Abstract
PPP1R13L was initially identified as a protein that binds to the NF-κB subunit p65/RelA and inhibits its transcriptional activity. It also binds p53 and inhibits its action. One set of experimental findings based on overexpression of PPP1R13L indicates that PPP1R13L blocks apoptosis. Another set of experiments, based on endogenous production of PPP1R13L, suggests that the protein may sometimes be pro-apoptotic. We have used primary mouse embryonic fibroblasts (MEFs), dually transformed by HRAS and adenovirus E1A and differing in their p53 status, to explore the effects of PPP1R13L overexpression, thus examining the ability of PPP1R13L to act as an oncoprotein. We found that overexpression of PPP1R13L strongly accelerated tumor formation by RAS/E1A. PPP1R13L overexpressing cells were depleted for both p53 and active p65/RelA and we found that both p53-dependent and -independent apoptosis pathways were modulated by PPP1R13L. Finally, studies with the proteasome inhibitor MG132 revealed that overexpression of PPP1R13L causes faster p53 degradation, a likely explanation for the depletion of p53. Taken together, our results show that increased levels of PPP1R13L can increase tumorigenesis and furthermore suggest that PPP1R13L can influence metastasis.
Keywords: PPP1R13L gene, malignant transformation, tumorigenesis, tumor cell migration, tumor suppressor p53
INTRODUCTION
The recently discovered apoptosis stimulating proteins of p53 (ASPP) family consists of three members, ASPP1, ASPP2, and the most evolutionary conserved PPP1R13L (iASPP), also known as RAI [1,2]. The three factors are encoded by the genes PPP1R13B, TP53BP2, and PPP1R13L, respectively. The family of proteins is characterized by a highly conserved carboxyl terminus-ankyrin repeats, Src homology 3 (SH3) domain and a proline-rich region. The proteins are thought to bind though these structures to several other proteins, including the tumor suppressor protein p53, BCL2, RelA/p65, protein phosphatase 1, YES-associated protein [1,2].
The best-known function of the ASPP1 and ASPP2 proteins is their ability to induce apoptosis via p53 and its family members, p63 and p73 [3]. ASPP1 and ASPP2 enhance the ability of p53 by causing it to specifically upregulate the expression of pro-apoptotic genes rather than genes involved in cell cycle arrest [3].
The third family member, PPP1R13L was initially identified and characterized as a novel RelA-associated inhibitor that binds to the nuclear factor-kappa B (NF-κB) subunit p65/RelA and inhibits its transcriptional activity [4]. Later it has become clear that PPP1R13L also binds to p53. In the nematode C. elegans the primary role of Ce-iASPP is to inhibit the pro-apoptotic activity of Ce-p53, which is normally stimulated in response to genotoxic stress. It is unknown if this finding can be generalized to other higher eukaryotes, as C. elegans lack NF-κB, so that the second major arm of the pathways is missing [5].
Two schools of thought exist regarding the primary role of PPP1R13L in mammals. The larger set of reports, which are mainly based on constitutive overexpression of PPP1R13L in transformed cells transfected with the relevant cDNA, indicate that the protein blocks apoptosis, presumably by binding and blocking p53 [3]. Another report, based on endogenous production of PPP1R13L, suggests that PPP1R13L may be pro-apoptotic [6]. Both set of findings could very well be true and reflect different roles at different concentration levels and putative activation levels in different cells.
Overexpression of PPP1R13L was detected in eight human breast carcinomas expressing wild-type p53 and normal levels of ASPP and in certain leukemias, underlining its potential importance in cancer [7]. Thus, if PPP1R13L has an anti-apoptotic role it might play role as an oncogene; in contrast if it is pro-apoptotic it might act as tumor suppressor.
To study the role of PPP1R13L in tumorigenesis we have used a combined in vitro and in vivo strategy, and used primary mouse embryonic fibroblasts (MEFs) as a model system. Transformation of the cells with a combination of oncogenic v-Ha-RAS Harvey rat sarcoma viral oncogene homologue (HRAS) and adenovirus E1A was used to obtain genetically defined malignant cells. Transformation through oncogenic ras requires either a cooperating oncogene or the inactivation of a tumor suppressor to abrogate senescence. The adenovirus E1A oncogene served this function.
MEFs expressing adenovirus E1A and activated RAS undergo p53-dependent apoptosis when treated with DNA-damaging or chemotherapeutic agents such as adriamycin (doxorubicin), or etoposide [8]. They also rapidly form tumors in nude mice. Utilizing these features we have explored the effects of PPP1R13L expression on dually transformed cells differing in their p53 status to examine the ability of PPP1R13L to act as an oncoprotein. We found that overexpression of PPP1R13L strongly accelerated tumor formation by RAS/E1A transformed cells and gave a phenotype with multiple tumor nodes, consistent with increased metastasis. At the same time the PPP1R13L overexpressing cells were depleted for both p53 and active p65/RelA. Through several different lines of investigation, we provide evidence that both p53-dependent and -independent apoptosis pathways are modulated by PPP1R13L. Finally, studies with the proteasome inhibitor MG132, suggest that overexpression of PPP1R13L causes faster p53 degradation, a likely explanation for the depletion of p53. The combined results indicate that overexpression of PPP1R13L will accelerate tumor growth and may be important for tumor metastasis.
MATERIALS AND METHODS
Cells, Plasmids and Gene Transfer
WT and p53−/− MEFs were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) and used between passages 3 and 5. p53−/− and wild-type MEFs were infected with retroviral vectors overexpressing both HRAS and E1A and retroviral vector expressing the PPP1R13L protein, to obtain transformed fibroblasts with defined p53 and PPP1R13L status. Cells infected with the respective empty retroviral vectors were used as a negative control. The retroviral vectors were as follows: LPC, control vector expressing puromycin phosphotransferase (puro). Control cycline D pLPC, a human cyclin D cDNA in pLPC puro. RAI pLPC (kindly donated by Kevin M. Ryan, Beatson Institute for Cancer Research, Glasgow, United Kingdom) was cloned into PCB6+ from cDNA with primers containing KpnI and NotI restriction sites. An EcoRI/EcoRV fragment from plasmid was then cloned into pLPC to generate RAI pLPC vector. Oncogenic RAS (HRASV12) and E1A were expressed by using WZL-Hygro-based retroviral vectors. E1A/HRASV12 was expressed by using a modified pBabe HRASV12 retroviral vector. Retroviruses were generated by transfection of Phoenix packaging cells (G. Nolan, Stanford University, Stanford, California). Infective supernatants were then administered to target cells followed by appropriate drug selection, puromycin (2 µg/mL, 2 d, Sigma Aldrich, St. Louis, MO) or hygromycin (75 µg/mL, 3 d, Sigma Aldrich). Due to the high efficiency of transfection and selection no cloning was performed. Calcium phosphate precipitation was used to transiently transfect cells.
Colony Formation Assay and Soft Agar Transformation Assay
Five thousand cells were plated in 60-mm dishes in complete culture medium. Triplicate experiments were performed for each cell type. The medium was changed on cells every 4 d. On the 10th day, plates were fixed with 4% formaldehyde, washed twice, and stained with 1% crystal violet, and the colonies were quantified. For soft agar transformation assays, cells were resuspended in 0.3% Noble agar (in DMEM supplemented with 10% FBS) at a density of 2 × 104 cells per well (in 6-well plates), and were plated onto solidified 0.5% Noble agar-containing bottom layer medium. Plated cells were fed weekly and were incubated at 37°C for 21 d. Plates were stained with 0.005% crystal violet for 1 h. Colonies were counted under a dissecting microscope. Experiments were repeated twice, and MEFs from two different infections were used in each assay.
DNA Content Analysis
Cell concentration for each sample was adjusted so that all samples were of the same concentration. Cells growing on10cmculture dishes were harvested and transferred to a 15-mL conical tube as a single cell suspension. Subsequently, they were centrifuged at 1000g for 5 min, and the supernatant was removed. Samples were washed twice in PBS without calcium or magnesium. At last wash, cell count of the total number of cells was performed, and recorded on the tubes. Pellets were resuspended in approximately 500 µL of PBS. Five milliliters of cold ethanol was added and samples were fixed at 4°C overnight. Next day, 5 × 106 cells were transferred to a 15-mL conical tube, centrifuged at 1000g to remove ethanol. After vortexing pellets were washed two times in 5 mL PBS + 1%bovine serum albumin (BSA) or calf serum. One hundred microliters of 10 × PI solution was added to each sample (500 µg/mL PI [Sigma, St. Louis, MO] in 3.8 × 10−2M sodium citrate, pH 7.0). Next, 100 µL of boiled RNase A (10 mg/mL prepared in 10 mM Tris–HCL, pH 7.5) was added and samples were incubated at 37°C for 30 min. Samples were analyzed by flow cytometry.
BrdU/DAPI Staining
The cells were grown on cover slips in a 6-well plate. After labeling with BrdU (10 mg/mL 5-bromo-2′-deoxyuridine [Sigma B-9285]) cells were incubated for 6 h at 37°C. At the end of incubation period, cells were washed twice with PBS. Cells were fixed with 3, 7% formaldehyde for 30 min at room temperature (RT). Next, cells were washed twice with PBS and permabilized with acetone −20°C for 5 min. After washing two times with PBS, 2 M HCL was added and samples were rotated for 10 min. Subsequently, samples were washed twice, blocked with 1% BSA in PBS containing 0.1% Tween for 1 h at RT. Anti-BrdU primary antibody (mouse, 1:4000) in blocking solution was added for 1 h at RT. Samples were washed four times with blocking solution, with 5 min rotation between washes. Rhodamine-conjugated anti-mouse secondary antibody was added for 1 h at RT. After incubation samples were washed four times with blocking solution with 5 min rotation between washes. Slides were mounted with media containing DAPI and analyzed under fluorescence microscopy.
Cell Viability and Apoptosis
The cells were plated in 96-well plates (2 × 104 cells per well) or 10 cm-culture dishes at concentration 7.5 × 105 cells per plate. Twenty-four hours after plating cells were washed and medium was replaced with culture medium containing appropriate drug at indicated concentrations. After drug addition cells were incubated for indicated time periods. Cell viability was measured using CellTiter-Blue® Cell Viability Assay (Promega Corporation, Madison, WI) according to the protocol provided by the manufacturer. Cells growing on 10 cm culture plates were stained with fluorescein isothiocyanate (FITC)-Annexin V. For flow cytometry, staining with FITC-Annexin V and propidium iodide were performed according to the manufacturer’s instructions (Pharmingen, BD Biosciences, San Jose, CA).
Western Blotting
Whole-cell lysates were derived by lysing cell pellets in SDS sample buffer (60 mM Tris–HCl at pH 6.8, 10% glycerol, 2%SDS, 5%2-mercaptoethanol). Samples corresponding to 30 µg of protein were separated on SDS–PAGE gels and transferred to Immobilon-P-membranes (Millipore, Billerica, MA). The primary antibodies used were as follows: Anti-p53 polyclonal antibody (CM5, or 505, 1:1000, NovaCastra, Newcastle Upon Tyne, UK), anti-HRAS antibody (C-20, sc-520, 1:1000 Santa Cruz Biotechnology, Santa Cruz, CA), anti-E1A antibody (sc-430 (13S-5) 1:500, Santa Cruz Biotechnology), anti-RAI (1:500, Sigma Aldrich), anti-cycline A antibody (c-19, 1:1000 Santa Cruz Biotechnology), anti-cdk4 antibody (c-22, 1:500 Santa Cruz Biotechnology), anti-cyclin D (c-20 1.1, 1:1000 Santa Cruz Biotechnology), anti-phospho NF-κB p65 antibody (1:1000 Santa Cruz Biotechnology), and anti-Tubulin antibody (1:1000, Abcam, Cambridge, UK).
Immunofluorescence
Cells growing at a confluence of 50% on coverslips in 6-well plates were treated with the indicated drugs and for the indicated time period. At the end of treatment, the cells were fixed in 3.7% formaldehyde in PBS followed by a 5 min permeabilization in 0.1% Triton X-100 in PBS and then incubated in PBS containing 0.1% BSA before staining with primary antibody. For caspase-3 expression, anti-cleaved caspase-3 monoclonal antibody (1:1000, R&D Systems, Minneapolis, MN) was used. Subsequently slides were counterstained with DAPI and analyzed under fluorescence microscopy. Cells were quantified by averaging three optical fields under 40× magnifications on two slides from two independent experiments.
Tumorigenicity Assays
Four- to 6-wk-old nude mice were obtained from Cold Spring Harbor Laboratory and maintained in the animal facility building at Cold Spring Harbor Laboratory in accordance with the institutional guidelines. Animals were kept in numbers no greater that 5/cage and had unlimited access to food and water. A total of 1.5 × 106 cells from each of the MEF cultures were suspended in PBS and injected subcutaneously into each flank of an animal, with four animals being used for each cell line. After injection, mice were monitored three times per week. Once apparent, the tumors were measured with calipers, and tumor volume was calculated using following formula: tumor volume (mm3) as length × width2 ×π/6. Animals with tumor measuring in excess of 1200 mm3 were sacrificed. For analysis of protein expression, tumors were snap–froze and pulverized in liquid nitrogen using a mortar and pestle. For Western blotting powdered tumors were lysed in Laemmli buffer (2% SDS, 10% glycerol, 5% 2-mercaptoethanol, 0.002% bromophenol blue, 0.075 M Tris–HCL). For histology analysis, tumors were fixed for 16 h in 4% formalin in PBS before embedding and sectioning. The Cold Spring Harbor Laboratory Animal Care and Use Committee approved all mouse experiments described in this work.
Histopathology
Fixed tissue samples were embedded in paraffin, and sections (7 µm) were stained with hematoxylin and eosin (H&E) according to standard protocols. For detection of KI67, representative sections were deparaffinized, rehydrated and processed in graded alcohols using the avidin–biotin immunoperoxidase method. Briefly, sections were submitted to antigen retrieval by microwave oven treatment for 15 min in TRYLOGY buffer (Cell Marque, Inc., Rocklin, CA). First, slides were incubated in 10% normal goat serum for 30 min and then overnight at 4°C with appropriate diluted primary antibody (rabbit antibody against Ki67, NovaCastra). Next, slides were incubated with biotinylated goat rabbit-specific immunoglobulins (Vector Laboratories, Burlingame, CA) at 1:500 dilution for 30 min and then with avidin–biotin peroxidase complexes (1:25; Vector Laboratories) for 30 min. As a chromogen, diaminobenzidine was used, and hematoxylin as the nuclear counterstained. Histopathological evaluation of carcinomas was performed by an experienced pathologist (L.Z.) using paraffin embedded tumor sections stained with hematoxylin/eosin.
Wound Healing Assay
Briefly, in vitro “scratch” wounds were created by scraping confluent cell monolayers in Petri dishes with a sterile pipette tip. After 24 h incubation in DMEM with 0.5% serum, migration was quantified by counting cells migration distance from the wound edge. Phase contrast microscopy images were taken using Axioskop (Zeiss, Thornwood, NY), at 40× magnification. Each determination represents the mean of five measurements from at least two individual experiments.
Assays for Cell Survival in Suspension and Cell Growth in Methylcellulose
Both assays were performed according to previously published protocols [9].
Proteosome Inhibition
Unless otherwise noted, the cells were treated with 100 µM MG132 (Sigma Aldrich) for 4h.The cells were washed with PBS and subsequently lysed for 30 min at 4°C in a buffer containing 120 mM NaCl, 50 mM HEPES pH 7.2, 1 mM EDTA, 0.1% NP-40, and complete protease inhibitor cocktail (Roche Diagnostics Corporation, Indianapolis, IN). The protein concentration was determined using Bio-Rad protein assay. The samples were separated on SDS–PAGE gels and transferred to Immobilon-P-membranes (Millipore). The primary antibodies used were as follows: Anti-p53 polyclonal antibody (CM5, or 505, 1:1000, NovaCastra), anti-human PPP1R13L antibody (1:1000, Invitrogen Corporation, Carlsbad, CA), activated anti-RelA/p65 subunit of NF-κB (NLS signal) (Chemicon MAB 3026, Rosemont, IL) and anti-Tubulin antibody (1:1000, Abcam).
RESULTS
PPP1R13L Suppresses Apoptosis Induced by Multiple Stimuli in RAS/E1A Transformed Mouse Embryonic Fibroblasts
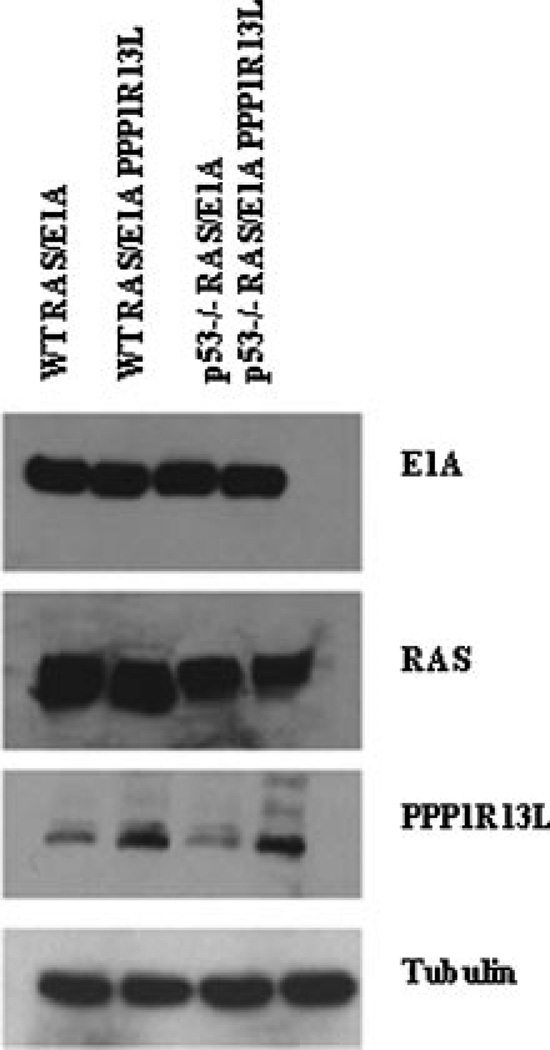
The tumor suppressor p53 is required for the efficient activation of apoptosis following treatment with chemotherapeutic compounds [10], and absence of p53 expression leads to a dramatic increase in cellular resistance to these agents. The hypothesis that the PPP1R13L protein is an inhibitor of p53, and that its expression enhances growth and tumorigenic potential of cells, led us to examine the effect of various anti-cancer agents on the apoptosis of our panel of four transformed cells types: wild-type and p53−/− RAS/E1A MEFs expressing PPP1R13L or empty vector. First, we quantified expression of the PPP1R13L mRNA and protein by qPCR and Western blot. Retroviral transduction increased the PPP1R13L mRNA expression about 100-fold and the protein expression approximately 3-fold compared to endogenous levels (Fig. 10; Supplementary Data). Then, the cells were treated with adriamycin or etoposide for 24 h using doses that have previously been reported to induce apoptosis in E1A/RASexpressing MEFs [8]. Figure 1a and b shows representative experiments from a series of four experiments in total. Adriamycin and Etoposide efficiently induced apoptosis in RAS/E1A wild-type MEFs but not in p53−/− RAS/E1A MEFs. Overexpression of PPP1R13L inhibited apoptosis induced by both compounds in MEFs RAS/E1A which are wild-type for p53 gene. Expression of PPP1R13L in cells, which were null for p53, had little effect.
Figure 1.
PPP1R13L suppresses apoptosis induced by multiple stimuli in RAS/E1A transformed MEFs and increases transformation in a way that is largely p53-dependent. (a) and (b) Cultures of RAS/E1A transformed wild-type or p53−/− MEFs, expressing PPP1R13L or vector alone were treated with 1 µg/mL adriamycin (a) or 100 µg/ mL etoposide (b) for 24 h (see Materials and Methods Section). Both adherent and floating cells where harvested and analyzed by flow cytometry. The percentage of apoptotic cells was determined by Annexin-V staining. (c) Cell transformation as quantified by colony forming assays. Colony numbers were estimated ~2 wk after transfection and represent the average ± SD determined from at least three transfections. (d) Cell transformation as quantified by soft agar assays. Cells expressing indicated gene combinations were plated in soft agar media and growth was monitored. Colonies were scored after 3 wk. Data represent the average ± SD determined from at least three independent experiments.
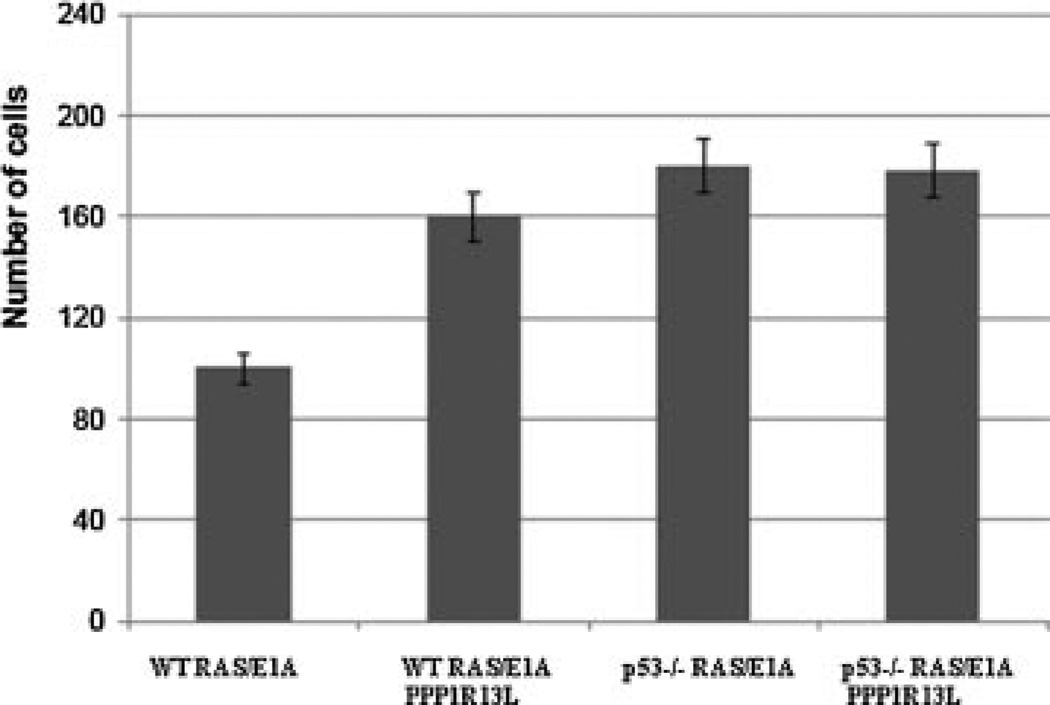
PPP1R13L Enhances Transformation In Vitro in a Manner That Is Largely P53 Dependent
Overexpression of the PPP1R13L protein could modulate RAS/E1A mediated transformation. Therefore, we plated the four cell types at low density and examined colony formation (Fig. 11; Supplemental Data). PPP1R13L-expressing p53-wild-type RAS/E1A MEFs cells grew significantly faster and formed more colonies than their control counterparts when plated at low density, but not quite as fast as the p53-null cells. PPP1R13L-expressing, oncogene-transduced MEFs, which were null for p53 tumor suppressor gene formed marginally more colonies as compared to p53−/− RAS/E1A MEFs control cells (Figure 1c).
The same infected mouse embryo cultures were also tested for their ability to form colonies in soft agar (Figure 1d). Wild-type MEFs and p53−/− MEFs transduced with both E1A and RAS formed anchorage-independent colonies, but with different efficiencies. In marked contrast to RAS/E1A transformed MEFs, those additionally overexpressing PPP1R13L formed two- to threefold more colonies in soft agarose. The RAS/E1A transduced MEFs null for p53 but overexpressing PPP1R13L possibly formed a marginally greater number of agar foci.
Thus, PPP1R13L was identified as a novel oncogene that cooperates with RAS and E1A to transform cells in vitro. One mechanism by which PPP1R13L potentiates the transforming activity of RAS/E1A oncogenes seems to be through its ability to inactivate p53 tumor suppressor gene.
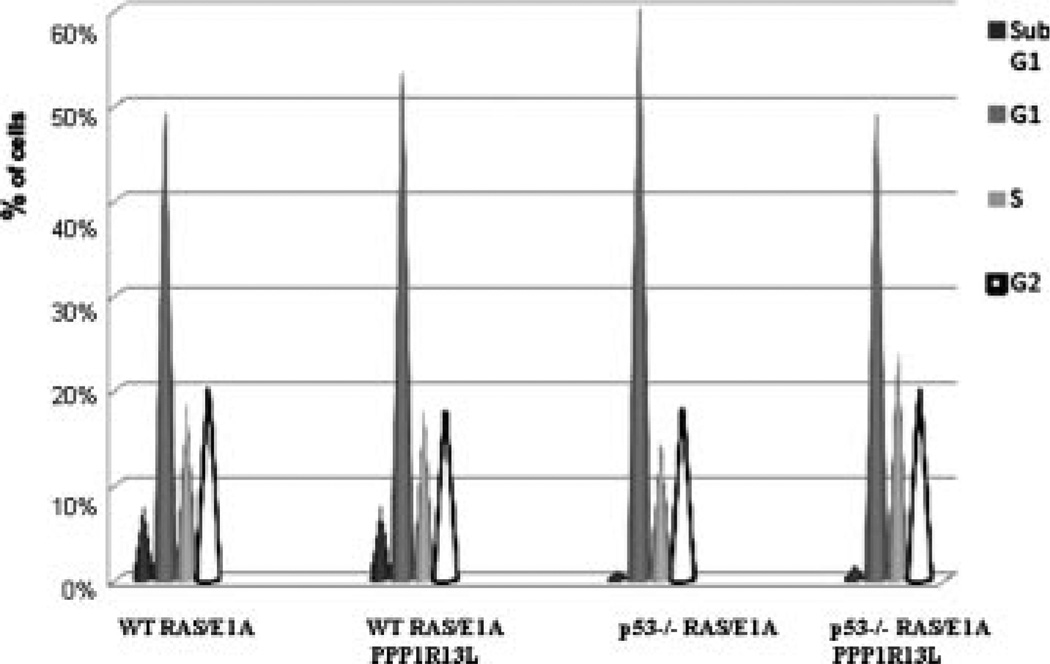
PPP1R13L Does Not Influence Cell Cycle Progression in RAS/E1A Transformed Mouse Embryonic Fibroblasts
We examined whether PPP1R13L overexpression had any effect on cell cycle progression in the studied cell types. FACS based cell-cycle analysis revealed no significant differences between RAS/E1A transduced MEFs, and their PPP1R13L expressing counterparts (Figure 2). Further, we confirmed these results by protein expression blotting for cyclines, which are involved in the progression of cells through the cell cycle (Fig. 12; Supplementary Data). No differences in the protein levels for cyclin A, cyclin-dependent kinase 4 (CDK4), cyclin D were observed between RAS/E1A transduced MEFs and their PPP1R13L overexpressing parallel lines. Taken together our results show that PPP1R13L overexpression inhibits p53-dependent apoptosis, but has no effect on cell cycle progression in RAS/E1A transformed MEFs.
Figure 2.
PPP1R13L does not influence cell cycle progression in RAS/E1A transformed MEFs. Cell cycle analysis of RAS/E1A transformed wild-type and p53−/− MEFs, expressing PPP1R13L or vector alone performed on FACS. Histogram of all cell cycle phases for all studied cell lines. Data are presented as the average from three independent experiments. The number of cells in each phase of cell cycle was determined as described in Materials and Methods Section.
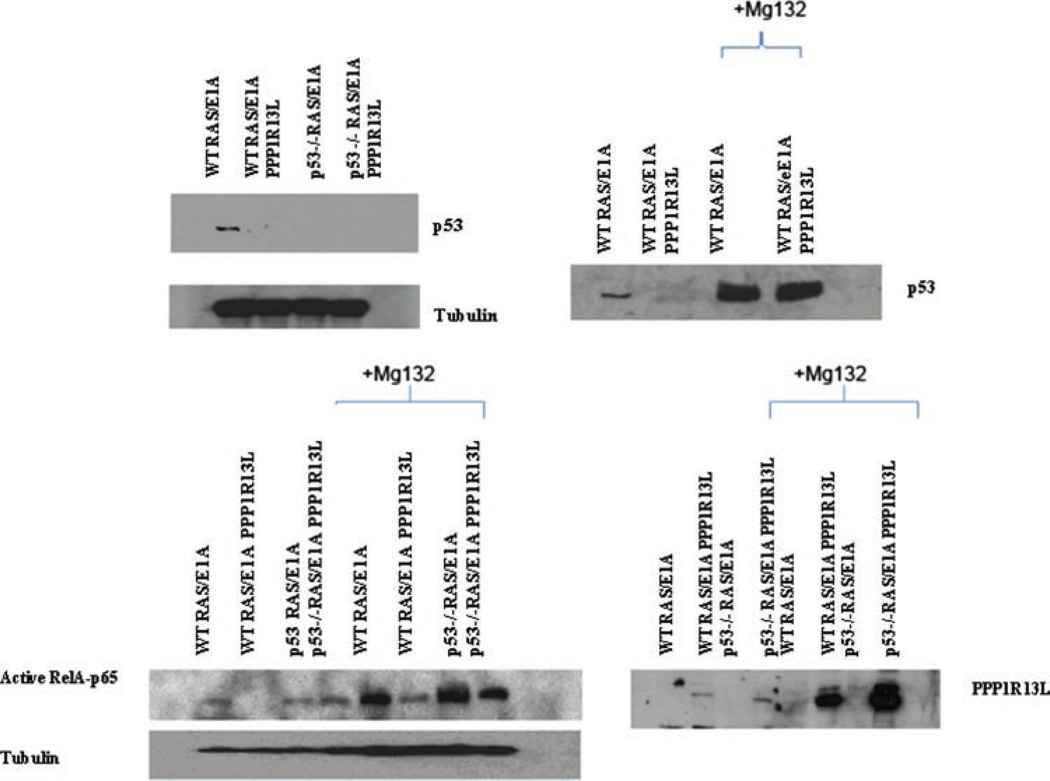
PPP1R13L Promotes p53 Turnover
PPP1R13L expression decreases p53 protein expression levels and PPP1R13L expression modulates p53-mediated apoptosis (Figure 3). We were interested whether PPP1R13L expression modulated p53 stability. Wild-type RAS/E1A transduced cells with or without PPP1R13L were incubated with or without the proteasome inhibitor MG132 and after the indicated time the level of p53 was determined. We found that overexpression of PPP1R13L depleted the cells of p53 (Figure 3). This effect was reversed by MG132. We observed a dramatic pile-up of p53 in the PPP1R13L-overexpressing, MG132-treated cells, consistent with the notion that p53 is rapidly degraded by the proteasomes in these cells. Overexpression of PPP1R13L also depleted the cells of the NF-κB subunit, RelA, which again could be reversed by MG132. Finally, MG132 caused a considerable pile-up of PPP1R13L itself, suggesting that this molecule also undergoes proteasomal degradation (Figure 3).
Figure 3.
PPP1R13L promotes p53 turnover. For p53 degradation RAS/E1A transduced wild-type MEFs expressing PPP1R13L/vector control were incubated with proteasome inhibitor 100 µM MG132 for 4 h. Lysates were prepared as described in Materials and Methods Section, and subjected for Western blotting with antip53 antibody. For PPP1R13L and RelA p65, proteasomal degradation ras/E1A transduced wild-type and p53−/− MEFs were incubated for 4 h with 100 µM MG132 inhibitor and treated as above. The membrane was probed with anti-PPP1R13L antibody and anti-phospho p65 antibody. Tubulin is shown as a loading control.
In Vivo, PPP1R13L Promotes Tumorigenesis Through Both p53-Dependent and -Independent Mechanisms
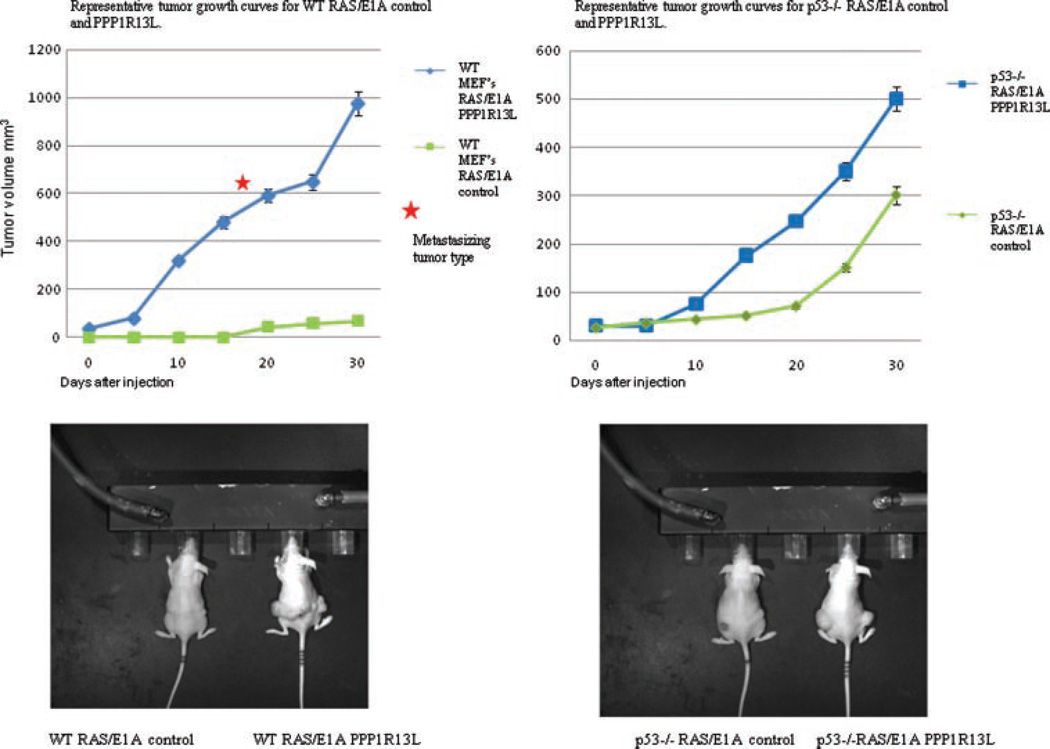
p53 tumor suppressor activity has been extensively studied in primary rodent cells, where p53 mutations can cooperate with the combinations of RAS and E1A oncogenes, in promoting oncogenic transformation [10]. Reduction of apoptosis provides a mechanism for this effect. We therefore investigated whether reduced susceptibility to apoptosis due to PPP1R13L overexpression would increase tumorigenic potential. We used cells harboring the same genetic lesions as described above to induce tumors in the rear flanks of immunocompromised mice.
After retroviral infection, cell populations were injected subcutaneously into nude mice and mice were continuously monitored for tumor formation at the sites of injection (Figure 4). Consistent with previous studies that E1A and RAS are sufficient for tumorigenic conversion of wild-type MEFs, tumor formed in animals injected with these cells within 20–30 d. E1A and RAS were also sufficient to cause tumorigenic conversion in p53−/− MEFs, with the tumors forming after 10–20 d. Importantly, in all cases tumors formed faster in animals injected with PPP1R13L expressing cells. Thus, p53-wild-type, PPP1R13L-overexpressing RAS/E1A cells were highly tumorigenic, with latency periods similar to the transformed p53−/− cells. Strikingly, RAS/E1A PPP1R13L expressing p53-wild-type MEFs developed aggressive subcutaneous tumors with local satellite tumors and also more distant metastasis in some cases. The RAS/E1A PPP1R13L p53−/− MEFs developed tumors faster than RAS/E1A p53−/− MEFs, and tumors formed by the former cells were invariably bigger (Figure 4). However, the metastasizing tumor type described above was not seen in the absence of p53. All in all, these results are in agreement with our in vitro results and support the idea that PPP1R13L overexpression favors tumorigenesis and progression. Histopathological evaluation revealed that the developing tumors could be classified as aggressive sarcomas with a high proliferation rate as assessed by the frequency of mitotic figures (Fig. 13; Supplemental Data, Figure 5).
Figure 4.
In vivo, PPP1R13L promotes tumorigenesis through both p53-dependent and -independent mechanisms. RAS/E1A-transformed wild-type or p53−/− MEFs of the indicated PPP1R13L genotypes (retroviral vector carrying PPP1R13L or only vector control) were injected into both flanks of athymic nude mice. The animals were checked periodically for tumor formation, and the dimensions of tumors were measured three times a week with calipers. The tumor size is plotted as tumor volume over time. The star indicates “metastasizing tumor type” observed in PPP1R13L RAS/E1A wild-type MEFs.
Figure 5.
Quantification of the incidence of mitotic cells in the histological sections of the tumors. Data represent average ± SD from three independent tumor sections. From each slide of each tumor at least six pictures were acquired and cells well counted.
Explanted Tumors Cells Retain Their In Vitro Phenotypes
The growth curves (Figure 6) show that in ex vivo cells that are p53-wild-type, PPP1R13L overexpression caused the cells to grow faster. The growth kinetics was similar to p53−/− RAS/E1A MEFs colonies. Western blot probing confirmed that each cell lines expressed RAS, E1A protein, and where indicated PPP1R13L protein (Figure 7). PPP1R13L suppressed apoptosis induced by multiple stimuli (adriamycin, etoposide) in tumor derived clones, which respond to treatment the same way as original clones (Fig. 14; Supplemental Data). This indicates that tumorigenicity did not result from additional mutations that might suppress apoptosis. In all experiments and under all tested conditions cells expressing PPP1R13L protein showed markedly higher viability, whether or not they were wild-type or p53−/−.
Figure 6.
Growth curves for explanted tumor cells. Cells growth curves were determined by plating 1 × 103 cells in a 6-well plate culture dishes and by culturing the cells in DMEM supplemented with 10% FBS under standard conditions. Cells were stained with trypan blue and counted every day for 7 d. The results are derived from at least three independent experiments with three replica dishes for every time points.
Figure 7.
Western blot of the explanted tumor cells. Cell extracts were prepared from cell cultured ex vivo from tumor mass and were subjected to Western blotting with the indicated antibodies. Tubulin served as a loading control.
We explored the use of the active form of caspase-3 for the detection of apoptotic events. Fluorescence images (Fig. 15; Supplementary Data) for PPP1R13L expressing cell, showed very few positive cells for activated caspase-3 in response to the apoptosis-inducing drug adrimaycin. In the case of p53−/− RAS/E1A PPP1R13L transduced clones almost no staining could be detected indicating high resistance of those cell to the above mentioned treatments. Additionally, no difference in BrdU incorporation could be detected between wild-type RAS/E1A, p53−/− RAS/E1A transduced cells and the paired PPP1R13L expressing cells (Fig. 16; Supplementary Data). Thus, PPP1R13L expression in tumor derived cells conferred cellular resistance to the cytotoxic effect of adriamycin and etoposide.
PPP1R13L Promotes Cell Migration and Modulates p53-Dependent Cell Death in Suspension
Enhanced cell migration is often a characteristic of invasive tumor cells [11]. Based on our demonstration that the PPP1R13L overexpressing wild-type tumors had a distinct phenotype we examined the effects of PPP1R13L modulation on migration. We tested the ability of the cells to migrate in a wound-healing assay. Cells were physically removed from a confluent monolayer and the ability of cells to fill the available space was quantified using phase-contrast microscopy. After 24 h, the p53−/− RAS/E1A transduced cells with or without PPP1R13L closed the wound completely. In comparison, the p53-wild-type PPP1R13L expressing transduced cells, moved significantly towards the gap, whereas wild-type RAS/E1A control cells migrated less (Figure 8). For phase contrast microscopy images see Figure 17; Supplementary Data.
Figure 8.
PPP1R13L promotes cell migration. Scratch wound assay on studied cells populations grown until confluence and on which a “mechanical” wound had been made. Quantitative determination (computer assisted phase contrast microscopy) of the ability of wild-type RAS/E1A and p53−/− RAS/E1A cells expressing PPP1R13L/vector control, to recolonize the wound. For phase contrast microscopy images see Supplementary Data (Fig. 17). The data are given as the means of five measurements from at least two independent experiments ± SEM.
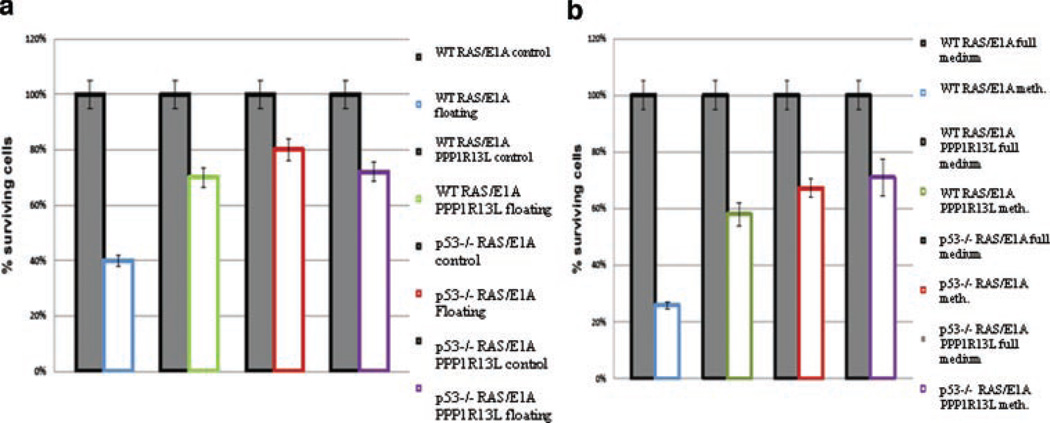
It has been shown that p53 promotes apoptosis in RAS/E1A transformed MEFs detached from substrate which could eliminate disseminated cells in vivo in the bloodstream [12]. To determine whether PPP1R13L expression might influence cell viability following disruption of cell–substrate interactions, we examined the viability of RAS/E1A transformed MEFs with known p53 and/or PPP1R13L status. Cell viability in floating assays and in methylcellulose was determined as illustrated in Figure 9a and b. RAS/E1A transduced p53−/− MEFs expressing PPP1R13L/vector control showed no significant reduction in viability after 24 h in suspension. These cells kept growing in suspension as if they were on solid substrate. However wild-type RAS/E1A MEFs when incubated in suspension showed a dramatic decrease in cell viability. The cell death of wild-type ras/E1a transformed MEFs was partially prevented by PPP1R13L overexpression. To further confirm our findings, we also studied cell survival in methylcellulose (Figure 9b). Again, survival rate of wild-type RAS/E1A transduced cells expressing PPP1R13L was strongly stimulated as compared to control cells (wild-type RAS/E1A). This increase in cell viability after incubation in methylcellulose was p53 dependent, since p53−/− RAS/E1A MEFs expressing PPP1R13L/vector control did not exhibit significantly altered cell growth. We conclude that in wild-type RAS/E1A transformed MEFs, abrogation of p53-dependent apoptosis by PPP1R13L overexpression promotes the survival of cells detached from substrate.
Figure 9.
PPP1R13L modulates p53-dependent cell death in suspension. (a) Viability of different cell types in suspension. Cell quantification was performed by MTT assay and/or CellTiter-Blue™ cell viability assay in 4 h after plating cells subjected to 24 h floating. All values were normalized to the values obtained in control cells growing 4 h on plate. Closed bars: viability of control cells, color bars: viability of cells after 24 h of floating. (b) Viability of cells subjected to incubation in methylcellulose. All values were normalized to the values obtained in control cells growing on the plate for a corresponding time (10 d). Cell quantification was performed by MTT assay and/or CellTiter-Blue™ cell viability assay.
DISCUSSION
In the present work, we have used RAS/E1A transformed mouse embryo fibroblasts (MEFs) as a model of tumor formation to analyze the role of PPP1R13L in tumor establishment and progression [13]. This model was chosen because transformation is induced in a simple and controlled way, avoiding the difficulties of analyzing the multiple and complex transformation mechanisms observed in cellular models derived from human tumors. In the present study, observations were made using both transformed p53 wild-type and transformed p53−/− MEFs making it possible to evaluate the role of p53 in the cells.
qPCR results showed that the level of mRNA encoding PPP1R13L protein increased about 100-fold after transfection. In contrast, the level of PPP1R13L protein was only moderately increased. This apparent discrepancy was most likely due to a fast turnover of the synthesized protein. This became apparent when degradation was blocked by the proteasome inhibitor MG132 and PPP1R13L accumulated to high levels.
Transfection with the PPP1R13L encoding vector led to depletion of active p65/RelA and p53 in the p53-wild-type cells. In the p53−/− cells active p65/RelA was not eliminated. It is not clear how p53 contributed to elimination of NF-κB. Again, the elimination seemed associated with rapid degradation, as the proteins piled up in cells treated with MG132. As it is known that PPP1R13L binds to these proteins, one can speculate that PPP1R13L-binding targets them for proteasome degradation. Although the mechanism whereby this occurs remains to be determined, it is noteworthy that PPP1R13L belongs to a group of proteins, acting as a “targeting proteins,” that is, facilitating the binding catalytic units of proteins to their targets. Thus, PPP1R13L may either act itself to direct p53 to the proteasome or guide a catalytic unit involved in protein modification to p53. Degradation would then follow as a consequence of the modification.
In agreement with previously published results [3], PPP1R13L-overexpressing cells were significantly more resistant to undergo an apoptosis process. We have found no changes in cell cycle progression of the cells. In conclusion, PPP1R13L is a potent p53 inhibitor, and inhibits apoptosis, but has no affect on cell-cycle arrest.
When we studied cell transformation with in vitro techniques it became apparent that transfection of wild-type p53 cells with the PPP1R13L-encoding vector led to a more transformed phenotype, reminiscent of that of the p53−/− cells. This was true for assays of cloning efficiency, soft agar growth and cell migration, all assays reflecting aspects of a transformed phenotype. The transfection had a limited effect on the p53−/− cells in these assays.
Our in vivo results demonstrated that the latency of tumors in nude mice was diminished and size was increased by transfection with the PPP1R13L encoding vector. In the case of p53 wild-type MEFs stably overexpressing PPP1R13L, we also observed a very characteristic “multifocal” phenotype, not observed in other settings. This phenotype suggests that the malignant cells migrated with greater ease in the animal. After injection of cells devoid of p53, those overexpressing of PPP1R13L produced tumors of greatly increased size, but the phenotype was still a solitary tumor.
Ex vivo studies indicated that the cell properties had not been changed during tumor growth. Thus, the in vivo studies allowed us to distinguish between absence of p53 and overexpression of PPP1R13L, and in particular the effect in p53−/− cells indicated that PPP1R13L must have other targets than p53. NF-κB was only depleted in p53 wild-type cells and one possibility is that depletion of p65/RelA led to the unusual multifocal phenotype in the p53 wild-type cells, but other targets for PPP1R13L could also cause this effect. We note with interest that Ryan et al. [14] have reported that loss of p65/RelA in these transformed cells accentuates their transformed character, but does not simulate the loss of p53. The multifocal phenotype could again reflect increased tumor migration and/or metastasis, a possibility that we currently pursue.
There are many possible reasons why the RAS/E1A p53 wild-type MEFs and RAS/E1A p53−/− MEFs cells respond differently, including the fact that the cells express different levels of NF-κB. The observation itself is however valid. We have corroborated the finding using the tail vein injection of the transformed cells, in which case the PPP1R13L overexpressing RAS/E1A p53 wild-type cells homed much better to internal organs than the other three cell types (data not shown).
To our knowledge, this study represents the first report that PPP1R13L functions as an oncoprotein, using both in vitro and in vivo approaches. However, PPP1R13L previously showed an ability to inhibit p53-mediated apoptosis in response to DNA damage in two human cell lines; U2OS and MCF7 [3]. Also, our findings are consistent with an early observation that the apoptotic, but not cell-cycle-arrest function of p53 is required to suppress the transforming activities of RAS and E1A [15].
There are two caveats with our experiments: First, the experiments deal with the expression of human PPP1R13L in mouse cells. Thus one must consider the possibility that species differences influence the results. We think this unlikely as PPP1R13L is known to be a highly conserved protein [3]. Secondly, the experiments deal exclusively with overexpression of PPP1R13L under the control of an external promoter without the putative regulation of the endogenous counterpart. Also, the experiments focus on transformed cells only. Thus, our experiments deal with the function of PPP1R13L in the context of a malignant setting and shed little light on its normophysiological role.
Although previous reports have suggest that PPP1R13L can function as an oncoprotein [3,16], we now provide a combination of in vitro and in vivo techniques to directly support this contention. Moreover, the finding that PPP1R13L can stimulate cell migration and metastasis though p53-independent mechanisms is surprising, and extend the interest in this protein beyond simply its antiapoptotic activity. How this occurs remains to be determined, but it is clear that PPP1R13L can signal through multiple targets. In principle, its ability to signal through distinct mechanisms may explain its different actions in the cell.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kevin Ryan for graciously providing the necessary vectors. We are grateful to Pamela Moody for helping with the FACS analysis, and Katherine McJunkin for assistance with the in vivo assays. This work was supported by The Lundbeck Foundation (grant number 43/05), The NovoNordisk Foundation (grant number 2007-11-30), Fabrikant Einar Willumsens Mindelegat, Foundation, The Boehringer Ingelheim Fond, The Danish Medical Research Council.
Abbreviations
- PPP1R13L
protein phosphatase 1, regulatory (inhibitor) subunit 13 like
- NF-κB
nuclear factor-kappa B
- MEFs
mouse embryonic fibroblasts
- HRAS
v-Ha-RAS Harvey rat sarcoma viral oncogene homologue
- E1A
adenovirus 5 early region 1A
- DMEM
Dulbecco’s modified Eagle’s medium
- BSA
bovine serum albumin
- BrdU
5-bromo-2′-deoxyuridine
- RT
room temperature
- PCR
polymerase chain reaction
Footnotes
SUPPLEMENTARY DATA
Are available at the Molecular Carcinogenesis website.
REFERENCES
- 1.Trigiante G, Lu X. The ASPPs and cancer. Nat Rev Cancer. 2006;6:217–226. doi: 10.1038/nrc1818. [DOI] [PubMed] [Google Scholar]
- 2.Slee EA, Lu X. The ASPP family: Deciding between life and death after DNA damage. Toxicol Lett. 2003;139:81–87. doi: 10.1016/s0378-4274(02)00421-6. [DOI] [PubMed] [Google Scholar]
- 3.Bergamaschi D, Samuels-Lev Y, O’Neil NJ, et al. iASPP oncoprotein is a key inhibitor of p53 conserved from worm to human. Nat Genet. 2003;33:162–167. doi: 10.1038/ng1070. [DOI] [PubMed] [Google Scholar]
- 4.Yang JP, Hori M, Sandra T, Okamoto T. Identification of a novel inhibitor of nuclear factor-kappaB, RelA-associated inhibitor. J Biol Chem. 1999;24:15662–15670. doi: 10.1074/jbc.274.22.15662. [DOI] [PubMed] [Google Scholar]
- 5.Ruvkun G, Hobert O. The taxonomy of developmental control in Caenorhabtidis elegans. Science. 1998;282:2033–2041. doi: 10.1126/science.282.5396.2033. [DOI] [PubMed] [Google Scholar]
- 6.Laska MJ, Strandbygard D, Kjeldgard A, et al. Expression of the RAI gene is conductive to apoptosis: Studies of induction and interference. Exp Cell Res. 2007;15:2611–2621. doi: 10.1016/j.yexcr.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, Wang M, Zhou C, Chen S, Wang J. The expression of iASPP in acute leukemias. Leuk Res. 2005;29:179–183. doi: 10.1016/j.leukres.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Lowe SW, Ruley HE. Stabilization of the p53 tumor suppressor is induced by adenovirus-5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 9.Nikiforov MA, Hagen K, Ossovskaya VS, et al. P53 modulation of anchorage independent growth and experimental metastasis. Oncogene. 1996;13:1709–1719. [PubMed] [Google Scholar]
- 10.Lowe SW, Ruley HE, Jacks T, Housman DE. P53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 11.Lowe SW, Jacks T, Housman DE, Ruley HE. Abrogation of oncogene-associated apoptosis allows transformation of p53-deficient cells. Proc Natl Acad Sci USA. 1994;91:2026–2030. doi: 10.1073/pnas.91.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samuelson AV, Lowe SW. Selective induction of p53 and chemosensitivity in RB-deficient cell by E1A mutants unable to bind RB-related proteins. Proc Natl Acad Sci USA. 1997;94:12094–12099. doi: 10.1073/pnas.94.22.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin HJ, Eviner V, Prendergast GC, White E. Activated H-ras rescues E1A-induced apoptosis and cooperates with E1A to overcome p53-dependent growth arrest. Mol Cell Biol. 1995;15:4536–4544. doi: 10.1128/mcb.15.8.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan KM, O’Prey J, Vousden KH. Loss of nuclear factor-κB is tumor promoting but does not substitute for loss of p53. Cancer Res. 2004;64:4415–4418. doi: 10.1158/0008-5472.CAN-04-1474. [DOI] [PubMed] [Google Scholar]
- 15.Ludwig RL, Bates S, Vousden KH. Differential activation of target cellular promotes by p53 mutants with impaired apoptotic function. Mol Cell Biol. 1996;16:4952–4960. doi: 10.1128/mcb.16.9.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan A, Lu X. ASPP: A new family of oncogenes and tumour suppressor genes. Br J Cancer. 2007;96:196–200. doi: 10.1038/sj.bjc.6603525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.