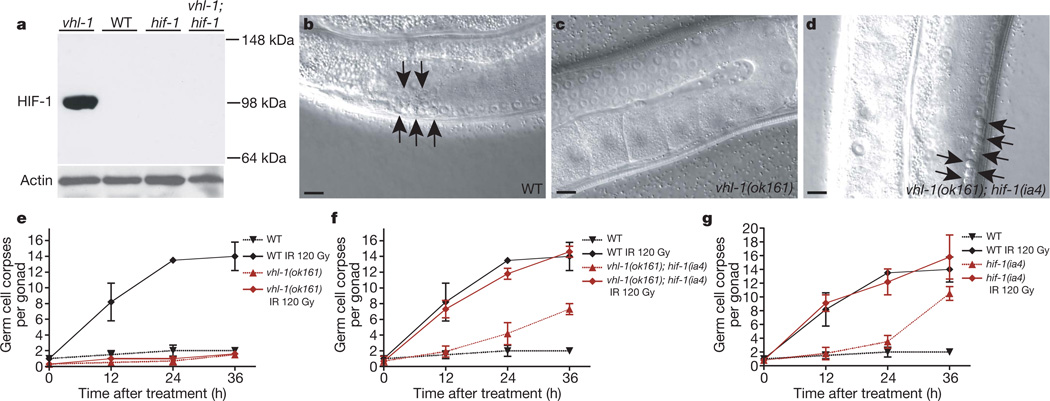
To determine whether HIF-1 alters DNA-damage-induced apoptosis, we assessed germ lines of wild-type and vhl-1(ok161) mutant animals. Loss of the negative regulator vhl-1 led to a marked increase in HIF-1 levels (Fig. 1a), as previously shown1. Although ionizing radiation (IR) and ultraviolet C (254 nm) induced an increase in the number of apoptotic germ cells in wild type, no such increase was seen in vhl-1(ok161) mutant animals (Fig. 1b, c, e and Supplementary Fig. 2a). Two lines of evidence indicate that the apoptotic defect in vhl-1(ok161) mutants is due to stabilized HIF-1, rather than to an alternative function of vhl-1. First, increasing HIF-1 levels via either hypoxia treatment or loss of egl-9 function also conferred resistance to IR (Supplementary Fig. 2b, d). Second, loss of HIF-1 function restored the sensitivity to IR in vhl-1(ok161) and egl-9(n586) mutant worms (Fig. 1d, f, g and Supplementary Fig. 2c). Taken together, these results indicate that HIF-1 antagonizes DNA-damage-induced apoptosis.
Figure 1. HIF-1 antagonizes DNA-damage-induced apoptosis.
a, HIF-1 western blot analysis of synchronized young adult hermaphrodites. All alleles used in this study are defined in Methods. b–d, Synchronized young adult hermaphrodites were exposed to ionizing radiation (IR) and germline apoptosis was analysed by DIC microscopy 12 h after treatment. Arrows indicate germ cell corpses. Scale bars, 10 µm. e–g, Quantification of germline apoptosis. Synchronized young adult hermaphrodites were exposed to IR and germline apoptosis was quantified at the indicated time points. Data shown represent the average of three to six independent experiments ± s.d. (n > 20 animals for each experiment and time point).
HIF-1 could antagonize apoptosis either by modulating the central apoptotic machinery or by interfering with the upstream signalling pathways that activate the apoptotic machinery in response to DNA damage. To distinguish between these two possibilities, we asked whether HIF-1 also affects other outputs of the DNA damage response pathway that are independent of apoptosis. We first monitored cell cycle arrest after IR in the mitotic germline compartment. Cell cycle arrest and apoptosis are induced via a common, conserved signalling cascade that branches into two distinct pathways upstream of CEP-1 (ref. 12; Supplementary Fig. 1). vhl-1(ok161) worms showed a normal cell cycle arrest upon IR, as assessed by the decrease in the number of proliferating cells in the stem cell compartment (Supplementary Fig. 2e). Moreover, embryonic lethality—an indirect measure of failures in DNA repair—was not affected by HIF-1, as we observed no significant change in vhl-1(ok161) or hif-1(ia4) mutants compared to wild-type worms after IR (Supplementary Table 1). Collectively, these findings indicate that the upstream DNA damage response pathway is fully functional in vhl-1(ok161) mutants. Therefore, HIF-1 must act either directly on the apoptotic machinery, or on the apoptosis-specific branch of the signalling cascade (Supplementary Fig. 1).
HIF-1 antagonizes CEP-1/p53 function
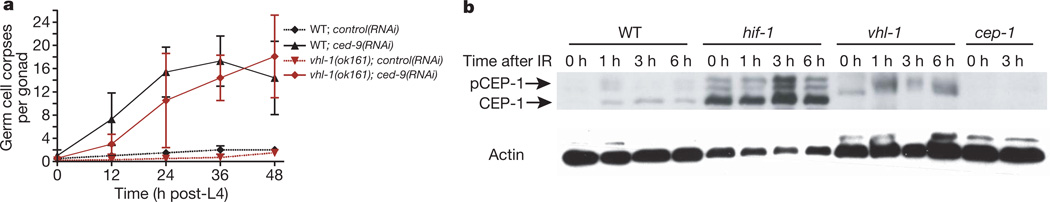
To refine the site of action of HIF-1 further, we next asked whether HIF-1 directly influences the core apoptotic machinery. In addition to mediating DNA-damage-induced apoptosis, CED-3, CED-4 and CED-9 participate in physiological germ cell death, a cep-1/p53-independent program that eliminates approximately 50% of all germ cells in the absence of exogenous stress11. Physiological germ cell death was still readily observed in vhl-1(ok161) and egl-9(n586) mutants, both of which overexpress HIF-1 (ref. 1) (Fig. 1a, e and Supplementary Fig. 2b), suggesting that the core machinery is fully functional in these mutants. Similarly, whereas mutations in ced-3 and ced-4 efficiently suppress ced-9(lf)-dependent cell death11, germ cell apoptosis triggered by RNA interference (RNAi)-mediated depletion of the Bcl-2 homologue CED-9 was not blocked in vhl-1(ok161) animals (Fig. 2a). Taken together, these results suggest that HIF-1 antagonizes apoptosis at the level of either CEP-1/p53 or the BH3-only domain protein EGL-1 (Supplementary Fig. 1) or that it acts in parallel to this pathway.
Figure 2. HIF-1 impedes DNA-damage-induced apoptosis by inhibiting CEP-1.
a, Wild-type or vhl-1(ok161) worms were grown on bacteria containing either an empty control RNAi vector or ced-9(RNAi). Germline apoptosis was quantified at the indicated time points beginning at the fourth larval (L4) stage. Data shown represent the average of three independent experiments ± s.d. (n > 20). b, CEP-1 western blot analysis of synchronized young adult animals. pCEP-1, phosphorylated CEP-1. β-actin was used as loading control.
In mammals, p53 is regulated via protein stability and post-translational modifications14. It was recently shown that C. elegans CEP-1 is also phosphorylated upon IR15. To determine whether HIF-1 alters CEP-1 activation, we compared CEP-1 levels by western blot analysis in IR-treated wild-type, hif-1(ia4) and vhl-1(ok161) worms. IR treatment of wild-type animals resulted in the appearance of two additional bands, representing phosphorylated CEP-1 (Fig. 2b)15. hif-1(ia4) mutants showed two very distinctive phenotypes. First, CEP-1 protein levels were much higher than in wild-type animals (Fig. 2b and Supplementary Figs 3 and 4a–c). Second, phosphorylated CEP-1 was readily detectable even without IR treatment (Fig. 2b). In vhl-1(ok161) mutants, which overexpress HIF-1, we observed a novel CEP-1 migration pattern (Fig. 2b) that is distinct from the wild-type and phosphorylated species reported previously15 and possibly represents a different type of CEP-1 modification. IR treatment of vhl-1(ok161) mutants resulted in a further shift in CEP-1 mobility (Fig. 2b). The strong changes in CEP-1 observed in vhl-1(ok161) and hif-1(ia4) mutants suggest that HIF-1 controls, directly or indirectly, CEP-1 activation.
HIF-1 inhibits apoptosis via TYR-2
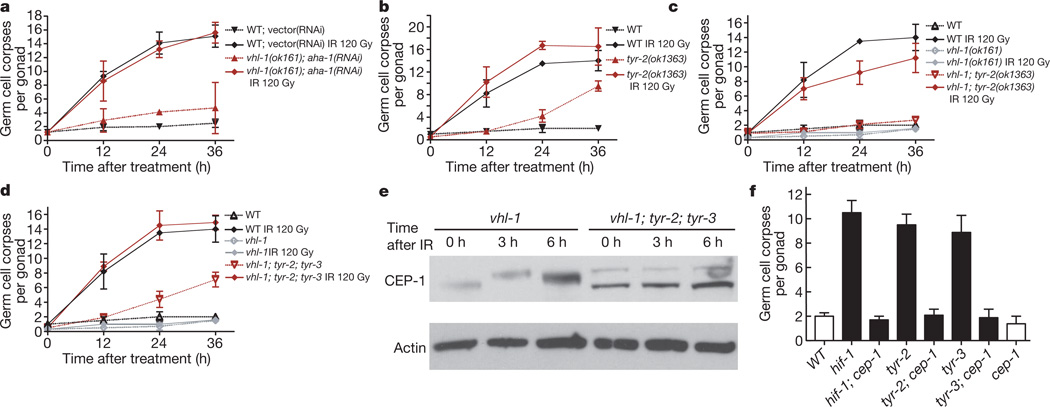
How could HIF-1 inhibit CEP-1? First, being a transcription factor, HIF-1 might promote the expression of one or more antiapoptotic genes that antagonize CEP-1. Alternatively, HIF-1 might directly interact with and regulate CEP-1. To discriminate between these two scenarios, we stabilized HIF-1 by means of the vhl-1(ok161) mutation and simultaneously knocked down the C. elegans HIFβ homologue aha-1 by RNAi to repress HIF-1 transcriptional activity (Supplementary Figs 2h and 5). We found that HIF-1 could not inhibit apoptosis in the absence of AHA-1: aha-1(RNAi); vhl-1(ok161) animals were as sensitive to IR as aha-1(RNAi) or wild-type control animals (Fig. 3a and Supplementary Fig. 4d). We thus conclude that HIF-1 antagonizes CEP-1 activation via transcriptional upregulation of one or more antiapoptotic target genes.
Figure 3. The antiapoptotic function of HIF-1 is mediated by a concerted action of TYR-2 and TYR-3.
a, Wild-type or vhl-1(ok161) worms were grown on bacteria containing either an empty control RNAi vector or the HIF-1β homologue aha-1(RNAi) beginning at the third larval stage (L3). Synchronized young adult animals were irradiated and germline apoptosis was quantified at the indicated time points. Data shown represent the average of three independent experiments ± s.d. (n > 20 per time point and experiment). b–d, Time-course analysis of germ cell apoptosis in synchronized control or irradiated young adult animals. Data shown represent the average of three to six independent experiments ± s.d. (n > 20). e, Western blot analysis of CEP-1 in synchronized young adult worms. β-actin was used as loading control. f, Analysis of germ cell apoptosis in unirradiated synchronized animals 48 h after L4 stage. Data shown represent the average of three independent experiments ± s.d. (n > 20).
Previous microarray studies of vhl-1(ok161) mutants and hypoxia-treated animals led to the identification of putative transcriptional targets of HIF-1 (refs. 16, 17). To determine whether any one of these targets might mediate the antiapoptotic activity of HIF-1, we tested whether RNAi depletion of any of these genes restores IR-induced apoptosis in vhl-1(ok161) mutants. Out of this set, only one gene, C. elegans tyrosinase tyr-2, was able to restore sensitivity to IR (Supplementary Fig. 6). To confirm the specificity of our tyr-2 RNAi depletion, we next analysed worms carrying the tyr-2(ok1363) deletion allele (Supplementary Fig. 7a–c). Whereas the tyr-2(ok1363) mutation did not alter IR response on its own (Fig. 3b), it could restore the IR response defect of vhl-1(ok161) animals close to wild-type levels (68–85%, Fig. 3c). We confirmed that tyr-2 expression is regulated by HIF-1 using quantitative RT–PCR: relative tyr-2 messenger RNA levels in vhl-1(ok161), wild-type and hif-1(ia4) worms were 1.74 ± 0.47 (s.d.), 1 and 0.30 ± 0.45, respectively. Our results indicate that HIF-1 antagonizes DNA-damage-induced apoptosis to a large part via transcriptional upregulation of the tyrosinase family member TYR-2.
TYR-2 is one of four annotated tyrosinase family members in the C. elegans genome. Given that loss of tyr-2 did not fully restore the apoptotic defect in vhl-1(ok161) mutants, we asked whether one of the other three tyrosinases might act in a partially redundant manner with TYR-2. We therefore used RNAi to deplete tyr-1, tyr-3 and tyr-4 in vhl-1(ok161) or vhl-1(ok161); tyr-2(ok1363) worms and analysed IR-induced apoptosis. Intriguingly, tyr-3(RNAi) led to an increase in apoptosis in both genetic backgrounds (Supplementary Fig. 8g, h). Consistent with the RNAi results, apoptosis was partially restored in tyr-3(ok1194); vhl-1(ok161) double mutants (Supplementary Fig. 4e, f), and essentially back to wild-type levels in the tyr-3(ok1194); tyr-2(ok1363); vhl-1(ok161) triple mutants (Fig. 3d). Deficiency in TYR-2 and TYR-3 also caused an increase in CEP-1 levels after IR (Fig. 3e and Supplementary Fig. 4b). Taken together, these findings lead us to propose a model in which HIF-1 acts via TYR-2 and TYR-3 to downregulate CEP-1 levels, thereby impeding apoptosis after DNA damage.
tyr-2(ok1363), tyr-3(ok1194) as well as hif-1(ia4) mutants showed increased basal levels of apoptosis in the absence of IR, most evidently at the 36 h after young adult stage (Figs 1g and 3b and Supplementary Fig. 4e). Because we observed increased CEP-1 levels in the absence of HIF-1 (Fig. 2b), we asked whether these elevated basal levels of germ cell apoptosis are dependent on CEP-1 function. Indeed, CEP-1 deficiency suppressed the increase in apoptosis in all three mutant backgrounds (Fig. 3f and Supplementary Fig. 2i). Thus, lack of TYR-2 and TYR-3 results in overactivation of CEP-1 and excessive germ cell death.
TYR-2 is expressed in ASJ neurons
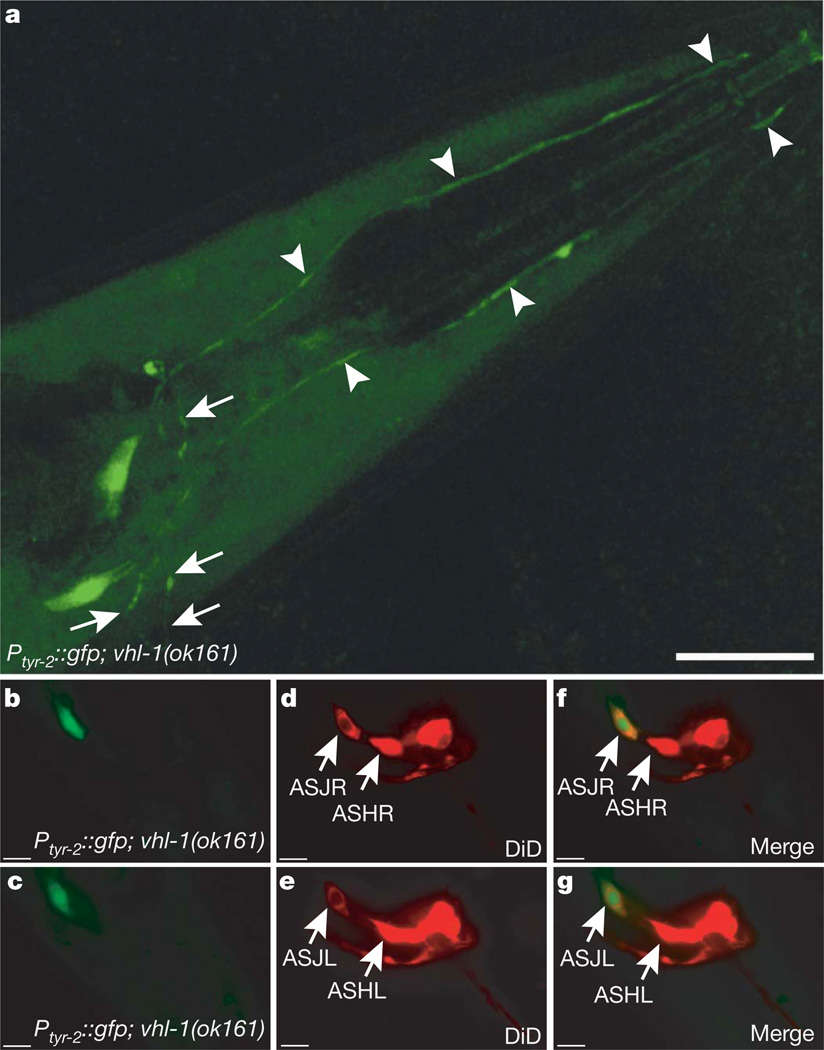
Because the antiapoptotic effect of HIF-1 is, to a large extent, mediated by TYR-2, we focused on the tyr-2 gene and asked how TYR-2 induction might inhibit CEP-1-dependent apoptosis. As a first step, we analysed the tyr-2 expression pattern using two transcriptional GFP reporter lines. Under normoxic conditions, tyr-2 was expressed in the uterine muscles and in the hypodermis (Supplementary Fig. 8e, f). However, HIF-1 stabilization by means of the vhl-1(ok161) mutation led to additional expression in two neurons in the head (Fig. 4a and Supplementary Fig. 8a–d). On the basis of their position and their ability to take up the dye 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate (DiD)18, we identified these two cells as the ASJL and ASJR amphid sensory neurons (Fig. 4b–g).
Figure 4. HIF-1 drives tyr-2 expression in ASJ neurons.
a, Transcriptional tyr-2 GFP-reporter opIs216[Ptyr-2::gfp] in a vhl-1(ok161) mutant background reveals HIF-1-dependent expression in two neurons in the head. Arrowheads indicate the dendritic extensions to the anterior end of the worm. Arrows indicate axons that are associated with the nerve ring. Scale bar, 20 µm. b–g, DiD staining in opIs216[Ptyr-2::gfp]; vhl-1(ok161) reveals that the GFP signal co-localizes with the most ventro-posterior amphid neurons, the ASJL and ASJR sensory neurons. Scale bars, 10 µm.
Because apoptosis was induced in the germ line rather than in these neurons, we investigated the possibility that HIF-1 acted cell non-autonomously. HIF-1 is ubiquitously expressed, but is not detectable in an adult animal under normoxia due to its rapid proteasome-dependent degradation. In vhl-1(ok161) mutants, stabilized HIF-1::GFP could be observed ubiquitously in the somatic tissues (Supplementary Figs 9c and 11g). However, HIF-1::GFP was not detectable in mitotic to pachytene zones of the germ line (Supplementary Fig. 9d–i) and was only present in mature, diakinesis stage oocytes, in the somatic gonadal sheath cells and in the distal tip cell (Supplementary Fig. 9f–i). The failure to detect HIF-1 in the cells undergoing apoptosis (Supplementary Figs 9–11) suggests that HIF-1 might regulate IR-induced germ cell apoptosis cell non-autonomously. Given the HIF-1-dependent expression of TYR-2 in the ASJ neurons, we hypothesized that these two neurons might be required for the inhibitory effect of HIF-1 on apoptosis.
To test this prediction, we specifically ablated the ASJ neurons in vhl-1(ok161) animals using laser microbeam surgery and tested whether this affected the rate of apoptosis after IR. The results unambiguously showed that the inhibitory effect of HIF-1 was eliminated by ablating both ASJ sensory neurons, but not by mock ablation (Fig. 5a). Moreover, ablating either ASJ neuron sensitized animals to IR and resulted in apoptotic levels intermediate between the vhl-1(ok161) and vhl-1(ok161); tyr-2(ok1363) animals (Fig. 5a). To confirm that the consequences of ASJ ablation were specific to the effect of HIF-1, rather than generally sensitizing worms to DNA-damage-induced apoptosis, we similarly ablated ASJ neurons in wild-type worms and did not find any significant increase in apoptosis (Supplementary Fig. 12a). Therefore, our results indicate that stabilized HIF-1 in ASJ sensory neurons results in TYR-2 expression, which in turn antagonizes DNA-damage-induced apoptosis in the germ line.
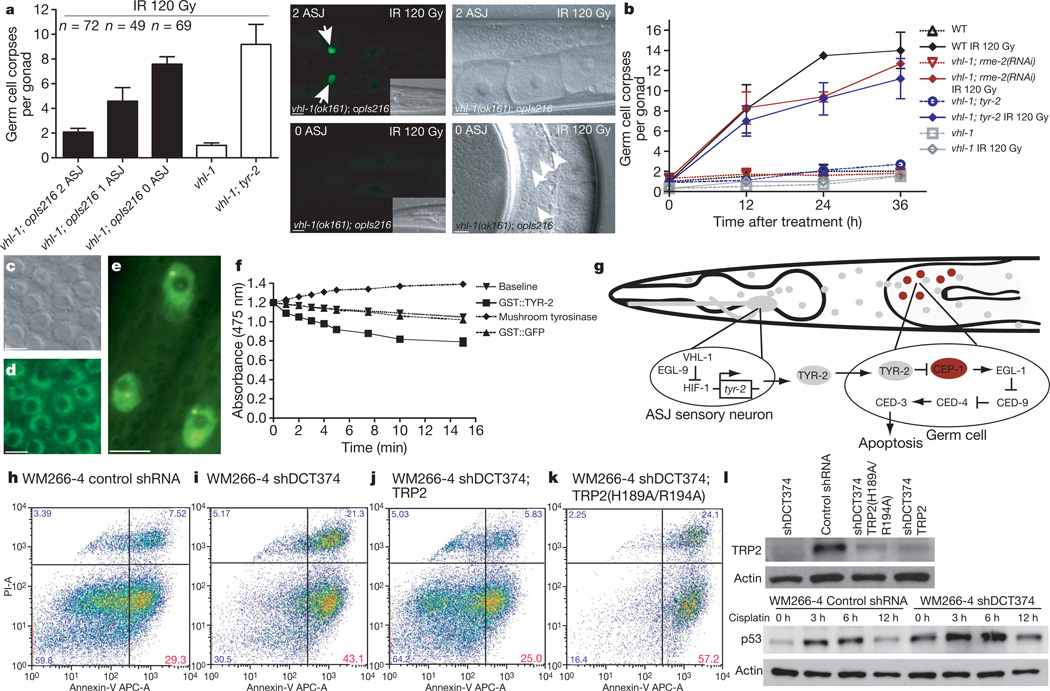
Figure 5. TYR-2 is a l-dopachrome tautomerase secreted by the ASJ neurons to inhibit apoptosis.
a, Laser microbeam ablation of ASJ neurons. Germline apoptosis of laser-ablated and mock-ablated animals was quantified 24 h after irradiation and the presence of GFP-positive ASJ neurons was determined subsequently. The number of intact ASJ neurons is indicated. Representative head and appendant germ lines are shown on the right; mock-ablated animals in the top row, ASJ-ablated animals are shown in the bottom row. Arrows indicate ASJ neurons; arrowheads apoptotic germ cells. Scale bars, 10 µm. b, Germline endocytosis was inhibited by rme-2(RNAi) and germ cell apoptosis quantified. Data shown represent the average of three independent experiments ± s.d. (n > 20 animals). c, d, DIC microscopy section and TYR-2::GFP nuclear localization in the germ line of Phus-1::tyr-2-SP::gfp::tyr-2(3′ region) animals, which express TYR-2 lacking the signal peptide (amino acids 1–22). Scale bars, 5 µm. e, Nuclear localization of TYR-2::GFP in Ptyr-2::tyr-2-SP::gfp::tyr-2(3′ region) animals. Hypodermal nuclei are depicted. Scale bar, 5 µm. f, l-dopachrome tautomerase activity of GST::TYR-2, GST::GFP and mushroom tyrosinase. g, Model for HIF-1-mediated inhibition of germline apoptosis. h–k, Loss of human tyrosinase-related protein 2 (TRP2; also called DCT) results in increased cisplatin-induced apoptosis in WM266-4 metastatic melanoma cells. Melanoma cells were treated with 50 µM cisplatin, stained with Annexin-V/propidium iodide (PI) 24 h after treatment and analysed by flow cytometry. TRP2 knock down (shDCT374) increases Annexin-V positive/PI negative fractions (early apoptotic cells) compared to control shRNA (Luciferase) in WM266-4 melanoma cells (h, i). The shDCT374 (targeting endogenous TRP2 5′ UTR) phenotype is rescued by expression of wild-type TRP2 (MSCV-DCT) (j), but not of the catalytically inactive point mutant TRP2(H189A/R194A) (MSCV-DCT)(k). Data shown are representative of four experiments. APC-A, allophycocyanin conjugate-Annexin-V. l, TRP2 and p53 western blot analysis in WM266-4 melanoma cells.
Our model predicts that constitutive expression of TYR-2 in ASJ neurons would inhibit apoptosis in germ cells. To test this prediction, we overexpressed TYR-2 in the ASJ neurons. As the model predicted, we found that expression of TYR-2, via the ASJ-specific trx-1 promoter, was sufficient to suppress the increased basal apoptotic levels arising from TYR-2 deficiency and inhibited DNA-damage-induced apoptosis almost completely (Table 1 and Supplementary Table 2).
Table 1.
I TYR-2 antagonizes DNA damage-induced apoptosis cell non-autonomously
| Genotype | Promoter | Control | IR 120 Gy | ||
|---|---|---|---|---|---|
| 24 h | 36 h | 24 h | 36 h | ||
| Wild type | – | 1.8 ± 0.7 | 1.8 ± 0.3 | 13.5 ± 0.4 | 14.0 ± 1.8 |
| vhl-1(ok161) | – | 0.7 ± 0.5 | 1.5 ± 0.2 | 1.0 ± 0.2 | 1.6 ± 0.3 |
| tyr-2(ok1363) | – | 4.2 ± 1.1 | 9.5 ± 0.9 | 16.7 ± 0.7 | 16.5 ± 3.3 |
| Ptrx-1::tyr-2::gfp; tyr-2(ok1363) | ASJ neuron | 2.4 ± 0.4 | 2.3 ± 0.2 | 4.3 ± 0.6 | 3.4 ± 0.1 |
| Ptyr-2::tyr-2::gfp; tyr-2(ok1363) | Endogenous | 2.0 ± 0.4 | 2.5 ± 0.6 | 3.1 ± 0.8 | 3.4 ± 0.9 |
| Ptyr-2::tyr-2-SP::gfp; tyr-2(ok1363) | Endogenous | 5.7 ± 1.0 | 5.5 ± 0.2 | 13.3 ± 0.9 | 14.2 ± 1.6 |
| Phus-1::tyr-2-SP::gfp; tyr-2(ok1363) | Germ line | 1.3 ± 0.6 | 1.5 ± 0.1 | 1.8 ± 0.4 | 1.9 ± 0.1 |
| Phus-1::tyr-2-SP::gfp; tyr-2(ok1363); rme-2(RNAi) | Germ line | 1.5 ± 0.4 | 1.6 ± 0.4 | 2.4 ± 0.1 | 2.5 ± 0.3 |
Quantification of germline apoptosis in transgenic animals after IR. Data shown represent the average of two to four independent experiments ±s.d. (n > 20 animals for each experiment and time point). SP indicates that the signal peptide (amino acids 1–22) has been deleted from the tyr-2 genomic region.
Secreted TYR-2 blocks germline apoptosis
How could HIF-1-dependent TYR-2 expressed in ASJ sensory neurons regulate apoptosis in the germ line? Tyrosinase family members possess a signal peptide that targets the protein either to melanosomes19 or the extracellular environment20. Analysis of the TYR-2 protein sequence revealed that its first 22 amino acids also form a putative signal peptide (Supplementary Figs 12b and 13a–d). This finding raised the possibility that HIF-1-induced TYR-2 could be secreted from the ASJ sensory neurons into the pseudocoelomic fluid to impede germ cell apoptosis.
We reasoned that TYR-2 could act at a distance in two ways. First, TYR-2 could be taken up by endocytosis and subsequently act in germ cells directly. Alternatively, a small molecule, for example, a melanin-related metabolite, produced by TYR-2 in the pseudocoelomic fluid could diffuse and act in germ cells. To distinguish between these possibilities, we knocked down rme-2, a gene required for endocytosis in the gonad21, and asked whether blockage of gonadal endocytosis restores germline apoptosis in vhl-1(ok161) mutants upon IR. We found that deficiency in RME-2 completely restored DNA-damage-induced apoptosis in vhl-1(ok161) mutants (Fig. 5b), but neither sensitized wild-type worms to IR nor resulted in an engulfment phenotype (Supplementary Fig. 14a). These findings are consistent with the notion that TYR-2 may be secreted and may enter the gonad by endocytosis.
If this model is correct, ectopic expression of TYR-2 in the germ line should be sufficient to prevent apoptosis caused by IR. We therefore expressed modified TYR-2 protein that lacks the signal peptide in the germ line (TYR-2-SP). TYR-2 expression as a result of this construct rescued tyr-2(ok1363) mutants and completely prevented DNA-damage-induced apoptosis, asserting again that TYR-2 acts in the germ line to antagonize apoptosis (Table 1 and Fig. 5c, d). In contrast, TYR-2-SP expressed under the control of the endogenous tyr-2 promoter did not protect against IR-induced apoptosis (Table 1, Fig. 5e and Supplementary Fig. 13a–d). To explore the possibility that other proteins in addition to TYR-2 could be taken up by endocytosis to assist TYR-2, we blocked endocytosis by rme-2(RNAi) in worms expressing TYR-2-SP in the germ line. We found that these worms were still completely defective in DNA-damage-induced apoptosis (Table 1). Collectively, these experiments indicate that TYR-2 within germ cells is necessary and sufficient to inhibit DNA-damage-induced apoptosis.
In humans, the tyrosinase family consists of tyrosinase, which converts l-tyrosine into l-DOPA, tyrosinase-related protein 1 (TRP1; also called TYRP1) and tyrosinase-related protein 2 (TRP2; also called DCT), which shows no oxidase activity but instead acts as a l-dopachrome tautomerase to catalyse l-dopachrome to 5,6-dihydroxyindole-2-carboxylic acid22,23. To determine TYR-2 enzymatic activity, we analysed recombinant, bacterially expressed GST::TYR-2. We found that like mammalian TRP2, GST::TYR-2 had no detectable tyrosinase activity (Supplementary Fig. 15), but showed l-dopachrome tautomerase activity (Fig. 5f).
On the basis of the antiapoptotic properties of C. elegans TYR-2 and its homology to human TRP2, we hypothesized that TRP2 might similarly represent a negative regulator of apoptosis. To test this hypothesis, we knocked down TRP2 by short hairpin RNA (shRNA) in human WM266-4 metastatic melanoma cells and analysed cisplatin-induced apoptosis (Supplementary Fig. 16). TRP2 knock down (shDCT374) increased cisplatin-induced apoptosis compared to control shRNA in WM266-4 cells (Fig. 5h, i, l and Supplementary Fig. 17). This phenotype was rescued by expression of wild-type TRP2 but not of the catalytically inactive point mutant TRP2(H189A/R194A) (Fig. 5j, k).
Three further traits were reminiscent of C. elegans TYR-2. First, shDCT374 and shDCT374;TRP2(H189A/R194A) melanoma cells exhibited both higher basal apoptotic fractions, compared to control shRNA cells, under untreated conditions (Supplementary Fig. 17). Second, TRP2, which normally localizes to melanosomes—specialized pigment organelles produced only in melanocytes—was found to localize to the nucleus in some (including WM266-4), but not all, melanoma cell lines (Supplementary Fig. 18). Last, in analogy to TYR-2-mediated CEP-1 inhibition, we asked whether human TRP2 impinges on p53 levels. We found that TRP2 knock down in WM266-4 melanoma cells increased basal and cisplatin-induced p53 levels (Fig. 5l). We conclude from this set of experiments that human TRP2 counteracts apoptotic cell death induction, possibly by means of its l-dopachrome tautomerase activity, and negatively affects the p53 pathway.
Discussion
Understanding the molecular events that contribute to resistance towards DNA-damage-induced apoptosis is of fundamental importance in tumorigenesis and cancer therapy9. We present evidence suggesting that stabilization of C. elegans HIF-1 in ASJ sensory neurons promotes expression of the tyrosinase family member TYR-2. We propose that TYR-2 acts as a pro-survival signalling molecule: after its secretion by the ASJ sensory neurons, TYR-2 acts on the germ line, directly or indirectly, as a critical negative regulator of CEP-1, thereby antagonizing DNA-damage-induced apoptosis (Fig. 5g). Our findings uncover a novel pathway by which two single neurons regulate apoptotic fate in another tissue.
Previous work has shown that behavioural responses of C. elegans to oxygen are controlled by the activity of the URX, AQR and PQR sensory neurons24,25. Here we present a second type of systemic response to hypoxia controlled by two different sensory neurons that have not been previously implicated in oxygen sensing. ASJ sensory neurons represent the major ultraviolet-sensitive cells mediating negative phototaxis and have been reported to control exit from the dauer stage26,27. C. elegans inhabits soil environments that vary from practically 0% to 21% oxygen. Worms are forced to move into hypoxic soil areas in search of bacteria, which consume oxygen faster than it diffuses. While feeding in such regions, the ASJ sensory neurons may protect germ cells from apoptosis, and thus may ensure that reproduction is not jeopardized. Notably, the role of the ASJ sensory neurons in hypoxia response is reminiscent of the mammalian carotid body, which transduces a fall in arterial oxygen partial pressure into systemic adaptation to hypoxia.
How could CEP-1 function be regulated by TYR-2? We found that in the absence of its signal peptide, TYR-2 localized to the nucleus. Nuclear localization of both TYR-2 and CEP-1 would be consistent with the possibility that TYR-2 directly interacts with, and perhaps modifies, CEP-1. Indeed, various tyrosinases have been shown to directly hydroxylate tyrosyl residues on proteins28,29. However, we failed to observe mass changes in CEP-1 peptides by MS/MS after in vitro incubation with TYR-2 (Supplementary Figs 19 and 20). Nonetheless, because tautomerization does not result in amass change, tautomerase activity cannot be excluded as a putative mechanism.
The antiapoptotic activity of TYR-2 is interesting in the light of reports that the human homologue TRP2 mediates resistance of melanomas to DNA-damaging agents30. Our data indicate that TRP2 counteracts apoptotic cell death induction, possibly by affecting p53 levels, and may therefore allow survival even in the absence of p53 mutations, which—unusual for such an aggressive cancer—are rarely present in melanomas31. Activating BRAF mutations in melanomas occur at a frequency of 50–70% and drive cancer progression by controlling the level of microphthalmia-associated transcription factor (MITF)32,33. Given that TRP2 in turn is transcriptionally regulated by MITF34, our findings suggest that TRP2 could act downstream of BRAF to affect tumour progression and therapy. The existence of such a potent antiapoptotic factor in melanocytes might be explained by its function: melanocytes must survive ultraviolet light to produce melanin; using the same pathway to promote both melanin synthesis and survival would be an elegant solution to this problem.
The antiapoptotic action of HIF-1 and TRP2 could also be present in other types of cancer. For instance, TRP2 was expressed in metastatic but not in non-metastatic human colorectal carcinoma cells35. We suggest that increased HIFα levels could induce tyrosinases such as TRP2 and thereby may provide an as yet unrecognized explanation for the correlation between increased HIFα and patient mortality.
Our discovery that TYR-2 acts cell non-autonomously is of particular interest, as it raises the possibility that HIFα stabilization in hypoxic areas of a tumour could increase viability of cancer cells in other parts of the tumour, thereby providing a paradigm for a cell non-autonomous regulation of p53-dependent apoptosis. Such a cell non-autonomous mechanism would introduce an additional layer of complexity within the tumour, as apoptotic cell fate would not only be dictated by the set of genetic alterations carried by a given tumour cell, but would also be influenced by the vicinity of hypoxic cells. If our model is correct, TRP2 might represent an intriguing target for therapeutic strategies.
METHODS SUMMARY
Strains
All strains were maintained and raised at 20 °C on NGM agar seeded with Escherichia coli OP50 (ref. 36). Bristol N2 strain was used as the wild-type strain. The mutations that were used are listed in the Methods.
C. elegans DNA damage response assay
Synchronized young adult worms (12 h post L4/adult moult stage) were exposed to indicated X-ray dosages (Gy). An Isovolt 160 HS X-ray machine (Rich. Seifert & Co.) was used to deliver the appropriate dosages. Germline apoptosis was quantified at indicated time points using differential interference contrast (DIC) microscopy, as previously described11. For RNAi experiments, synchronized L1 larvae were transferred onto plates seeded with bacteria expressing the respective RNAi clone37. Germline apoptosis was quantified as described above, starting from the 12 h post L4/adult moult stage. For cell cycle arrest quantification, staged young adult worms (12 h post L4/adult moult) were irradiated, and cell cycle arrest was assessed 12 h after treatment by counting the number of mitotic nuclei present in the area within 50 µm from the distal tip cell.
Laser ablation
ASJ neurons in L1/L2 animals were identified using DiD staining (20 µM for 2 h) as previously described18. Synchronized L1/L2 animals were ablated, or mock-ablated in parallel, using a laser microbeam as described previously38. Worms were allowed to recover on plates, and irradiated upon reaching the young adult stage (12 h post L4/adult). Germline apoptosis was quantified 24 h after treatment followed by ASJ neuron survey. Ablation of ASJ neurons was confirmed by either absence of GFP signal in opIs216[Ptyr-2::gfp]; vhl-1(ok161) animals or absence of DiD staining of the ASJ neurons in wild-type animals.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
METHODS
Strains
All strains were maintained and raised at 20 °C on NGM agar seeded with Escherichia coli OP50 (ref. 36). Bristol N2 strain was used as the wild-type strain.
C. elegans DNA damage response assay
Synchronized young adult worms (12 h post L4/adult moult stage) were exposed to indicated X-ray dosages (Gy). An Isovolt 160 HS X-ray machine (Rich. Seifert & Co.) was used to deliver the appropriate dosages. Germline apoptosis was quantified at indicated time points using differential interference contrast (DIC) microscopy, as previously described11. For RNAi experiments, synchronized L1 larvae were transferred onto plates seeded with bacteria expressing the respective RNAi clone37. Germline apoptosis was quantified as described above, starting from the 12 h post L4/adult moult stage. For cell cycle arrest quantification, staged young adult worms (12 h post L4/adult moult) were irradiated, and cell cycle arrest was assessed 12 h after treatment by counting the number of mitotic nuclei present in the area within 50 mm from the distal tip cell.
Laser ablation
ASJ neurons in L1/L2 animals were identified using DiD staining (20 µM for 2 h) as previously described18. Synchronized L1/L2 animals were ablated, or mock-ablated in parallel, using a laser microbeam as described previously38. Worms were allowed to recover on plates, and irradiated upon reaching the young adult stage (12 h post L4/adult). Germline apoptosis was quantified 24 h after treatment followed by ASJ neuron survey. Ablation of ASJ neurons was confirmed by either absence of GFP signal in opIs216[Ptyr-2::gfp]; vhl-1(ok161) animals or absence of DiD staining of the ASJ neurons in wild-type animals.
Strains
The following mutations were used: vhl-1(ok161), egl-9(n586), hif-1(ia4), tyr-2(ok1363), tyr-3(ok1194), cep-1(gk138), cat-2(e1112).
Transgenic lines
The following transgenic lines were used: opIs206[Phif-1::hif-1::gfp::hif-1(3′ UTR)], opIs216[Ptyr-2::gfp::let-858(3′ UTR)], opIs217[Ptyr-2::gfp::let-858(3′ UTR)], opIs233[Punc-119::tyr-2::gfp::tyr-2(3′ UTR)], opIs240[Ptyr-2::tyr-2:: gfp::tyr-2(3′ UTR)], opIs241[Pdpy-7::tyr-2::gfp::tyr-2(3′ UTR)], opIs245[Phus-1::tyr-2::gfp::tyr-2(3′ UTR)], opEx1321[Ptyr-2::tyr-2-SP::gfp::tyr-2(3′ UTR)], opEx1322 [Ptyr-2::tyr-2-SP::gfp::tyr-2(3′ UTR)], opEx1323[Ptyr-2::tyr-2-SP::gfp::tyr-2(3′ UTR)], opEx1350[Phus-1::tyr-2-SP::gfp::tyr-2(3′ UTR)], opEx1351[Phus-1::tyr-2-SP::gfp::tyr-2(3′ UTR)], opIs316[Ptrx-1::tyr-2::gfp::tyr-2(3′ UTR)], opIs271[Ptyr-3::tyr-3::gfp::tyr-3(3′ UTR)], opIs34[Phus-1::hus-1::gfp::let858(3′ UTR)] (ref. 39), opIs425[Ptrx-1::vhl-1(hairpin)::let-858(3′ UTR)]. Schematic representation of the constructs can be found in Supplementary Fig. 21.
Relative quantification of transcripts
Synchronized young adults were collected for total RNA extraction followed by cDNA synthesis (SuperScript III Platinum) and quantitative real-time PCR (qRT–PCR) as described previously39. Transcript levels were normalized to tbp-1 and pgk-1.
The forward and reverse primer sequences were: tbp-1 5′-TTGGATTTGAAGAAGATTGCATTG-3′, 5′-AATGACTGCTGCGAAACGTTT-3′; pgk-1 5′-GCGATATTTATGTCAATGATGCTTTC-3′, 5′-TGAGTGCTCGACTCCAACCA-3′; W07A12.6 5′-AAGAATACAACATCATGTCTTACAAGC-3′, 5′-TGGAAGTAATGGATCCAAGGATTA-3′; nhr-57 5′-CCGGGCTTCTGATGTTACTG-3′, 5′-CGCTTTGACTTCCATCGTTT-3′; tyr-1 5′-TCACAGGCTCCTGATTCAGAC-3′, 5′-CTTCCGTGCAGCACTGTC-3′; tyr-2 5′-TGAATCAGAAGCGGTGGAG-3′, 5′-GGTCCATTCGACGGATCA-3′; tyr-3 5′-TGGATATCGCTTGTCTTTGTG-3′, 5′-AGTGGACGACCTTGAACCAC-3′; tyr-4 5′-AGATGGGATGATGGAGCAAG -3′, 5′-CATTCCTGGTGGTAATGCAA-3′; egl-1 5′-CAGGACTTCTCCTCGTGTGAAGATTC-3′, 5′-CGAAGTCATCGCACATTGCTGCTA -3′; ced-13 5′-ACGGTGTTTGAGTTGCAAGC-3′, 5′-GCCAATATTATATTCAACCGTGTTTGAGT-3′.
RNAi screen for HIF-1 target genes mediating antiapoptotic function
Staged vhl-1(ok161) L1 were transferred to plates seeded with bacteria expressing the respective RNAi clone37. Worms were grown to the young adult stage (12 h after the L4/adult moult) and were irradiated with 120 Gy as described above. Germline apoptosis was quantified 24 h after treatment.
Western blot analysis
Synchronized young adult worms were exposed to 120 Gy ionizing radiation. Worms were collected at indicated time points in lysis buffer (250 mM sucrose, 10 mM HEPES, 0.1 mM EDTA, 1 mM PMSP, phosphatase inhibitor). Proteins were extracted by adding 212–300 mm glass beads (Sigma) and treating the worms with a BIO101/Savant FastPrep FP120 cell disrupter for 3 times for 45 s. Extracts were centrifuged for 10 min and the supernatant was heated to 90 °C for 10 min in Laemmli buffer. Proteins were separated by SDS–PAGE, transferred to PVDF (Amersham) membrane, and immunoblotted with CEP-1 antibody (gift from A. Gartner). To reduce nonspecific staining, CEP-1 antibody was pre-absorbed with OP50 acetone-powder and cep-1(gk138) acetone worm powder over night at 4 °C. β-Actin antibody (Sigma) was used as loading control.
Immunohistochemistry
For CEP-1 immunostaining, dissected gonads from synchronized young adult worms (12 h post L4 stage) were fixed with 4% formaldehyde, 100 mM K2HPO4 for 30 min and treated with methanol for 20 min. Antibody incubations and washes were performed as described40. Affinity-purified polyclonal anti-CEP-1 antibody was used at 1:50 dilution and Alexa 488 secondary antibody at 1:300 dilution.
Melanoma cells were fixed in 4% formaldehyde and incubated with anti-Pep8 (1:400) TRP2 antibody (provided by V. Hearing) and Alexa 488 secondary antibody (1:400). Cells were mounted in Vectashield mounting medium.
l-Dopachrome tautomerase activity
Enzymatic activity was determined spectrophotometrically by monitoring the decrease in absorbance at 475 nm. Fresh solutions of l-dopachrome were prepared by oxidation of l-dopa with silver (I) oxide, as previously described41. l-Dopa (0.5 mM) in 0.05 M sodium phosphate buffer, pH 6.8, was shaken vigorously with silver (I) oxide for 5 min. The final suspension was filtered through a 0.45 µm Millipore filter. Solutions were immediately used for kinetic experiments.
Tyrosinase activity
Enzymatic activity was determined spectrophotometrically by monitoring absorbance at 475 nm. 0.5 mM l-tyrosine solution in 0.05 M sodium phosphate buffer, pH 6.8, was incubated with the enzyme. Mushroom tyrosinase was obtained from Sigma (specific activity 2,085 U mg−1).
Embryonic survival assay
Synchronized L4 larvae were exposed to different dosages of IR, transferred to fresh plates 24 h after treatment and were allowed to lay eggs for 8 h. Adults were then removed and eggs were counted. Unhatched eggs were quantified 24 h later and percentage of embryonic survival was calculated.
Antibody production
HIF-1 rabbit polyclonal antibody was raised against the HIF-1 peptides DDTRDYIGRQPEIVEC and DRSPPAKRMHQSGPS (Eurogentech). Antibody was affinity purified.
Multiple alignment
Alignments were performed using MUSCLE software42.
Protein analysis
Gel bands were cut in small pieces and washed twice with 100 µl of 100 mM NH4HCO3/50% acetonitrile, then washed once with 50 µl acetonitrile. All three supernatants were discarded. Ten microlitres trypsin was added (0.01 mg µl−1 in 10 mM Tris/2 mM CaCl2, pH 8.2) with 10 µl buffer (10 mM Tris/2 mM CaCl2, pH 8.2), and incubated over night at 37 °C. Supernatant was removed and gel pieces were extracted twice with 100 µl 0.1% TFA/50% acetonitrile. All three supernatants were combined and dried. Samples were dissolved in 15 µl 0.1% TFA. The samples were desalted by using Ziptip C18 and were mixed 1:1 with matrix solution (5 mg ml−1 HCCA in 0.1% TFA/50% ACN). Samples were measured by MALDI/MS and MALDI/MS/MS.
Cell culture
Human cell lines were cultured in Dulbecco’s modified Eagle medium (DMEM, Invitrogen-Gibco), supplemented with 10% fetal bovine serum, 50 U ml−1 penicillin, 50 µg ml−1 streptomycin at 37 °C with 5% CO2.
Vectors
All retroviruses were generated using the murine stem cell virus (MSCV) backbone (Clontech). For the expression of miR-30 based shRNAs (shRNA-mirs), we used the retroviral vector MSCV-LTRmiR30-PIG (LMP) described previously43. The shRNAs were cloned into LMP as 110-bp XhoI–EcoRI fragments obtained by annealing oligonucleotides generating sticky ends. Guide strands were designed to target both isoforms of the human dopachrome tautomerase TRP2, and had the following sequences (5′–3′): shDCT197, UGUGAUUCAAACAACUAACAGA; shDCT223, UACAUGAACGUGCACACACAAU; shDCT374, UUACUUUCCUUGUCUCUGUCGU; shDCT1180, UGUACACACAUCACACUCGUUC; shDCT1182, UCUGUACACACAUCACACUCGU; shDCT1371, AAGGUUGGCAAUUUCAUGCUGU; shDCT1372, UAAGGUUGGCAAUUUCAUGCUG.
An shRNA efficiently targeting firefly luciferase (shLuc1309) was used as control44. The coding sequence of human TRP2 was amplified by PCR from the IMAGE clone 4705935 (imaGenes, clone IRATp970E0539D) and XhoI–MfeI ligated into MSCV-LTR-PGK-Hygro. A catalytically inactive TRP2 mutant (H189A/R194A) was obtained by site-directed mutagenesis.
Retroviral transduction
Cells were transduced as previously described45; however, target cells were infected only once without centrifugation. Retroviral co-transduction was carried out as previously described46. Efficiency of retroviral transduction of GFP expressing constructs was assessed 48 h after infection by flow cytometry (Guava EasyCyte, Guava Technologies). Transduced cell populations were selected 48 h after infection, using either 2.5 mg ml−1 puromycin (Sigma) or 400 mg ml−1 hygromycin B (Roche).
Antibodies for western blot analysis of human cells
Blots were probed with antibodies against tyrosinase-related protein 2 (TRP2) (Pep8, 1:4,000, gift from V. Hearing), p53 (DO1, 1:500, Abcam) or β-actin (AC-15, 1:15000, Sigma).
FACS analysis using Annexin-V/PI staining
Human melanoma cells were treated with cisplatin for 24 h. Both attached and floating cells were collected. Cells were washed twice in phosphate-buffered saline and re-suspended in Annexin-V binding buffer (Invitrogen). Thereafter, 5 µl Annexin-V-APC (BD Biosciences) and 1 µl propidium iodide (PI) (1 mg ml−1) were added to 100 µl cell suspension and incubated for 20 min in the dark. Finally, 400 µl Annexin-V binding buffer was added. Cells were analysed on a BD FACSCanto II flow cytometer.
Supplementary Material
Acknowledgements
We thank Y. Lazebnik, A. Gartner, A. Hajnal, J. Jiricny, R. Wenger and C. Mosimann for critical reading of the manuscript. We are grateful to S. Schrimpf, L. Stergiou, E. Bogan, S. Egger, O. Georgiev, M. Moser and Hengartner, Hajnal and Lowe laboratory members for help and discussions. We thank Y. Auchli and P. Hunziker from the Functional Genomics Center Zurich for protein analysis. We are grateful to V. Hearing and J. Valencia for the TRP2 antibody and help. This work was supported by the Swiss National Science Foundation, the Kanton of Zurich and the Josef-Steiner Foundation. M.O.H. is Ernst Hadorn endowed Professor of Molecular Biology. A.S. was supported by Oncosuisse and the Swiss National Science Foundation. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions A.S. designed experiments, performed most of the experiments and analysed data; I.K. generated opIs425 transgenic animals and performed experiments; C.F. established shRNA melanoma cell lines and helped to perform melanoma experiments; S.W.L. contributed to the design of melanoma experiments and analysed data; M.O.H. designed experiments and analysed the data. A.S. and M.O.H. wrote the paper.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Epstein AC, et al. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 2.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 3.Ivan M, et al. Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc. Natl Acad. Sci. USA. 2002;99:13459–13464. doi: 10.1073/pnas.192342099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu F, White SB, Zhao Q, Lee FS. HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl Acad. Sci. USA. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Latif F, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 6.Höckel M, Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J. Natl Cancer Inst. 2001;93:266–276. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 7.Zhong H, et al. Overexpression of hypoxia-inducible factor 1a in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- 8.Semenza GL. Targeting HIF-1 for cancer therapy. Nature Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 9.Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: a link between cancer genetics and chemotherapy. Cell. 2002;108:153–164. doi: 10.1016/s0092-8674(02)00625-6. [DOI] [PubMed] [Google Scholar]
- 10.Jiang H, Guo R, Powell-Coffman JA. The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc. Natl Acad. Sci. USA. 2001;98:7916–7921. doi: 10.1073/pnas.141234698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- 12.Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- 13.Derry WB, Putzke AP, Rothman JH. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science. 2001;294:591–595. doi: 10.1126/science.1065486. [DOI] [PubMed] [Google Scholar]
- 14.Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 15.Quevedo C, Kaplan DR, Derry WB. AKT-1 regulates DNA-damage-induced germline apoptosis in C. elegans. Curr. Biol. 2007;17:286–292. doi: 10.1016/j.cub.2006.12.038. [DOI] [PubMed] [Google Scholar]
- 16.Bishop T, et al. Genetic analysis of pathways regulated by the von Hippel-Lindau tumor suppressor in Caenorhabditis elegans. PLoS Biol. 2004;2:e289. doi: 10.1371/journal.pbio.0020289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen C, Nettleton D, Jiang M, Kim SK, Powell-Coffman JA. Roles of the HIF-1 hypoxia-inducible factor during hypoxia response in Caenorhabditis elegans. J. Biol. Chem. 2005;280:20580–20588. doi: 10.1074/jbc.M501894200. [DOI] [PubMed] [Google Scholar]
- 18.Hedgecock EM, Culotti JG, Thomson JN, Perkins LA. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev. Biol. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- 19.Wang N, Hebert DN. Tyrosinase maturation through the mammalian secretory pathway: bringing color to life. Pigment Cell Res. 2006;19:3–18. doi: 10.1111/j.1600-0749.2005.00288.x. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y, Setaluri V, Takechi Y, Houghton AN. Sorting and secretion of a melanosome membrane protein, gp75/TRP1. J. Invest. Dermatol. 1997;109:788–795. doi: 10.1111/1523-1747.ep12340971. [DOI] [PubMed] [Google Scholar]
- 21.Grant B, Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell. 1999;10:4311–4326. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.del Marmol V, Beermann F. Tyrosinase and related proteins in mammalian pigmentation. FEBS Lett. 1996;381:165–168. doi: 10.1016/0014-5793(96)00109-3. [DOI] [PubMed] [Google Scholar]
- 23.Tsukamoto K, Jackson IJ, Urabe K, Montague PM, Hearing VJ. A second tyrosinase-related protein, TRP-2, is a melanogenic enzyme termed DOPAchrome tautomerase. EMBO J. 1992;11:519–526. doi: 10.1002/j.1460-2075.1992.tb05082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray JM, et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- 25.Chang AJ, Bargmann CI. Hypoxia and the HIF-1 transcriptional pathway reorganize a neuronal circuit for oxygen-dependent behavior in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 2008;105:7321–7326. doi: 10.1073/pnas.0802164105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward A, Liu J, Feng Z, Xu XZ. Light-sensitive neurons and channels mediate phototaxis in C. elegans. Nature Neurosci. 2008;11:916–922. doi: 10.1038/nn.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bargmann CI, Horvitz HR. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science. 1991;251:1243–1246. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- 28.Ito S, Kato T, Shinpo K, Fujita K. Oxidation of tyrosine residues in proteins by tyrosinase. Formation of protein-bonded 3,4-dihydroxyphenylalanine and 5-S-cysteinyl-3,4-dihydroxyphenylalanine. Biochem. J. 1984;222:407–411. doi: 10.1042/bj2220407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freddi G, et al. Tyrosinase-catalyzed modification of Bombyx mori silk fibroin: grafting of chitosan under heterogeneous reaction conditions. J. Biotechnol. 2006;125:281–294. doi: 10.1016/j.jbiotec.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Chu W, et al. Tyrosinase-related protein 2 as a mediator of melanoma specific resistance to cis-diamminedichloroplatinum(II): therapeutic implications. Oncogene. 2000;19:395–402. doi: 10.1038/sj.onc.1203315. [DOI] [PubMed] [Google Scholar]
- 31.Montano X, Shamsher M, Whitehead P, Dawson K, Newton J. Analysis of p53 in human cutaneous melanoma cell lines. Oncogene. 1994;3:1455–1459. [PubMed] [Google Scholar]
- 32.Wellbrock C, et al. Oncogenic BRAF regulates melanoma proliferation through the lineage specific factor MITF. PLoS ONE. 2008;3:e2734. doi: 10.1371/journal.pone.0002734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 34.Bertolotto C, et al. Different cis-acting elements are involved in the regulation of TRP1 and TRP2 promoter activities by cyclic AMP: pivotal role of M boxes (GTCATGTGCT) and of microphthalmia. Mol. Cell. Biol. 1998;18:694–702. doi: 10.1128/mcb.18.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ying-Tao Z, Yi-Ping G, Lu-Sheng S, Yi-Li W. Proteomic analysis of differentially expressed proteins between metastatic and non-metastatic human colorectal carcinoma cell lines. Eur. J. Gastroenterol. Hepatol. 2005;17:725–732. doi: 10.1097/00042737-200507000-00006. [DOI] [PubMed] [Google Scholar]
- 36.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 38.Avery L, Horvitz HR. Pharyngeal pumping continues after laser killing of the pharyngeal nervous system of C. elegans. Neuron. 1989;3:473–485. doi: 10.1016/0896-6273(89)90206-7. [DOI] [PubMed] [Google Scholar]
References
- 39.Hofmann ER, et al. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr. Biol. 2002;12:1908–1918. doi: 10.1016/s0960-9822(02)01262-9. [DOI] [PubMed] [Google Scholar]
- 40.Schumacher B, et al. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120:357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 41.Palumbo A, d’Ischia M, Misuraca G, Prota G. Effect of metal ions on the rearrangement of dopachrome. Biochim. Biophys. Acta. 1987;925:203–209. doi: 10.1016/0304-4165(87)90110-3. [DOI] [PubMed] [Google Scholar]
- 42.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nature Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 44.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genome. Nature Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 45.Serrano M, et al. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 46.Zuber J, et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009;23:877–889. doi: 10.1101/gad.1771409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.