Cells from Cockayne syndrome patients and a mouse model of the disease show increased metabolism as a result of impaired autophagy-mediated removal of damaged mitochondria.
Abstract
Cockayne syndrome (CS) is a devastating autosomal recessive disease characterized by neurodegeneration, cachexia, and accelerated aging. 80% of the cases are caused by mutations in the CS complementation group B (CSB) gene known to be involved in DNA repair and transcription. Recent evidence indicates that CSB is present in mitochondria, where it associates with mitochondrial DNA (mtDNA). We report an increase in metabolism in the CSBm/m mouse model and CSB-deficient cells. Mitochondrial content is increased in CSB-deficient cells, whereas autophagy is down-regulated, presumably as a result of defects in the recruitment of P62 and mitochondrial ubiquitination. CSB-deficient cells show increased free radical production and an accumulation of damaged mitochondria. Accordingly, treatment with the autophagic stimulators lithium chloride or rapamycin reverses the bioenergetic phenotype of CSB-deficient cells. Our data imply that CSB acts as an mtDNA damage sensor, inducing mitochondrial autophagy in response to stress, and that pharmacological modulators of autophagy are potential treatment options for this accelerated aging phenotype.
Neurodegeneration and cachectic dwarfism are cardinal symptoms of the autosomal recessive segmental progeria Cockayne syndrome (CS; Nance and Berry, 1992). Average life expectancy is 12 yr; however, the phenotype shows substantial variation in severity. 80% of CS cases are caused by mutations in the group B gene (CSB), with the remaining 20% caused by mutations in CSA. CSB is involved in transcription-coupled nucleotide excision DNA repair (TC-NER; Anindya et al., 2010) and has been proposed to carry out chromatin remodeling (Citterio et al., 2000), act as a transcription factor (Le May et al., 2010), and function as a co-regulator of base excision repair (BER; Stevnsner et al., 2008).
The CSBm/m mouse displays a mild neurological phenotype, with an increased susceptibility to UV-induced cancer (van der Horst et al., 1997). This hypersensitivity to UV is a hallmark of CSB-deficient cells in culture and is related to the lack of TC-NER. However, CSB-deficient cells are also sensitive to alkylating and oxidizing agents (Stevnsner et al., 2008) and display defective repair of the DNA lesions 8-oxoguanine (Dianov et al., 1999) and 8-hydroxyadenine (Tuo et al., 2002). Oxidative lesions, repaired by BER, are particularly pertinent because the accumulation of oxidized proteins, lipids, and/or nucleic acids have been proposed to be an underlying cause of aging (Balaban et al., 2005). Endogenous oxidizing radicals originate predominantly from the mitochondria, where the mitochondrial BER machinery acts as the primary protector. Recent findings indicate that CSB is present in the mitochondria, suggesting a role for CSB in mitochondrial DNA (mtDNA) repair (Aamann et al., 2010; Kamenisch et al., 2010). We now report that CSB participates in mitochondrial maintenance by inducing autophagy in response to mitochondrial stress. Lack of CSB leads to mitochondrial dysfunction and increased metabolism, both at the organismal and cellular level. Accordingly, we show that pharmacological activation of autophagy reverses the bioenergetic profile. Based on this, we propose that CSB acts as a sensor of mtDNA damage and regulates mitochondrial autophagy and that treatment with rapamycin or lithium chloride could potentially attenuate some symptoms related to CS.
RESULTS
CSBm/m mice show age-related and universal loss of fat
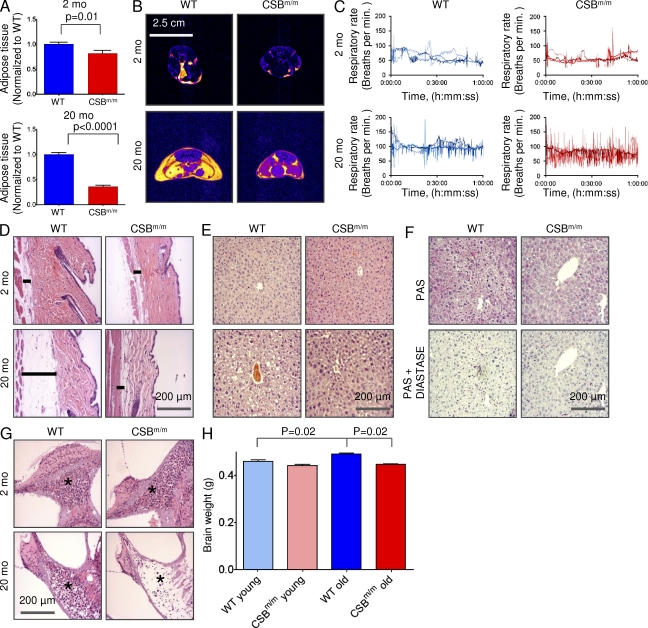
Because mitochondrial dysfunction can have an effect on overall metabolism, we investigated the distribution and volume of adipose tissue, using T1-weighted MRI, in old and young WT and CSBm/m mice. The combined volume of the abdominal and epididymal fat depots in old CSBm/m mice was only 36% of the amount in old WT mice (Fig. 1 A). Interestingly, this difference was already present in the young group, which showed 82% of the amount of fat compared with age-matched controls. The loss of fat was not only subcutaneous but also visceral fat (Fig. 1 B and Video 1). Curiously, during the MRI scans the old CSBm/m mice were found to be particularly difficult to maintain at a constant respiration frequency under anesthesia, as evident from the large fluctuations in respiration frequency (Fig. 1 C).
Figure 1.
The old CSBm/m mice show signs of neurodegeneration and universal loss of fat. (A) Quantification of adipose tissue using T1-weighted MRI in young (2 mo) and old (20 mo) WT and CSBm/m mice (n = 3; data are represented as mean ± SEM). (B) Representative axial T1-weighted MRI scans of the CSBm/m and WT mice. Yellow denotes fat. (C) Respiration rates of mice during the MRI scans. (D) HE staining of abdominal skin in young and old WT and CSBm/m mice. The bar denotes the subcutaneous fat layer (n = 3). (E) HE staining of the liver. (F) Periodic acid-Schiff staining of the liver of the 20-mo-old mice with and without diastase to show glycogen content. (G) HE staining of the inner ear. The star denotes the spiral ganglion (n = 3). (H) Brain weight of WT and CSBm/m mice (n = 3; data are represented as mean ± SEM).
Skin histology of old CSBm/m mice also showed loss of subcutaneous fat by HE staining (Fig. 1 D) as previously reported (Kamenisch et al., 2010). Liver histology from the old WT mice, but not CSBm/m animals, showed vacuolization of the cytoplasm (Fig. 1 E) caused either by an accumulation of lipids or glycogen. Using periodic acid-Schiff staining, we found no difference in the amount of glycogen in the liver tissue (Fig. 1 F), indicating a decrease in liver lipids in old CSBm/m.
The CSBm/m mouse shows a CS-like phenotype
To further investigate a possible phenotype in the CSB-deficient mice, we performed extensive histology, hematology, and serology on young and old CSBm/m and WT mice. Table S1 shows a list of serum chemistry, hematology, and organ weights examined. The only overall significant difference uncovered when comparing the young WT and CSBm/m mice was an ∼50% increase in thymus weight (31 mg, WT vs. 48 mg, CSBm/m; P = 0.03), indicating a delay in organ involution; however, no histological disparity was observed (unpublished data).
For the old CSBm/m mice, subtle signs of neurodegeneration were present. A substantial loss of spiral gangliocytes was found in the inner ear of the old CSBm/m mice (Fig. 1 G), consistent with the sensorineural hearing loss associated with CS (Nance and Berry, 1992). The CSBm/m mice displayed significantly smaller brain weights compared with WT (Fig. 1 H), although the cerebrum, cerebellum, spinal cord, and ischiatic nerve showed no sign of pathology (unpublished data). Several mitochondrial diseases show skeletal or heart myopathy, yet no histological changes were found in the CSBm/m mice (unpublished data), despite a trend toward increased plasma creatine kinase in the CSB-deficient mice (Table S1).
CSBm/m mice show increased metabolism
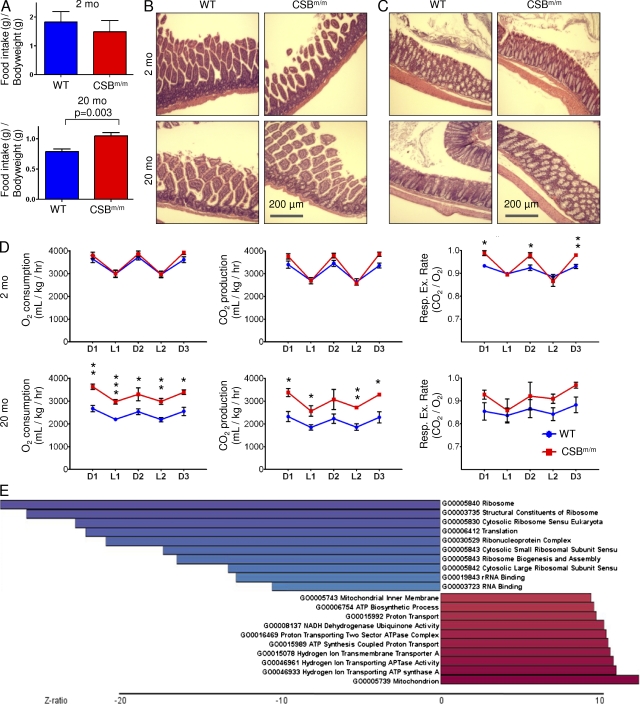
The observed fat loss could be the result of a decrease in food consumption, gastrointestinal malabsorption, or increased energy expenditure. Surprisingly, the old CSBm/m mice consumed ∼33% more food than the age-matched WT mice (Fig. 2 A, n = 3). No steatorrhea was observed, and histology of the jejunum, ileum, and colon did not reveal pathology that could indicate malabsorption in the mutant animals (Fig. 2, B and C). We therefore investigated the overall metabolism of the mice. Housing the animals in metabolic cages revealed increased O2 consumption and CO2 production without an increase in movement in the old CSBm/m mice (Fig. 2 D; movement data not depicted). Interestingly, there was a trend toward increased respiratory exchange rates in the old CSBm/m mice, indicating that the loss of fat was not the result of a preference for β-oxidation (Fig. 2 D). Serum chemistry showed no difference in blood glucose or cholesterol (Table S1); however, a decrease in circulating triglycerides was observed in old CSBm/m mice, reflecting the increased metabolism.
Figure 2.
CSB deficiency increases whole body metabolism. (A) 1 wk food intake CSBm/m compared with age-matched WT mice (n = 3; data are represented as mean ± SEM). (B and C) HE staining of the small (B) and large (C) intestines (n = 3). (D) Mice were placed in metabolic cages for 72 h. O2 consumption and CO2 production were measured. D1= dark phase 1, L1 = light phase 1, etc. (n = 3–8; data are represented as mean ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001). (E) Gene ontology terms of a microarray comparing MDFs from WT and CSBm/m mice (n = 3–4; blue is down-regulated and red is up-regulated).
Intriguingly, by data-mining a previously published and validated microarray of mouse dermal fibroblasts (MDFs) from the CSBm/m animals (Garinis et al., 2009), we found a strong correlation between metabolism and CSB deficiency. When comparing the expression profiles of MDFs from WT and CSBm/m mice, we found 90 genes that were significantly differentially regulated (±1.5 SD, 32 genes down- and 58 up-regulated; Fig. S1 A). Interrogating the gene ontology terms in greater detail, we saw a clear increase in genes involved in energy metabolism in the CSB-deficient fibroblasts. All of the top 10 up-regulated gene ontology terms and a total of 22 out of 94 up-regulated terms were ATP production related, indicating an attempt by the cells to up-regulate energy production (Fig. 2 E and Fig. S1 B). When examining the down-regulated gene ontology terms, the list is dominated by terms involved in protein synthesis, in accordance with previous literature of organisms under energy deprivation (Hansen et al., 2007).
Cellular metabolism is increased in CSB-deficient cultured cells
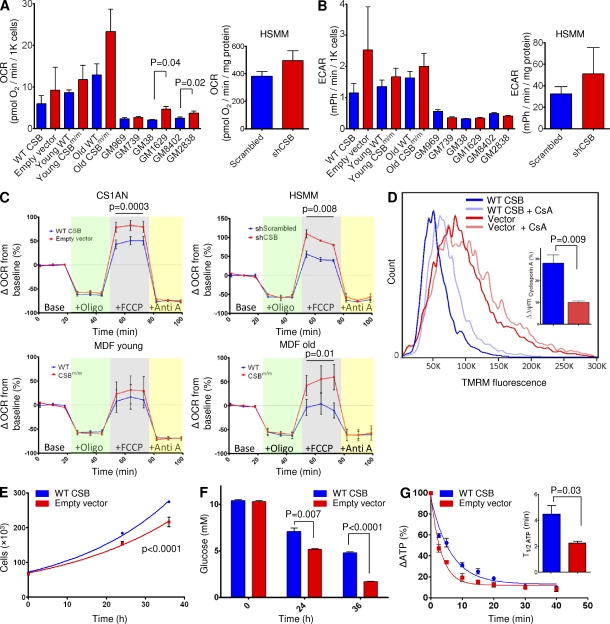
To explore if the hypermetabolic condition of the mouse was present at the cellular level, we investigated the respiration in various CSB-deficient cell lines. MDFs from young and old CSBm/m and WT mice, as well as fibroblasts derived from the CS patient CS1AN complemented with WT CSB or an empty vector, were examined. Additionally, we performed lentiviral knockdown of CSB in primary human myotubes (human skeletal muscle myotube [HSMM]). Finally, we also examined three primary CSB-deficient patient cell lines (GM739, GM1629, and GM2838) with age- and gender-matched control lines (GM969, GM38, and GM8402, respectively). Oxygen consumption and extracellular acidification were measured using the Seahorse XF24 analyzer. Every CSB-deficient cell line tested showed increased basal respiration, although only two cell lines reached significance when compared with the controls (GM38 vs. GM1629 and GM8402 vs. GM2838; n = 3, paired Student’s t test; Fig. 3 A). All the CSB-deficient cells that were compared with isogenic controls showed increased extracellular acidification rates, reflecting not only an increase in oxygen consumption but also increased glycolysis (Fig. 3 B). Additionally, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP)–uncoupled respiration was found to be increased in CSB-deficient CS1AN cells, myotubes, and MDFs from old CSBm/m mice (Fig. 3 C; CS1AN: n = 7, paired Student’s t test; HSMM: n = 3, paired Student’s t test; old MDF: n = 3, paired Student’s t test). An increase in FCCP-uncoupled respiration could be the result of an increased mitochondrial membrane potential. Indeed, we found a robust increase in tetramethyl rhodamine methyl ester (TMRM) staining by flow cytometry (Fig. 3 D). Additionally, we observed a large variation in membrane potential within the CSB-deficient cells, as evident by the broadened peak, perhaps reflecting a significantly more heterogeneous population of mitochondria in CSB. Remarkably, addition of cyclosporin A, which increased the membrane potential in the WT cells, did not affect the membrane potential in CSB-deficient CS1AN cells, indicating a decreased activity of the membrane permeability transition pore (mPTP) in CSB-deficient cells (Fig. 3 D).
Figure 3.
CSB deficiency increases cellular metabolism. (A and B) Oxygen consumption rates (OCRd; A) and extracellular acidification rates (ECARd; B) were increased in CSB-deficient cells. CS1AN: CSB patient cell line expressing an empty vector or a vector containing WT CSB. HSMM: HSMMs expressing scrambled shRNA or shRNA targeted to CSB. MDF: MDFs isolated from young and old CSBm/m mice. Primary CSB patient fibroblasts (GM739, GM1629, and GM2838) and healthy age-/gender-matched controls (GM969, GM38, and GM 8402) are shown. Blue are cells expressing WT CSB. Red are CSB-deficient cells (n = 3–7; data are represented as mean ± SEM). (C) FCCP-uncoupled respiration was measured by sequential addition of oligomycin (green background), FCCP (gray background), and antimycin A (yellow background; n = 3–7; data are represented as mean ± SEM). (D) Mitochondrial membrane potential in CS1AN cells measured by 20 nM TMRM staining and flow cytometry and its responsiveness to treatment with cyclosporin A (CsA). Inset: quantification of the response to cyclosporin A (n = 3; data are represented as mean ± SEM). (E) Growth curve of CS1AN cells expressing either an empty vector or WT CSB. (n = 3; data are represented as mean ± SEM). (F) Glucose concentration was measured in the media of CS1AN cells expressing WT CSB or an empty vector (n = 3; data are represented as mean ± SEM). (G) ATP consumption of CS1AN cells expressing WT CSB or an empty vector after inhibition of ATP production with 100 mM 2-deoxyglucose and 1 µM oligomycin. Inset: half-life of ATP (n = 3; data are represented as mean ± SEM).
In cultured cells, the main carbon donor for metabolism is glucose. To support the respiratory data, we monitored the glucose concentration and number of CS1AN cells over 36 h. Cell number data were fitted to the exponential growth curve (y = y0 × ekX; WT CSB, r2 = 0.99; empty vector, r2 = 0.96). Interestingly, CSB-deficient cells showed a significantly slower growth rate and consumed around twofold more glucose than cells expressing WT CSB (Fig. 3, E [n = 3, F-test] and F [n = 3]). When measuring ATP levels, however, we found no difference between the CS1AN cells expressing WT CSB and the empty vector (unpublished data). Based on these observations, we would predict that the CSB-deficient cells would consume more ATP and, indeed, that is what we found when we monitored the ATP levels after inhibiting both glycolysis (using 2-deoxyglucose) and oxidative phosphorylation (using oligomycin; Fig. 3 G).
CSB-deficient cultured cells show an increased mitochondrial content
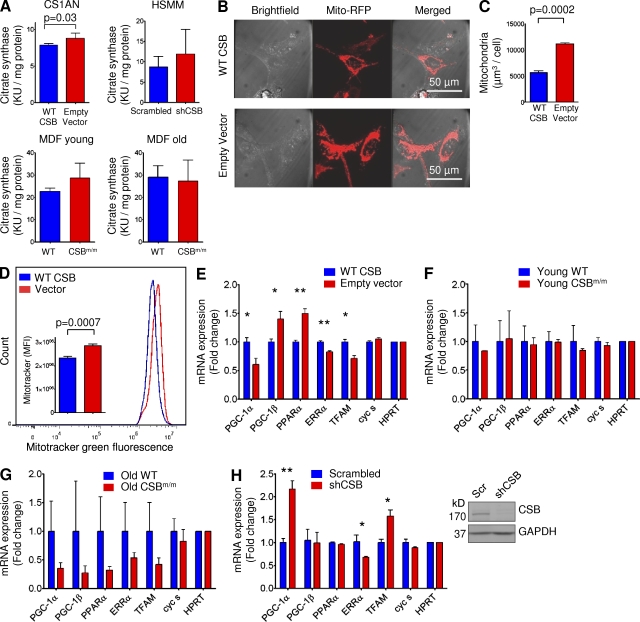
We next explored if the increase in cellular metabolism was reflected by an increase in mitochondrial content. Citrate synthase is a marker for mitochondrial content (Holloszy et al., 1970) and CSB-deficient CS1AN cells showed an ∼10% increase in citrate synthase activity compared with cells expressing WT CSB (Fig. 4 A, n = 5). This increase was also found in HSMM subjected to CSB knockdown, yet failed to reach significance. To further investigate mitochondrial content, we transfected CS1AN cells expressing WT CSB or empty vector with RFP-tagged mitochondrial protein. Live cells were then imaged on a confocal microscope and z-stacks were quantified. Using this approach, CSB-deficient CS1AN cells showed a twofold increase in mitochondrial content (Fig. 4, B and C, n = 3). We substantiated these results by flow cytometry, observing a clear increase in MitoTracker green staining intensity in CSB-deficient CS1AN cells compared with cells expressing WT CSB (Fig. 4 D, n = 5).
Figure 4.
CSB-deficient cells show increased mitochondrial content and decreased mitochondrial biogenesis. (A) Citrate synthase activity in CS1AN cells expressing WT CSB or an empty vector or in MDF from young or old CSBm/m or WT mice or in HSMM expressing scrambled or CSB-specific shRNA (n = 3–5; data are represented as mean ± SEM). (B) Representative confocal image of CS1AN cells expressing WT CSB or an empty vector transfected with the mitochondrial RFP-tagged protein using organelle lights. (C) Quantification of the mitochondrial RFP signal normalized to cell number (n = 3; data are represented as mean ± SEM). (D) Representative flow cytometry experiment of CS1AN cells expressing WT CSB or empty vector treated with 50 nM MitoTracker green. Inset: quantification of the flow cytometry data (n = 3; data are represented as mean ± SEM). (E–H) Quantitative PCR analysis of genes involved in mitochondrial biogenesis in CS1AN cells expressing WT CSB or empty vector (E), MDFs from young (F) and old (G) mice, and HSMM subjected to lentiviral CSB knockdown (H). The inset in H shows efficiency of lentiviral shRNA knockdown of CSB (n = 3; data are represented as mean ± SEM; *, P < 0.05; **, P < 0.01).
The amount of mitochondria in a cell must be a balance between synthesis and destruction. We therefore evaluated if changes in mitochondrial biogenesis could account for the altered mitochondrial content and function. In cooperation with estrogen-related receptor α (ERR-α), peroxisome proliferator-activated receptor γ co-activator 1α (PGC-1α) potently stimulates mitochondrial biogenesis. Although young CSBm/m MDFs showed no changes in the gene expression of the transcriptional regulators ERR-α, PGC-1α, and transcription factor A, mitochondrial (TFAM), CSB-deficient CS1AN, and old CSBm/m MDFs revealed specific decreases in these factors (Fig. 4, E–G). In the CSB knockdown HSMM, we observed a decrease in ERR-α, consistent with the other CSB-deficient cell lines. PGC-1α and TFAM, however, were increased in the knockdown myotubes (Fig. 4 H). The reason for this discrepancy is at present unclear, but perhaps the full phenotype of CSB deficiency has not developed in the HSMM during the 3 d that passed from shRNA transfection to RNA isolation. Considering the consistent mitochondrial phenotype we see both across species and cell types with no consistent changes in these transcription factors leads us to believe that biogenesis was not causal in the phenotype.
CSB-deficient cells show decreased autophagy and an accumulation of damaged mitochondria
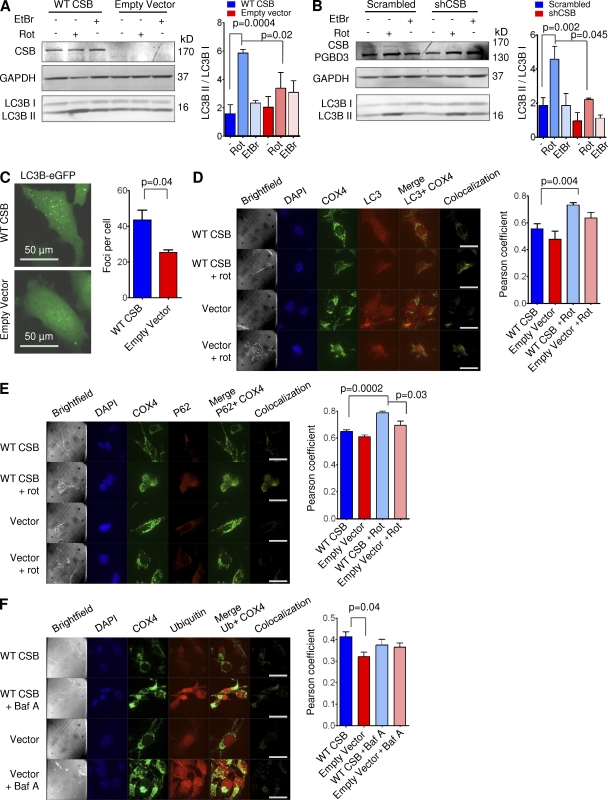
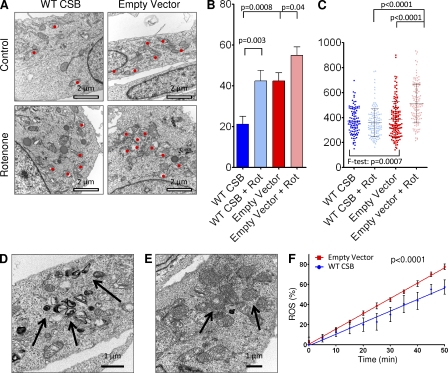
The observation mentioned in the previous section prompted us to investigate if macro-autophagy (hereon autophagy), the primary degradation route of mitochondria (Levine and Kroemer, 2008), might be altered in CSB-deficient cells. Upon formation of autophagic vacuoles, cytosolic LC3B (isoform-I, LC3B-I) is cleaved and inserted into the autophagic membrane (isoform-II, LC3B-II), allowing autophagy to be monitored by measuring the LC3B-II/LC3B-I ratio (Levine and Kroemer, 2008). Previous literature has shown that the mitochondrial toxicant rotenone induces autophagy (Chen et al., 2007). Remarkably, we saw a significant defect in rotenone-induced autophagy in CSB-deficient CS1AN cells and HSMM after 24 h of treatment (Fig. 5, A and B; CS1AN, n = 3; HSMM, n = 2–5). Even basal autophagy was decreased in the CSB-deficient HSMM. Interestingly, treatment with ethidium bromide did not increase the LC3II/LC3I ratio, indicating that mtDNA replication is independent of autophagy. This difference was not the result of a change in autophagic flux as measured by LC3B levels after treatment with bafilomycin A and chloroquine (unpublished data). To investigate if a defect in autophagy could be observed in the CSB-deficient mice, we investigated whether LC3 levels were altered in whole brain lysates. Correlating well with the defect being specifically mitochondrial, we found no change in LC3B levels between WT and CSB-deficient animals (unpublished data). Accordingly, we did not find any CSB-dependent difference in the accumulation of ubiquitinated protein in the mice. We did, however, observe an increase in ubiquitinated protein with age as previously described (Ohtsuka et al., 1995; unpublished data). To corroborate our findings in the CSB-deficient cells, we transfected CS1AN cells expressing WT CSB or the empty vector with a plasmid encoding eGFP-tagged LC3. The amount of eGFP-LC3 foci correlates with the formation of autophagic vacuoles (Jackson et al., 2005) and, upon quantification, we saw a significant decrease in foci formation in the CSB-deficient cells, indicating a decrease in autophagy (Fig. 5 C, n = 3). To investigate whether the autophagic vacuoles contained mitochondria, we performed confocal microscopy examining the colocalization of endogenous LC3B and mitochondrial cytochrome c oxidase subunit 4 (COX4). Interestingly, when measuring the unbiased global Pearson’s coefficient we saw a significant increase in the colocalization of LC3B and mitochondria in the CSB-proficient cells in response to rotenone treatment (Fig. 5 D). This data could indicate a defect in the recruitment of factors that have been proposed to specifically induce mitochondrial degradation. Indeed, when measuring the colocalization between P62 and COX4 we saw a significant decrease in the global Pearson’s coefficient in the CSB-deficient cells (Fig. 5 E). Ubiquitination of proteins in the outer mitochondrial membrane have been suggested to selectively lead to the degradation of damaged mitochondria (Geisler et al., 2010). Interestingly, when investigating this in the CS1AN cells we observed a decrease in colocalization of ubiquitin and COX4 in cells lacking CSB (Fig. 5 F). There was neither a difference in the amount of ubiquitin staining at base line nor in the amount after inhibition of autophagy by bafilomycin A, supporting the notion of normal autophagic flux in the CSB-deficient cells. To further corroborate these data, we performed electron microscopy on the CS1AN cell lines after 24 h of rotenone treatment. In agreement with the previous findings, the CSB-deficient CS1AN cells had an increased amount of damaged mitochondria both at baseline and after rotenone treatment compared with cells expressing WT CSB. This suggested a failure to clear the damaged mitochondria (Fig. 6, A and B). When measuring the mean mitochondrial diameter, we found that the CSB-deficient cells displayed a much greater variation in mitochondrial width than those from WT, demonstrating a significantly more heterogeneous mitochondrial population (Fig. 6 C) correlating well with our TMRM staining. In addition, a subset of CSB-deficient cells showed an accumulation of hyperdense membranous material (Fig. 6 D) and formation of multiple lamellar mitochondrial cristae (onion formation), indicative of loss of mtDNA (Kukat et al., 2008) or altered complex five assembly (Paumard et al., 2002; Fig. 6 E). Decreased autophagy could lead to an accumulation of damaged mitochondria and increased reactive oxygen species (ROS) production. Indeed, using dihydroethidium (Aykin-Burns et al., 2009) we found a significant increase in ROS formation in CSB-deficient CS1AN cells compared with cells expressing WT CSB (Fig. 6 F, n = 10).
Figure 5.
Mitochondrial autophagy is decreased in CSB-deficient cells. (A and B) Western blots of CS1AN cells expressing WT CSB or an empty vector (A) or HSMM subjected to lentiviral knockdown of CSB treated with vehicle, 5 µM rotenone (autophagy inducer), or 500 ng/ml ethidium bromide for 24 h (B; n = 3, data are represented as mean ± SEM). (C) Representative image of CS1AN cells expressing WT CSB or an empty vector transfected with eGFP-tagged LC3 and quantification of the foci per cell (n = 3; data are represented as mean ± SEM). (D) Representative image of CS1AN cells expressing WT CSB or an empty vector treated with vehicle or rotenone and stained for LC3B and COX4 (mitochondria; bars, 50 µm) and quantification of the Pearson’s coefficient of these cells (n = 4; data are represented as mean ± SEM). (E) Representative image of CS1AN cells expressing WT CSB or an empty vector treated with vehicle or rotenone and stained for P62 and COX4 (bars, 50 µm) and the quantification of the Pearson’s coefficient of these cells (n = 4; data are represented as mean ± SEM). (F) Representative image of CS1AN cells expressing WT CSB or an empty vector treated with vehicle or bafilomycin A and stained for ubiquitin and COX4 (bars, 50 µm) and the quantification of the Pearson’s coefficient of these cells (n = 4; data are represented as mean ± SEM).
Figure 6.
CSB-deficient cells show accumulation of damaged mitochondria. (A) Representative electron microscopy images of CS1AN cells lines after treatment with vehicle or 5 µM rotenone. Red dots denote swollen/damaged mitochondria (n = 12). (B) Quantification of damaged mitochondria in vehicle and rotenone-treated CS1AN cells expressing WT CSB or an empty vector (n = 12; data are represented as mean ± SEM). (C) Mitochondrial diameter in CS1AN cells expressing WT CSB or an empty vector after vehicle or rotenone treatment. Each point represents one mitochondria and cross bar represents mean ± SD. (D) Arrows marking hyperdense membranous material in CS1AN cells expressing an empty vector. (E) Arrows marking mitochondrial onion formation in CS1AN cells expressing an empty vector. (F) ROS formation measured by dihydroethidium in CS1AN cells expressing WT CSB or an empty vector (n = 10; data are represented as mean ± SD).
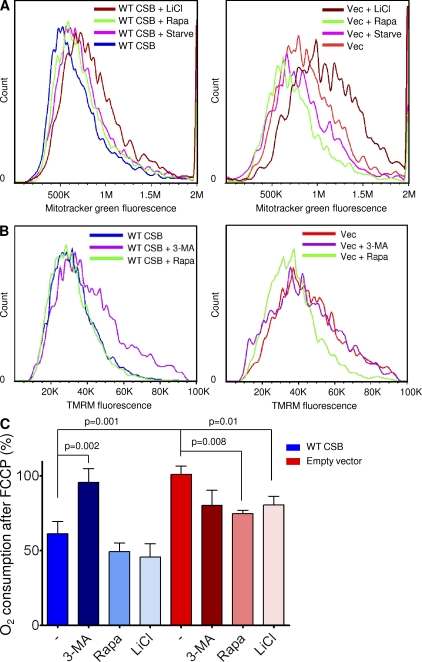
Serum starvation, as well as the FDA-approved drugs rapamycin and lithium chloride, is known to induce autophagy in cells. We therefore tested if these conditions could alter the bioenergetic profile of the CSB-deficient cells. Interestingly, both starvation and rapamycin were able to decrease the mitochondrial content in CSB-deficient cells (Fig. 7 A). Furthermore, by treating the CSB-deficient cells with rapamycin for 24 h we were able to decrease the membrane potential in CSB-deficient cells, whereas inhibiting autophagy using 3-methyladenine (3-MA) led to an increase in membrane potential only in the cell expressing WT CSB (Fig. 7 B). Finally, using the Seahorse XF24 analyzer we found that treatment with either rapamycin or lithium chloride led to a decrease in FCCP-uncoupled respiration in the CSB-deficient cells, whereas treatment with 3-MA led to an increase in FCCP-uncoupled respiration essentially phenocopying the CSB-deficient cells (Fig. 7 C).
Figure 7.
Rapamycin rescues the bioenergetic profile of CSB-deficient cells. (A) Mitochondrial content of CS1AN cells expressing WT CSB (left) or an empty vector (right) was measured using flow cytometry and 50 nM MitoTracker green staining after treatment with the autophagy inducer 1 µM rapamycin (rapa), serum starvation (starve), or 10 mM of the autophagy inducer lithium chloride (LiCl; n = 3 representative experiments). (B) The membrane potential of CS1AN cells expressing WT CSB (left) or an empty vector (right) was measured using flow cytometry and 20 nM TMRM staining after treatment with 1 µM rapamycin (rapa) or 10 mM of the autophagy inhibitor 3-MA for 24 h (n = 6 representative experiments). (C) The bioenergetics profile of CS1AN cells expressing WT CSB or an empty vector after treatment with 10 mM lithium chloride, 1 µM rapamycin, or 3-MA for 24 h (n = 3; data are represented as mean ± SEM).
DISCUSSION
We describe herein a dramatic age-related fat loss in the CSBm/m mouse. We show that this phenotype is caused by an increase in baseline metabolism, as seen by increased food intake and increased oxygen consumption. On the cellular level, CSB deficiency led to increased oxygen consumption, hyperpolarized mitochondria, and increased mitochondrial volume that does not seem to be explained by increased mitochondrial biogenesis. The elevated mitochondrial metabolism appears to instead stem from decreased mitochondrial degradation. Indeed, we find defective autophagy in CSB-deficient cells in response to mitochondrial stress, possibly caused by a defect in the recruitment of P62 to mitochondria as well as decreased mitochondrial ubiquitination. Consistent with an accumulation of damaged mitochondria, free radical formation is also increased in the CSB-deficient cells. Remarkably, using pharmacological activators of autophagy we are able to revert the bioenergetic profile of the CSB-deficient cells, indicating a possible treatment for this disease.
Based on our findings, we propose the following model. CSB localizes to the mitochondria and interacts with the nucleoid, especially under conditions of oxidative stress (Aamann et al., 2010; Kamenisch et al., 2010). We hypothesize that CSB plays a role as a sensor of mtDNA damage, either by directly binding to mtDNA or as an indirect signaling protein. DNA damage triggers a response, perhaps leading to the formation of the mPTP. Induction of the mPTP has been shown to signal autophagy (Carreira et al., 2010). Accordingly, mtDNA damage will selectively induce degradation of damaged mitochondria. Interestingly, inhibition of the mPTP by cyclosporin A protects against ischemia-reperfusion injury (Gomez et al., 2009). Based on our model, we would therefore predict increased resistance to ischemia-reperfusion injury in CSB-deficient cells as the result of a decrease in overall induction of the mPTP. Indeed, increased survival after kidney artery clamping has recently been reported in the CSBm/m mouse (Susa et al., 2009).
Although a direct role of CSB in mitochondrial maintenance through autophagy seems to be the simplest explanation, we could propose a connection to some of the characterized functions of CSB. It is well known that nuclear DNA damage accumulates in cells and tissues deficient in CSB not only as a result of the nucleotide excision repair defect in transcription-coupled repair but, presumably, also because of a defect in BER. Accordingly, this nuclear DNA repair defect could lead to increased poly ADP-ribose polymerase activation and, consequently, increased poly ADP ribosylation. This is a process with a high energy cost because: (a) loss of NAD+ leads to increased NADH/NAD+ and, thus, an inhibition of glycolysis/oxidative phosphorylation; and (b) NAD+ recycling costs two ATP molecules for every ADP-ribose moiety. An increase in energy expenditure, as we observe, could be countered by decreasing inner mitochondrial membrane uncoupling which will lead to more ATP produced per oxygen consumed. This would lead to an increased membrane potential that would directly lead to increased ROS formation. The autophagic defect could be a consequence of increased membrane potential because depolarization of the inner membrane is thought to lead to autophagy.
Previous studies have shown increased autophagy in several models of longevity (Levine and Klionsky, 2004), and life extension by caloric restriction or daf-2-mutation in Caenorhabditis elegans is dependent on autophagy (Meléndez et al., 2003; Jia and Levine, 2007). mTOR is a master inhibitor of autophagy (Díaz-Troya et al., 2008), and mice treated with the mTOR inhibitor rapamycin show increased life span (Harrison et al., 2009). Several autophagy gene (ATG) knockout mouse models display CS like phenotype. For example, the ATG5−/− mouse shows normal embryonic development but early neonatal death, presumably from energy deprivation (Kuma et al., 2004). Brain-specific knockout of ATG7 or ATG5 leads to neurodegeneration with loss of purkinje cells in the cerebellum (Hara et al., 2006; Komatsu et al., 2006). In humans, decreased autophagy has been implicated in several neurodegenerative diseases (Martinez-Vicente and Cuervo, 2007), and increased autophagy attenuates aspects of neurodegeneration (Chen et al., 2008; Rangaraju et al., 2009). Based on the evidence that increased autophagy may lead to life extension and neuroprotection, we are speculating that a deficiency in autophagy could lead to a premature aging phenotype such as CS. Interestingly, we do not observe any severe neurological phenotype in our CSB-deficient mice. As has been pointed out, mirroring human neurodegenerative diseases in other organisms seems to be difficult (Jucker, 2010). The Parkinson’s disease models are particularly interesting because a defect in mitochondrial degradation has been proposed to underlie the human condition (Geisler et al., 2010). However, like the CSB mutant mouse, little phenotype is seen in transgenic mouse models of this disease (Hatano et al., 2009), suggesting that rodents may have redundant mitophagic pathways or display increased resistance to mitochondrial degradation defects.
According to our model, lack of CSB induces a stochastic accumulation of damaged mitochondria, increased oxygen consumption, and ROS production. Oxidative stress has been observed in several neurodegenerative diseases (Trushina and McMurray, 2007), providing a link between our findings of increased ROS production and decreased autophagy to the neuropathology seen in CS. These findings could also explain the accumulation of lesions found in mitochondria from CSB-deficient cells (Muftuoglu et al., 2009; Osenbroch et al., 2009; Aamann et al., 2010; Kamenisch et al., 2010). The increased oxygen and glucose consumption we observe indicates a state of cellular energy deprivation. The microarray analysis we present further corroborates this conclusion. Increased energy consumption could explain the cachexia seen in humans with this disease. Accordingly, a recent paper by Kamenisch et al. (2010) reported subcutaneous fat loss in aged CSBm/m mice and ascribed this to increased mitochondrial DNA deletions. The increase in mitochondrial deletions is of particular interest because mtDNA deletions have been reported to stimulate autophagy (Prigione and Cortopassi, 2007). In agreement with our model, a deficiency in autophagy caused by loss of CSB would be predicted to lead to an accumulation of mitochondrial deletions.
In addition to the recent evidence of the presence of CSB in the mitochondria (Aamann et al., 2010; Kamenisch et al., 2010), indirect evidence of a mitochondrial role of CSB comes from biochemical analysis. Berquist and Wilson (2009) showed that Ca2+ stimulates and Mg2+ inhibits the strand annealing activity of CSB. Interestingly, Ca2+ is believed to accumulate in mitochondria (Rizzuto and Pozzan, 2006), and several mitochondrial proteins are activated by Ca2+ and inhibited by Mg2+ (Panov and Scarpa, 1996). Further evidence comes from our observation that old CSBm/m mice were difficult to maintain at stable respiration rates under isoflurane anesthesia. Increased ROS production and decreased mitochondrial respiration are observed after inhalation anesthesia (Stowe and Kevin, 2004). It seems likely that an underlying mitochondrial dysfunction would be aggravated by the exposure to isoflurane. Further support for a mitochondrial involvement in CS is the considerable similarity of the phenotype to mitochondrial diseases. Mitochondrial neurogastrointestinal encephalomyopathy (Nishino et al., 1999), Leigh syndrome (Finsterer, 2008), and NARP syndrome (Gelfand et al., 2011) bear substantial resemblance to CS, whereas a subset of cases of CoQ10 deficiency (Rötig et al., 2000), Leber’s hereditary optical atrophy (Funalot et al., 2002), and chronic progressive external ophthalmoplegia (Zeviani et al., 1990) show overlapping phenotypical characteristics. Additional evidence for a role of CSB outside nuclear DNA repair comes from the observation that CS patients do not show increased risk of cancer.
Currently, only palliative treatment is offered to CS patients, although one compound is currently under clinical trial (www.clinicaltrials.gov). Our study opens up the idea of stimulating autophagy as a potential therapeutic avenue. We show here that rapamycin reverses the bioenergetic profile of the CSB-deficient cells. This compound is currently approved by the FDA and could be a natural choice, although the adverse effects are severe. Interestingly, the demyelination seen in CS has been related to microglial activation (Weidenheim et al., 2009) and the immunosuppressant effect of rapamycin could, therefore, perhaps attenuate these symptoms as well. Lithium chloride is known to induce autophagy and could be another interesting treatment option. Besides the well known neuroprotective properties of this drug, a common adverse effect is weight gain (Grandjean and Aubry, 2009), which would benefit these patients as well. A different strategy could be to target downstream effects of the mitochondrial dysfunction. Antioxidants have shown little or even detrimental effect in long term population studies (Bjelakovic et al., 2008). However, they are used to treat stroke by attenuating ROS formation during ischemia-reperfusion injury (Wilson and Gelb, 2002) and could perhaps be beneficial to CS patients. A more pragmatic approach could be to increase food intake in these patients, perhaps lessening some symptoms. However, this could be a double-edged sword because increased caloric intake could decrease autophagy.
In closing, we have uncovered a new possible mechanism underlying CS. We propose a model for the disease that opens up new avenues of therapeutic remediation. However, the implications of this study extend beyond CS into basic cellular mechanisms and breathe new life into the mitochondrial theory of aging.
MATERIALS AND METHODS
Animals.
Male homozygous CSBm/m mice were compared with sex- and age-matched C57BL/6 mice. 2-mo-old (young group) or 20–24-mo-old (old group) mice were used. Mice were housed at the National Institute on Aging (NIA) under standard conditions and fed standard animal chow. All experiments were approved by the NIA Animal Care and Use Committee and performed in accordance with National Institutes of Health Guidelines for the Care and Use of Laboratory Animals (National Institutes of Health Publication 85-23, National Institutes of Health, Bethesda, MD). Food consumption was determined by administering 100 g of standard chow to single-housed animals and measuring the remaining amount 1 wk later.
MRI.
Comparison of fat distribution was performed using magnetic resonance imaging (MRI). Mice (n = 3) were anesthetized with ∼1.5% isoflurane in O2 administered via a face mask and were maintained at a body temperature of 36 ± 1°C during data acquisition. Respiration was monitored via an MR-Compatible monitoring and gating system (Model 1025; SA Instruments Inc.) responsive to thoracic excursion. During the experiment, isoflurane concentration was adjusted ±0.5% to maintain a respiration rate of 60–80/min. Imaging was performed on a 9.4T/105-mm Avance III spectrometer (Bruker BioSpin Corp.) using a 30-mm MicroMouse 2.5 imaging probe for the young mouse group, and a 7T/30-cm Bruker spectrometer using a 72-mm inner diameter whole body resonator for the old mouse group. Multi-slice axial and coronal images were acquired using a moderately T1-weighted spin echo sequence. MRI parameters included TE/TR = 25 ms /500 ms, NEX = 2, FOV = 5.0 × 5.0 cm (axial imaging) or 6.0 × 5.0 cm (coronal imaging), slice thickness = 1.0 mm, and matrix size 256 × 128 for both mouse groups. Adipose tissue was quantified using ImageJ (Abrámoff et al., 2004) within each slice of the stacked images to permit measurement of the abdominal adipose tissue volume.
Serology, hematology, and histopathology.
Mice (n = 3) were anesthetized, weighed, and bled. Blood from each mouse was processed into anti-coagulated whole blood and sera for hematology evaluation (HV 1700; Drew Scientific) and serum chemistry evaluation (Vet/Ace 39–10700-14; Alfawassermann), respectively. After phlebotomy, mice were euthanized with CO2, necropsied, organs weighed, and tissues (adrenal gland, bladder, cerebellum, cerebrum, colon, ductus deferens, esophagus, epididymis, femur bone marrow, heart, liver, inner ear, ischiatic nerve, kidney, knee joints, lens, lung, lymph nodes, pancreas, pituitary, prostate, retina, skeletal muscle, skin, skull, sinuses, small/large intestines, spinal cord, spleen, stomach, testis, thymus, thyroid, parathyroid, and trachea) collected for histopathologic evaluation. The tissues were fixed in 10% neutral-buffered formalin for a minimum of 48 h before paraffin embedding. Paraffin-embedded tissues were sectioned 5 µm thick, placed on glass slides, and stained with hematoxylin and eosin for histopathologic evaluation by a veterinary pathologist.
Metabolic and physical activity.
Metabolic rate of the mice (n = 3–8) was assessed by indirect calorimetry in open-circuit oxymax chambers using the Comprehensive Laboratory Animal Monitoring System (CLAMS; Columbus Instruments). Mice were single housed and maintained at ∼24°C under a 12/12-h light/dark cycle (light period 0600–1800). Water and food were available ad libitum. All mice were acclimatized to monitoring cages for 3–6 h, after which twice-hourly automated recordings of gas exchange were taken over the next 60 h. The concentrations of O2 and CO2 were monitored at the inlet and outlet of the sealed chambers to calculate respiration. The sensors were calibrated against a standard gas mix containing defined quantities of O2, CO2, and N2. Constant airflow (0.6 liter/min) was drawn through the chamber and monitored by a mass-sensitive flow meter. Each chamber was measured for 30 s at 30-min intervals. Movement (both horizontal and vertical) was monitored by beams 0.5 in apart on horizontal and vertical planes providing a high-resolution grid covering the X, Y, and Z planes. Beam breaks representing mouse movement were recorded in 30-s epochs. Mice were weighed before and after each trial. The respiratory exchange ratio (RER) is simply the ratio between the carbon dioxide production and the oxygen consumption.
Microarray.
Gene expression of CSBm/m MDFs compared with WT from a previously published microarray (E-MEXP-1968; ArrayExpress) was performed. Raw hybridization intensity data were log-transformed and normalized to yield z-scores. One dataset was found to be an outlier by scatter plot, sample clustering, and principal component analysis (Fig. S1, C–E) and was excluded from further analysis. The z-ratio was calculated as the difference between the observed gene z-scores for the experimental and the control comparisons, and dividing by the SD associated with the distribution of these differences. Z-ratio values +2.0 or −2.0 were chosen as cut-off values, defining increased and decreased expression, respectively. A complete set of 522 cellular pathways was obtained from the Molecular Signatures Database (MSigDB, Broad Institute, Massachusetts Institute of Technology, Cambridge, MA). The complete set was tested for gene set enrichment using parametric analysis of gene set enrichment. For each pathway z-score, a p-value was computed using JMP 6.0 software to test for the significance of the z-score obtained. These tools were part of DIANE 1.0 (see http://www.grc.nia.nih.gov/branches/rrb/dna/diane_software.pdf for information).
Cell culture.
SV40-transformed CS1AN cells stably transfected with either CSB (WT CSB) or empty vector were cultured in DME supplemented with 10% FBS, 1% pen-strep, and 400 µg/ml geneticin and grown in 20% O2/5% CO2 at 37°C. Primary human skeletal muscle myoblasts (HSMM; Lonza) were cultured in SkGM-2 medium (Lonza) according to company protocols in humidified 20% O2/5% CO2 at 37°C. To differentiate myoblasts into myotubes, the medium was switched to DME:F-12 (Lonza) supplemented with 3% fetal horse serum. Only passage five fully differentiated myotubes were used in experiments. HEK293T cells (American Type Culture Collection) were grown in DME supplemented with 10% Hyclone FBS (Thermo Fisher Scientific) at 37°C in 20% O2/5% CO2. Primary patient and control cells (Coriell Cell Repository) were grown in DME supplemented with 10% FBS, 1% pen-strep. The use of the commercially available primary cell lines were conducted in accordance with the National Institutes of Health guidelines.
Whole cell extracts were produced by incubating cells in lysis buffer (LB; 50 mM Tris-HCL, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 1× protease inhibitor cocktail; Roche) for 30 min on ice. To ensure complete mitochondrial lysis, the lysate was subjected to sonication at 5 W for 5 s, three times performed on ice. The extract was then cleared by centrifugation at 16,000 g for 10 min. Protein concentrations were measured using the Bradford assay with bovine serum albumin as a standard.
Isolation of MDFs.
MDFs from old and young CSBm/m mice and WT age-matched (n = 3) controls were isolated as follows: tail skin biopsies were minced using a scalpel, placed into a 35-mm dish with 1.5 ml complete medium (DME, 10% FBS, 100 U ml−1 penicillin, 100 U ml−1 streptomycin, and 0.25 µg ml−1 fungizone) and 1 ml collagenase solution (1,000 U ml−1), and then incubated at 37°C in a 20% O2/5% CO2 incubator. After 24 h, the cells were triturated, passed through netting to remove large particles, and centrifuged for 5 min at 200 g. The cell pellet was resuspended in 3 ml complete medium and transferred to a 25-cm2 flask and cultured to 90% confluency. At this point, 0.75 × 106 cells were subcultured into 75-cm2 flasks; this is considered passage 1. The cell culture was passaged at 0.75 × 106 cells per 75-cm2 flask every 4–6 d. At passage 3, cells were harvested and frozen in 10% DMSO, 70% FBS, and 20% DME and stored at −150°C. Upon thawing, cells were cultured in AmnioMAX II medium (Invitrogen) in humidified 3% O2 and 5% CO2. All cells used for experiments were at passage 5.
Electron microscopy.
Electron microscopy of CS1AN cells were performed by Electron Microscopy Bioservices. In brief, cells untreated or treated with 5 µM rotenone for 24 h grown in 150 mm plates were fixed for 5 min at room temperature in 2.5% glutaraldehyde and 2% paraformaldehyde in Millonig’s buffer. Cells were then scraped, spun down, and allowed to fix for an additional 2–3 h at 4°C. The cell pellet was washed twice carefully in Millonig’s buffer. The cell pellet was then minced post-fixed in 1.0% OsO4 and washed in ultrapure water. Samples were then en bloc stained with 2.0% aqueous uranyl acetate, serially dehydrated in ethanol, embedded in Spurr’s plastic resin, and allowed to polymerize overnight at 70°C. Ultrathin 60–80-nm-thick sections were made using an ultramicrotome (UC7; Leica), mounted onto 200 mesh copper grids, and post-stained with uranyl acetate and Reynold’s lead citrate. Transmission electron microscopy was then performed on an electron microscope (Tecnai Spirit Twin Transmission; FEI) at 80 kV. Images were quantified by counting the percentage of damaged mitochondria per image (n = 12–14) or by measuring the mitochondrial diameter using ImageJ (n > 100).
Lentivirus generation and infections.
shRNA vectors against CSB were purchased from Santa Cruz Biotechnology, Inc. A cocktail of three vectors containing the following shRNA sequences were used: 5′-GTCTTACGAGATACCATAATTCAAGAGATTATGGTATCTCGTAAGACTTTTT-3′, 5′-CCAGAAGCAAGACAGTGAATTCAAGAGATTCACTGTCTTGCTTCTGGTTTTT-3′, and 5′-GTCTTCCGAGAACTATTGATTCAAGAGATCAATAGTTCTCGGAAGACTTTTT-3′. As a control, a plasmid containing a scrambled shRNA was used (Plasmid-A; Santa Cruz Biotechnology, Inc.). Lentiviral production was done by co-transfection of pCMV delta R8.2 (plasmid 12263 deposited by Didier Trono, Addgene Inc.) backbone, pCMV-VSV-G (plasmid 8454, deposited by Robert Weinberg, Addgene Inc.), and the vectors expressing the shRNA into 293T cells by Fugene 6 transfection as per manufacturer’s protocol (Roche). The media containing the virus was harvested after 48 h, filtered through 0.45-µm pore filter, flash frozen, and stored at −80°C. Cells were seeded at a density of 2 × 105 in 10-cm plates, 24 h before the infection. For infection, the media was removed and the cells were washed once with PBS. The frozen virus was thawed quickly and added to the plates. The plates were rocked every 15 min by hand and incubated at 37°C for 2 h. Fresh media was subsequently added.
Cellular oxygen consumption.
Oxygen consumption and extracellular acidification rate measurements were performed using the Seahorse XF-24 instrument (Seahorse Biosciences). Cells were seeded into a Seahorse tissue culture plate at a density according to Table S1, and 16 h later media was changed to unbuffered XF assay media at pH 7.4 (Seahorse Biosciences), supplemented with 25 mM glucose (Sigma-Aldrich), 1 mM sodium-pyruvate, and 1 mM glutamax (Invitrogen). Cells were incubated for 1 h at 37°C at ambient O2 and CO2 concentration before measurements were taken. Respiration was measured in four blocks of 3 × 3 min. The first block measured the basal respiration rate. Next, oligomycin (EMD) was added to inhibit complex V and the second block was measured. Then, FCCP (Sigma-Aldrich) was added to uncouple respiration and the third block was measured. Finally, antimycin A (Sigma-Aldrich) was added to inhibit complex 3 and the last measurements were performed. See Table S2 for compound concentrations used. All compound concentrations used had been optimized for that cell line. Immediately after finishing the measurements, cells were trypsinized and counted using a Coulter counter (Beckman Coulter). For HSMM, cells were plated in Seahorse tissue culture plates and allowed to grow to 90% confluency. The media was then changed to DME:F-12 (Lonza) supplemented with 3% fetal horse serum and the myoblasts were allowed to differentiate for 3 d. Lentiviral knockdown was then performed by addition of virus directly to each well. 3 d later, respiratory measurements were performed as described. After the experiment, myotubes were lysed in LB, the protein content was measured, and respiration was normalized to protein concentration. Lysates were subsequently used for immunoblotting to investigate knockdown efficiency.
Mitochondrial membrane potential.
In brief, 0.4 × 106 CS1AN cells expressing WT CSB or empty vector were harvested by trypsin, washed, and resuspended in DME without phenol indicator (Invitrogen). Cells were then stained with 20 nM TMRM for 15 min at 37°C with or without 1 µM cyclosporin A. Fluorescence was measured by a flow cytometer (C6; Accuri). Treatments with 10 mM lithium chloride (Sigma-Aldrich) or 1 µM rapamycin (Sigma-Aldrich) were done for 24 h before membrane potential.
Glucose consumption.
Glucose consumption was measured by plating 50,000 CS1AN cells expressing WT CSB or an empty vector in a 24-well dish (n = 3). 24 h later, media was changed to DME containing 10 mM glucose and supplemented with 1% pen-strep, 10% FBS, and 400 µg/ml geneticin. At the given time points, the glucose concentration was measured in the media using the GlucCell glucose monitoring system as per manufacturer’s instruction (Cesco Bioengineering). The cells were washed twice in PBS, trypsinized, and counted.
ATP concentration.
In brief, 10,000 cells per well were plated in a 96-well dish. 24 h later, ATP levels were measured in using the ATPlite assay according to manufacturer’s protocol (PerkinElmer). For the ATP consumption assay, 100 mM 2-deoxyglucose and 1 µM oligomycin was added at time 0. Cells were subsequently lysed at the indicated time points and the ATP concentration was measured. Luminescence was measured on the 1450 MicroBeta TriLux Microplate Scintillation and Luminescence Counter (PerkinElmer).
Mitochondrial content.
Citrate synthase activity (n = 3) was measured spectrophotometrically as described in Srere (1969) with modifications. Two different dilutions of the sample were loaded in duplicates into a 96-well dish. The linear increase in absorbance was measured at 412 nm over 5 min and the concentration was determined against a standard curve of purified citrate synthase (Sigma-Aldrich). Transfection with RFP-tagged mitochondrial protein (Organelle Lights; Invitrogen) was further used to investigate the amount of mitochondria according to manufacturer’s protocol (n = 3). In brief, 105 cells were plated in glass-bottom 20-mm plates. The next day, cells were washed in PBS and 500 µl of the transduction solution was added. Cells were incubated 2 h in 37°C before the transduction solution was removed and the enhancer solution was added. After 2 h of incubation at 37°C, the enhancer solution was removed and normal growth media was added. 24 h later, live cells were imaged at 40× on a confocal microscope (Eclipse TE-2000e; Nikon), with a 60 ms brightfield and 200 ms rfp exposure time. 0.2 µm z-stacks were used to image entire cells and, with an average of at least 25 cells per transfection, were used per replica experiment. Quantification was performed by normalizing voxel count to cell number using the Volocity software (PerkinElmer). Mitochondrial content was additionally investigated using MitoTracker green FM (Invitrogen) as previously described (Pendergrass et al., 2004) with slight modification (n = 3). In brief, 0.75 × 106 CS1AN cells expressing WT CSB or empty vector were harvested by trypsin, washed, and resuspended in DME without phenol indicator (Invitrogen). Half the cells were then stained with 50 nM MitoTracker green FM for 30 min at 37°C. Cells were washed twice and resuspended in PBS before fluorescence was measured by a flow cytometer (C6). Unstained cells were used to gate for stained cells.
PCR.
Total RNA was isolated from cells using Trizol reagent (Invitrogen) per the manufacturer’s protocol (n = 3). Reverse transcription reactions were performed using Iscript (Bio-Rad Laboratories). Primers were designed using the Primer Express 2.0 (Applied Biosystems). The sequences of primers are shown in Table S3. Real-time PCR was performed using SYBR green PCR mix (Applied Biosystems) with a primer concentration of 500 nM. Hypoxanthine phosphoribosyltransferase (HPRT) served as an internal control to normalize samples.
Western blots.
Western blots were performed according to standard procedure. 30–100 µg proteins per lane were used. Samples were analyzed using 4–20% tris-glycine SDS-PAGE (Invitrogen). The following antibodies were used: rabbit anti-CSB (Santa Cruz Biotechnology, Inc.), rabbit anti-LC3B (Novus Biologicals), mouse anti-GAPDH (Santa Cruz Biotechnology, Inc.), rabbit anti-lamin A/C (Santa Cruz Biotechnology, Inc.), mouse anti-P62 (Abcam), goat anti-COX4 (Santa Cruz Biotechnology, Inc.), and rabbit anti-ubiquitin (Santa Cruz Biotechnology, Inc.). Secondary HRP-conjugated antibodies and ECL plus (GE Healthcare) were used to visualize the protein bands on a Typhoon scanner. Quantification was performed using ImageQuant (GE Healthcare).
For autophagy, 1 × 106 CS1AN cells were plated in 100 mm dishes (n = 3). 24 h later, cells were treated with vehicle, 5 µM rotenone, or 500 ng/ml ethidium bromide. Cells were incubated 24 h and harvested. Cells were lysed in LB. For HSMM, 10 × 105 myoblasts were plated in 100 mm dishes and differentiated as described. Lentiviral knockdown was performed and, 3 d later, cells were treated with vehicle, 5 µM rotenone, or 500 ng/ml ethidium bromide before they were harvested.
Microscopy.
To investigate colocalization, 5 ×104 cells were plated in 4-well chamber slides and grown overnight. The next day, the media was changed and vehicle, 5 µM rotenone, or 100 nM bafilomycin was added. Cells were treated for 24 h before being fixed for 15 min in 3.7% paraformaldehyde in PBS. Cells were then washed before being permeabilized in 0.25% Triton X-100 for 10 min on ice. Subsequently, cells were washed in PBS and blocked overnight in PBS containing 5% FBS. Primary antibodies were added at a concentration of 1:100 and incubated for 1 h at 37°C. After being washed, secondary antibodies were added at 1:1,000 and incubated for 1 h at 37°C. Cells were then washed 6 × 10 min in PBS before being mounted in prolong antifade gold with DAPI (Invitrogen). Images were acquired at 60× on a confocal microscope (Eclipse TE-2000e), 50 ms brightfield, 300 ms exposure time for COX4 at 488 nm, and 900 ms exposure time for P62 or LC3B at 647 nm. Quantification was done by averaging the unbiased global Pearson’s coefficients of at least 10 unselected images per experiment using Volocity software. For live cell imaging of LC3B-eGFP, 105 cells were plated in glass-bottom 20-mm plates. The next day, cells were transfected with LC3-EGFP plasmid (plasmid 11546, deposited by K. Kirkegaard, Addgene Inc.) using Fugene-6 transfection according to the manufacturer’s protocol (Roche). 2 d later, live cell imaging was performed at 40× on a confocal microscope (Eclipse TE-2000e), with a 60 ms brightfield and 600 ms gfp exposure time. 0.2 µm z-stacks were used to image entire cells, and the means of at least 20 cells per transfection were used per replica experiment (n = 3). Quantification was performed by automatic unbiased spot finding using the Volocity software.
ROS production.
The ROS marker dihydroethidium (DHE; Invitrogen) was used to measure intracellular ROS, predominantly superoxide (n = 10). 4 µM DHE induced no visible cellular toxicity after 2 h of incubation and was subsequently used to measure ROS. Target cells were plated into 96-well plates 12 h before the experiment. Before commencing the assay, the medium was removed and cells washed twice with PBS before the addition of DHE probe diluted in prewarmed DME without phenol indicator (Invitrogen). Fluorescence was measured using 520 nm excitation/610 nm emission at 5-min intervals for 60 min using a FLUOstar Optima (BMG Labtech) with bottom reading capability. The mitochondrial ETC blocker antimycin A was used as a positive control and to ensure that cellular dye uptake was not rate limiting. After completion of the assay, the cells in each well were counted and used to normalize the results.
Statistics.
Prism 4.0 (GraphPad Software) was used to plot data, make graphs, and statistical analysis. Data are plotted as mean ± SEM. Unless otherwise stated, two-tailed unpaired Student’s t test was performed.
Online supplemental material.
Fig. S1 shows the list of genes significantly changed in the analyzed microarray as well as the quality control we performed on the data. Video 1 shows a representative axial MRI scan of the young and old WT and CSBm/m mice. Table S1 shows serum chemistry, hematology, and organ weights of the old and young WT and CSBm/m mice. Table S2 shows a list of cells and compound concentrations used in the oxygen consumption experiments. Table S3 shows a list of primers used for quantitative PCR. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20111721/DC1.
Acknowledgments
We would like to thank Dr. Mark Bryant at the Division of Veterinary Resources, National Institutes of Health (NIH), for his invaluable help regarding mouse pathology. Further thanks should go to Dr. Kevin Becker, Dr. Yongqing Zhang, and Dr. Elin Lehrmann for their help with reanalyzing the microarray. Finally, we would like to thank Chris Dunn for his invaluable technical support.
This research was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- 3-MA
- 3-methyladenine
- BER
- base excision repair
- COX4
- cytochrome c oxidase subunit 4
- CS
- Cockayne syndrome
- ERR-α
- estrogen-related receptor α
- FCCP
- carbonyl cyanide-p-trifluoromethoxyphenylhydrazone
- HSMM
- human skeletal muscle myotube
- MDF
- mouse dermal fibroblast
- mPTP
- membrane permeability transition pore
- mtDNA
- mitochondrial DNA
- PGC-1α
- peroxisome proliferator-activated receptor γ co-activator 1α
- ROS
- reactive oxygen species
- TFAM
- transcription factor A, mitochondrial
- TMRM
- tetramethyl rhodamine methyl ester
References
- Aamann M.D., Sorensen M.M., Hvitby C., Berquist B.R., Muftuoglu M., Tian J., de Souza-Pinto N.C., Scheibye-Knudsen M., Wilson D.M., III, Stevnsner T., Bohr V.A. 2010. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 24:2334–2346 10.1096/fj.09-147991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrámoff M.D., Magalhaes P.J., Ram S.J. 2004. Image processing with imageJ. Biophotonics International. 11:36–41 [Google Scholar]
- Anindya R., Mari P.O., Kristensen U., Kool H., Giglia-Mari G., Mullenders L.H., Fousteri M., Vermeulen W., Egly J.M., Svejstrup J.Q. 2010. A ubiquitin-binding domain in Cockayne syndrome B required for transcription-coupled nucleotide excision repair. Mol. Cell. 38:637–648 10.1016/j.molcel.2010.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aykin-Burns N., Ahmad I.M., Zhu Y., Oberley L.W., Spitz D.R. 2009. Increased levels of superoxide and H2O2 mediate the differential susceptibility of cancer cells versus normal cells to glucose deprivation. Biochem. J. 418:29–37 10.1042/BJ20081258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban R.S., Nemoto S., Finkel T. 2005. Mitochondria, oxidants, and aging. Cell. 120:483–495 10.1016/j.cell.2005.02.001 [DOI] [PubMed] [Google Scholar]
- Berquist B.R., Wilson D.M., III 2009. Nucleic acid binding activity of human Cockayne syndrome B protein and identification of Ca(2+) as a novel metal cofactor. J. Mol. Biol. 391:820–832 10.1016/j.jmb.2009.06.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelakovic G., Nikolova D., Gluud L.L., Simonetti R.G., Gluud C. 2008. Antioxidant supplements for prevention of mortality in healthy participants and patients with various diseases. Cochrane Database Syst. Rev. 2:CD007176. [DOI] [PubMed] [Google Scholar]
- Carreira R.S., Lee Y., Ghochani M., Gustafsson A.B., Gottlieb R.A. 2010. Cyclophilin D is required for mitochondrial removal by autophagy in cardiac cells. Autophagy. 6:462–472 10.4161/auto.6.4.11553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Steele A.D., Hutter G., Bruno J., Govindarajan A., Easlon E., Lin S.J., Aguzzi A., Lindquist S., Guarente L. 2008. The role of calorie restriction and SIRT1 in prion-mediated neurodegeneration. Exp. Gerontol. 43:1086–1093 10.1016/j.exger.2008.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.Q., McMillan-Ward E., Kong J.M., Israels S.J., Gibson S.B. 2007. Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J. Cell Sci. 120:4155–4166 10.1242/jcs.011163 [DOI] [PubMed] [Google Scholar]
- Citterio E., Van Den Boom V., Schnitzler G., Kanaar R., Bonte E., Kingston R.E., Hoeijmakers J.H.J., Vermeulen W. 2000. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 20:7643–7653 10.1128/MCB.20.20.7643-7653.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov G., Bischoff C., Sunesen M., Bohr V.A. 1999. Repair of 8-oxoguanine in DNA is deficient in Cockayne syndrome group B cells. Nucleic Acids Res. 27:1365–1368 10.1093/nar/27.5.1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Troya S., Pérez-Pérez M.E., Florencio F.J., Crespo J.L. 2008. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 4:851–865 [DOI] [PubMed] [Google Scholar]
- Finsterer J. 2008. Leigh and Leigh-like syndrome in children and adults. Pediatr. Neurol. 39:223–235 10.1016/j.pediatrneurol.2008.07.013 [DOI] [PubMed] [Google Scholar]
- Funalot B., Reynier P., Vighetto A., Ranoux D., Bonnefont J.P., Godinot C., Malthièry Y., Mas J.L. 2002. Leigh-like encephalopathy complicating Leber’s hereditary optic neuropathy. Ann. Neurol. 52:374–377 10.1002/ana.10299 [DOI] [PubMed] [Google Scholar]
- Garinis G.A., Uittenboogaard L.M., Stachelscheid H., Fousteri M., van Ijcken W., Breit T.M., van Steeg H., Mullenders L.H.F., van der Horst G.T.J., Brüning J.C., et al. 2009. Persistent transcription-blocking DNA lesions trigger somatic growth attenuation associated with longevity. Nat. Cell Biol. 11:604–615 10.1038/ncb1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S., Holmström K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. 2010. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12:119–131 10.1038/ncb2012 [DOI] [PubMed] [Google Scholar]
- Gomez L., Li B., Mewton N., Sanchez I., Piot C., Elbaz M., Ovize M. 2009. Inhibition of mitochondrial permeability transition pore opening: translation to patients. Cardiovasc. Res. 83:226–233 10.1093/cvr/cvp063 [DOI] [PubMed] [Google Scholar]
- Grandjean E.M., Aubry J.M. 2009. Lithium: updated human knowledge using an evidence-based approach: part III: clinical safety. CNS Drugs. 23:397–418 10.2165/00023210-200923050-00004 [DOI] [PubMed] [Google Scholar]
- Hansen M., Taubert S., Crawford D., Libina N., Lee S.J., Kenyon C. 2007. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 6:95–110 10.1111/j.1474-9726.2006.00267.x [DOI] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Harrison D.E., Strong R., Sharp Z.D., Nelson J.F., Astle C.M., Flurkey K., Nadon N.L., Wilkinson J.E., Frenkel K., Carter C.S., et al. 2009. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 460:392–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano T., Kubo S., Sato S., Hattori N. 2009. Pathogenesis of familial Parkinson’s disease: new insights based on monogenic forms of Parkinson’s disease. J. Neurochem. 111:1075–1093 10.1111/j.1471-4159.2009.06403.x [DOI] [PubMed] [Google Scholar]
- Holloszy J.O., Oscai L.B., Don I.J., Molé P.A. 1970. Mitochondrial citric acid cycle and related enzymes: adaptive response to exercise. Biochem. Biophys. Res. Commun. 40:1368–1373 10.1016/0006-291X(70)90017-3 [DOI] [PubMed] [Google Scholar]
- Jackson W.T., Giddings T.H., Jr, Taylor M.P., Mulinyawe S., Rabinovitch M., Kopito R.R., Kirkegaard K. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:e156 10.1371/journal.pbio.0030156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K.L., Levine B. 2007. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 3:597–599 [DOI] [PubMed] [Google Scholar]
- Jucker M. 2010. The benefits and limitations of animal models for translational research in neurodegenerative diseases. Nat. Med. 16:1210–1214 10.1038/nm.2224 [DOI] [PubMed] [Google Scholar]
- Kamenisch Y., Fousteri M., Knoch J., von Thaler A.K., Fehrenbacher B., Kato H., Becker T., Dollé M.E., Kuiper R., Majora M., et al. 2010. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J. Exp. Med. 207:379–390 10.1084/jem.20091834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 441:880–884 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Kukat A., Kukat C., Brocher J., Schäfer I., Krohne G., Trounce I.A., Villani G., Seibel P. 2008. Generation of rho0 cells utilizing a mitochondrially targeted restriction endonuclease and comparative analyses. Nucleic Acids Res. 36:e44 10.1093/nar/gkn124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. 2004. The role of autophagy during the early neonatal starvation period. Nature. 432:1032–1036 10.1038/nature03029 [DOI] [PubMed] [Google Scholar]
- Le May N., Mota-Fernandes D., Vélez-Cruz R., Iltis I., Biard D., Egly J.M. 2010. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol. Cell. 38:54–66 10.1016/j.molcel.2010.03.004 [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D.J. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 6:463–477 10.1016/S1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- Levine B., Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell. 132:27–42 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M., Cuervo A.M. 2007. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 6:352–361 10.1016/S1474-4422(07)70076-5 [DOI] [PubMed] [Google Scholar]
- Meléndez A., Tallóczy Z., Seaman M., Eskelinen E.L., Hall D.H., Levine B. 2003. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 301:1387–1391 10.1126/science.1087782 [DOI] [PubMed] [Google Scholar]
- Muftuoglu M., de Souza-Pinto N.C., Dogan A., Aamann M., Stevnsner T., Rybanska I., Kirkali G., Dizdaroglu M., Bohr V.A. 2009. Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J. Biol. Chem. 284:9270–9279 10.1074/jbc.M807006200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance M.A., Berry S.A. 1992. Cockayne syndrome: review of 140 cases. Am. J. Med. Genet. 42:68–84 10.1002/ajmg.1320420115 [DOI] [PubMed] [Google Scholar]
- Nishino I., Spinazzola A., Hirano M. 1999. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science. 283:689–692 10.1126/science.283.5402.689 [DOI] [PubMed] [Google Scholar]
- Ohtsuka H., Takahashi R., Goto S. 1995. Age-related accumulation of high-molecular-weight ubiquitin protein conjugates in mouse brains. J. Gerontol. A Biol. Sci. Med. Sci. 50A:B277–B281 10.1093/gerona/50A.5.B277 [DOI] [PubMed] [Google Scholar]
- Osenbroch P.O., Auk-Emblem P., Halsne R., Strand J., Forstrøm R.J., van der Pluijm I., Eide L. 2009. Accumulation of mitochondrial DNA damage and bioenergetic dysfunction in CSB defective cells. FEBS J. 276:2811–2821 10.1111/j.1742-4658.2009.07004.x [DOI] [PubMed] [Google Scholar]
- Panov A., Scarpa A. 1996. Mg2+ control of respiration in isolated rat liver mitochondria. Biochemistry. 35:12849–12856 10.1021/bi960139f [DOI] [PubMed] [Google Scholar]
- Paumard P., Vaillier J., Coulary B., Schaeffer J., Soubannier V., Mueller D.M., Brèthes D., di Rago J.P., Velours J. 2002. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 21:221–230 10.1093/emboj/21.3.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergrass W., Wolf N., Poot M. 2004. Efficacy of MitoTracker Green and CMXrosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry A. 61A:162–169 10.1002/cyto.a.20033 [DOI] [PubMed] [Google Scholar]
- Prigione A., Cortopassi G. 2007. Mitochondrial DNA deletions and chloramphenicol treatment stimulate the autophagic transcript ATG12. Autophagy. 3:377–380 [DOI] [PubMed] [Google Scholar]
- Rangaraju S., Hankins D., Madorsky I., Madorsky E., Lee W.H., Carter C.S., Leeuwenburgh C., Notterpek L. 2009. Molecular architecture of myelinated peripheral nerves is supported by calorie restriction with aging. Aging Cell. 8:178–191 10.1111/j.1474-9726.2009.00460.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R., Pozzan T. 2006. Microdomains of intracellular Ca2+: molecular determinants and functional consequences. Physiol. Rev. 86:369–408 10.1152/physrev.00004.2005 [DOI] [PubMed] [Google Scholar]
- Rötig A., Appelkvist E.L., Geromel V., Chretien D., Kadhom N., Edery P., Lebideau M., Dallner G., Munnich A., Ernster L., Rustin P. 2000. Quinone-responsive multiple respiratory-chain dysfunction due to widespread coenzyme Q10 deficiency. Lancet. 356:391–395 10.1016/S0140-6736(00)02531-9 [DOI] [PubMed] [Google Scholar]
- Srere P.A. 1969. [ 1] Citrate Synthase. Methods Enzymol. 13:3–11 10.1016/0076-6879(69)13005-0 [DOI] [Google Scholar]
- Stevnsner T., Muftuoglu M., Aamann M.D., Bohr V.A. 2008. The role of Cockayne Syndrome group B (CSB) protein in base excision repair and aging. Mech. Ageing Dev. 129:441–448 10.1016/j.mad.2008.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe D.F., Kevin L.G. 2004. Cardiac preconditioning by volatile anesthetic agents: a defining role for altered mitochondrial bioenergetics. Antioxid. Redox Signal. 6:439–448 10.1089/152308604322899512 [DOI] [PubMed] [Google Scholar]
- Susa D., Mitchell J.R., Verweij M., van de Ven M., Roest H., van den Engel S., Bajema I., Mangundap K., Ijzermans J.N., Hoeijmakers J.H.J., de Bruin R.W. 2009. Congenital DNA repair deficiency results in protection against renal ischemia reperfusion injury in mice. Aging Cell. 8:192–200 10.1111/j.1474-9726.2009.00463.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trushina E., McMurray C.T. 2007. Oxidative stress and mitochondrial dysfunction in neurodegenerative diseases. Neuroscience. 145:1233–1248 10.1016/j.neuroscience.2006.10.056 [DOI] [PubMed] [Google Scholar]
- Tuo J.S., Jaruga P., Rodriguez H., Dizdaroglu M., Bohr V.A. 2002. The cockayne syndrome group B gene product is involved in cellular repair of 8-hydroxyadenine in DNA. J. Biol. Chem. 277:30832–30837 10.1074/jbc.M204814200 [DOI] [PubMed] [Google Scholar]
- van der Horst G.T., van Steeg H., Berg R.J., van Gool A.J., de Wit J., Weeda G., Morreau H., Beems R.B., van Kreijl C.F., de Gruijl F.R., et al. 1997. Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell. 89:425–435 10.1016/S0092-8674(00)80223-8 [DOI] [PubMed] [Google Scholar]
- Weidenheim K.M., Dickson D.W., Rapin I. 2009. Neuropathology of Cockayne syndrome: Evidence for impaired development, premature aging, and neurodegeneration. Mech. Ageing Dev. 130:619–636 10.1016/j.mad.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Wilson J.X., Gelb A.W. 2002. Free radicals, antioxidants, and neurologic injury: possible relationship to cerebral protection by anesthetics. J. Neurosurg. Anesthesiol. 14:66–79 10.1097/00008506-200201000-00014 [DOI] [PubMed] [Google Scholar]
- Zeviani M., Bresolin N., Gellera C., Bordoni A., Pannacci M., Amati P., Moggio M., Servidei S., Scarlato G., DiDonato S. 1990. Nucleus-driven multiple large-scale deletions of the human mitochondrial genome: a new autosomal dominant disease. Am. J. Hum. Genet. 47:904–914 [PMC free article] [PubMed] [Google Scholar]