Abstract
The MA (matrix) domain of the retroviral Gag polyprotein plays several critical roles during virus assembly. Although best known for targeting the Gag polyprotein to the inner leaflet of the plasma membrane for virus budding, more recently studies have revealed that MA also contributes to selective packaging of the genomic RNA (gRNA) into virions. In this perspective, we will summarize recent progress in understanding how MA participates in genome incorporation. We will compare the mechanisms by which the MA domains of different retroviral Gag proteins influence gRNA packaging, highlighting variations and similarities in how MA directs the subcellular trafficking of Gag, interacts with host factors, and binds to nucleic acids. A deeper understanding of how MA participates in these diverse functions at different stages in the virus assembly pathway will require more detailed information about the structure of the MA domain within the full-length Gag polyprotein. In particular, it will be necessary to understand the structural basis of the interaction of MA with gRNA, host transport factors, and membrane phospholipids. A better appreciation of the multiple roles MA plays in genome packaging and Gag localization may guide the development of novel antiviral strategies in the future.
Introduction
All infectious retroviruses contain two copies of positive-stranded genomic RNA (gRNA) packaged into virions as noncovalently linked dimers 1-4. During the assembly of virus particles, the Gag polyprotein specifically packages gRNA, although a small amount of other viral RNAs and cellular RNAs are also found in virus particles 1, 5, 6. What makes the process of gRNA packaging so challenging to decipher is the highly specific and selective binding of Gag to gRNA in preference to other RNAs.
Viral RNAs undergo processing in the nucleus, just like cellular mRNAs, with the addition of a 5′ methylguanine cap and a 3′ polyA tail 7. A portion of the viral RNA undergoes splicing to yield mRNAs that encode Env glycoproteins and accessory viral factors. The viral gRNA remains unspliced and is indistinguishable from viral mRNAs that are exported into the cytoplasm to synthesize the structural Gag and GagPol proteins. Both the viral gRNA and mRNA contain the psi (ψ) sequence, a highly structured cis-acting element located within the 5′-untranslated region (UTR) and/or upstream coding regions of the gag gene 1,2. For some retroviruses, ψ is also present on spliced viral mRNAs, providing evidence that the ψ sequence itself is not sufficient to explain how gRNA is preferentially incorporated into virions 1,8.
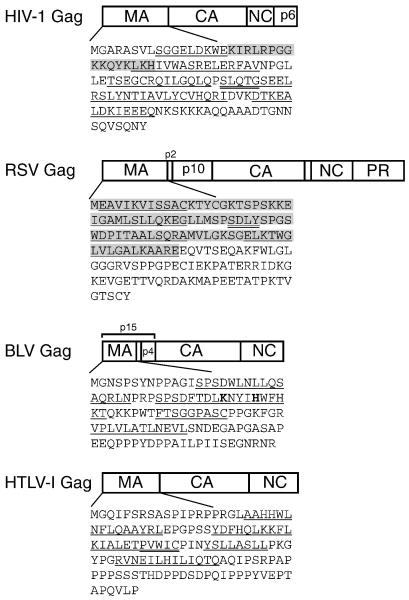
Efforts to identify the trans-acting factors involved in gRNA packaging have centered on understanding the RNA-binding activity of the Gag polyprotein. The Gag protein possesses general nucleic acid binding activity but also specifically recognizes the ψ sequence in the context of the genome-length viral RNA. Gag is synthesized as a polyprotein precursor, and in this form Gag directs genome packaging and particle assembly within the infected cell. Once virions are released, the Gag precursor is cleaved into MA (matrix), CA (capsid), and NC (nucleocapsid) proteins plus additional peptide sequences that vary for each retrovirus 1. A comparison of the Gag polyproteins and MA domain sequences of human immunodeficiency virus type 1 (HIV-1), Rous sarcoma virus (RSV), bovine leukemia virus (BLV), and human T cell lymphotropic/leukemia virus type 1 (HTLV-I) are depicted in Figure 1.
Figure. 1. Domain structure of Gag and sequences of the MA domains of RSV, HIV-1, BLV, and HTLV-1.
The organization of the domains in retroviral Gag polyproteins discussed in this review (MA, matrix; CA, capsid; NC, nucleocapsid); PR (protease). The amino acid sequences of each MA domain are indicated, with the α-helices underlined and the 310 helices underlined with double lines based on coordinates defined by three-dimensional structural analyses 106, 120-122. Residues in bold-face type represent residues that are required for interactions with nucleic acids. In HIV-1 MA, the gray box denotes the highly basic region (HBR) which is involved in nucleic acid binding, PIP(4,5)2 interaction, and plasma membrane targeting. For RSV MA, the gray box delineates the membrane-binding domain and the NLS in MA. The boundaries of the helices shown for HTLV-I MA are predicted from the three-dimensional structure solved for the homologous HTLV-II MA protein106. Residues indicated in bold-type in the BLV MA sequence have been implicated in genome encapsidation102.
Despite the fact that the Gag precursor selects and packages the viral genome, many biochemical and biophysical studies to date have focused on the nucleic acid binding activities of the mature MA and NC proteins. One reason for this approach is that MA and NC are more amenable to in vitro biochemical methods than the full-length Gag protein due to technical challenges in purifying full-length Gag from recombinant expression systems 9-11. Additionally, relatively pure MA and NC proteins could be readily isolated in high quantities from virions and subjected to biochemical analyses.
Studies focusing on NC demonstrate that it possesses high affinity for the ψ packaging sequence in vitro. Moreover, the NC domain of Gag determines whether its cognate genome is preferentially packaged over a competitor gRNA bearing the ψ sequence of a different retrovirus 2, 2-17. However, the relatively low affinity of NC for viral RNA sequences outside of the ψ region 16-19 implies that cooperative protein-protein interactions may be stimulated by the nonspecific RNA binding of the Gag NC domain during virus particle morphogenesis. The observation that viral RNA serves a scaffolding role during virus biogenesis5 highlights the importance of interactions between the NC domain in Gag and the viral genome in providing structural stability to the virus particle. However, it is possible that other regions of Gag, in particular the MA domain, also make contact with the gRNA during the process of particle assembly, as discussed below. Thus, despite the wealth of data pointing to NC as the critical determinant of specific genome packaging in most retroviruses, studies have revealed a complementary role for MA in specific encapsidation of gRNA.
The notion that MA might influence genome incorporation arose from early studies which reported that the mature MA proteins from different retroviruses has nucleic acids binding activity, with evidence of binding to both single stranded and double stranded RNA and DNA, although in most cases there was a lack of specificity (see Table 1)17, 18, 20-23. The biological relevance of the nonspecific nucleic acid binding activity of retroviral MA proteins was questioned for many years because MA is located near the lipid envelope of the virion, away from the ribonucleic acid core and crosslinking of virions did not show MA-RNA interactions 1. The examination of mature virions is potentially misleading, however, because gRNA packaging is mediated by the immature Gag polyprotein inside the cell, and intracellular Gag-gRNA interactions may be disrupted prior to particle release. Thus, the role of the MA domain of Gag in gRNA interactions is likely to be most crucial during immature particle formation.
Table 1.
In vitro binding affinities for retroviral MA proteins and nucleic acids
| Retrovirus MA Protein |
Binding Affinity Kd (M) | Nucleic acid | Reference |
|---|---|---|---|
| HIV | 3-30 × 10−9 | RNA 50mer (SELEX) | 56 |
| HIV | 5 ×10−7 | RNA with homology to the pol region |
54 |
| HIV | 1.5 × 10−5 | ssDNA 30mer | 51 |
| HIV | 5.42 × 10−7 | ssDNA 20mer | 50 |
| HIV | 3.25 ×10−7 | minihelixLys | 50 |
| RSV | 3.6 × 10−12 | Viral RNA | 17,18 |
| RSV | 1 × 10−7* | Viral RNA | 22 |
Intrinsic Kd estimated at 2.9 × 10−3 to 9.1 × 10−4 M.
In this article, we will summarize the current understanding of the role of the Gag MA domain in gRNA packaging, emphasizing the data available for HIV-1, RSV, BLV, and HTLV-I. One intriguing feature of this comparative approach is that the precise mechanisms by which the MA domains influence genome encapsidation appear to vary for different viruses. Perhaps future experiments will reveal more common themes as further information about the contribution of the Gag MA domain in selective gRNA packaging becomes available.
Direct and indirect roles of MA in genome packaging
HIV-1 MA binding to nucleic acids influences gRNA packaging and intracellular trafficking of Gag
Since the discovery of HIV-1 in 1983 24, identifying the determinants of genome encapsidation has been the focus of intense study. Several investigators found that the NC domain binds directly to the ψ sequence by virtue of the zinc knuckle domains and basic residues 13, 25-33 and mediates Gag multimerization in conjunction with the CA domain of Gag 29, 34-37. Genetic and imaging studies permitted the dissection of the sequence of events from Gag:gRNA binding to particle assembly, revealing that HIV-1 Gag-Gag interactions are initiated in the cytoplasm 38-41;. Efforts to determine the subcellular location of Gag:gRNA recognition yielded different results, with data demonstrating the formation of viral ribonucleoprotein complexes near the nucleus at a pericentriolar site 42, within the cytoplasm 41, and at the plasma membrane 43. Irrespective of where Gag:gRNA complexes form initially, they are subsequently targeted to the plasma membrane through a bipartitie signal consisting of an N-terminal myristic acid moiety and a cluster of basic residues in the Gag MA domain 44. Studies suggest that the myristate is buried within the hydrophobic MA globular domain until Gag reaches the membrane, triggering a conformational change in MA and exposing the myristic acid for insertion into the lipid bilayer 45-47. Binding of key basic residues in MA to the acidic phospholipid PI(4,5)P2, which is enriched in the plasma membrane, may account for the specificity of Gag being targeted to the plasma membrane rather than to internal membranes 48-50.
Beyond its role in plasma membrane targeting and binding, HIV-1 MA also has nucleic acid binding activity with affinity for both RNA and DNA 51-53. Although specific binding of HIV-1 MA to the ψ sequence has not been demonstrated, a basic-rich region of MA does bind with high affinity (Kd = 5 × 10−7 M) to an RNA molecule highly homologous to a segment of the pol sequence 54. Viral mutants that disrupt this MA:RNA interaction exhibit a delay in replication, although the level of gRNA packaging in these mutant viruses was not examined. The region of MA that binds to this RNA was mapped to the N-terminal basic sequence, and substitution of two or more basic residues disrupted RNA binding. In support of the idea that the MA domain makes direct contact with the viral genome, basic residues in HIV-1 MA can substitute for the RNA-induced assembly functions of NC 52, 55. In vitro RNA binding data suggest that Gag contains two independent RNA-binding sites, one in MA and the other in NC, that appear to contact RNA simultaneously 56.
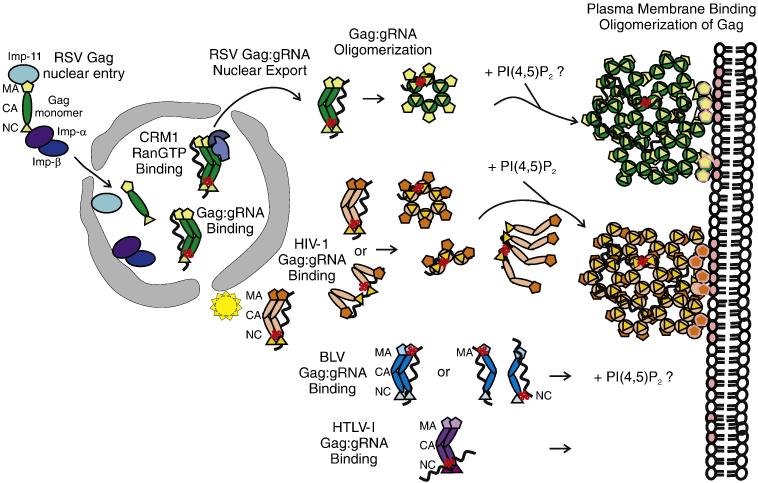
Supporting this finding, structural studies reveal that the Gag protein adopts a U-shaped conformation in solution whereby the MA and NC domains are in close proximity 57, 58. The conformation of Gag remains “folded over” when bound to RNA, but in the presence of both nucleic aids and membranes a structural change is triggered, resulting in extension of the protein 58. Thus, it appears that basic residues in both MA and NC each bind to the gRNA prior to membrane binding, although it is possible that other cellular RNAs present in HIV-1 virions may also interact with MA or NC 6. This collection of experiments suggest that the HIV-1 Gag MA domain may directly play a role in genome encapsidation by interacting with gRNA, likely by binding to regions outside of ψ. As illustrated in Figure 2, the current model shows that the Gag MA and/or NC domains bind to gRNA in the cytoplasm, inducing dimers or small oligomers of Gag to nucleate on the viral RNA. As the Gag:gRNA complex approaches the plasma membrane, PI(4,5)P2 competes with RNA for binding to MA, causing a conformational change in Gag. Gag-Gag interactions are strengthened cooperatively at the membrane, forming a dense aggregation of Gag:gRNA complexes that form an incomplete hexameric lattice that forms the immature virus particle 59-61.
Fig. 2. Model illustrating the role of the Gag MA domain in regulating gRNA binding, subcellular trafficking, genome encapsidation and particle assembly.
After synthesis in the cytoplasm, Gag proteins destined to bind the gRNA for packaging are transported to specific subcellular locations. The MA domain of Gag is represented by pentagons, CA by ovals, and NC by triangles. The gRNA is a wavy black line and the ψ sequence is depicted as a red cloverleaf. For RSV Gag (green), NLSs in the MA and NC domains interact with host import factors importin-11 and the importin-α/β complex to direct Gag into the nucleus where Gag interacts with the ψ sequence on the gRNA. RSV Gag:gRNA binding induces a conformation change in RSV Gag that promotes binding to CRM1:RanGTP, facilitating nuclear export. RSV Gag forms oligomers that are transported to the plasma membrane, possibly through an interaction with the phosphoinositol PIP(4,5)2 (denoted by pink ovals in the inner leaflet of the lipid bilayer). A discontinuous hexameric lattice of RSV Gag proteins bound to gRNA assembles at the membrane, encapsidating the genome into the assembling particle. For HIV (orange), Gag interacts with its gRNA at a pericentriolar location (illustrated as a yellow star) or in the cytoplasm, inducing Gag dimer formation. The model illustrates the MA and NC domains interacting with gRNA in an extended conformation (top) or in a folded conformation (bottom), with the NC domain binding to ψ and the MA domain binding to the gRNA at a different location. It is possible that the MA domain is bound to a cellular RNA rather than to the gRNA. HIV-1 Gag:gRNA oligomers form and are transported toward the periphery of the cell. Upon binding to PIP(4,5)2, the MA domain releases the gRNA and Gag adopts an extended conformation with MA facing the membrane and NC binding to the gRNA. The hexamers of Gag associate with the plasma membrane and assemble into a hexameric lattice. For BLV (blue), the site of Gag:gRNA complex formation is not known. The MA domain of BLV Gag is shown binding to the ψ sequence, although the NC domain also contacts the gRNA through nonspecific interactions. Further details regarding the mechanism of BLV Gag assembly are not well understood. For HTLV-I, the model depicts the ψ sequence on the gRNA binding to the NC domain of Gag because it is not known whether MA plays a role in genome encapsidation. It has been shown that nucleic acid binding does not influence membrane binding, and PIP(4,5)2 is not required for membrane targeting119
RSV MA functions in nuclear localization of Gag
The alpharetrovirus RSV was discovered in 191062 and is the basis for many seminal discoveries, including the initial identification of the ψ RNA packaging element63. Further studies defined the minimal ψ packaging signal as a 160-nucleotide sequence that resides almost exclusively in the 5′ leader sequence of the genome, with its 3′ border just upstream of the splice donor site 6,64,65. Thus, both unspliced (genome-length) and spliced viral RNAs contain the ψ site, raising the question of how the Gag protein can preferentially package the gRNA over viral mRNAs. In retroviruses like HIV-1, important portions of the packaging signal continue beyond the major 5′ splice site (several hundred bases into the gag coding region), making differentiation of gRNA from spliced viral mRNAs more straightforward because ψ is present only on the gRNA 11, 25, 28, 66-72. Because of this variation in the location of ψ, it is conceivable that RSV and HIV-1 may rely on somewhat different strategies for specific, ψ-mediated gRNA selection.
The NC region of RSV Gag is the primary domain required for gRNA encapsidation. RNA interactions are mediated by basic residues flanking the two zinc-finger motifs in NC whereas residues within the zinc-fingers themselves bind directly to the minimal ψ sequence 6,64,65,73-76. Interestingly, an RSV Gag mutant that contained the NC domain of murine leukemia virus in place of its own resulted in reduced, but not absent, packaging specificity 77. This result, in addition to others, suggest that other regions of Gag may influence packaging efficiency in concert with NC 77,78. In support of this possibility, mutants involving the N-terminal region of MA including deletions, basic residues substitutions, and alteration of the membrane-targeting domain are associated with defects in gRNA dimerization and selective encapsidation 79. The decrease in selective gRNA packaging is due to changes in the protein sequence of MA rather than as a result of mutations at the RNA level that impair genome recognition or packaging. Because monomeric gRNA is packaged in these viral mutants, dimerization of the genome does not appear to be absolutely required for RSV encapsidation, unlike some other retroviruses. In vitro binding experiments support this finding, as the RSV NC protein binds tightly to ψ with 1:1 stoichiometry 73.
Recent studies have elucidated a mechanism by which the RSV MA domain influences gRNA packaging indirectly by virtue of its role in regulating Gag subcellular localization (Fig. 2) 80,82. After its synthesis on cytosolic ribosomes, the RSV Gag polyprotein undergoes transient nuclear localization, a step that is required for efficient genome encapsidation 80,8. Nuclear import occurs by virtue of nuclear localizations signals (NLS) in the MA and NC domains. The NLS in MA is a complex and nonclassical nuclear targeting signal that binds directly to importin-11, an unusual import receptor, to facilitate nuclear entry of Gag 83. Another import factor, known as transportin SR or transportin-3, was also identified as a mediator of MA nuclear entry, although its role in Gag nuclear targeting has not been investigated. A second NLS resides in the Gag NC domain and consists of a highly basic region that also is involved in nucleic acid binding 81. This classical NLS binds directly to the canonical import receptor importin-α, which then recruits its cofactor importin-β to translocate Gag into the nucleus. Dissection of the molecular mechanism underlying nuclear import of Gag revealed that nucleic acids are effective competitors for binding of importin-α to NC. An RNA molecule containing the minimal ψ sequence was a much more effective competitor for binding to the NC domain of Gag compared to a nonviral RNA or DNA, demonstrating highly specific binding of ψ to the recombinant full-length RSV Gag protein in vitro.
RSV MA also possesses nucleic acid binding activity, with early experiments reporting an apparent disassociation constant of 1-10 nM for RSV RNA, and much lower affinity binding to ribosomal RNA (rRNA) 15,16. Subsequent studies disputed the claim that there was any specificity of MA for viral RNA, instead finding that MA bound to viral RNA, ribosomal RNA, and DNA equally and with low affinity (Table 1) 21,84. However, evidence for specific nucleic acid binding activity was demonstrated by the finding that RNA competes for RSV MA:importin-11 binding better than DNA does, although there is no specificity for ψ 81. Surprisingly, the ψ sequence is a much more effective competitor than nonviral RNA for importin-11 binding to the full-length Gag protein, suggesting that the MA domain may contribute to specificity of the Gag:ψ RNA binding. This intriguing result can be explained by either (a) an indirect mechanism in which Gag NC:ψ binding induces a global structural change that disrupts MA:importin-11 interaction or (b) a direct mechanism in which Gag MA itself contacts the ψ sequence, displacing importin-11. Additional experimentation is needed to determine what direct role, if any, the specific interaction of the Gag MA domain plays in gRNA selection and packaging; however, it does seem clear that the MA and NC domains work together to spatially and temporally regulate RSV Gag nucleocytoplasmic localization, thereby influencing genome incorporation.
A combination of genetic and biochemical evidence suggest that once in the nucleus, RSV Gag recognizes and binds to the gRNA, inducing a conformational change that reveals a leucine-rich nuclear export signal (NES) within the p10 domain in Gag 80,81. The Gag p10 NES binds directly to CRM1-RanGTP, a major exporter of RNA-binding proteins from the nucleus 81,82. The Gag-gRNA complex is exported through the nuclear pore, shedding CRM1, and then travels to the plasma membrane where multimerization of Gag and budding occur. The nuclear localization of Gag proteins from other retroviruses (HIV-185, murine leukemia virus86, Mason-Pfizer monkey virus87 and foamy viruses 88-91), retrotransposons (Ty192), and retroelements 93,94 have also been reported (reviewed in 95). In the case of HIV-1, the original report that the HIV-1 MA protein contains a NLS as well as a CRM1-dependent NES has been disputed 96-98. Thus, whether HIV-1 Gag undergoes a nuclear trafficking step that is linked to gRNA packaging85 is controversial and will require further investigation. For the human foamy virus Gag protein, nuclear localization is mediated by the RNA binding domain, leading investigators to hypothesize that foamy virus Gag picks up its gRNA in the nucleus88. Recently, a CRM1-dependent NES was identified and the authors again speculated that Gag:gRNA binding might occur in the nucleus, although no experiments were shown that directly test this hypothesis 89. Thus, it is possible that directing Gag into the nucleus via the MA and NC sequences for selection of gRNA for packaging is a mechanism not unique to RSV, but may be shared by genetically diverse groups of retroviruses.
The BLV Gag MA domain plays a major role in selective gRNA packaging
Identification of the cis and trans factors required for genome packaging of the deltaretroviruses BLV led to some surprises. Rather than consisting of a concise, continuous sequence limited to the 5′ untranslated region, the ψ sequence was found to be composed of two distinct segments of gRNA, one in the leader sequence and the other within coding sequence of gag 99-101. Furthermore, although the BLV Gag NC domain contains two zinc finger domains and basic amino acids that are important for gRNA packaging102, the mature BLV NC protein possesses only nonspecific RNA-binding activity, with no selectivity for viral RNA sequences containing ψ 22. Instead of NC, the MA region of Gag plays a predominant role in specific selection and packaging of the genome 22,102, 103. The BLV MA(p15) protein, which is further cleaved into MA(p10) and MA(p4) upon completion of virus maturation, binds specifically to two segments of viral RNA derived from the 5′ end of the genome: the first segment contains the viral RNA dimerization domain 22, 102, 103, while the second segment encompasses the ψ sequence, extending from the 5′ leader sequence through a portion of the gag gene 103. Interestingly, the fully mature MA protein (p10) lacks specific RNA binding, implying that the immature form of MA(p15) present within the Gag precursor is involved in packaging of the gRNA by binding selectively to ψ 22. Moreover, the MA(p15) protein, but not MA(p10), binds preferentially to a dimer of the 5′ leader viral RNA compared to the denatured viral RNA species. The MA(p15) protein:RNA complex apparently forms first, then dimerization of the 5′ viral RNA sequence ensues. Thus, binding of the MA(p15) region of BLV Gag to the 5′ leader viral RNA sequence containing both the dimerization signal and the ψ encapsidation signal may provide a mechanism to ensure that two genomes are specifically packaged into virions 22, 103.
In support of these in vitro studies, experiments that examined the role of the BLV MA and NC domains in gRNA encapsidation concluded that basic residues within both regions of Gag are required for optimal genome packaging102. The basic residues in BLV MA that are most important for gRNA packaging (K41 and H45, Fig. 1) are not involved in plasma membrane targeting of Gag. Furthermore, the codons for K41 and H45 lie outside of the ψ sequence, indicating that the deleterious effects of mutating K41 and H45 are likely mediated at the level of MA protein domain rather than due to an effect on gRNA structure or sequence 100-102. Thus, for BLV the composite data suggest that MA and NC may both bind to ψ itself or to nearby RNA sequences that contribute to selective gRNA incorporation102. Alternatively, it is feasible that the MA and NC domains within a single BLV Gag molecule might bind to separate gRNA molecules as an alternative mechanism to incorporate two genomes into one virus particle (Fig. 2). Further structural studies will be needed to investigate how MA and NC act together to contribute to genome recognition and encapsidation.
Because mutants involving basic residues in the MA domain of the related deltaretrovirus HTLV-I MA 22, 102, 104 also impair infectivity but maintain plasma membrane localization, it has been suggested that HTLV-I MA 22, 102, 104 might also play a role in specific packaging of the gRNA102. This possibility is especially intriguing because of the functional conservation of several basic residues in the BLV and HTLV-I MA 22, 102, 104 sequences and the similarity of the three dimensional structures of the deltaretrovirus MA domains 105, 106. While it is possible that all of the deltaretroviruses use similar mechanisms for genome encapsidation, there are no published studies to date that address this interesting possibility.
The Power of Comparative Retrovirology: Studies for the Future
Packaging of the retroviral gRNA into virus particles is essential for productive replication, hence disruption of Gag:gRNA binding is an attractive target for antiviral therapy. A critical step toward designing optimal packaging inhibitors is to understand the mechanism of encapsidation at the molecular level. The NC domain of Gag is certainly a critical determinant of in gRNA packaging, but it has become increasingly apparent that the Gag MA region contributes to the specificity of genome recognition, influences the compartment of the cell where Gag:gRNA binding is initiated, and regulates the location and timing of the final encapsidation of gRNA into the assembling virus particle. The critical role of the Gag MA domain in genome packaging must be considered when developing antiretroviral agents that interfere with gRNA incorporation, rather than focusing solely on abrogating NC:ψ interactions 107-117.
Many of the regulatory activities of MA require the participation of host co-factors that mediate subcellular trafficking and compete with nucleic acids for binding to MA. In the case of RSV, the nuclear import factor importin-11 binds to the MA domain of Gag in the absence of nucleic acids to facilitate import of Gag into the nucleus where gRNA binding occurs 81. Once the gRNA binds to Gag, importin-11 can no longer associate with the MA sequence, presumably to ensure that the Gag:gRNA complex has a one-way ticket out of the nucleus. It is perplexing that the RSV ψ sequence competes with MA:importin-11 binding better than nonviral RNA or DNA. Does this result imply that the RSV MA domain recognizes the ψ sequence specifically during genome binding or is the interaction due to a structural element in ψ that happens to bind to MA better than other RNAs? Further experiments will be needed to determine whether there is a significant biological role for the apparent preference of the MA domain for the ψ packaging element. In addition, the recent discovery that the foamy virus Gag protein contains an NES similar to that of RSV Gag has raised speculation that foamy virus might also select its genome genome in the nucleus89. Future investigations will reveal whether additional Gag proteins undergo transient nuclear trafficking for the purposes of gRNA encapsidation. If so, these Gag proteins may utilize different mechanisms for export other than CRM-1.
In contrast to RSV, the HIV-1 Gag MA domain appears to bind selectively to a sequence in the gRNA at a location other than ψ 54. Apparently, the MA and NC domains of HIV-1 Gag both make contact with the gRNA in the cytoplasm. It is possible that the MA domain binds to a segment of the gRNA near ψ, and this interaction may enhance specificity by (a) altering the conformation of ψ, (b) by promoting genome dimerization, or (c) by inducing tighter binding of NC to ψ via an allosteric effect. Alternatively, if MA binds to a segment outside of ψ with high affinity, then Gag has two “handles” (the other being NC bound to ψ) to hold on more tightly to the gRNA. Once the HIV-1 Gag:gRNA complex approaches the periphery of the cell, the plasma membrane specific phosphoinositol PI(4,5)P2 competes successfully with nucleic acids for binding to the MA domain 50,188. The Gag MA:PI(4,5)P2 association induces a conformational change in Gag that results in elongation of the protein, with the N terminus buried in the lipid bilayer of the plasma membrane and the C terminal region bound to the gRNA 57. In this way, the MA domain regulates the timing of the extension of Gag and ensures that the switch in conformation occurs at the correct location—the plasma membrane. These carefully orchestrated events guarantee that the genome becomes encapsidated into the emerging virus particle during the budding process.
The mechanism of gRNA selection and packaging is less clear for the deltaretroviruses BLV and HTLV-I, but it appears that the MA and NC domains share in facilitating specific genome binding. Whether the MA and NC regions of BLV Gag bind to the same or different gRNA molecules is not known (Figure 2); this question is worth pursuing because it may provide a key insight into how retroviruses ensure that two genomes are incorporated into every virion. The observation that the BLV Gag MA domain has specific affinity for the ψ packaging signal also raises the question of whether MA remains associated with the gRNA within the immature and mature virus particles. Many of the details regarding the mechanism of specific RNA packaging for HTLV-I remain poorly defined, although a recent report suggests it does that HTLV-I and HIV-1 MA have fundamentally different mechanisms of interaction with both nucleic acids and membranes119.
It is curious that different retroviruses appear to have evolved distinct mechanisms to govern genomic RNA encapsidation even though the outcome of the process is remarkably conserved throughout the retrovirus family. The Gag MA domain has either leading or supportive roles in genome selection and encapsidation depending on the virus. Although the mechanistic details may differ, comparative studies across retroviral genera have provided compelling support for the involvement of MA in genomic RNA packaging. The value of these comparative studies has elevated the importance of the observations made in each individual virus and has emphasized the need for continuing to study the properties of multiple retroviruses.
In the future, studies investigating the role of the Gag MA domain in gRNA packaging should focus on determining where in the host cell Gag initially binds the genome, how MA facilitates subcellular trafficking of the viral ribonucleoprotein complex, and the influence of MA:gRNA interactions on Gag multimerization. It is hopeful that determining the ultrastructural properties of Gag bound to the gRNA at high resolution will reveal whether MA contacts the RNA in an immature virus particle or whether this interaction is limited to the intracellular environment. Perhaps the most difficult yet enormously informative experiments will be to solve high-resolution structures of Gag (with and without viral RNA) in complex with its cellular binding partners, including protein co-factors and membrane components. Defining alternative structures of Gag may yield clues about the dynamic conformational changes induced by transient interactions with RNA, proteins, and lipids that are needed to complete the complicated journey from the ribosome to the site of assembly on the plasma membrane. Successful outcomes of these experimental approaches will be the critical next steps in elucidating common themes and uncovering distinct roles for the MA domain of retroviral Gag proteins in encapsidation of the viral RNA genome.
Acknowledgements
The authors gratefully acknowledge support from the National Institutes of Health (R01 CA76534 to L.J.P) and the Penn State College of Medicine (Graduate Student Research Award to N.G.) using the Pennsylvania State Department of Health Tobacco Settlement Funds. The Department of Health specifically disclaims responsibility for any analyses, interpretations or conclusions. We thank the anonymous reviewers for their constructive criticisms that improved the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 2.Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr.Top.Microbiol.Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 3.Rein A. Retroviral RNA packaging: a review. Arch.Virol.Suppl. 1994;9:513–522. doi: 10.1007/978-3-7091-9326-6_49. [DOI] [PubMed] [Google Scholar]
- 4.Jewell NA, Mansky LM. In the beginning: genome recognition, RNA encapsidation and the initiation of complex retrovirus assembly. J Gen Virol. 2000;81(Pt 8):1889–1899. doi: 10.1099/0022-1317-81-8-1889. [DOI] [PubMed] [Google Scholar]
- 5.Muriaux D, Mirro J, Harvin D, Rein A. RNA is a structural element in retrovirus particles. Proc Nat Acad Sci USA. 2001;98:5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rulli SJ, Jr., Hibbert CS, Mirro J, Pederson T, Biswal S, Rein A. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J Virol. 2007;81:6623–31. doi: 10.1128/JVI.02833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogt VM. Retroviral virions and genomes. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1997. pp. 27–69. [PubMed] [Google Scholar]
- 8.Aronoff R, Hajjar AM, Linial ML. Avian retroviral RNA encapsidation: reexamination of functional 5′ RNA sequences and the role of nucleocapsid Cys-His motifs. J Virol. 1993;67:178–188. doi: 10.1128/jvi.67.1.178-188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell S, Vogt VM. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell S, Vogt VM. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berkowitz RD, Ohagen A, Hoglund S, Goff SP. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–56. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Souza V, Summers MF. How retroviruses select their genomes. Nat Rev Microbiol. 2005;3(8):643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 14.Poon DTK, Li G, Aldovini A. Nucleocapsid and Matrix Protein Contributions to Selective Human Immunodeficiency Virus Type 1†Genomic RNA Packaging. J Virol. 1998;72:1983–1993. doi: 10.1128/jvi.72.3.1983-1993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol. 1995;69:5716–22. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karpel RL, Henderson LE, Oroszlan S. Interactions of retroviral structural proteins with single-stranded nucleic acids. J Biol Chem. 1987;262:4961–7. [PubMed] [Google Scholar]
- 17.Leis JP, McGinnis J, Green RW. Rous sarcoma virus p19 binds to specific double-stranded regions of viral RNA: effect of p19 on cleavage of viral RNA by RNase III. Virology. 1978;84:87–98. doi: 10.1016/0042-6822(78)90220-9. [DOI] [PubMed] [Google Scholar]
- 18.Leis JP, Scheible P, Smith RE. Correlation of RNA binding affinity of avian oncornavirus p19 proteins with the extent of processing of virus genome RNA in cells. J Virol. 1980;35:722–31. doi: 10.1128/jvi.35.3.722-731.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luban J, Goff SP. Binding of human immunodeficiency virus type 1 (HIV-1) RNA to recombinant HIV-1 gag polyprotein. J Virol. 1991;65:3203–12. doi: 10.1128/jvi.65.6.3203-3212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darlix JL, Spahr PF. Binding sites of viral protein P19 onto Rous sarcoma virus RNA and possible controls of viral functions. J Mol Biol. 1982;160:147–61. doi: 10.1016/0022-2836(82)90172-3. [DOI] [PubMed] [Google Scholar]
- 21.Meric C, Spahr P. Rous sarcoma virus nucleic-acid binding protein p12 is necessary for viral 70S dimer formation and packaging. J Virol. 1986;60:450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steeg CM, Vogt VM. RNA-binding properties of the matrix protein (p19gag) of avian sarcoma and leukemia viruses. J Virol. 1990;64:847–855. doi: 10.1128/jvi.64.2.847-855.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoh I, Kyushiki H, Sakamoto Y, Ikawa Y, Yoshinaka Y. Bovine leukemia virus matrix-associated protein MA(p15): further processing and formation of a specific complex with the dimer of the 5′-terminal genomic RNA fragment. J Virol. 1991;65:6845–6855. doi: 10.1128/jvi.65.12.6845-6855.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 25.Berkowitz RD, Goff SP. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–46. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 26.Harrison GP, Lever AM. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–53. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cimarelli A, Sandin S, Hoglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J Virol. 2000;74:3046–57. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–8. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 29.Lever AM. HIV-1 RNA packaging. Adv Pharmacol. 2007;55:1–32. doi: 10.1016/S1054-3589(07)55001-5. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence DC, Stover CC, Noznitsky J, Wu Z, Summers MF. Structure of the intact stem and bulge of HIV-1 Psi-RNA stem-loop SL1. J Mol Biol. 2003;326:529–42. doi: 10.1016/s0022-2836(02)01305-0. [DOI] [PubMed] [Google Scholar]
- 31.Hagan N, Fabris D. Direct mass spectrometric determination of the stoichiometry and binding affinity of the complexes between nucleocapsid protein and RNA stem-loop hairpins of the HIV-1 Psi-recognition element. Biochemistry. 2003;42:10736–45. doi: 10.1021/bi0348922. [DOI] [PubMed] [Google Scholar]
- 32.Shubsda MF, Paoletti AC, Hudson BS, Borer PN. Affinities of packaging domain loops in HIV-1 RNA for the nucleocapsid protein. Biochemistry. 2002;41:5276–82. doi: 10.1021/bi016045+. [DOI] [PubMed] [Google Scholar]
- 33.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J Mol Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 34.Bennett RP, Nelle TD, Wills JW. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derdowski A, Ding L, Spearman P. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J Virol. 2004;78:1230–42. doi: 10.1128/JVI.78.3.1230-1242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorfman T, Luban J, Goff SP, Haseltine WA, Gottlinger HG. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–69. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Accola MA, Strack B, Gottlinger HG. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J Virol. 2000;74:5395–402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowzard JB, Bennett RP, Krishna NK, Ernst SM, Rein A, Wills JW. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J Virol. 1998;72:9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin J, Sturgeon T, Chen C, Watkins SC, Weisz OA, Montelaro RC. Distinct intracellular trafficking of equine infectious anemia virus and human immunodeficiency virus type 1 Gag during viral assembly and budding revealed by bimolecular fluorescence complementation assays. J Virol. 2007;81:11226–35. doi: 10.1128/JVI.00431-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milev MP, Brown CM, Mouland AJ. Live cell visualization of the interactions between HIV-1 Gag and the cellular RNA-binding protein Staufen1. Retrovirology. 2010;7:41. doi: 10.1186/1742-4690-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kutluay SB, Bieniasz PD. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 2010;6:e1001200. doi: 10.1371/journal.ppat.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poole E, Strappe P, Mok HP, Hicks R, Lever AM. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and FRET. Traffic. 2005;6:741–755. doi: 10.1111/j.1600-0854.2005.00312.x. [DOI] [PubMed] [Google Scholar]
- 43.Kemler I, Meehan A, Poeschla EM. Live-cell coimaging of the genomic RNAs and Gag proteins of two lentiviruses. J Virol. 2010;84:6352–66. doi: 10.1128/JVI.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou W, Parent LJ, Wills JW, Resh MD. Identification of a membrane-binding domain within the amino-terminal region of Human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of Human immunodeficiency virus type 1 Matrix protein in vivo supports a conformational myristyl switch mechanism. J Virol. 1997;71:6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc.Natl.Acad.Sci.U.S.A. 2006;103(30):11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang C, Loeliger E, Luncsford P, Kinde I, Beckett D, Summers MF. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc Natl Acad Sci U S A. 2004;101:517–22. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci U S A. 2004;101:14889–94. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. J Virol. 2008;82:2405–17. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones CP, Datta SA, Rein A, Rouzina I, Musier-Forsyth K. Matrix Domain Modulates HIV-1 Gag’s Nucleic Acid Chaperone Activity via Inositol Phosphate Binding. J Virol. 2011;85:1594–603. doi: 10.1128/JVI.01809-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alfadhli A, Still A, Barklis E. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J Virol. 2009;83:12196–203. doi: 10.1128/JVI.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ott DE, Coren LV, Gagliardi TD. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J Virol. 2005;79:13839–47. doi: 10.1128/JVI.79.22.13839-13847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramalingam D, Duclair S, Datta SA, Ellington A, Rein A, Prasad VR. RNA aptamers directed to human immunodeficiency virus type 1 Gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J Virol. 2011;85:305–14. doi: 10.1128/JVI.02626-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purohit P, Dupont S, Stevenson M, Green MR. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA. 2001;7:576–84. doi: 10.1017/s1355838201002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burniston MT, Cimarelli A, Colgan J, Curtis SP, Luban J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J Virol. 1999;73:8527–40. doi: 10.1128/jvi.73.10.8527-8540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lochrie MA, Waugh S, Pratt DG, Jr., Clever J, Parslow TG, Polisky B. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 1997;25:2902–10. doi: 10.1093/nar/25.14.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Datta SA, Curtis JE, Ratcliff W, Clark PK, Crist RM, Lebowitz J, Krueger S, Rein A. Conformation of the HIV-1 Gag protein in solution. J Mol Biol. 2007;365:812–824. doi: 10.1016/j.jmb.2006.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Datta SA, Heinrich F, Raghunandan S, Krueger S, Curtis JE, Rein A, Nanda H. HIV-1 Gag Extension: Conformational Changes Require Simultaneous Interaction with Membrane and Nucleic Acid. J Mol Biol. 2011;406:205–14. doi: 10.1016/j.jmb.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Briggs JA, Johnson MC, Simon MN, Fuller SD, Vogt VM. Cryo-electron microscopy reveals conserved and divergent features of gag packing in immature particles of Rous sarcoma virus and human immunodeficiency virus. J Mol Biol. 2006;355(1):157–168. doi: 10.1016/j.jmb.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 60.Carlson LA, de Marco A, Oberwinkler H, Habermann A, Briggs JA, Krausslich HG, Grunewald K. Cryo electron tomography of native HIV-1 budding sites. PLoS Pathog. 2010;6:e1001173. doi: 10.1371/journal.ppat.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. Structure and assembly of immature HIV. Proc Natl Acad Sci U S A. 2009;106:11090–5. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rous P. A transmissible avian neoplasm (sarcoma of the common fowl) J Exp Med. 1910;12:696–705. doi: 10.1084/jem.12.5.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Linial M, Medeiros E, Hayward WS. An avian oncovirus mutant (SE 21Q1b) deficient in genomic RNA: biological and biochemical characterization. Cell. 1978;15:1371–81. doi: 10.1016/0092-8674(78)90062-4. [DOI] [PubMed] [Google Scholar]
- 64.Banks JD, Linial ML. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. The Journal of Virology. 2000;74:456–464. doi: 10.1128/jvi.74.1.456-464.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Banks JD, Yeo A, Green K, Cepeda F, Linial ML. A minimal avian retroviral packaging sequence has a complex structure. J Virol. 1998;72:6190–6194. doi: 10.1128/jvi.72.7.6190-6194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clever J, Sassetti C, Parslow TG. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–9. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clever JL, Miranda D, Jr., Parslow TG. RNA structure and packaging signals in the 5′ leader region of the human immunodeficiency virus type 1 genome. J Virol. 2002;76:12381–7. doi: 10.1128/JVI.76.23.12381-12387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clever JL, Taplitz RA, Lochrie MA, Polisky B, Parslow TG. A heterologous, high-affinity RNA ligand for human immunodeficiency virus Gag protein has RNA packaging activity. J Virol. 2000;74:541–6. doi: 10.1128/jvi.74.1.541-546.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–7. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McBride MS, Panganiban AT. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–73. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buchschacher GL, Jr., Panganiban AT. Human immunodeficiency virus vectors for inducible expression of foreign genes. J Virol. 1992;66:2731–9. doi: 10.1128/jvi.66.5.2731-2739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parolin C, Dorfman T, Palu G, Gottlinger H, Sodroski J. Analysis in human immunodeficiency virus type 1 vectors of cis-acting sequences that affect gene transfer into human lymphocytes. J Virol. 1994;68:3888–95. doi: 10.1128/jvi.68.6.3888-3895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou J, McAllen JK, Tailor Y, Summers MF. High affinity nucleocapsid protein binding to the muPsi RNA packaging signal of Rous sarcoma virus. J.Mol.Biol. 2005;349(5):976–988. doi: 10.1016/j.jmb.2005.04.046. [DOI] [PubMed] [Google Scholar]
- 74.Lee EG, Alidina A, May C, Linial ML. Importance of basic residues in binding of rous sarcoma virus nucleocapsid to the RNA packaging signal. J of Virol. 2003;77:2010–2020. doi: 10.1128/JVI.77.3.2010-2020.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee E. g., Linial ML. Basic Residues of the Retroviral Nucleocapsid Play Different Roles in Gag-Gag and Gag-{Psi} RNA Interactions. J Virol. 2004;78:8486–8495. doi: 10.1128/JVI.78.16.8486-8495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aronoff R, Linial M. Specificity of retroviral RNA packaging. J Virol. 1991;65:71–80. doi: 10.1128/jvi.65.1.71-80.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dupraz P, Spahr P. Specificity of Rous sarcoma virus nucleocapsid protein in genomic RNA packaging. J Virol. 1992;66:4662–4670. doi: 10.1128/jvi.66.8.4662-4670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sakalian M, Wills JW, Vogt VM. Efficiency and selectivity of RNA packaging by Rous sarcoma virus Gag deletion mutants. J Virol. 1994;68:5969–5981. doi: 10.1128/jvi.68.9.5969-5981.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garbitt RA, Albert JA, Kessler MD, Parent LJ. trans-acting inhibition of genomic RNA dimerization by Rous sarcoma virus matrix mutants. J Virol. 2001;75:260–268. doi: 10.1128/JVI.75.1.260-268.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garbitt-Hirst R, Kenney SP, Parent LJ. Genetic evidence for a connection between Rous sarcoma virus gag nuclear trafficking and genomic RNA packaging. J Virol. 2009;83:6790–7. doi: 10.1128/JVI.00101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gudleski N, Flanagan JM, Ryan EP, Bewley MC, Parent LJ. Directionality of nucleocytoplasmic transport of the retroviral gag protein depends on sequential binding of karyopherins and viral RNA. Proc Natl Acad Sci U S A. 2010;107:9358–63. doi: 10.1073/pnas.1000304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Scheifele LZ, Garbitt RA, Rhoads JD, Parent LJ. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc.Natl.Acad.Sci.U.S.A. 2002;99:3944–3949. doi: 10.1073/pnas.062652199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Butterfield-Gerson KL, Scheifele LZ, Ryan EP, Hopper AK, Parent LJ. Importin beta family members mediate alpharetrovirus Gag nuclear entry via interactions with MA and NC. J Virol. 2006;80:1798–806. doi: 10.1128/JVI.80.4.1798-1806.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meric C, Darlix JL, Spahr PF. It is Rous sarcoma virus protein P12 and not P19 that binds tightly to Rous sarcoma virus RNA. J Mol Biol. 1984;173:531–8. doi: 10.1016/0022-2836(84)90396-6. [DOI] [PubMed] [Google Scholar]
- 85.Dupont S, Sharova N, DeHoratius C, Virbasius CM, Zhu X, Bukrinskaya AG, Stevenson M, Green MR. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature. 1999;402(6762):681–685. doi: 10.1038/45272. [DOI] [PubMed] [Google Scholar]
- 86.Nash MA, Meyer MK, Decker GL, Arlinghaus RB. A subset of Pr65gag is nucleus associated in murine leukemia virus-infected cells. The Journal of Virology. 1993;67(3):1350–1356. doi: 10.1128/jvi.67.3.1350-1356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bohl CR, Brown SM, Weldon RA., Jr. The pp24 phosphoprotein of Mason-Pfizer monkey virus contributes to viral genome packaging. Retrovirology. 2005;2:68. doi: 10.1186/1742-4690-2-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schliephake AW, Rethwilm A. Nuclear localization of foamy virus Gag precursor protein. The Journal of Virology. 1994;68:4946–4954. doi: 10.1128/jvi.68.8.4946-4954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Renault N, Tobaly-Tapiero J, Paris J, Giron ML, Coiffic A, Roingeard P, Saib A. A nuclear export signal within the structural Gag protein is required for prototype foamy virus replication. Retrovirology. 2011;8:6. doi: 10.1186/1742-4690-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Saib A, Puvion-Dutilleul F, Schmid M, Peries J, de The H. Nuclear targeting of incoming human foamy virus Gag proteins involves a centriolar step. The Journal of Virology. 1997;71:1155–1161. doi: 10.1128/jvi.71.2.1155-1161.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu SF, Edelmann K, Strong RK, Moebes A, Rethwilm A, Linial ML. The carboxyl terminus of the human foamy virus Gag protein contains separable nucleic acid binding and nuclear transport domains. The Journal of Virology. 1996;70:8255–8262. doi: 10.1128/jvi.70.12.8255-8262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dang VD, Levin HL. Nuclear import of the retrotransposon Tf1 is governed by a nuclear localization signal that possesses a unique requirement for the FXFG nuclear pore factor Nup124p. Mol.Cell Biol. 2000;20:7798–7812. doi: 10.1128/mcb.20.20.7798-7812.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Casacuberta E, Marin FA, Pardue ML. Intracellular targeting of telomeric retrotransposon Gag proteins of distantly related Drosophila species. Proc.Natl.Acad.Sci U.S.A. 2007;104(20):8391–8396. doi: 10.1073/pnas.0702566104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haoudi A, Kim MH, Champion S, Best-Belpomme M, Maisonhaute C. The Gag polypeptides of the Drosophila 1731 retrotransposon are associated to virus-like particles and to nuclei. FEBS Lett. 1995;377(1):67–72. doi: 10.1016/0014-5793(95)01305-9. [DOI] [PubMed] [Google Scholar]
- 95.Parent L. New insights into the nuclear localization of retroviral Gag proteins. Nucleus. 2011;2:92–97. doi: 10.4161/nucl.2.2.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Depienne C, Roques P, Creminon C, Fritsch L, Casseron R, Dormont D, Dargemont C, Benichou S. Cellular distribution and karyophilic properties of matrix, integrase, and Vpr proteins from the human and simian immunodeficiency viruses. Exp Cell Res. 2000;260:387–95. doi: 10.1006/excr.2000.5016. [DOI] [PubMed] [Google Scholar]
- 97.Fouchier RA, Meyer BE, Simon JH, Fischer U, Malim MH. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hearps AC, Wagstaff KM, Piller SC, Jans DA. The N-terminal basic domain of the HIV-1 matrix protein does not contain a conventional nuclear localization sequence but is required for DNA binding and protein self-association. Biochemistry. 2008;47:2199–210. doi: 10.1021/bi701360j. [DOI] [PubMed] [Google Scholar]
- 99.Mansky LM, Gajary LC. The primary nucleotide sequence of the bovine leukemia virus RNA packaging signal can influence efficient RNA packaging and virus replication. Virology. 2002;301:272–80. doi: 10.1006/viro.2002.1578. [DOI] [PubMed] [Google Scholar]
- 100.Mansky LM, Krueger AE, Temin HM. The bovine leukemia virus encapsidation signal is discontinuous and extends into the 5′ end of the gag gene. J Virol. 1995;69:3282–9. doi: 10.1128/jvi.69.6.3282-3289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mansky LM, Wisniewski RM. The bovine leukemia virus encapsidation signal is composed of RNA secondary structures. J Virol. 1998;72:3196–204. doi: 10.1128/jvi.72.4.3196-3204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang H, Norris KM, Mansky LM. Involvement of the matrix and nucleocapsid domains of the bovine leukemia virus Gag polyprotein precursor in viral RNA packaging. J Virol. 2003;77:9431–8. doi: 10.1128/JVI.77.17.9431-9438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Katoh I, Yasunaga T, Yoshinaka Y. Bovine leukemia virus RNA sequences involved in dimerization and specific gag protein binding: close relation to the packaging sites of avian, murine, and human retroviruses. J Virol. 1993;67:1830–1839. doi: 10.1128/jvi.67.4.1830-1839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Le Blanc I, Rosenberg AR, Dokhelar MC. Multiple functions for the basic amino acids of the human T-cell leukemia virus type 1 matrix protein in viral transmission. J Virol. 1999;73:1860–7. doi: 10.1128/jvi.73.3.1860-1867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Matthews S, Mikhailov M, Burny A, Roy P. The solution structure of the bovine leukaemia virus matrix protein and similarity with lentiviral matrix proteins. EMBO J. 1996;15:3267–74. [PMC free article] [PubMed] [Google Scholar]
- 106.Christensen AM, Massiah MA, Turner BG, Sundquist WI, Summers MF. Three-dimensional structure of the HTLV-II matrix protein and comparative analysis of matrix proteins from the different classes of pathogenic human retroviruses. J Mol Biol. 1996;264:1117–31. doi: 10.1006/jmbi.1996.0700. [DOI] [PubMed] [Google Scholar]
- 107.Rice WG, Baker DC, Schaeffer CA, Graham L, Bu M, Terpening S, Clanton D, Schultz R, Bader JP, Buckheit RW, Jr., Field L, Singh PK, Turpin JA. Inhibition of multiple phases of human immunodeficiency virus type 1 replication by a dithiane compound that attacks the conserved zinc fingers of retroviral nucleocapsid proteins. Antimicrob Agents Chemother. 1997;41:419–26. doi: 10.1128/aac.41.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pannecouque C, Szafarowicz B, Volkova N, Bakulev V, Dehaen W, Mely Y, Daelemans D. Inhibition of HIV-1 replication by a bis-thiadiazolbenzene-1,2-diamine that chelates zinc ions from retroviral nucleocapsid zinc fingers. Antimicrob Agents Chemother. 2010;54:1461–8. doi: 10.1128/AAC.01671-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dietz J, Koch J, Kaur A, Raja C, Stein S, Grez M, Pustowka A, Mensch S, Ferner J, Moller L, Bannert N, Tampe R, Divita G, Mely Y, Schwalbe H, Dietrich U. Inhibition of HIV-1 by a peptide ligand of the genomic RNA packaging signal Psi. Chem Med Chem. 2008;3:749–55. doi: 10.1002/cmdc.200700194. [DOI] [PubMed] [Google Scholar]
- 110.Huang M, Maynard A, Turpin JA, Graham L, Janini GM, Covell DG, Rice WG. Anti-HIV agents that selectively target retroviral nucleocapsid protein zinc fingers without affecting cellular zinc finger proteins. J Med Chem. 1998;41:1371–81. doi: 10.1021/jm9708543. [DOI] [PubMed] [Google Scholar]
- 111.McDonnell NB, De Guzman RN, Rice WG, Turpin JA, Summers MF. Zinc ejection as a new rationale for the use of cystamine and related disulfide-containing antiviral agents in the treatment of AIDS. J Med Chem. 1997;40:1969–76. doi: 10.1021/jm970147+. [DOI] [PubMed] [Google Scholar]
- 112.Rice WG, Turpin JA, Huang M, Clanton D, Buckheit RW, Jr., Covell DG, Wallqvist A, McDonnell NB, DeGuzman RN, Summers MF, Zalkow L, Bader JP, Haugwitz RD, Sausville EA. Azodicarbonamide inhibits HIV-1 replication by targeting the nucleocapsid protein. Nat Med. 1997;3:341–5. doi: 10.1038/nm0397-341. [DOI] [PubMed] [Google Scholar]
- 113.Druillennec S, Dong CZ, Escaich S, Gresh N, Bousseau A, Roques BP, Fournie-Zaluski MC. A mimic of HIV-1 nucleocapsid protein impairs reverse transcription and displays antiviral activity. Proc Natl Acad Sci U S A. 1999;96:4886–91. doi: 10.1073/pnas.96.9.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schito ML, Goel A, Song Y, Inman JK, Fattah RJ, Rice WG, Turpin JA, Sher A, Appella E. In vivo antiviral activity of novel human immunodeficiency virus type 1 nucleocapsid p7 zinc finger inhibitors in a transgenic murine model. AIDS Res Hum Retroviruses. 2003;19:91–101. doi: 10.1089/088922203762688595. [DOI] [PubMed] [Google Scholar]
- 115.Warui DM, Baranger AM. Identification of specific small molecule ligands for stem loop 3 ribonucleic acid of the packaging signal Psi of human immunodeficiency virus-1. J Med Chem. 2009;52:5462–73. doi: 10.1021/jm900599v. [DOI] [PubMed] [Google Scholar]
- 116.Chung J, Ulyanov NB, Guilbert C, Mujeeb A, James TL. Binding characteristics of small molecules that mimic nucleocapsid protein-induced maturation of stem-loop 1 of HIV-1 RNA. Biochemistry. 2010;49:6341–51. doi: 10.1021/bi100660r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jenkins L. M. Miller, Ott DE, Hayashi R, Coren LV, Wang D, Xu Q, Schito ML, Inman JK, Appella DH, Appella E. Small-molecule inactivation of HIV-1 NCp7 by repetitive intracellular acyl transfer. Nat Chem Biol. 2010;6:887–9. doi: 10.1038/nchembio.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chukkapalli V, Oh SJ, Ono A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc Natl Acad Sci U S A. 2010;107:1600–5. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Inlora J, Chukkapalli V, Derse D, Ono A. Gag Localization and Virus-Like Particle Release Mediated by the Matrix Domain of Human T-Lymphotropic Virus Type 1 Gag Are Less Dependent on Phosphatidylinositol-(4,5)-Bisphosphate than Those Mediated by the Matrix Domain of HIV-1 Gag. J Virol. 2011;85:3802–10. doi: 10.1128/JVI.02383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Massiah MA, Staricg MR, Paschall C, Summers MF, Christensen AM, Sundquist WI. The three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J.Mol.Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 121.Matthews S, Mikhailov M, Burny A, Roy P. The solution structure of the bovine leukaemia virus matrix protein and similarity with lentiviral matrix proteins. EMBO Jl. 1996;15:3267–3274. [PMC free article] [PubMed] [Google Scholar]
- 122.McDonnell JM, Fushman D, Cahill SM, Zhou W, Wolven A, Wilson CB, Nelle TD, Resh MD, Wills J, Cowburn D. Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus. J.Mol.Biol. 1998;279:921–928. doi: 10.1006/jmbi.1998.1788. [DOI] [PubMed] [Google Scholar]