Abstract
Epigenetic pathways can regulate gene expression by controlling and interpreting chromatin modifications. Cancer cells are characterized by altered epigenetic landscapes, and commonly exploit the chromatin regulatory machinery to enforce oncogenic gene expression programs1. Although chromatin alterations are, in principle, reversible and often amenable to drug intervention, the promise of targeting such pathways therapeutically has been limited by an incomplete understanding of cancer-specific dependencies on epigenetic regulators. Here we describe a non-biased approach to probe epigenetic vulnerabilities in acute myeloid leukaemia (AML), an aggressive haematopoietic malignancy that is often associated with aberrant chromatin states2. By screening a custom library of small hairpin RNAs (shRNAs) targeting known chromatin regulators in a genetically defined AML mouse model, we identify the protein bromodomain-containing 4 (Brd4) as being critically required for disease maintenance. Suppression of Brd4 using shRNAs or the small-molecule inhibitor JQ1 led to robust antileukaemic effects in vitro and in vivo, accompanied by terminal myeloid differentiation and elimination of leukaemia stem cells. Similar sensitivities were observed in a variety of human AML cell lines and primary patient samples, revealing that JQ1 has broad activity in diverse AML subtypes. The effects of Brd4 suppression are, at least in part, due to its role in sustaining Myc expression to promote aberrant self-renewal, which implicates JQ1 as a pharmacological means to suppress MYC in cancer. Our results establish small-molecule inhibition of Brd4 as a promising therapeutic strategy in AML and, potentially, other cancers, and highlight the utility of RNA interference (RNAi) screening for revealing epigenetic vulnerabilities that can be exploited for direct pharmacological intervention.
AML represents a paradigm for understanding how complex patterns of cooperating genetic and epigenetic alterations lead to tumorigenesis3,4. Although this complexity poses a challenge for the development of targeted therapies, diverse gene mutations in AML generally converge functionally to deregulate similar core cellular processes. One key event in AML initiation is the corruption of cell-fate programs to generate leukaemia stem cells that aberrantly self-renew and thereby maintain and propagate the disease5. Although it is incompletely understood, this process has been linked to changes in regulatory chromatin modifications2. For example, common AML oncogenes such as those encoding the AML1-ETO and MLL fusion proteins induce self-renewal programs at least in part through reprogramming of epigenetic pathways6,7. In addition, several genes encoding epigenetic regulators have been identified as targets of somatic mutation in AML8,9. Because epigenetic alterations induced by oncogenic stimuli are potentially reversible, chromatin regulators are being explored as candidate drug targets1.
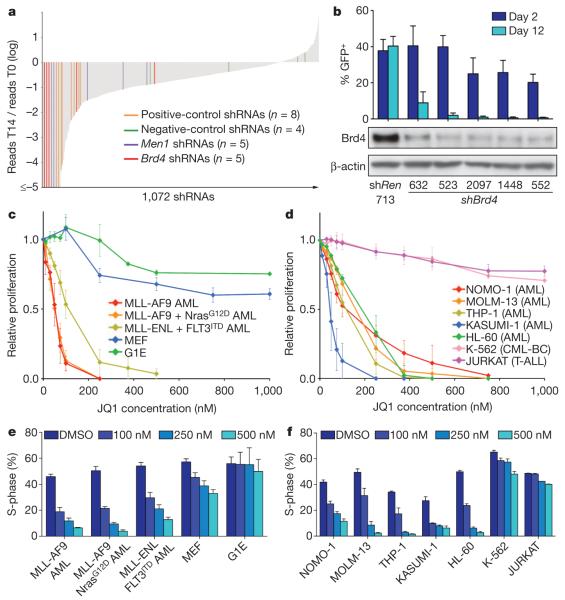
To probe epigenetic pathways required for AML maintenance systematically, we built a custom shRNA library targeting 243 known chromatin regulators, including most ‘writers’, ‘readers’ and ‘erasers’ of epigenetic marks (Supplementary Fig. 1 and Supplementary Table 1). This library of 1,094 shRNAs (3–6 per gene) was constructed in TRMPV-Neo, a vector optimized for negative-selection RNAi screening, and was transduced as one pool into an established Tet-on-competent AML mouse model driven by MLL-AF9 and NrasG12D (ref. 10). After drug selection, shRNA expression was induced by addition of doxycycline, and changes in library representation after 14 days of culture were monitored using deep sequencing of shRNA guide strands amplified from genomic DNA (Fig. 1a and Supplementary Fig. 2). Using the scoring criterion of more than twenty-fold depletion in each of two independent replicates, 177 shRNAs were strongly depleted. These included all eight positive-control shRNAs targeting essentialgenes (Rpa1, Rpa3, Pcna and Polr2b), as well as several shRNAs targeting two known MLL-AF9 cofactors (Men1 and Psip1)11,12. Genes for which at least two independent shRNAs scored were subjected to extensive one-by-one validation using an independent MLL-AF9/NrasG12D AML cell line and vector system (Supplementary Fig. 3a). In both the primary screen and validation stages, several shRNAs targeting Brd4 were among the most strongly depleted, identifying this gene as the top scorer in the screen (Fig. 1a and Supplementary Fig. 3b).
Figure 1. AML growth is sensitive to Brd4 inhibition.
a, Pooled negative-selection screening in MLL-AF9/NrasG12D leukaemia, depicting changes in representation of 1,072 informative shRNAs during 14 days of culture. shRNA abundance ratios were calculated as the number of reads after 14 days of culture on doxycycline (T14) divided by the number of reads before doxycycline treatment (T0), and plotted as the mean of two replicates in ascending order. Completely depleted shRNAs (0 reads at T14, n = 71) were plotted as a ratio of 10−5; highlighted shRNAs in this group are shown with even spacing in alphabetical order. Positive controls included shRNAs targeting Rpa1, Rpa3, Pcna and Polr2b. Negative-control shRNAs targeted renilla luciferase or Braf. b, Percentage of GFP-positive MLL-AF9/NrasG12D leukaemia cells 2 days and 12 days after transduction with LMN constructs expressing the indicated shRNAs (top panel). Western blotting of whole-cell lysates prepared from MEF cultures transduced with the indicated TtTMPV-shRNAs and induced with doxycycline for 5 days (bottom panel). shREN, renilla luciferase shRNA. Representative experiments are shown. c, d, Proliferation rates of JQ1-treated cells, calculated by measuring the increase in viable cell number after 72 h in culture and fitting data to an exponential growth curve. Results are normalized to the proliferation rate of vehicle/DMSO-treated cells, set to 1 (n = 3). CML-BC, chronic myeloid leukaemia blast crisis; T-ALL, T-cell acute lymphoblastic leukaemia. e, f, Percentage of cells in S-phase (bromodeoxyuridine (BrdU)+) after JQ1 treatment for 48 h at the indicated concentrations (n = 3). BrdU was pulsed for 30 min in all experiments shown. All error bars represent s.e.m.
Brd4 is a member of the BET family of bromodomain-containing proteins that bind to acetylated histones to influence transcription13. BRD4 is also a proto-oncogene that can be mutated via chromosomal translocation in a rare form of squamous-cell carcinoma14, although a role in leukaemia has not been described. The recent development of small-molecule BET bromodomain inhibitors15,16, together with our screening results, prompted us to investigate the suitability of Brd4 as an AML drug target. Five independent Brd4 shRNAs showed a close correspondence between knockdown efficiency and growth inhibition, indicating on-target effects (Fig. 1b). Suppression of Brd4 led to cell-cycle arrest and apoptosis of leukaemia cells, whereas the equivalent knockdown in immortalized murine embryonic fibroblasts (MEFs) led to only modest cell-cycle inhibition without cytotoxicity (Supplementary Fig. 4a–d). Brd4 knockdown also failed to influence the growth of non-transformed G1E erythroblast cells (Supplementary Fig. 4e). In addition, shRNAs targeting BRD4 were sufficient to induce cell-cycle arrest in two MLL-AF9+ human AML lines (Supplementary Fig. 5). Together, these results indicate that Brd4 is a critical requirement in MLL-AF9-induced AML.
Next, we examined the sensitivity of leukaemia cells to JQ1, a first-in-class small-molecule inhibitor of BET bromodomains with highest affinity for the first bromodomain of Brd4 (ref. 15). Proliferation of mouse MLL-fusion leukaemia cells was notably sensitive to sub-micromolar JQ1 concentrations, as compared to proliferation of fibroblasts and G1E cells (Fig. 1c), correlating with the relative impact of Brd4 shRNAs on proliferation of these different cell types. We also examined growth-inhibitory effects of JQ1 in a series of established human leukaemia cell lines as well as in adult and paediatric primary leukaemia samples. We observed broad growth-suppressive activity of JQ1 (IC50 < 500 nM) in 13 out of 14 AML cell lines (Fig. 1d and Supplementary Fig. 6a, b) and in 12 out of 15 primary human AML samples, representing diverse disease subtypes (Supplementary Fig. 7, 8). In addition, all three primary MLL-rearranged infant leukaemias tested were sensitive to JQ1 (Supplementary Fig. 8), whereas other non-AML-leukaemia and solid-tumour cell lines showed minimal sensitivity to the compound (Fig. 1d and Supplementary Fig. 6c). In all AML lines examined, JQ1 treatment triggered cell-cycle arrest and apoptosis, similar to the effects seen upon shRNA-mediated Brd4 knockdown (Fig. 1e, f and Supplementary Figs 7–9). Together, these data indicate a critical requirement for Brd4 in AML proliferation that can be effectively inhibited using the bromodomain inhibitor JQ1.
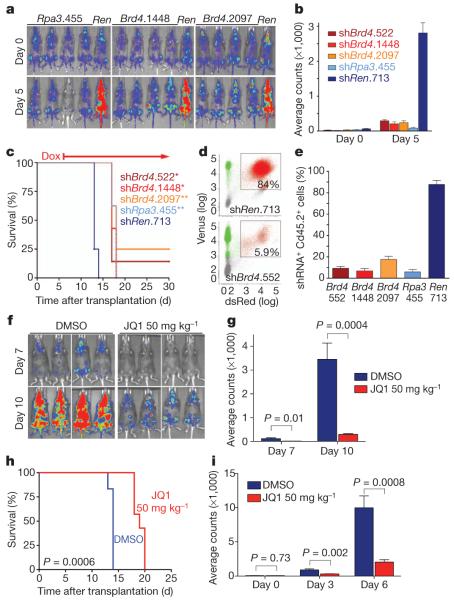
We next investigated the relevance of Brd4 to AML progression in vivo. To suppress Brd4 in established AML in mice, Tet-on-competent MLL-AF9/NrasG12D leukaemia cells were transduced with TRMPV-Neo constructs containing Brd4 shRNAs or control shRNAs, and transplanted into secondary recipient mice (Supplementary Fig. 10). After disease onset, confirmed by bioluminescent imaging, shRNA expression was induced by doxycycline administration. Subsequent monitoring showed that Brd4 knockdown resulted in a marked delay in leukaemia progression and a survival benefit (Fig. 2a–c and Supplementary Fig. 11). Taking advantage of the dsRed reporter linked to shRNA expression in the TRMPV-Neo vector10, flow-cytometry analysis verified that Brd4-shRNA-positive cells were depleted in the terminal leukaemia burden as compared to controls, indicating that the mice succumbed to an outgrowth of Brd4-shRNA-negative cells (Fig. 2d, e). Together, these data indicate that RNAi-mediated suppression of Brd4 inhibits leukaemia progression in vivo.
Figure 2. Brd4 is required for AML progression in vivo.
a, Bioluminescent imaging of mice transplanted with MLL-AF9/NrasG12D leukaemia cells harbouring the indicated TRMPV-shRNAs. Doxycycline was administered upon disease onset, 6 days after transplant. Day 0 indicates the time of doxycycline treatment. b, Quantification of bioluminescent imaging responses after doxycycline treatment. Mean values of four replicate mice are shown. c, Kaplan-Meier survival curves of recipient mice transplanted with the indicated TRMPV-shRNA leukaemia lines. The interval of doxycycline treatment is indicated by the arrow. Each shRNA group contained 6–8 mice. Statistical significance compared to shRen was calculated using a log-rank test; *, P = 0.0001; **, P < 0.0001. d, Representative flow cytometry plots of donor-derived (Cd45.2+) bone marrow cells in terminally diseased doxycycline-treated mice. The gate shown includes dsRed+/shRNA+ cells. e, Percentage of dsRed+/shRNA+ cells in the Cd45.2+ terminal leukaemia burden. Mean values of four replicate mice are shown. f, Bioluminescent imaging of MLL-AF9/NrasG12D leukaemia recipient mice at the indicated day after initiation of treatment with JQ1 (50 mg−1 kg−1 d−1) or DMSO carrier. g, Quantification of bioluminescent imaging responses to JQ1 treatment. Mean values of six DMSO- and seven JQ1-treated mice are shown. P values were calculated using a two-tailed Student’s t-test. h, Kaplan–Meier survival curves of control and JQ1-treated mice. Statistical significance was calculated using a log-rank test. In f, g and h, JQ1 treatment was initiated on day 1 after transplant of 50,000 leukaemia cells. i, Quantification of bioluminescent imaging responses to JQ1 treatment in established disease. Treatment of leukaemic mice was initiated 6 days after transplant, when disease first became detectable by imaging. Mean values of six DMSO- and seven JQ1-treated mice are shown. P values were calculated using a two-tailed Student’s t-test. All error bars represent s.e.m.
To examine whether JQ1 has single-agent activity in AML, mice transplanted with MLL-AF9/NrasG12D leukaemia cells were treated either with daily injections of JQ1 (50 mg kg−1) or with vehicle. JQ1 administration led to a marked delay in disease progression and significantly extended survival (Fig. 2f–h). JQ1 also showed single-agent activity in an intervention setting, in which treatment was initiated only after disease was detected by bioluminescent imaging (Fig. 2i and Supplementary Fig. 12). Comparable effects were observed in an independent AML mouse model based on expression of AML1-ETO9a and NrasG12D and loss of p53 (ref. 17), which is known to be insensitive to conventional chemotherapy (Supplementary Fig. 13). Consistent with previous findings15, JQ1 treatment was well tolerated in mice, with little if any impact on normal haematopoiesis (Supplementary Figs 14–16). Collectively, these findings demonstrate that JQ1 has potent and leukaemia-specific effects as a single agent in vivo.
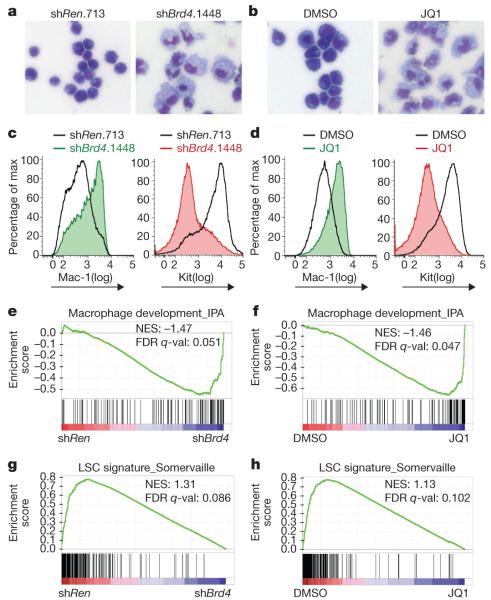
AML is characterized by an expanded self-renewal capacity linked with an inability to complete terminal myeloid differentiation. Therefore, we next considered whether Brd4 influences the differentiation state of leukaemia cells. Both expression of Brd4 shRNA and JQ1 treatment altered the morphology of MLL-AF9/NrasG12D leukaemia from myelomonocytic blasts to cells with a macrophage-like appearance (Fig. 3a, b and Supplementary Fig. 17a). Upon Brd4 inhibition, leukaemia cells showed increased surface expression of integrin αM (Itgam, also known as Mac-1), a myeloid differentiation marker, and decreased expression of Kit, a marker associated with leukaemia stem cells (LSCs) in mouse models of MLL-rearranged leukaemia (Fig. 3c, d and Supplementary Fig. 17b, c)18,19. In addition, JQ1 treatment induced morphological signs of maturation phenotypes in most of the primary leukaemia samples tested, albeit to varying degrees (Supplementary Figs 7 and 8).
Figure 3. Brd4 inhibition leads to myeloid differentiation and leukaemia stem-cell depletion.
a, b, Light microscopy of May–Grünwald/Giemsa-stained MLL-AF9/NrasG12D leukaemia cells after 2 days of doxycycline-induced shRNA expression or 2 days of JQ1 treatment (100 nM). Expression of shRNA was induced in TRMPV-transduced leukaemia cells. Imaging was performed with a ×40 objective. Representative images of three biological replicates are shown. c, d, FACS analysis of Mac-1 and Kit surface expression after 4 days of shRNA expression or 2 days of JQ1 treatment (100 nM). A representative experiment of three biological replicates is shown. e–h, GSEA plots evaluating changes in macrophage and LSC gene signatures upon Brd4 inhibition. In e and g, RNA for expression arrays was obtained from sorted dsRed+/shRNA+ cells (shRen versus three different Brd4 shRNAs) after 2 days of doxycycline induction. In f and h, microarray data were obtained from leukaemia cells treated for 2 days with DMSO or 100 nM JQ1. NES, normalized enrichment score; FDR q-val, false discovery rate q-value (the probability that a gene set with a given NES represents a false-positive finding).
To further investigate whether suppression of Brd4 affects the LSC compartment, we performed gene set enrichment analysis (GSEA) of expression microarray data obtained from Brd4-shRNA-treated and JQ1-treated leukaemia cells20. GSEA revealed a marked upregulation of macrophage-specific gene expression after Brd4 inhibition (Fig. 3e, f), as well as global downregulation of a gene expression signature previously shown to discriminate LSCs from non-self-renewing leukaemia cell populations (Fig. 3g, h)21. A similar profile of gene expression changes was seen after JQ1 treatment of THP-1 cells, a human MLL-AF9-expressing AML cell line (Supplementary Fig. 18). Although we cannot exclude the involvement of additional cellular targets, the strong concordance between phenotypes induced by Brd4 shRNAs and JQ1 supports Brd4 as the relevant target of JQ1 in AML. Together, these findings indicate that Brd4 is critically required to maintain LSC populations and prevent terminal myeloid differentiation.
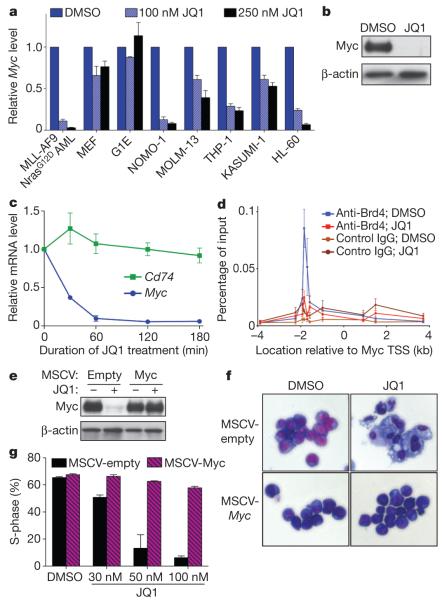
Recent evidence indicates that the Myc transcriptional network has an important role in LSC self-renewal22,23. Because previous studies also implicate Myc as a potential downstream target of Brd4 (refs 24, 25), we examined whether this regulatory function was relevant to the antileukaemic effects of JQ1. Brd4 inhibition with shRNAs or JQ1 led to a marked reduction of mRNA and protein levels of Myc in MLL-AF9/NrasG12D leukaemia, whereas these effects were minimal in MEF and G1E cells (Fig. 4a, b and Supplementary Fig. 19a–c). Downregulation of Myc mRNA levels occurred within 60 min of JQ1 exposure, qualitatively preceding the increased expression of genes related to macrophage differentiation, such as Cd74 (Fig. 4c). Supporting a direct role in Myc transcriptional regulation, chromatin immunoprecipitation (ChIP) experiments identified a region of focal Brd4 occupancy about 2 kilo-bases upstream of the Myc promoter and this was eliminated after exposure to competitive JQ1 (Fig. 4d). As expected, RNAi- or JQ1-induced suppression of Brd4 led to a global reduction in expression of Myc target genes22,26 (Supplementary Figs 19d and 20). Notably, JQ1 treatment triggered Myc downregulation in a broad array of mouse and human leukaemia cell lines examined (Fig. 4a, b and Supplementary Fig. 21), indicating that JQ1 may provide a means to suppress the Myc pathway in a range of AML subtypes.
Figure 4. JQ1 suppresses the Myc pathway in leukaemia cells.
a, Quantitative reverse transcription PCR (qRT–PCR) of relative Myc mRNA levels in the indicated mouse or human cells after a 48-h treatment with JQ1. Results were normalized to Gapdh, with the relative mRNA level in untreated cells set to 1 (n = 3). b, Western blotting of whole-cell lysates prepared from MLL-AF9/NrasG12D leukaemia cells treated for 48 h with DMSO or 250 nM JQ1. A representative experiment of three biological replicates is shown. c, qRT–PCRtime course at indicated time points after treatment of MLL-AF9/NrasG12D leukaemia cells with 250 nM JQ1. Results were normalized toGapdh, with the relative mRNA level in untreated cells set to 1 (n = 3). d, ChIP–qPCR performed in MLL-AF9/NrasG12D leukaemia cells with the indicated antibodies and PCR primer locations (n = 6 for DMSO; n = 4 for JQ1-treated). TSS, transcription start site. e, Western blotting of whole-cell lysates prepared from MLL-AF9/NrasG12D leukaemia cells transduced with empty vector or a Myc-cDNA-containing MSCV retrovirus. Cells were treated for 48 h with DMSO or 250 nM JQ1. A representative experiment of three biological replicates is shown. f, Light microscopy of May–Grünwald/Giemsa-stained MLL-AF9/NrasG12D leukaemia cells transduced with an empty vector or with the Myc cDNA. Cells were treated for 5 days with 50 nM JQ1 and imaged using a×40 objective. A representative image of three biological replicates is shown. g, Quantification of BrdU incorporation after a 30-min pulse in MLL-AF9/NrasG12D leukaemia cells transduced with empty control vector or the Myc cDNA. Cells were treated with JQ1 for 5 days at the indicated concentrations (n = 3). All error bars shown represent s.e.m.
To evaluate whether suppression of Myc confers the growth-inhibitory effects of JQ1, we generated MLL-AF9/NrasG12D leukaemia cultures in which the Myc cDNA was ectopically expressed from a retroviral promoter, which resulted in Myc expression levels that were only slightly elevated but entirely resistant to JQ1-induced suppression (Fig. 4e and Supplementary Fig. 22a). Notably, ectopic Myc expression conferred nearly complete resistance to cell-cycle arrest and macrophage differentiation induced by JQ1 and Brd4 shRNA (Fig. 4g, h and Supplementary Figs 22b and 23). Furthermore, global gene-expression profiling revealed that most of the JQ1-induced transcriptional changes are probably secondary effects of Myc downregulation (Supplementary Fig. 24). Notably, ectopic Myc expression was unable to prevent JQ1-induced cell death, indicating that Brd4 has Myc-independent roles in regulating cell survival (Supplementary Fig. 22c, d). These findings collectively support a role for Brd4 in maintaining Myc expression to preserve an undifferentiated cellular state in AML.
By taking a non-biased screening approach targeting epigenetic regulators, our study has identified Brd4 as a critical factor required for AML disease maintenance. Because Brd4 is not evidently mutated or overexpressed in AML (Supplementary Fig. 25), the exquisite sensitivity of leukaemia cells to Brd4 inhibition would not have been revealed simply through genetic or transcriptional characterization of this disease. We further show that the bromodomain inhibitor JQ1 has broad activity in diverse AML contexts and, by comparing its effects to those induced by Brd4 shRNAs, we provide evidence that Brd4 is the primary target for the antileukaemic activity of JQ1. Of note, JQ1 is a first-generation chemical inhibitor yet to be optimized for in vivo delivery, with a half-life of only about 1 h in rodents (Supplementary Fig. 26 and ref. 15). The more robust antileukaemic effects seen using Brd4 shRNAs in vivo indicate that second-generation derivatives of this compound may have greater clinical activity. Regardless, our results unambiguously highlight the utility of RNAi screening for revealing candidate drug targets in cancer.
As a competitive inhibitor of the acetyl-lysine binding domain, JQ1 interferes with the ability of Brd4 to ‘read’ histone acetylation marks that facilitate transcriptional activation15. When applied to leukaemia cells, JQ1 interferes with transcriptional circuits supporting self-renewal, thus targeting LSCs and inducing terminal differentiation. In a parallel study, we identified the transcription factor Myb as a critical mediator of addiction to the MLL-AF9 oncogene (Zuber et al., in press). Notably, global gene-expression changes observed after genetic or pharmacological inhibition of Brd4 are remarkably similar to those seen upon suppressing Myb (Zuber et al., in press), indicating that Myb and Brd4 may intersect functionally in a common transcriptional circuit that is essential for malignant self-renewal. A key downstream effector of both Myb and Brd4 is the oncoprotein Myc (ref. 27), which has been validated as an attractive therapeutic target but has thus far escaped efforts at pharmacological inhibition28,29. Although the precise mechanism remains to be further defined, targeting Brd4 abolishes Myc expression and limits self-renewal, with selectivity for the malignant context, thus avoiding the haematopoietic toxicities that may be associated with systemic Myc inhibition30. As such, our study may define a general strategy to disarm oncogenic pathways through the direct modulation of the epigenetic machinery.
METHODS SUMMARY
Pooled negative-selection RNAi screening
A customized shRNA library targeting 243 chromatin-regulating mouse genes was designed using miR30-adapted BIOPREDsi predictions, and was generated by PCR-cloning a pool of oligonucleotides synthesized on 55k arrays (Agilent Technologies). Pools of shRNAs were subcloned into the TRMPV-Neo vector together with control shRNAs, and transduced into Tet-on MLL-AF9/NrasG12D leukaemia cells for negative-selection screening, essentially as described previously10. All shRNA sequences as well as primary screening data are provided in Supplementary Table 1.
Animal studies
All mouse experiments were approved by the Cold Spring Harbor animal care and use committee. For conditional RNAi experiments in vivo, Tet-on MLL-AF9/NrasG12D leukaemia cells were transduced with TRMPV-Neo-shRNA constructs, followed by transplantation into sub-lethally irradiated recipient mice, as described previously10. For shRNA induction, animals were treated with doxycycline in both drinking water (2 mg ml−1 with 2% sucrose; Sigma-Aldrich) and food (625 mg kg−1, Harlan laboratories). For JQ1 treatment trials, a stock of 100 mg ml−1 JQ1 in DMSO was diluted 20-fold by dropwise addition of a 10% 2-hydroxypropyl-β-cyclodextrin carrier (Sigma) under vortexing, yielding a 5 mg ml−1 final solution. Mice were intraperitoneally injected daily with freshly diluted JQ1 (50 or 100 mg kg−1) or a similar volume of carrier containing 5% DMSO.
Microarray analysis
Expression microarrays were performed using Affymetrix ST 1.0 GeneChips. Raw microarray data can be accessed at Gene Expression Omnibus, GSE29799. Pathway analysis was performed using GSEA v2.07 software with 1,000 phenotype permutations20. All gene sets used for GSEA are provided in Supplementary Table 2.
Supplementary Material
Acknowledgements
We thank B. Ma, S. Muller and M. Weissenboeck for technical assistance; J. Simon, E. Earl and L. Bianco for support with mouse work; C. dos Santos for assistance with LSK FACS analysis; S. Hearn for microscopy support; G. Hannon, K. Chang, and E. Hodges for shRNA technology support; A. Gordon and M. Hammell for bioinformatics support; L. Dow for assistance with mouse pathology sample preparation; and G. Blobel for comments on the manuscript. We thank the Don Monti Memorial Research Foundation and Laurie Strauss Leukemia Foundation for research support. J.Z. was supported by a research fellowship from the German Research Foundation (DFG) and by the Andrew Seligson Memorial Clinical Fellowship at CSHL; A.R.R. was supported by an NIH traineeship and the Barbara McClintock fellowship; J.E.B and J.Q. are supported by the Damon-Runyon Cancer Research Foundation and Smith Family Foundation. P.B. is supported by a Damon Runyon-Lilly Clinical Investigator Award and a Leukemia and Lymphoma Society (LLS) Translational Research Program Grant, and is an LLS Scholar in Clinical Research. S.W.L. is supported by a Specialized Center of Research (SCOR) grant from the Leukemia and Lymphoma Society of America, and by the Howard Hughes Medical Institute; C.R.V., J.S. and E.W. are supported by the CSHL President’s Council and the SASS Foundation for Medical Research.
Appendix
METHODS
Pooled negative-selection RNAi screening
A custom shRNA library targeting 243 chromatin-regulating mouse genes was designed using miR30-adapted BIOPREDsi predictions (six shRNAs per gene) and constructed by PCR-cloning a pool of oligonucleotides synthesized on 55k customized arrays (Agilent Technologies) as previously described10. After sequence verification, 1,094 shRNAs (3–6 per gene) were combined with several positive- and negative-control shRNAs at equal concentrations in one pool. This pool was subcloned into TRMPV-Neo and transduced into Tet-on MLL-AF9/NrasG12D leukaemia cells using conditions that predominantly lead to a single retroviral integration and represent each shRNA in a calculated number of >500 cells (a total of thirty million cells at infection, 2% transduction efficiency). Transduced cells were selected for 5 days using 1 mg ml−1 G418 (Invitrogen); at each passage more than twenty million cells were maintained to preserve library representation throughout the experiment. After drug selection, T0 samples were obtained (~twenty million cells per replicate) and cells were subsequently cultured with 0.5 mg ml−1 G418 and 1 μg ml−1 doxycycline to induce shRNA expression. After 14 days (12 passages, T14), about fifteen million shRNA-expressing (dsRed+/Venus+) cells were sorted for each replicate using a FACSAriaII (BD Biosciences). Genomic DNA from T0 and T14 samples was isolated by two rounds of phenol extraction using PhaseLock tubes (5prime) followed by isopropanol precipitation. Deep-sequencing template libraries were generated by PCR amplification of shRNA guide strands as previously described10. Libraries were analysed on an Illumina Genome Analyser at a final concentration of 8 pM; 18 nucleotides of the guide strand were sequenced using a custom primer (miR30EcoRISeq, TAGCCCCTTGAATTCCGAGGCAGTAGGCA). To provide a sufficient baseline for detecting shRNA depletion in experimental samples, we aimed to acquire >500 reads per shRNA in the T0 sample, which required more than ten million reads per sample to compensate for disparities in shRNA representation inherent in the pooled plasmid preparation or introduced by PCR biases. With these conditions, we acquired T0 baselines of >500 reads for 1,072 (97% of all) shRNAs. Sequence processing was performed using a customized Galaxy platform31. For each shRNA and condition, the number of matching reads was normalized to the total number of library-specific reads per lane and imported into a database for further analysis (Access 2003, Microsoft). All shRNA sequences are provided in Supplementary Table 1.
Animal studies
All mouse experiments were approved by the Cold Spring Harbor animal care and use committee. Leukaemia cells were transplanted by tail-vein injection of 1 × 106 cells into sub-lethally (5.5 Gy) irradiated B6/SJL(CD45.1) recipient mice. For whole-body bioluminescent imaging, mice were intraperitoneally injected with 50 mg kg−1 D-Luciferin (Goldbio), and after 10 min, analysed using an IVIS Spectrum system (Caliper LifeSciences). Quantification was performed using Living Image software (Caliper LifeSciences) with standardized rectangular regions of interests covering the mouse trunk and extremities. For shRNA induction, animals were treated with doxycycline in both drinking water (2 mg ml−1 with 2% sucrose; Sigma-Aldrich) and food (625 mg kg−1, Harlan laboratories). For JQ1 treatment trials, a stock of 100 mg ml−1 JQ1 in DMSO was 20-fold diluted by dropwise addition of a 10% 2-hydroxypropyl-β-cyclodextrin carrier (Sigma) under vortexing, yielding a 5 mg ml−1 final solution. Mice were intraperitoneally injected daily with freshly diluted JQ1 (50 or 100 mg kg−1) or a similar volume of carrier containing 5% DMSO.
Plasmids
For conditional RNAi experiments, shRNAs were expressed from either the TRMPV-Neo vector or the TtTMPV-Neo vector, which have been described previously10. For screen validation, shRNAs were cloned into LMN(MSCV-miR30-PGK-NeoR-IRES-GFP), which was generated from LMP32 by replacing the PuroR transgene with a NeoR cassette. For Myc rescue experiments, the wild-type mouse Myc cDNA was subcloned into MSCV-PGK-Puro-IRES-GFP (MSCV-PIG)33.
Cell culture
All mouse MLL-leukaemia cell lines were derived from bone marrow obtained from terminally ill recipient mice, and were cultured in RPMI 1640 (Gibco-Invitrogen) supplemented with 10% FBS, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin. MLL-AF9(alone), MLL-AF9/NrasG12D, Tet-on MLL-AF9/NrasG12D and MLL-ENL/FLT3ITD cell cultures were derived as described previously10,17. Tet-on immortalized MEF cultures were described previously10. G1E cells were provided by M. Weiss. MEF cells were grown in DMEM with 10% FBS, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and 1% glutamine (GIBCO). G1E cells were grown in IMDM with 15% FBS, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin, 2 U ml−1 erythropoietin (Sigma) and 10% Kit-ligand-conditioned medium. All human leukaemia cell lines were cultured in RPMI-1640 with 10% FBS, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin, except KASUMI-1 cells, which were cultured in 20% FBS. NOMO-1, MOLM-13, EoL-1, NB4, HNT-34 and CMK were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ). KASUMI-1, HL-60, MV4-11, KG1, HEL, THP-1, B16-F10 and IMR-90 were obtained from ATCC. K-562 cells were provided by M. Carroll. U2OS, HeLa and Jurkat were provided by the CSHL tissue culture facility. CD34.MA9.NRAS and CD34.MA9.FLT3 cells were generated by retroviral transduction of umbilical-cord-blood CD34+ cells as described previously34,35. All retroviral packaging was performed using ecotropic Phoenix cells according to established protocols (http://www.stanford.edu/group/nolan/tutorials/retpkg_1_packlines.html).
Western blotting
For Brd4 immunoblotting, 30 μg of whole-cell lysate RIPA extracts (25 mM Tris (pH 7.6), 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) were loaded into each lane. For Myc immunoblotting, cells were lysed directly in Laemmli buffer and about 50,000 cell-equivalents were loaded into each lane. Protein extracts were resolved by SDS polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to nitrocellulose for blotting.
Proliferation assay
Competitive proliferation assays using shRNAs in LMN or TRMPV-/TtRMPV-Neo vectors were performed as outlined in Supplementary Fig. 3a and as described previously10, respectively. Proliferation assays for JQ1 in vitro testing were performed by counting the increase in viable cell numbers over 72 h in the presence of different JQ1 concentrations. Dead cells were excluded using propidium iodide (PI) staining. Measurements of cell concentration were performed on a Guava Easycyte (Millipore), gating only viable cells (FSC/SSC/PI−). Proliferation rates were calculated using the equation ln(cell concentration at 72 h/cell concentration at 0 h)/72. Relative proliferation rates were calculated by normalizing to the rate of DMSO-treated cells.
May–Grünwald–Giemsa cytospin staining
MLL-AF9/NrasG12D leukaemia cells were treated with 1 ug ml−1 doxycycline for 2 days to induce shRNA expression from TRMPV-Neo or TtTMPV vectors, or treated with 100 nM JQ1 for 2 days. 50,000 cells were resuspended in 100 μl FACS buffer (5% FBS, 0.05% NaN3 in PBS) and cytospun onto glass slides using a Shandon Cytospin 2 Centrifuge at 500 rpm for 5 min. May–Grünwald (Sigma) and Giemsa (Sigma) stainings were performed according to manufacturer’s protocols. Images were collected using a Zeiss Observer Microscope with a 340 objective.
BrdU cell-cycle analysis and annexin V flow cytometry
BrdU incorporation assays were performed according to the manufacturer’s protocol (BD, APC BrdU flow kit), with cells pulsed with BrdU for 30 min. Cells were co-stained with 7-aminoactinomycin D or 4′,6-diamidino-2-phenylindole (DAPI) for DNA content measurement. For all conditional shRNA experiments, the analysis was gated on Venus+/dsRed+ (shRNA+) cell populations. Annexin V apoptosis staining was performed according to the manufacturer’s protocol (BD, APC annexin V). To analyse shRNA-mediated induction of apoptosis specifically, annexin V was quantified in viable shRNA-expressing cells (FSC/SSC; Venus+/dsRed+). Notably, this gating selectively evaluates early apoptotic cells (Annexin V+, DAPI−), excluding accumulated dead cells (Annexin V+, DAPI+). All analyses were performed using FlowJo software (Tree Star).
shRNA experiments in human AML cell lines
THP-1 and MOLM-13 cells were modified to express the ecotropic receptor and rtTA3 using retroviral transduction of MSCV-RIEP (MSCV-rtTA3-IRES-EcoR-PGK-Puro) followed by drug selection (0.5 and 1 ug ml−1 puromycin for 1 week, respectively). The resulting cell lines were transduced with ecotropically packaged TRMPV-Neo-shRNA retroviruses, selected with 400 μg ml−1 G418 for 1 week and treated with 1 μg ml−1 doxycycline to induce shRNA expression. The relative change in Venus+/dsRed+ (shRNA++) cells was monitored on a Guava Easycyte (Millipore). BrdU cell-cycle analysis was performed as described above.
Adult primary leukaemia sample analysis
The study was approved by the Institutional Review Board (ethics committee) of the Medical University of Vienna. Primary leukaemic cells were obtained from peripheral blood or bone marrow aspirate samples. Informed consent was obtained before blood donation or bone marrow puncture in each case. Diagnoses were established according to criteria provided by the French-American-British (FAB) cooperative study group36,37 and the World Health Organization (WHO)38. Mononuclear cells were prepared using Ficoll and stored in liquid nitrogen until used. HL60 and MOLM13 cell lines (obtained from DSMZ) were included as controls. After thawing, the viability of AML cells ranged from 70% to 99% as assessed by trypan blue exclusion. Primary cells (thawed mononuclear cells, 5–10 × 104 cells per well) and cell lines (1-5 × 104 cells per well) were cultured in 96-well microtitre plates (TPP) in RPMI-1640 medium (PAA laboratories) with 10% fetal calf serum (FCS, Pasching) in the absence or presence of JQ1 (10–5,000 nM) at 37 °C (5% CO2) for 48 h. In selected experiments, primary AML cells were incubated with JQ1 in the presence or absence of a cocktail of proliferation-inducing cytokines: recombinant human (rh) G-CSF, 100 ng ml−1 (Amgen), rhSCF, 100 ng ml−1 (Peprotech) and rhIL-3, 100 ng ml−1 (Novartis). After 48 h, 0.5 μCi 3H-thymidine was added (16 h). Cells were then harvested on filter membranes in a Filtermate 196 harvester (Packard Bioscience). Filters were air-dried and the bound radioactivity was measured in a β-counter (Top-Count NXT, Packard Bioscience). All experiments were performed in triplicates. Proliferation was calculated as a percentage of control (cells kept in control medium), and the inhibitory effects of JQ1 were expressed as IC50 values. In 7 out of 12 patients, drug-exposed cells were analysed for morphologic signs of differentiation by Wright-Giemsa staining on cytospin slides. The thymidine incorporation assay was chosen as a proliferation assay because of its superior sensitivity and ease of implementation for suspension cells, as compared to other proliferation assays, such as MTT.
Paediatric primary leukaemia sample analysis
Diagnostic bone marrow samples were collected, under protocols approved by an institutional review board, from newly diagnosed children with acute leukaemia. Informed consent was obtained in accordance with the Helsinki protocol. At the time of collection, primary leukaemic cells were enriched by density centrifugation using Ficoll-Paque PLUS (GE Healthcare) and subsequently stored in liquid nitrogen. Vials of cryopreserved cells were thawed, resuspended in media, and live leukaemic cells were enriched by density centrifugation. Cells were maintained in supplemented media with 20% FBS. All leukaemia cell cultures were incubated at 37 °C in 5% CO2. Primary leukaemia samples were treated with dose-ranges of JQ1 and vehicle control for 72 h in 96-well plates. For the annexin binding assays, cells were harvested and stained with Annexin V-PEand 7-AAD (BD Pharmingen), read on a FACSCalibur and analysed with FlowJo software (Tree Star). For the WST-1 assays, WST-1 reagent (Roche Diagnostics) was added to the culture medium (1:10 dilution) and absorbance was measured at 450 nm using a Bio-Rad model 680 microplate reader (Bio-Rad Laboratories). WST-1 assays were performed in triplicate. Primary leukaemia samples were treated with 250 nM JQ1 and vehicle control for 48 h in 96-well plates. Cytospins were prepared at baseline, 24 h and 48 h and stained with Wright-Giemsa solution (Sigma-Aldrich). Images were acquired using a Nikon Eclipse E600 microscope system (Nikon). Although similar to other metabolic assays measuring cell proliferation (for example, MTT), WST-1 has superior sensitivity for the assessment of cytotoxicity in primary leukaemia samples.
Histological analysis of bone marrow
Paraffin-embedded sections were stained with haematoxylin and eosin (H&E). Photographs were taken on a Nikon Eclipse 80i microscope with a Nikon Digital Sight camera using NIS-Elements F2.30 software at a resolution of 2560 × 1920. Using Adobe Photoshop CS2, images were re-sized and set at a resolution of 300 pixels inch−1, autocontrast was applied and unsharp mask was used to improve image clarity.
FACS evaluation of normal haematopoiesis
Bone marrow cells were obtained by flushing mouse femurs and tibias, followed by erythrocyte lysis with ACK buffer (150 mM NH4Cl, 10 mM KHCO3 and 0.1 mM EDTA). Samples were washed in FACS buffer (5% FBS, 0.05% NaN3 in PBS), followed by staining (two million cells in 100 μl of FACS buffer) for 1 h. Antibody dilutions used were: mouse haematopoietic lineage eFluor 450 cocktail (1:100), PE-Cy7 anti-mouse Kit (1:50), APC anti-mouse Sca-1 (1:100), APC anti-mouse B220 (1:100), APC anti-mouse Cd11b (1:100), APC anti-mouse TER-119 (1:100) and APC anti-mouse Gr-1 (1:100). Stained samples were analysed on an LSRII flow cytometer. Data analysis was performed using FlowJo software (Treestar).
Expression microarrays
Microarrays were performed through the CSHL microarray shared resource. RNA was isolated from 107 cells using RNeasy Mini Kit (Qiagen). RNA quality was assessed on an Agilent 2100 Bioanalyser, RNA 6000 Pico Series II Chips (Agilent) and samples with a RIN score of 7.0 or greater passed. RNA was amplified by a modified Eberwine technique, amplified antisense RNA was then converted to cDNA using a WT Expression kit (Ambion). The cDNA was then fragmented and terminally labelled with biotin, using the Affymetrix GeneChip WT Terminal Labelling kit (Affymetrix). Samples were then prepared for hybridization, hybridized, washed and scanned according to the manufacturer’s instructions on Mouse Gene ST 1.0 GeneChips (Affymetrix). Affymetrix Expression Console QC metrics were used to pass the image data. Raw data was processed by Affymetrix and Limma packages in R-based Bioconductor. Heatmaps were generated using GenePattern software39; RMA-processed microarray data was converted into a log2 scale, selected gene lists were row-normalized and visualized using the HeatMapImage module on GenePattern. All raw microarray data files are available from the Gene Expression Omnibus (GSE29799).
GSEA analysis
Gene set enrichment analysis20 was performed using GSEA v2.07 software with 1,000 phenotype permutations. Leukaemia-stem-cell and Myc gene sets were obtained from the indicated publications21,22,26. The macrophage-development gene set was obtained from the Ingenuity Pathway Analysis (IPA) software (Ingenuity). To perform GSEA on human microarray data, mouse gene sets were converted into human gene names using bioDBNet dbWalk (http://biodbnet.abcc.ncifcrf.gov/db/dbWalk.php) or manually using the NCBI database. A detailed description of GSEA methodology and interpretation is provided at http://www.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html. In brief, the normalized enrichment score (NES) provides ‘the degree to which a gene set is overrepresented at the top or bottom of a ranked list of genes’. The false discovery rate q-value (FDR q-val) is ‘the estimated probability that a gene set with a given NES represents a false positive finding’. ‘In general, given the lack of coherence in most expression datasets and the relatively small number of gene sets being analyzed, an FDR cutoff of 25% is appropriate.’ Gene sets used in this study are included in Supplementary Table 2.
Chromatin immunoprecipitation
ChIP assays were performed exactly as described40. Crosslinking was performed with sequential EGS (Pierce) and formaldehyde41. All results were quantified by quantitative PCR performed using SYBR green (ABI) on an ABI 7900HT. Each immunoprecipitate signal was referenced to an input standard-curve dilution series (immunoprecipitate/input) to normalize for differences in starting cell number and for primer amplification efficiency.
qRT–PCR
RNA was prepared using TRIzol reagent (Invitrogen). Synthesis of cDNA was performed using qScript cDNA SuperMix (Quanta Biosciences). Quantitative PCR analysis was performed on an ABI 7900HT with SYBR green (ABI). All signals were quantified using the ΔCt method. All signals were normalized to the levels of Gapdh.
Antibodies
The anti-Brd4 antibody used for western blotting was a gift from G. Blobel. The anti-Brd4 antibody used for ChIP was purchased from Sigma (HPA015055). The anti-Myc antibody was purchased from Epitomics (1472-1). Antibodies used in FACS were: APC anti-mouse CD117/Kit (Biolegend, 105811), APC anti-mouse CD11b (Biolegend, 101211), Pacific Blue anti-mouse CD45.2 (Biolegend, 109820), mouse haematopoietic lineage eFluor 450 cocktail (eBioscience, 88-7772-72), APC anti-mouse CD45R/B220 (Biolegend, 103212), APC anti-mouse TER-119/erythroid cells (Biolegend, 116212), APC anti-mouse Ly-6G/Gr-1 (eBioscience, 17-5931), PE-Cy7 anti-mouse CD117/Kit (eBioscience, 25-1171-82) and APC anti-mouse Sca-1 (eBioscience, 17-5981-81). The anti-β-actin HRP antibody was purchased from Sigma (A3854).
- 31.Taylor J, Schenck I, Blankenberg D, Nekrutenko A. Using galaxy to perform large-scale interactive data analyses in. Curr. Protoc. Bioinformatics Ch. 2007;10 doi: 10.1002/0471250953.bi1005s19. Unit 10.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nature Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 33.Hemann MT, et al. An epi-allelicseries of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nature Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- 34.Wei J, et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell. 2008;13:483–495. doi: 10.1016/j.ccr.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wunderlich M, Mulloy JC. Model systems for examining effects of leukemia-associated oncogenes in primary human CD34+ cells via retroviral transduction. Methods Mol. Biol. 2009;538:263–285. doi: 10.1007/978-1-59745-418-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bennett JM, et al. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br. J. Haematol. 1976;33:451–458. doi: 10.1111/j.1365-2141.1976.tb03563.x. [DOI] [PubMed] [Google Scholar]
- 37.Bennett JM, et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British cooperative group. Ann. Intern. Med. 1985;103:620–625. doi: 10.7326/0003-4819-103-4-620. [DOI] [PubMed] [Google Scholar]
- 38.Vardiman JW, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 39.Reich M, et al. GenePattern 2.0. Nature Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 40.Steger DJ, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell. Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng PY, Vakoc CR, Chen ZC, Blobel GA, Berger SL. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques. 2006;41:694–698. doi: 10.2144/000112297. [DOI] [PubMed] [Google Scholar]
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Author Contributions J.Z., J.S. and C.R.V. designed research, performed experiments and analysed data. E.W., A.R.R. and M.J.T. performed experiments and analysed data. C.J. performed microarray experiments. A.C. assisted with shRNA library design. H.H., E.A.S., D.M., K.B., P.B. and P.V. performed experiments with primary leukaemia specimens. J.C.M. and M.W. generated and provided engineered AML lines. S.C.K. performed histological analysis. J.Q. and J.E.B. designed research and synthesized and supplied JQ1. J.Z., S.W.L. and C.R.V. co-wrote the paper. S.W.L. and C.R.V. supervised the research.
Author Information Reprints and permissions information is available at www.nature.com/reprints. Raw microarray data can be accessed at Gene Expression Omnibus, GSE29799. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Chi P, Allis CD, Wang GG. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nature Rev. Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nature Rev. Cancer. 2010;10:23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilliland DG, Jordan CT, Felix CA. The molecular basis of leukemia. Hematology (Am. Soc. Hematol. Educ. Program) 2004:80–97. doi: 10.1182/asheducation-2004.1.80. [DOI] [PubMed] [Google Scholar]
- 4.Figueroa ME, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl Acad. Sci. USA. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krivtsov AV, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 9.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuber J, et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nature Biotechnol. 2011;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yokoyama A, et al. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 13.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 14.French CA, et al. BRD4–NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 2003;63:304–307. [PubMed] [Google Scholar]
- 15.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuber J, et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009;23:877–889. doi: 10.1101/gad.1771409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 19.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 20.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Somervaille TC, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong P, et al. The miR-17-92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res. 2010;70:3833–3842. doi: 10.1158/0008-5472.CAN-09-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol. Cell. Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuhmacher M, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt M, Nazarov V, Stevens L, Watson R, Wolff L. Regulation of the resident chromosomal copy of c-myc by c-Myb is involved in myeloid leukemogenesis. Mol. Cell. Biol. 2000;20:1970–1981. doi: 10.1128/mcb.20.6.1970-1981.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soucek L, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 30.Wilson A, et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.