Gain- and loss-of-function analyses reveal that the transcription factor EBF1 is required for normal differentiation and function of marginal zone, B-1, follicular, and germinal center B cells in mice.
Abstract
The transcription factor EBF1 is essential for lineage specification in early B cell development. In this study, we demonstrate by conditional mutagenesis that EBF1 is required for B cell commitment, pro–B cell development, and subsequent transition to the pre–B cell stage. Later in B cell development, EBF1 was essential for the generation and maintenance of several mature B cell types. Marginal zone and B-1 B cells were lost, whereas follicular (FO) and germinal center (GC) B cells were reduced in the absence of EBF1. Activation of the B cell receptor resulted in impaired intracellular signaling, proliferation and survival of EBF1-deficient FO B cells. Immune responses were severely reduced upon Ebf1 inactivation, as GCs were formed but not maintained. ChIP- and RNA-sequencing of FO B cells identified EBF1-activated genes that encode receptors, signal transducers, and transcriptional regulators implicated in B cell signaling. Notably, ectopic expression of EBF1 efficiently induced the development of B-1 cells at the expense of conventional B cells. These gain- and loss-of-function analyses uncovered novel important functions of EBF1 in controlling B cell immunity.
Hematopoietic stem cells (HSCs) in the bone marrow give rise to all mature B cell types in peripheral lymphoid organs, which provide humoral immunity for protection against foreign pathogens. HSCs first differentiate to lymphoid-primed multipotent progenitors (LMPPs) and common lymphoid progenitors (CLPs), which consist of Ly6D− all-lymphoid progenitors (ALPs) and Ly6D+ B cell–biased lymphoid progenitors (BLPs; Inlay et al., 2009). BLPs initiate the B cell gene expression program and differentiate via the prepro–B cell stage to pro–B cells, which undergo B lineage commitment (Inlay et al., 2009). Pro–B cells subsequently develop via pre–B cells into immature B lymphocytes that emigrate from the bone marrow to the spleen, where they differentiate into distinct mature B cell types (Hardy et al., 2007; Allman and Pillai, 2008).
The entry of lymphoid progenitors into the B cell pathway depends on several transcription factors, including the helix-loop-helix protein E2A, the early B cell factor EBF1, and the paired domain transcription factor Pax5 (Nutt and Kee, 2007; Medvedovic et al., 2011). These three regulators act in the genetic hierarchy E2A→EBF1→Pax5, as early B cell development is sequentially arrested at the ALP, prepro–B cell, or earliest pro–B cell stage in the absence of E2A, EBF1, and Pax5, respectively (Bain et al., 1994; Lin and Grosschedl, 1995; Nutt et al., 1997; Inlay et al., 2009). Moreover, the transcription factor E2A directly activates the Ebf1 gene by binding to the distal Ebf1 promoter (Smith et al., 2002; Roessler et al., 2007), which results in the initiation of Ebf1 expression at the CLP stage (Zandi et al., 2008; Inlay et al., 2009). EBF1 in turn binds to and activates the Pax5 promoter region (Decker et al., 2009), which gives rise to maximal Pax5 expression in pro–B cells (Fuxa and Busslinger, 2007). Finally, Pax5 further increases Ebf1 expression through a positive feedback loop by binding to the proximal Ebf1 promoter (Fuxa et al., 2004; Roessler et al., 2007), which leads to completion of the B cell commitment process in pro–B cells (Medvedovic et al., 2011).
At the molecular level, EBF1 is known to collaborate with E2A in the activation of B cell–specific genes, such as the surrogate light chain genes Igll (λ5) and Vpreb1 (Sigvardsson et al., 1997; O’Riordan and Grosschedl, 1999). Consistent with this finding, B cell–specific genes are not activated at the CLP stage in Ebf1 mutant mice (Zandi et al., 2008). EBF1 also represses B lineage–inappropriate genes, which may restrict the developmental options of lymphoid progenitors to the B cell lineage (Pongubala et al., 2008), similar to the B cell commitment factor Pax5 (Medvedovic et al., 2011). EBF1 controls gene activity as an epigenetic regulator, as it can induce DNA demethylation, nucleosome remodeling, and active chromatin modifications at its target genes (Maier et al., 2004; Decker et al., 2009; Treiber et al., 2010). Genome-wide analyses have recently identified a large spectrum of regulated EBF1 target genes in pro–B cells, which revealed an important role for EBF1 in pre–B cell receptor (pre–BCR) and phosphoinositide 3-kinase (PI3K) signaling, as well as in cell adhesion and migration during early B lymphopoiesis (Lin et al., 2010; Treiber et al., 2010). Hence, the function of EBF1 at the onset of B cell development has been fairly well characterized. EBF1 is expressed throughout B lymphopoiesis from the pro–B to the mature B cell stage (Hagman et al., 1993). However, nothing is yet known about the role of EBF1 in late B cell development.
Here, we have performed conditional loss-of-function experiments to demonstrate that EBF1 is essential for the generation of all mature B cell types. Marginal zone (MZ) and B-1 B cells were lost upon conditional Ebf1 inactivation, whereas follicular (FO) and germinal center (GC) B cells were generated in reduced numbers, but tolerated the loss of EBF1 for some time. EBF1 was, however, required for the maintenance of GC B cells during an immune response as well as for intracellular calcium signaling, proliferation, and survival of activated FO B cells in response to BCR stimulation. Genome-wide ChIP- and RNA-sequencing of FO B cells identified activated EBF1 target genes and indirectly EBF1-regulated genes that code for cell surface receptors, intracellular signal transducers and transcription factors implicated in different B cell signaling pathways. Notably, gain-of-function experiments revealed that ectopic Ebf1 expression from the Rosa26 locus efficiently induced the development of bona fide B-1 cells at the expense of MZ, FO, and GC B cells. Together, these data identified novel important roles of EBF1 in the generation, maintenance, and function of distinct mature B cell types.
RESULTS
Nonredundant in vivo functions of EBF1 and Pax5 at the onset of B cell development
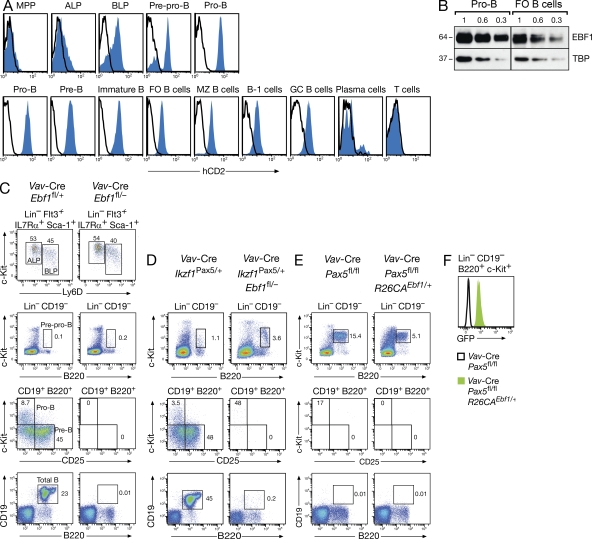
To investigate the lymphoid expression of Ebf1 at the single-cell level, we generated an Ebf1ihCd2 allele by inserting an internal ribosome entry sequence (IRES)-hCd2 (ihCd2) reporter gene into the 3′ untranslated region of the Ebf1 gene, which resulted in normal B cell development of Ebf1ihCd2/ihCd2 mice (Fig. S1 and unpublished data). As shown by flow cytometric analysis of Ebf1ihCd2/+ mice (Fig. 1 A), Ebf1 expression is weakly activated in ALPs, increases in BLPs and prepro–B cells, remains high in pro–B, pre–B, and immature B cells of the bone marrow, is moderately down-regulated in FO, MZ, B-1, and GC B cells of the spleen, and is subsequently lost in terminally differentiated plasma cells of the bone marrow. Furthermore, Ebf1 is not expressed in other hematopoietic lineages, as exemplified for T cells (Fig. 1 A and unpublished data). Semiquantitative immunoblot analysis revealed a threefold lower expression of the EBF1 protein in FO B cells compared with pro–B cells (Fig. 1 B), which is consistent with the moderate down-regulation of expression of the Ebf1ihCd2 allele that is observed in all mature B cell types (Fig. 1 A).
Figure 1.
Nonredundant functions of EBF1 and Pax5 in early B cell development. (A) Ebf1ihCd2/+ (blue) and WT (black line) mice were analyzed by flow cytometry for human (h) CD2 expression in different progenitors, B cell types, and T cells, which were defined as described in Materials and methods. (B) Expression of EBF1 protein in cultured pro–B cells and MACS-sorted FO B cells from lymph nodes. One of two immunoblot experiments is shown. Numbers indicate the relative proportion of nuclear extracts analyzed. The EBF1 protein abundance is normalized to expression of the TBP, and the size (kilodaltons) of the two proteins is indicated to the left. (C and D) B cell development in Vav-Cre Ebf1fl/− mice with or without ectopic Pax5 expression from the Ikzf1Pax5 allele. Progenitor and B cell types were investigated by flow cytometry of bone marrow cells isolated from mice of the indicated genotypes. The relative percentage of each cell type is indicated in the respective quadrant, and the gating is shown above the FACS plot. The different cell types were defined as described in Materials and methods. (E and F) B cell development in Vav-Cre Pax5fl/fl mice with or without ectopic EBF1 expression from the R26CAEbf1 allele (E). GFP expression, indicating EBF1 expression, in Pax5-deficient progenitors of Vav-Cre R26CAEbf1/+ Pax5fl/fl mice (F). Number of mice of each genotype analyzed: n = 8 (C); n = 5 (D); and n = 3 (E).
To study the function of Ebf1 throughout B cell development, we generated a floxed (fl) Ebf1 allele, which resulted in Cre-mediated deletion of the last 429 codons of the Ebf1 gene and subsequent expression of an EBF1-GFP fusion protein from the deleted Ebf1Δ allele (Fig. S2, A–C). Surprisingly, however, Ebf1Δ/+ mice expressed GFP at a significant level only in pro–B cells, but not at later developmental stages (Fig. S2 D), suggesting that the presence of the N-terminal 148 aa of EBF1 appeared to destabilize the GFP fusion protein. Importantly, B cell development was normal in homozygous Ebf1fl/fl mice, indicating that the floxed Ebf1 allele provided WT EBF1 function (unpublished data). Pan-hematopoietic deletion of the floxed Ebf1 allele with the Vav-Cre transgene (de Boer et al., 2003) did not affect the differentiation of HSCs to ALPs and BLPs, but resulted in a stringent differentiation arrest at the prepro–B cell stage in Vav-Cre Ebf1fl/– mice (Fig. 1 C), which therefore recapitulated the early developmental block of Ebf1−/− mice (Lin and Grosschedl, 1995).
Although Ebf1 is upstream of Pax5 in the genetic hierarchy of B cell development (Medvedovic et al., 2011), the loss of either transcription factor results in a similar block at the transition to the committed pro–B cell stage in adult mice (Lin and Grosschedl, 1995; Nutt et al., 1997). The question therefore arises whether EBF1 and Pax5 fulfill different or largely redundant functions during B cell commitment. This issue has been addressed by retroviral rescue experiments using in vitro cultured progenitors, which demonstrated that ectopic Pax5 expression could not rescue the EBF1 deficiency (Medina et al., 2004) and that retroviral EBF1 expression was unable to overcome the Pax5 mutant developmental arrest (Pongubala et al., 2008). In a similar situation, however, retroviral Pax5 expression in E2A (Tcf3)-deficient progenitors failed to rescue pro–B cell development in vitro (Seet et al., 2004), and yet transgenic Pax5 expression under the control of the Ikaros (Ikzf1) locus from the Ikzf1Pax5 allele (Souabni et al., 2002) restored pro–B cell development in vivo in the bone marrow of Vav-Cre Tcf3fl/fl Ikzf1Pax5/+ mice (Kwon et al., 2008). As shown by flow cytometric analysis (Fig. 1 D), B cell development was still arrested at the prepro–B cells in the bone marrow of Vav-Cre Ebf1fl/– Ikzf1Pax5/+ mice like in control Vav-Cre Ebf1fl/– mice, demonstrating that ectopic Pax5 expression in vivo is unable to compensate for the loss of EBF1 in early B cell development.
To perform the reciprocal experiment, we inserted a C-terminally tagged Ebf1 minigene into the ubiquitously expressed Rosa26 (R26) locus between upstream sequences consisting of a CMV enhancer-actin promoter (CA) region and floxed Neo-Stop cassette and a downstream region containing an IRES-Gfp gene and polyadenylation signal (Fig. S3, A and B). Upon Cre-mediated excision of the Neo-Stop cassette, the CA promoter drives expression of both EBF1 and GFP from the R26CAEbf1/+ allele (Fig. S3, C and D). As shown by flow cytometric analysis (Fig. 1 E), B cell development was still arrested at the same c-Kithi (pre)pro–B cell stage in Vav-Cre Pax5fl/fl R26CAEbf1/+ mice as in Vav-Cre Pax5fl/fl littermates. However, the R26CAEbf1 allele gave rise to GFP, and thus EBF1 expression in Vav-Cre Pax5fl/fl R26CAEbf1/+ (pre)pro–B cells (Fig. 1 F) and could overcome the Ebf1 mutant differentiation arrest by promoting pro–B cell development in Vav-Cre Ebf1fl/– R26CAEbf1/+ mice (Fig. S3 C). We therefore conclude that ectopic EBF1 expression from the Rosa26 locus is unable to rescue the Pax5 deficiency in early B cell development. Together, these data indicate that Pax5 and EBF1 fulfill nonredundant in vivo functions during B cell commitment.
Essential function of EBF1 in early B cell development
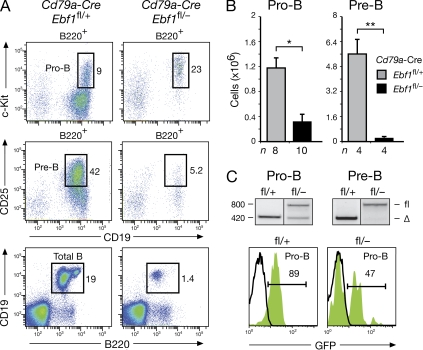
To investigate the role of EBF1 during B lymphopoiesis, we used the Cd79a-Cre and Cd23-Cre lines to delete the floxed Ebf1 allele of control Ebf1fl/+ and experimental Ebf1fl/– littermates at different stages of B cell development. This experimental strategy was validated by our observation that similar numbers of all B cell subtypes were present in the bone marrow and spleen of heterozygous Ebf1+/− mice compared with WT mice (unpublished data). The Cd79a-Cre line (also known as mb1-Cre line; Hobeika et al., 2006) initiates gene deletion at the transition from prepro–B to committed pro–B cells (Kwon et al., 2008), and was thus used to study the effect of EBF1 in early B cell development (Fig. 2). Pro–B cells (CD19+c-Kit+) were fourfold reduced in the bone marrow of Cd79a-Cre Ebf1fl/– mice compared with control Cd79a-Cre Ebf1fl/+ littermates (Fig. 2, A and B). PCR genotyping and GFP expression demonstrated that about half of the remaining pro–B cells in Cd79a-Cre Ebf1fl/– mice had undergone Ebf1 deletion in contrast to complete deletion in all pro–B cells of control littermates. Hence, pro–B cells do not tolerate the loss of EBF1 and thus stringently depend on this transcription factor (Fig. 2 C). Pre–B cells (CD19+CD25+) were decreased by 28-fold in Cd79a-Cre Ebf1fl/– mice and contained only the undeleted floxed allele contrary to control Cd79a-Cre Ebf1fl/+ littermates (Fig. 2, A–C). Consequently, only pro–B cells without Ebf1 deletion could differentiate to pre–B cells, indicating that EBF1 is required for the pro–B to pre–B cell transition.
Figure 2.
Function of EBF1 in early B cell development. (A and B) Relative percentages (A) and absolute numbers (B) of pro–B and pre–B cells were determined by flow cytometric analysis of bone marrow from Cd79a-Cre Ebf1fl/+ mice (gray bars; fl/+) and Cd79a-Cre Ebf1fl/− littermates (black bars; fl/−). n, number of mice analyzed. Statistical data (B) are shown with SEM and were analyzed by Student’s t test. *, P < 0.05; **, P < 0.01. (C) Deletion of the floxed Ebf1 allele in pro–B and pre–B cells was analyzed by PCR and GFP expression. The PCR fragments corresponding to the deleted (Δ) or intact (fl) floxed allele are indicated to the right and their size (base pairs) to the left of the gel.
Role of EBF1 in the generation of all mature B cell types
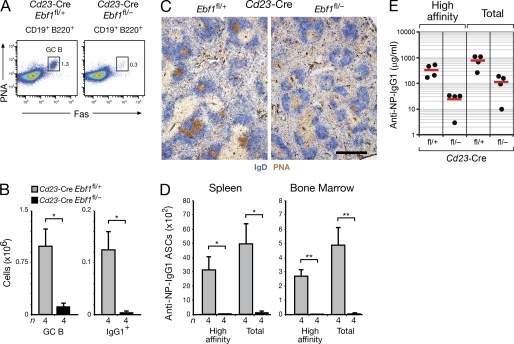
As the Cd23-Cre line initiates Cre-mediated deletion in immature B cells and leads to efficient deletion in MZ and FO B cells of the spleen (Kwon et al., 2008), we used this Cre line to study the role of EBF1 in late B cell development. Total B cells were consistently reduced by 40% in the spleen of Cd23-Cre Ebf1fl/– mice compared with control Cd23-Cre Ebf1fl/+ littermates at 8 and 16 wk of age (Fig. 3, A and B). FO B cells of Cd23-Cre Ebf1fl/– mice were similarly decreased (Fig. 3, A and B), underwent full deletion of the floxed Ebf1 allele (Fig. 3 C), and, as a result, lost most of their EBF1 protein compared to control FO B cells, as shown by semiquantitative immunoblot analysis of CD23+ splenic B cells (Fig. 3 D). Hence, FO B cells seem to tolerate the loss of EBF1 for quite some time. Competitive bone marrow reconstitution experiments revealed, however, that the development of EBF1-deficient FO B cells of the Cd23-Cre Ebf1fl/fl genotype was more strongly impaired in the presence of competing WT FO B cells (unpublished data). Notably, EBF1-deficient FO B cells significantly down-regulated only the expression of the surface protein CD21 (Fig. 3 E and not depicted) in contrast to the radical change of cell surface phenotype observed in Pax5-deficient FO B cells (Horcher et al., 2001). Consistent with this notion, the Pax5 protein is normally expressed in EBF1-deficient FO B cells (Fig. 3 D), demonstrating that Pax5 expression is no longer under the control of EBF in late B cell development.
Figure 3.
Role of EBF1 in late B lymphopoiesis. (A and B) Relative percentages and absolute numbers of B-1, MZ, and FO B cells were analyzed by flow cytometry of the spleen from Cd23-Cre Ebf1fl/+ mice (gray bars) and Cd23-Cre Ebf1fl/− littermates (black bars) at the age of 8 (A) and 16 (B) weeks. n, number of mice analyzed. Statistical data are shown with SEM and were analyzed by the Student’s t test. **, P < 0.01; ***, P < 0.001. (C) PCR genotyping of sorted FO, MZ, and B1 B cells of the indicated genotypes. The size (base pairs) and identity of the PCR fragments is shown to the left and right of the gel, respectively. C, genomic Ebf1fl/fl or Ebf1fl/Δ control DNA. (D) CD23+ splenic B cells of the indicated genotypes were isolated by MACS sorting before immunoblot analysis of serially diluted nuclear extracts (indicated by wedges) with EBF1, TBP, and Pax5 antibodies. The size (kilodaltons) of the proteins is indicated to the left. One of two immunoblots is shown. (E) Flow cytometric analysis of the cell surface phenotype of mature B cells (IgMloIgDhi) from the spleen of Cd23-Cre Ebf1fl/+ mice (gray) and Cd23-Cre Ebf1fl/− littermates (black line). (F) Analysis of MZ B cells as CD1dhiCD21hi splenic B cells in 8-mo-old Cd23-Cre Ebf1fl/− and control Cd23-Cre Ebf1fl/+ littermates.
In comparison to FO B cells, the MZ and B-1 B cells of the spleen were more strongly (2.5–4 fold) reduced in Cd23-Cre Ebf1fl/– mice relative to control Cd23-Cre Ebf1fl/+ littermates (Fig. 3, A, B and F). In this context, it is important to note that the CD19+B220lo/– B-1 cells of the spleen exhibited the classical B-1 cell surface phenotype, including no or low expression of CD23 (see below). Our observation that the Cd23-Cre line gave rise to the loss of CD23lo/– B-1 cells in Cd23-Cre Ebf1fl/– mice indicates that B-1 cells also develop from transitional CD23+ B cells similar to MZ B cells. Interestingly, the floxed Ebf1 allele was not deleted in the residual MZ and B-1 B cells of Cd23-Cre Ebf1fl/– mice in contrast to control littermates (Fig. 3 C). These data therefore demonstrated that the differentiation of MZ and B-1 B cells stringently depends on EBF1 in contrast to FO B cells.
EBF1-dependent maintenance of GC B cell development
We next immunized Cd23-Cre Ebf1fl/– and control Cd23-Cre Ebf1fl/+ mice with sheep red blood cells (SRBCs) to study the role of EBF1 in the development of GC B cells at day 7 after immunization (Fig. 4, A–E). As shown by flow cytometry, the number of GC B cells was 4.5-fold reduced in the spleen of Cd23-Cre Ebf1fl/– mice compared with control littermates (Fig. 4, A and B). Although most GC B cells of Cd23-Cre Ebf1fl/– mice efficiently deleted the floxed Ebf1 allele (Fig. 4 C), they behaved like control GC B cells as they down-regulated CD23 and CD38 expression and normally proliferated as shown by their increased cell size (Fig. 4 D). Moreover, the loss of EBF1 only minimally affected class switch recombination to IgG1 and had no effect on switching to IgG3, as GC B cells expressing IgG1 (9.5×) and IgG3 (4×) were similarly reduced like total GC B cells (4.5×; Fig. 4 B). Histological analysis revealed that GCs with PNA+ GC B cells (brown) were present in similar numbers but had a significantly reduced size in the spleen of Cd23-Cre Ebf1fl/– mice relative to control Cd23-Cre Ebf1fl/+ littermates (Fig. 4 E). At day 14 after SRBC immunization, GC B cells were 10-fold and thus more severely reduced (Fig. 4, F and G). Moreover, only rare GCs of small size could be detected by histological analysis in the spleen of Cd23-Cre Ebf1fl/– mice compared with control Cd23-Cre Ebf1fl/+ littermates (Fig. 3 H). We conclude therefore that EBF1 is required for the maintenance, but not initiation of GC B cell development.
Figure 4.
Impaired maintenance of GC B cells in the absence of EBF1. (A and B) The abundance and immunoglobulin class switching of splenic GC B cells were determined by flow cytometry as relative percentages (A) and absolute cell numbers (B). 7 d after immunization of Cd23-Cre Ebf1fl/+ mice (gray bars) and Cd23-Cre Ebf1fl/− mice (black bars) with SRBCs. n, number of mice analyzed. (C) PCR genotyping of sorted GC B cells 7 d after immunization with SRBCs. The size (base pairs) and identity of the PCR fragments is shown to the left and right of the gel, respectively. C, genomic Ebf1fl/Δ control DNA. (D) Characterization of EBF1-deficient GC B cells. Splenic GC B cells (PNA+Fas+B220+CD19+; gray) of Cd23-Cre Ebf1fl/+ and Cd23-Cre Ebf1fl/– mice were analyzed for their cell size and expression of CD23 and CD38 at day 7 after immunization with SRBCs. Non–B cells (B220–CD19–; dashed line) and FO B cells (CD19+CD21intCD23hi; black line) served as controls. (E) Size of GCs in the spleen of Cd23-Cre Ebf1fl/− and Cd23-Cre Ebf1fl/+ mice at day 7 after immunization with SRBCs. Cryosections of the spleen were stained for peanut agglutinin (PNA, brown) and IgD (blue) expression as described in Materials and methods. The black scale bar corresponds to 500 µm. (F–H) GC B cells in the spleen of Cd23-Cre Ebf1fl/− mice and control Cd23-Cre Ebf1fl/+ littermates at day 14 after immunization with SRBCs, as shown by flow cytometry (F and G) and histological analysis (H). Statistical data (B and G) are shown with SEM and were analyzed by Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
We next investigated the immune response to the T cell–dependent antigen 4-hydroxy-3-nitrophenylacetyl-conjugated keyhole limpet hemocyanin at day 14 after immunization (Fig. 5). GC B cells were eightfold decreased and IgG1+ GC B cells were reduced by 33-fold in the spleen of Cd23-Cre Ebf1fl/– mice compared with control Cd23-Cre Ebf1fl/+ littermates (Fig. 5, A and B) consistent with the identification of very few GCs of small size by immunohistochemical analysis (Fig. 5 C). Moreover, very little anti–NP-IgG1 antibody-secreting cells (ASCs) could be detected by ELISPOT assay in the spleen and bone marrow of Cd23-Cre Ebf1fl/– mice in contrast to control Cd23-Cre Ebf1fl/+ littermates (Fig. 5 D). Analysis of serum antibody titers by ELISA additionally revealed that high-affinity and total anti–NP-specific IgG1 antibodies were reduced by 14- and 7.5-fold, respectively, in Cd23-Cre Ebf1fl/– mice relative to Cd23-Cre Ebf1fl/+ littermates (Fig. 5 E). Together these data demonstrate that EBF1 is required for maintaining the GC B cell and plasma cell responses to foreign antigens.
Figure 5.
Impaired immune responses in the absence of EBF1. (A–E) NP-specific immune responses at day 14. Cd23-Cre Ebf1fl/+ mice (fl/+; gray bars) and Cd23-Cre Ebf1fl/– littermates (fl/−; black bars) were immunized with 4-hydroxy-3-nitrophenylacetyl-conjugated keyhole limpet hemocyanin (in Alum) and analyzed after 14 d by flow cytometric analysis of splenic B cells (A and B), immunohistochemical staining of the spleen (C), ELISpot assay (D), and ELISA (E). (D) The total number of anti–NP-IgG1 ASCs in the spleen or bone marrow (femur and tibia of the two hind legs) was determined by ELISpot assay using NP4-BSA– or NP23-BSA–coated plates for detecting cells secreting high-affinity or total anti–NP-IgG1 antibodies, respectively. (E) The serum titers of anti–NP-specific IgG1 antibodies were analyzed by ELISA using NP4-BSA– or NP23-BSA–coated plates for detecting high-affinity or total IgG1 antibodies, respectively. Bars, 500 µm. n, number of mice analyzed. Statistical data (B and D) are shown with SEM and were analyzed by Student’s t test. *, P < 0.05; **, P < 0.01.
EBF1-dependent survival and proliferation of FO B cells in response to BCR signaling
To investigate the role of EBF1 in BCR signaling, we purified splenic B cells from Cd23-Cre Ebf1fl/– and control Cd23-Cre Ebf1fl/+ mice to measure intracellular calcium mobilization in response to stimulation with anti-IgM antibodies. Intracellular calcium signaling was strongly impaired in Cd23-Cre Ebf1fl/– B cells in contrast to control Cd23-Cre Ebf1fl/+ B cells (Fig. 6 A), indicating that the loss of EBF1 results in a severe BCR signaling defect. We next performed a time-course analysis of anti-IgM stimulation with FO B cells, which were purified from lymph nodes by T cell depletion. Cd23-Cre Ebf1fl/– FO B cells were already preferentially lost at 12 h after BCR stimulation, and the remaining cells contained an increasingly larger fraction of Annexin V–positive cells, reaching 83% of all FO B cells after 48 h, in contrast to control Cd23-Cre Ebf1fl/+ B cells (Fig. 6 B). As signaling through the transmembrane protein tyrosine phosphatase CD45 cooperates with BCR stimulation in controlling cell survival (Huntington et al., 2006), we repeated the time-course analysis with FO B cells of the same genotypes, which were this time positively selected by MACS sorting with anti-B220 beads. The number of Cd23-Cre Ebf1fl/– B cells significantly increased and Annexin V+ cells decreased at all time points of BCR stimulation as a result of continuous activation of the CD45 isoform B220 by the presence of anti-B220 beads (Fig. 6 B). Hence, CD45 signaling does not depend on EBF1, and can thus partially rescue the survival defect of EBF1-deficient FO B cells in response to BCR stimulation. Finally, we labeled B220-sorted FO B cells with the CellTrace Violet reagent to study their proliferation in response to stimulation with anti-IgM and IL-4 for 4 d. As revealed by the dilution of the CellTrace Violet dye, 30% of the control Cd23-Cre Ebf1fl/+ B cells proliferated under these conditions in contrast to 3% of the Cd23-Cre Ebf1fl/– B cells (Fig. 6 C). These data therefore revealed an essential role of EBF1 in the regulation of B cell survival and proliferation in response to BCR signaling.
Figure 6.
B cell signaling defects in the absence of EBF1. (A) FO B cells were purified from the spleen of Cd23-Cre Ebf1fl/+ mice (black line; n = 2) and Cd23-Cre Ebf1fl/– mice (red line; n = 4) by MACS depletion of non–B cells with anti-PE beads after staining with PE-labeled TCRβ, CD4, CD8a, Mac1, and DX5 (Lin) antibodies. Intracellular Ca2+ fluxes were recorded as the fluorescence 405/485 nm ratio of Indo-1 emission after addition of anti-IgM antibody (arrow). (B) Partial rescue of activated EBF1-deficient B cells from apoptosis by CD45 stimulation. FO B cells from lymph nodes of Cd23-Cre Ebf1fl/– mice (open triangle and gray surface; n = 4) and Cd23-Cre Ebf1fl/+ littermates (black square and line; n = 4) were purified by MACS depletion of T cells with anti-PE beads after staining with PE-labeled TCRβ, CD4, and CD8a antibodies or were positively selected with anti-B220 MACS beads. The number of B cells and their Annexin V staining were determined by flow cytometry by gating on live cells at the indicated time points after anti-IgM stimulation. (C) FO B cells from lymph nodes of the indicated genotypes were positively selected with anti-B220 MACS beads and stained with CellTrace Violet reagent before stimulation with anti-IgM and IL-4 or with anti-CD40 and IL-4 for 4 d. The relative percentages of proliferating and IgG1+ B cells were determined by flow cytometric analysis. n = 3 (both genotypes) for anti-IgM plus IL-4 stimulation; n = 8 (Cd23-Cre Ebf1fl/+) and n = 10 (Cd23-Cre Ebf1fl/–) for anti-CD40 plus IL-4 stimulation. (D) FO B cells were purified from lymph nodes of Cd23-Cre Ebf1fl/– and Cd23-Cre Ebf1fl/+ mice, as described in B, before stimulation with anti-CD40 and IL-4 for up to 4 d. The number and Annexin V staining of live B cells were determined by flow cytometry. n = 4 for each genotype. (E) PCR genotyping of T cell–depleted B cells from lymph nodes of the indicated genotypes after 4 d of anti-CD40 plus IL-4 stimulation. C, genomic Ebf1fl/Δ control DNA.
Similar to BCR stimulation, treatment with anti-CD40 and IL-4 resulted in increased apoptosis of T cell–depleted FO B cells from the lymph nodes of Cd23-Cre Ebf1fl/– mice, whereas positive selection and stimulation with anti-B220 beads largely rescued the survival defect of EBF1-deficient FO B cells compared with control Cd23-Cre Ebf1fl/+ B cells (Fig. 6, D and E). In contrast to BCR signaling, the B220-sorted FO B cells from Cd23-Cre Ebf1fl/– mice efficiently proliferated upon anti-CD40 plus IL-4 treatment and underwent class switch recombination to IgG1 at a similar frequency compared with control Cd23-Cre Ebf1fl/+ FO B cells (Fig. 6 C). We conclude therefore that EBF1 plays no or only a minimal role in controlling B cell proliferation and IgG1 class switch recombination in response to CD40 and IL-4 signaling.
EBF1 regulates genes involved in different signaling pathways of FO B cells
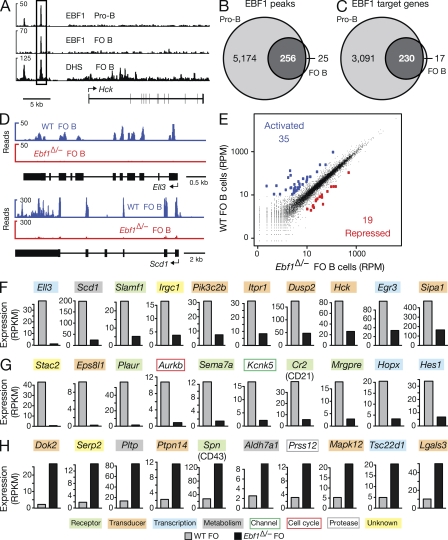
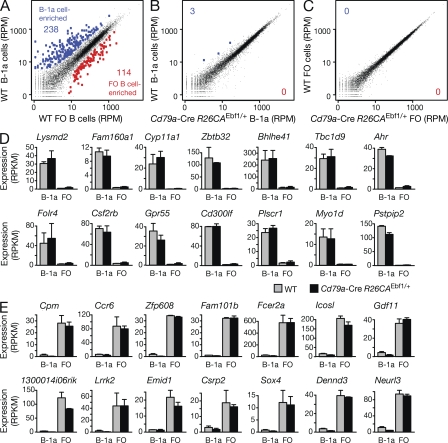
To gain insight into the molecular function of EBF1 in mature B cells, we next determined the genomic binding pattern and target genes of EBF1 in FO B cells, which were purified from lymph nodes of WT C57BL/6 mice by MACS depletion of non-B cells. The purified FO B cells were analyzed by chromatin immunoprecipitation (ChIP) with an EBF1 antibody combined with deep sequencing (ChIP-seq). Using a p-value of <10−10 for peak calling, we identified only 281 EBF1 peaks in FO B cells (Fig. 7, A and B; Table S1). Importantly, the EBF1 consensus recognition sequence was present in 95% of all EBF1 peaks (Fig. S4 A), which defined 247 EBF1 target genes in FO B cells (Fig. 7 C and Fig. S4 B). In contrast, ChIP-sequencing with the same EBF1 antibody detected 5,430 EBF1 peaks corresponding to 3,321 EBF1 target genes in Rag2−/− pro–B cells (Fig. 7, A-C; and Fig. S4 B). These numbers are similar to the 5,071 EBF1 peaks and 3,138 EBF1 target genes that were recently identified by ChIP-sequencing of A-MuLV-transformed pro–B cells (Treiber et al., 2010). Interestingly, 91% of all EBF1 peaks in FO B cells are already present in pro–B cells, thus defining 230 common EBF1 target genes (Fig. 7, B and C). In summary, our ChIP-sequencing data revealed few EBF1 peaks and target genes in FO B cells.
Figure 7.
Genome-wide analysis of regulated EBF1 target genes in FO B cells. (A) EBF1 binding at the Hck locus. EBF1-binding sites were identified by ChIP-sequencing in short-term cultured Rag2−/− pro–B cells and mature FO B cells, which were MACS-sorted from lymph nodes of WT C57BL/6 mice. DNase I hypersensitive (DHS) sites of FO B cells are additionally shown together with the exon–intron structure of the Hck gene and a scale bar indicated in kilobases. (B and C) Identification of EBF1 peaks and target genes in FO B cells compared with pro–B cells. EBF1 peaks (B) were identified by peak calling using a p-value of <10−10 and were assigned to target genes (C), if they were located from −50 kb upstream of the transcription start site (TSS) to +50 kb downstream of the transcription end site (TES). A second ChIP-seq experiment (Table S2) yielded similar results. (D) Identification of Ell3 and Scd1 as EBF1-activated genes in FO B cells by RNA-sequencing of polyA+ RNA from WT and Cd23-Cre Ebf1fl/− (Ebf1Δ/−) FO B cells (sorting strategy in Fig. S5, A and C). (E) Scatter plot of gene expression differences observed between WT and Ebf1Δ/− FO B cells. The normalized expression values of each gene in the two B cell types were plotted as reads per gene per million mapped sequence reads (RPM). Genes are highlighted in blue or red color, if they were expressed >10 RPMs and regulated at least fourfold between the two cell types. The data of one RNA-sequencing experiment for each cell type was analyzed (Table S2). (F–H) Expression of activated EBF1 target genes (F), as well as indirectly EBF1-activated (G) and EBF1-repressed (H) genes. The expression of each gene in WT (gray bar) and Ebf1Δ/− (black bar) FO B cells is shown as normalized expression value, which was determined as reads per kilobase of exon per million mapped sequence reads (RPKM). Different colors indicated genes of distinct functional categories.
We next investigated to what extent these EBF1 target genes depend on EBF1 for their expression in FO B cells. To this end, we used RNA-sequencing to determine and compare the gene expression level of each gene in FO B cells isolated from the spleen of WT and Cd23-Cre Ebf1fl/– (Ebf1Δ/–) mice (Fig. S5, A and C), as exemplified for the EBF1-activated genes Ell3 and Scd1 (Fig. 7 D). Scatter plot analysis of normalized expression values (RPM) of all genes in WT and Ebf1 mutant FO B cells demonstrated that the transcription of most genes (indicated in black) was not or was only minimally affected in the absence of EBF1 (Fig. 7 E). A small number (35) of genes were more than fourfold activated (blue), and even fewer genes (19) were similarly repressed (red) by EBF1 in WT FO B cells compared with Ebf1Δ/– FO B cells (Fig. 7 E). When we also considered EBF1 binding at the regulated genes, we identified only 10 activated EBF1 target genes that were more than threefold activated by EBF1 and were additionally expressed at a level of >10 RPKM in WT FO B cells (Fig. 7 F and Fig. S4 B). Using the same criteria, we could, however, not identify a single EBF1-repressed target gene in FO B cells. In addition, EBF1 indirectly activated 10 genes by a factor of >5 (Fig. 7 G) and indirectly repressed 10 genes by a factor of >4 (Fig. 7 H), whereby these genes were further selected for an expression level of >10 RPKM in WT FO B cells (activated genes) or Ebf1Δ/– FO B cells (repressed genes). The differential expression of the Cr2 (CD21) and Spn (CD43) genes was furthermore confirmed by flow cytometric analysis of control and Ebf1Δ/– mature B cells (Fig. 3 E). Importantly, the EBF1-activated genes code for cell surface and adhesion receptors (Slamf7, Cr2; Plaur, Sema7a, and Mrgpre), signal transducers (Pik3c2b, Hck; Dusp2, Sipa1, Eps8l, and Itpr1), and transcription factors (Ell3, Egr3, Hes1, and Hopx) that have been implicated in different B cell signaling pathways (Fig. 7, F and G; see Discussion).
Ectopic EBF1 expression promotes differentiation of B-1 cells at the expense of B-2 cells
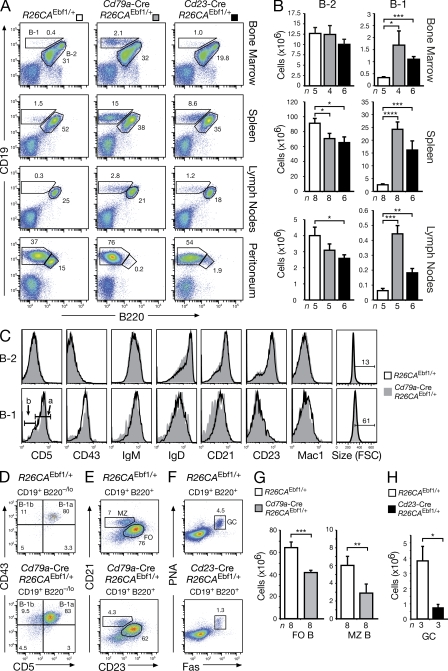
We next performed gain-of-function experiments to further investigate the role of EBF1 in late B cell development. Although ectopic expression of EBF1 from the R26CAEbf1 allele strongly impaired T lymphopoiesis in the thymus of Vav-Cre R26CAEbf1/+ mice (unpublished data), similar to retroviral EBF1 expression (Zhang et al., 2003), it did not appear to affect B cell development in the bone marrow of Vav-Cre R26CAEbf1/+ or Cd79a-Cre R26CAEbf1/+ mice (unpublished data). However, B-1 cells (CD19+B220lo/–) were strongly increased at the expense of B-2 cells (CD19+B220+) in the spleen, lymph nodes, and peritoneum and, to a lower degree, in the bone marrow of Cd79a-Cre R26CAEbf1/+ and Cd23-Cre R26CAEbf1/+ mice in contrast to control R26CAEbf1/+ mice (Fig. 8, A and B). This increase in B-1 cells was more pronounced upon activation of the R26CAEbf1 allele with the earlier deletion Cd79a-Cre line compared with the later deletion Cd23-Cre line, which likely induced Cre-mediated deletion only transiently in transitional CD23+ B cells during B-1 cell differentiation (Fig. 8, A and B). Comparison by flow cytometric analysis indicated that the CD19+B220lo/– B-1 cells in the spleen of Cd79a-Cre R26CAEbf1/+ mice were proliferating according to their larger cell size, exhibited the classical B-1 cell surface phenotype (CD43+IgMhiIgDloCD21loCD23lo/–Mac1–), and consisted predominantly of CD5+ B-1a cells (83%) with a smaller contribution of CD5– B-1b cells (9.5%), which is similar to the splenic B-1 cells of control R26CAEbf1 littermates (Fig. 8, C and D). Moreover, the B-1a and B-1b cells in the peritoneum of both Cd79a-Cre R26CAEbf1/+ and control R26CAEbf1 mice were also comparable with regard to their proliferation rate and cell surface phenotype (unpublished data) and, in contrast to the splenic B-1 cells, they also expressed the myeloid marker Mac1, as previously described (Berland and Wortis, 2002).
Figure 8.
Increased B-1 and reduced B-2 cell differentiation upon ectopic EBF1 expression. (A and B) B-1 (CD19+B220lo/–) and B-2 (CD19+B220+) cells were analyzed by flow cytometry in the bone marrow, spleen, lymph nodes, and peritoneum of R26CAEbf1/+ (white bars), Cd79a-Cre R26CAEbf1/+ (gray bars), and Cd23-Cre R26CAEbf1/+ (black bar) mice (A). Absolute cell numbers were determined for B-1 and B-2 cells in the bone marrow, spleen, and lymph nodes (B). n, number of mice analyzed. (C) Cell surface phenotype of B-1 and B-2 cells from the spleen of R26CAEbf1/+ mice (black line) and Cd79a-Cre R26CAEbf1/+ (gray) littermates. The B-1a (CD5+) and B-1b (CD5–) B cells are indicated. (D) Flow cytometric identification of B-1a cells (CD5+CD43+CD19+B220lo/–) and B-1b cells (CD5–CD43+CD19+B220lo/–) in the spleen of Cd79a-Cre R26CAEbf1/+ and control R26CAEbf1/+ littermates. The relative percentage of each B1 cell type is shown in the respective quadrant. (E–H) The relative percentages and absolute numbers of MZ and FO B cells (E and G), as well as GC B cells (F and H) were determined by flow cytometric analysis of the spleen from mice of the indicated genotypes. GC B cells were analyzed 7 d after immunization with SRBCs. Statistical data (B, G, and H) are shown with SEM and were analyzed by Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
In contrast to the B-1 cells, the FO and MZ B cell numbers were reduced by 30 and 50%, respectively, in the spleen of Cd79a-Cre R26CAEbf1/+ mice compared with control R26CAEbf1/+ littermates (Fig. 8, E and G). Moreover, mature B cells from the lymph nodes of both genotypes gave rise to identical calcium mobilization in response to BCR stimulation, suggesting that BCR signaling was not affected by ectopic EBF1 expression (unpublished data). Surprisingly, GC B cells were decreased by fivefold, and thus more strongly reduced in the spleen of Cd23-Cre R26CAEbf1/+ mice compared with control R26CAEbf1/+ mice at day 7 after immunization with sheep red blood cells (Fig. 8, F and H), which was confirmed by immunohistochemical analysis (unpublished data). Hence, both ectopic EBF1 expression and EBF1 loss resulted in impaired MZ, FO, and GC B cell development, quite in contrast to the observed opposite effects of these genetic manipulations on B-1 cell differentiation. In summary, we conclude that ectopic EBF1 expression promotes the differentiation of B-1 cells at the expense of mature B-2 cell types.
Normal gene expression and VH gene usage in EBF1-induced B-1 cells
To further characterize the EBF1-induced B-1 cells, we defined the gene expression signatures characteristic of splenic B-1a and FO B cells. To this end, we compared the expression profiles of sorted WT B-1a and FO B cells (Fig. S5 A) by RNA sequencing. As shown by scatter plot analysis (Fig. 9 A), 238 genes (blue) were significantly up-regulated in splenic B-1a cells, whereas 114 genes (red) were significantly up-regulated in FO B cells. In contrast, only three significant gene expression changes could be detected by comparing WT and Cd79a-Cre R26CAEbf1/+ B-1a or FO B cells (Fig. 9, B and C; and Fig. S5 B). Importantly, the analysis of individual B-1a cell-enriched transcripts revealed that they were highly expressed in both WT and Cd79a-Cre R26CAEbf1/+ B-1a cells in contrast to their low expression in FO B cells of both genotypes (Fig. 9 D). An inverse picture was observed for FO B cell–enriched transcripts, which were highly expressed in WT and Cd79a-Cre R26CAEbf1/+ FO B cells, but not in B-1a cells of either genotype (Fig. 9 E). Hence, ectopic EBF1 expression minimally affected gene expression in both B-1a and FO B cells.
Figure 9.
RNA-sequencing identified the EBF1-induced B-1a cells as bona fide B-1a cells. (A–C) Scatter plots of gene expression differences between WT and EBF1-overexpressing B-1a and FO B cells. Two independent RNA-sequencing experiments (Table S2) were performed for each FACS-sorted B-1a or FO B cell type isolated from the spleen of WT or Cd79a-Cre R26CAEbf1/+ mice (sorting strategy in Fig. S5, A and B). The average of the normalized expression values (RPM) for each gene were plotted to indicate the gene expression differences between the different cell types. Genes are highlighted in blue or red if they were called as differentially expressed genes with an adjusted p-value of <0.1. (D and E) Expression of B-1a cell–enriched (D) and FO B cell–enriched (E) transcripts, which were identified by comparison of WT B-1a and FO B cells (A). The expression of the indicated genes in B-1a and FO B cells of WT (gray bars) and Cd79a-Cre R26CAEbf1/+ (black bars) mice is shown as average of the normalized expression value (RPKM) together with the SEM.
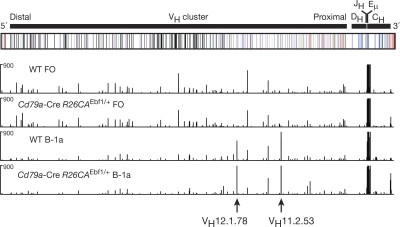
The B-1 cells differ from B-2 cells in their antigen specificity, as they express antibodies that weakly recognize self-antigens, including plasma membrane phospholipids such as phosphatidylcholine (Berland and Wortis, 2002; Baumgarth, 2011). In particular, phosphatidylcholine-binding B-1 cells predominantly express one of two immunoglobulin heavy- and light-chain combinations encoded by either the VH11 and Vκ9 or VH12 and Vκ4 gene segments, which determine the antibody specificity for phosphatidylcholine (Reininger et al., 1987; Hardy et al., 1989; Seidl et al., 1997). To analyze the VH gene repertoire in splenic B-1a and FO B cells, we mapped, by RNA sequencing, the transcripts of functionally rearranged VHDJH genes to the Igh locus in B-1a and FO B cells of WT and Cd79a-Cre R26CAEbf1/+ mice. As shown in Fig. 10, the Cμ gene segments in the proximal Igh region were similarly transcribed in B-1a and FO B cells of both genotypes. In contrast, the expressed VH sequences of the rearranged VHDJH genes were skewed toward the use of the VH11.2.53 and VH12.1.78 gene segments (Johnston et al., 2006) in B-1a cells of WT and Cd79a-Cre R26CAEbf1/+ mice, whereas these two VH genes were not expressed in FO B cells of either genotype (Fig. 10). Similar analysis of Vκ gene expression identified the Vκ4-91 and Vκ9-128 genes (Brekke and Garrard, 2004) to be uniquely expressed in B-1a cells of both WT and Cd79a-Cre R26CAEbf1/+ mice in contrast to FO B cells (unpublished data). Consequently, the EBF1-induced B-1a cells generated phosphatidylcholine-specific antibodies at a similar frequency compared with WT B-1a cells. Hence, the comparable VH and Vκ gene usage and similar gene expression pattern of WT and Cd79a-Cre R26CAEbf1/+ B-1a cells unequivocally demonstrates that ectopic EBF1 expression induces the generation of bona fide B-1a cells.
Figure 10.
Normal VH gene repertoire in EBF1-induced B-1a cells. Analysis of the rearranged and expressed VH gene repertoire of B-1a and FO B cells by RNA-sequencing. The abundance of the 76-nt-long sequence reads, which correspond to the RNA transcripts mapping to the variable (VH), diversity (DH), joining (JH), and constant (CH) gene segments of the Igh locus, are shown for B-1a and FO B cells of WT and Cd79a-Cre R26CAEbf1/+ mice. A schematic diagram of the Igh locus based on the C57BL/6 VH gene assembly (Johnston et al., 2006) indicates members of the distinct VH gene families in different colors. Arrows denote the positions of the VH11.2.53 and VH12.1.78 gene segments.
DISCUSSION
The transcription factor EBF1, like Pax5, is exclusively expressed from early B cell progenitors to mature B cells in the B lymphoid lineage of the hematopoietic system, and yet its function has so far only been explored at the initiation of B lymphopoiesis. In this study, we have uncovered novel important functions of EBF1 in early and late B cell development by performing conditional gain- and loss-of-function experiments. EBF1 is first expressed in BLPs at the onset of B cell development (Inlay et al., 2009; this study), activates Pax5 transcription by binding to the Pax5 promoter region (Decker et al., 2009), and then participates in a positive cross-regulatory loop with Pax5 to maintain each other’s expression at the pro–B cell stage (Fuxa et al., 2004; Roessler et al., 2007). By expressing Pax5 and EBF1 independently of this feedback loop, we have now shown that ectopic Pax5 expression from the Ikaros locus is unable to rescue the Ebf1 mutant phenotype at the start of B lymphopoiesis and that EBF1 expression from the Rosa26 locus cannot overcome the Pax5 deficiency in vivo. These data therefore demonstrate that EBF1 and Pax5 fulfill nonredundant functions during B cell commitment in vivo, despite the fact that both transcription factors often bind to the same regulatory elements of common target genes in pro–B cells (Lin et al., 2010; Treiber et al., 2010; unpublished data). EBF1 is furthermore essential for maintaining pro–B cell development and promoting the transitions to pre–B cells, as pro–B cells were reduced and pre–B cells were lost upon Ebf1 inactivation with the early deleting Cd79a-Cre line. The strict EBF1 dependence of pro–B cell development and pre–BCR signaling is best explained by the multitude of activated EBF1 target genes that code for essential signaling molecules including the surrogate light chains λ5 and VpreB1/2 and the signal-transducing proteins Igα and Igβ (Treiber et al., 2010).
In mature B cells, Ebf1 and Pax5 are regulated independently of each other, as Ebf1 is normally transcribed in Pax5 mutant FO B cells (unpublished data), whereas Pax5 is normally expressed in EBF1-deficient FO B cells (this study). Although FO B cells are reduced by 40% upon conditional Ebf1 inactivation with the late deleting Cd23-Cre line, the remaining FO B cells tolerate the loss of EBF1 for some time. Similarly, GC B cells in the absence of EBF1 are initially formed, but are later lost during the immune reaction, indicating that EBF1 is required for maintaining the GC B cell and plasma cell responses to foreign antigen. Interestingly, EBF1 indirectly activates the Cr2 gene, coding for the complement receptor CD21, in mature B cells. Notably, GC B cell development and B cell responses to T cell–dependent antigens are severely impaired in Cr2 mutant mice (Ahearn et al., 1996; Molina et al., 1996), as the survival of GC B cells is critically dependent on the expression of CD21 (Fischer et al., 1998). Hence, the strong down-regulation of CD21 expression in EBF1-deficient FO B cells may partly explain the loss of GC B cells and impaired immune responses in Cd79a-Cre Ebf1fl/– mice. The transmembrane proteins CD21 and CD19 form a co-stimulatory receptor complex on mature B cells that cooperates with the BCR to efficiently bind and respond to complement C3d-fixed antigens (Fearon and Carroll, 2000). Although CD21 recognizes the complement fragment C3d, CD19 functions as the signaling molecule of the complex to recruit Vav, PI3K, and phospholipase Cγ2 (PLCγ2) to the plasma membrane, which leads to increased calcium signaling and cell survival in response to BCR stimulation (Tuveson et al., 1993; O’Rourke et al., 1998; Buhl and Cambier, 1999; Roberts and Snow, 1999; Brooks et al., 2000). Intracellular calcium mobilization, cell survival, and proliferation are strongly reduced in EBF1-deficient FO B cells upon BCR activation, which may reflect impaired co-stimulation by the CD19–CD21 complex caused by the down-regulation of CD21 expression in the absence of EBF1. Interestingly, EBF1 directly activates the expression of the type 1 IP3 receptor (Itpr1), which functions as an intracellular calcium release channel in response to the second messenger inositol 1,4,5-trisphosphate (IP3; Scharenberg et al., 2007). Hence, the reduced expression of Itpr1 is likely to contribute to the calcium signaling defect of EBF1-deficient FO B cells.
Genome-wide analysis of EBF binding unexpectedly revealed only 281 EBF1 peaks and 247 EBF1 target genes in FO B cells, although the EBF1 protein is expressed only at a threefold lower level in FO B cells compared with pro–B cells. As we analyzed EBF1 binding in quiescent FO B cells, it remains a possibility that EBF1 may interact with more target genes in activated B cells. However, the observed small number of EBF1 peaks is unlikely to result from technical difficulties to detect EBF1 binding, as ChIP-sequencing with the same EBF1 antibody identified 5,071 peaks corresponding to 3,321 EBF1 target genes in pro–B cells, similar to recently published results obtained by ChIP-sequencing of A-MuLV-transformed pro–B cells (Treiber et al., 2010). In contrast to EBF1, Pax5 binds to 15,468 genomic sites in mature cells (unpublished data), which results in the colocalization of Pax5 peaks with two-thirds (184) of all EBF1-binding sites in FO B cells. Surprisingly, EBF1 efficiently activates only 10 (4%) of its target genes in FO B cells. One of these target genes, Ell3, is commonly activated in pro–B and mature B cells, and yet the other activated target genes are also already bound by EBF1 in pro–B cells, suggesting that transcription factors expressed in late B cell development cooperate with EBF1 to regulate these genes specifically in FO B cells (Treiber et al., 2010; this study). EBF1 directly activates a single transcription factor gene coding for Egr3, which has been implicated in the control of T cell proliferation and survival in response to (pre)TCR signaling (Xi and Kersh, 2004; Safford et al., 2005; Carter et al., 2007). As Egr3 is only threefold activated by EBF1 in FO B cells, it is unlikely able to mediate the much stronger indirect regulation of several EBF1-dependent genes. In contrast, the direct target gene Ell3 is 36-fold down-regulated in the absence of EBF1 and codes for a transcription elongation factor and component of the super-elongation complex (SEC), which controls the release of promoter-proximal stalled RNA polymerase II into productive elongation of transcription (Miller et al., 2000; Smith et al., 2011). It is thus conceivable that the near loss of Ell3 may interfere with the transcription elongation of indirectly EBF1-activated genes.
Compared with Cr2, other EBF1-activated genes coding for cell surface receptors or signal transducers have been less well characterized with regard to their function in late B cell development. The cell surface receptor Slamf1 (CD150) is activated by homophilic Slamf1 interactions between B cells or B and T cells within lymphoid follicles, and it enhances B cell proliferation as a co-stimulatory receptor in concert with BCR or CD40 signaling by activating the PI3K and MAPK pathways (Punnonen et al., 1997; Mikhalap et al., 2004; Yurchenko et al., 2010). The Src family kinase Hck has been implicated in cell proliferation in response to IL-6 signaling (Podar et al., 2004), whereas the phosphatase Dusp2 negatively regulates BCR-mediated cell proliferation by dephosphorylating the MAP kinase Erk2 upon co-ligation of the BCR and low-affinity IgG receptor FcγRIIb by IgG-containing immune complexes (Brown et al., 2004; Zhang et al., 2005). The GTPase-activating protein Sipa1 promotes signaling from the small G protein Rap1, which has been implicated in B-1a cell development by controlling the generation of the skewed Vκ gene repertoire (Ishida et al., 2006), and in lymphocyte adhesion and trafficking by enhancing integrin function (Katagiri et al., 2004; Sebzda et al., 2002). The GPI-anchored urokinase receptor Plaur is also involved in signaling lymphocyte adhesion, migration, and proliferation during immune responses, in addition to its proteolytic activation of the urokinase plasminogen activator (uPA) leading to extracellular matrix degradation (Blasi and Carmeliet, 2002). The cell surface protein semaphorin 7A (Sema7a) is known for its important function in T cell–mediated inflammatory responses, as its expression on activated T cells promotes integrin α1β1 binding and recruitment of macrophages to sites of inflammation (Suzuki et al., 2007). The PI3K C2β (Pik3c2b) is a member of the poorly characterized class II PI3K family and has been implicated in cell migration by associating with the trimeric Eps8–Abi–Sos1 complex, which functions as a guanine nucleotide exchange factor to activate the small G protein Rac, leading to actin cytoskeleton remodeling (Maffucci et al., 2005; Katso et al., 2006). Finally, the adaptor molecule Eps8L1 interacts with the signaling chain CD3ε of the TCR (Kesti et al., 2007) and, like its related protein Eps8, promotes actin reorganization by controlling the activity of Rac (Offenhäuser et al., 2004). Although the function of some of the EBF1-activated genes in mature B cells remains to be investigated, their role in the related T cell lineage suggests that the reduced expression of these genes in response to EBF1 loss also contributes to the Ebf mutant phenotype in late B cell development.
EBF1 is particularly important for the generation of B-1 and MZ B cells, as these two innate-like mature B cell types do not tolerate the loss of EBF1. Although B-1 cells are predominantly located in peritoneal and pleural cavities and MZ B cells in the antigen-filtrating MZ of the spleen, they share similar properties and constitute the first line of defense against blood-borne microorganisms by rapidly differentiating into short-lived plasma cells (Martin and Kearney, 2002; Hardy et al., 2007; Baumgarth, 2011). B-1 and MZ B cells have self-renewal potential and express a selected BCR repertoire recognizing bacterial cell wall constituents and self-antigens such as phospholipids (Hao and Rajewsky, 2001; Martin and Kearney, 2002; Baumgarth, 2011; Montecino-Rodriguez and Dorshkind, 2012). Although the loss of B-1 and MZ B cells upon conditional Ebf1 inactivation precludes the identification of regulated EBF1 target genes in these two cell types, it is worth noting that B-1a cells are reduced in Cr2 (CD21)-null mice (Ahearn et al., 1996), which is consistent with the idea that strong autoantigen-driven BCR signaling is required for the maturation and self-renewal of B-1a cells (Casola et al., 2004; Baumgarth, 2011; Montecino-Rodriguez and Dorshkind, 2012). In this context, it is important to emphasize that the loss of B-1 cells upon Ebf1 mutation may be best explained by the impaired proliferation of EBF1-deficient mature B cells in response to BCR stimulation.
Furthermore, we have shown that ectopic expression of EBF1 from the Rosa26 locus efficiently induces the development of B-1 cells at the expense of MZ and FO B cells. Importantly, the EBF1-induced B-1a cells exhibit the same gene expression signature and preference for immunoglobulin rearrangements, creating phosphatidylcholine-specific antibodies, as WT B-1a cells, which identifies EBF1 as the first transcription factor capable of diverting conventional B cells in the B-1 cell fate. The B-1 cell lineage is thought to develop from B-1 cell–specific progenitors in early B lymphopoiesis (Montecino-Rodriguez et al., 2006). In agreement with this idea, B-1 cell development is most efficiently induced if EBF1 expression from the Rosa26 locus is activated by early deletion with the Cd79a-Cre line. However, activation of EBF1 expression by the Cd23-Cre line, which initiates Cre expression only in CD23+ transitional B cells of the spleen (Kwon et al., 2008), is still able to generate B-1 cells at an advanced stage of B cell development. This latter finding is more in line with the activation-induced model postulating that B-1 cells develop in response to the recognition of certain self-antigens and common bacterial cell wall constituents by the BCR (Berland and Wortis, 2002). It remains to be seen whether the EBF1-induced B-1 cells originate predominantly in fetal B lymphopoiesis, like most B-1 cells of WT mice (Montecino-Rodriguez and Dorshkind, 2012). Investigating the developmental timing and critical EBF1 target genes involved in the B-2 to B-1 lineage diversion are interesting and challenging questions for future studies.
MATERIALS AND METHODS
Mice.
The following mice were maintained on the C57BL/6 genetic background: Ebf1+/− (Lin and Grosschedl, 1995), Pax5fl/fl (Horcher et al., 2001), Ikzf1Pax5/+ (Souabni et al., 2002), Cd79aCre/+ (Hobeika et al., 2006), Mox2Cre/+ (Tallquist and Soriano, 2000), transgenic Vav-Cre (de Boer et al., 2003), transgenic Cd23-Cre (Kwon et al., 2008), and transgenic FLPe (Rodríguez et al., 2000) mice. Throughout this article, the heterozygous Cd79aCre/+ genotype is referred to as Cd79a-Cre (also known as mb1-Cre; Hobeika et al., 2006). All animal experiments were performed according to valid project licenses, which were approved and regularly controlled by the Austrian Veterinary Authorities.
Generation of the Ebf1ihCd2 allele.
The targeting vector for generating the Ebf1ihCd2 allele was obtained by first inserting the following sequences in the 5′ to 3′ direction (Fig. S1 A) into the Ebf1 bacterial artificial chromosome (BAC) RP23-14018 by recombineering in Escherichia coli: 233-bp MfeI–AscI fragment containing C-terminal tag sequences (fused in frame to the last Ebf1 codon), 1.8-kb AscI–SalI fragment containing the IRES-hCd2 (ihCd2) reporter gene (flanked by frt sites), and 1.9-kb SalI–XhoI fragment containing the mouse phosphoglycerate kinase promoter linked to the neomycin resistance gene (flanked by loxP sites). The tag sequences contained cleavage sites for the PreScission (PreSc) and TEV proteases, epitopes for Flag and V5 antibodies, and a biotin acceptor sequence (Biotin) for biotinylation by the E. coli biotin ligase BirA. In a second step, the Ebf1ihCd2 targeting vector was generated by excising and inserting the integrated sequences together with the flanking 5′ (1.9 kb) and 3′ (5.0 kb) homology regions by recombineering from the modified BAC into the pBV-DTA-pA plasmid containing an HSV-TK gene (for negative selection). 15 µg SgrA1-linearized DNA was electroporated into cells (107) of the hybrid C57BL/6 × 129SV ES cell line A9, followed by selection with 250 µg/ml G418. PCR-positive clones were verified by Southern blot analysis of BamHI-digested DNA (Fig. S1 B) before injection into C57BL/6 blastocysts and the generation of Ebf1ihCd2−Neo/+ mice. The Ebf1ihCd2 allele was obtained by crossing Ebf1ihCd2−Neo/+ mice with the Mox2-Cre line (Tallquist and Soriano, 2000). The following primers were used for PCR genotyping of Ebf1ihCd2/+ mice: (a) 5′-TCTCACTATTTTGCAAAGTCTGAGG-3′; (b) 5′-GGGCTTGCTTTTTATCAATACAGA-3′; and (c) 5′-TTGTAGTCAGGTCCCTGGAACA-3′. The WT Ebf1 allele was identified as a 624-bp PCR fragment with the primer pair a/b, and the Ebf1ihCd2 allele as a 410-bp PCR fragment with the primer pair a/c (Fig. S1 C).
Generation of the floxed Ebf1 allele.
The conditional Ebf1 targeting vector was assembled in the pBV-DTA-pA plasmid containing an HSV-TK gene (for negative selection), as well as a polylinker with appropriate restriction sites for insertion of the following sequences in the 5′ to 3′ direction (Fig. S2 A): a 5.0-kb NotI–AscI fragment (cloned as long 5′ homology region from BAC RP23-82015 by recombineering in E. coli); a loxP site present in the polylinker; a 3.2-kb XhoI–SalI fragment (containing the 3′ end of Ebf1 intron 5, exon 6 fused in frame to Ebf1 cDNA sequences from exon 7 to the 3′ untranslated region, 6 copies of the SV40 polyA region and a second loxP site); a 1.7-kb SalI–RsrII fragment (containing the 3′ end of Ebf1 intron 5 and the first 14 codons of exon 6 linked in frame to a Gfp gene, polyadenylation signal, frt site, and the 3′ end of the neomycin [Neo] resistance gene [in opposite orientation to Ebf1]); 1.35-kb RsrII–ClaI fragment (containing the middle and 5′ end sequences of the Neo gene, the mouse phosphoglycerate kinase promoter, and the second frt site); and a 1.9-kb ClaI–BsiWI fragment (cloned as short 3′ homology region from BAC RP23-82015 by recombineering). 15 µg SgrA1-linearized DNA was electroporated into cells (107) of the hybrid C57BL/6 × 129SV ES cell line A9, followed by selection with 250 µg/ml G418. PCR-positive clones were verified by Southern blot analysis of ApaI-digested DNA (Fig. S2 B) before injection into C57BL/6 blastocysts and the generation of Ebf1fl−neo/+ mice. The Ebf1fl allele was obtained by crossing Ebf1fl−neo/+ mice with the FLPe line (Rodríguez et al., 2000) and the Ebf1Δ allele by crossing Ebf1fl/+ mice with the Mox2-Cre line (Tallquist and Soriano, 2000). The following primers were used for PCR genotyping of Ebf1fl/+ mice: (a) 5′-CTGTCCCTAGGATCCGTTGA-3′; (b) 5′-AGATGGATTACGTCCAACAGAA-3′; and (c) 5′-ACACCACGACCTGGAATCTC-3′. The WT Ebf1 allele was identified as a 647-bp PCR fragment with the primer pair a/b, and the floxed Ebf1fl allele as a 363-bp PCR fragment with the primer pair a/c (Fig. S2 C). A second set of primers was used for simultaneous PCR genotyping of the floxed and deleted Ebf1 alleles: (d) 5′-CCCAAATAGAGTTTTCTGTCCCTA-3′; (e) 5′-AGATGAACTTCAGGGTCAGCTTG-3′; and (f) 5′-AGGAATGACCTTCTGTAACCTCTG-3′. The floxed Ebf1fl allele gave rise to an 800-bp PCR fragment with the primer pair d/e, and the deleted Ebf1Δ allele to a 420-bp PCR fragment with the primer pair d/f.
Generation of the R26CAEbf1 allele.
Full-length Ebf1 cDNA linked in frame to the tag sequences shown in Fig. S3 A was cloned into the AscI site of the targeting vector CAG-STOP-eGFP-ROSA26TV (www.addgene.org). The AsiSI-linearized targeting vector was electroporated into cells (107) of the hybrid C57BL/6 × 129SV ES cell line A9, followed by selection with 250 µg/ml G418. Targeted ES cell clones were identified by Southern blot analysis (Fig. S3 B). The following primers were used for PCR genotyping of R26CAEbf1/+ mice: (a) 5′-AAGGGAGCTGCAGTGGAGTA-3′; (b) 5′-TAAGCCTGCCCAGAAGACTC-3′; and (c) 5′-CGCCGCCGGGATCACTCTCG-3′. The WT Rosa26 (R26) allele was identified as a 209-bp PCR fragment with primers a/b, and the R26CAEbf1 allele as a 508-bp PCR fragment with primers c/b.
Flow cytometry.
Mice at 4–5 or 6–10 wk of age were used for FACS analyses of lymphoid progenitors and mature B cell types, respectively. Single-cell suspensions of bone marrow, spleen, and lymph nodes were incubated with CD16/CD32 Fc block (BD) to inhibit unspecific antibody binding. For flow cytometry, cells were stained with the following antibodies: anti-B220/CD45R (RA3-6B2), CD3ε (145-2C11), CD4 (GK1.5), CD8a (53–6.7), CD5 (53–7.3), CD11b/Mac1 (M1/70), CD11c (HL3), CD19 (1D3), CD20 (AISB12), CD21/CD35 (7G6), CD22 (Cy34.1), CD23 (B3B4), CD24/HSA (M1/69), CD25/IL-2Rα (PC61), CD28 (37.51), CD38 (90), CD40 (3/23), CD43 (S7), CD44 (IM781), CD49b (DX5), CD86 (GL1), CD93/AA4.1 (PB493), CD95/Fas (Jo2), CD117/c-Kit (ACK4), CD127/IL-7Rα (A7R34), CD135/Flt3 (A2F10.1), CD138 (281–2), F4/80 (CI:A3-1), Gr1 (RB6-8C5), MHCII (M5-114), IgD (1.19), IgG1 (A85-1), IgG3 (R40-82), IgM (M41.42), IgMa (Igh-6a/DS-1), IgMb (AF6-78), NK1.1 (PK136), Ly5.1 (A20), Ly5.2 (104), Ly6C (6C3), Ly6D (49H4.3), Sca1/Ly6A (D7), TCRβ (H57-597), Ter119 (TER119), Thy1.2 (53–2.1), and human CD2 (RPA-2.10) antibodies.
FACS sorting and definition of hematopoietic cell types.
The different hematopoietic cell types were identified by flow cytometry or sorted with a FACSAria machine (BD), as follows: LSK MPPs (Lin–IL-7Rα–Sca1hic-Kithi), CLPs (Lin–IL-7Rα+Flt3+Sca1loc-Kitlo), ALP (Ly6D– CLPs), BLP (Ly6D+ CLPs), prepro–B (DX5–Ly6C–CD19–B220+c-Kit+), pro–B (CD19+c-Kit+CD25–IgM–IgD–), pre–B (CD19+CD25+c-Kit–IgM–IgD–), immature B (CD19+IgMhiIgDloCD93+), mature B (CD19+ IgMlo IgDhi CD93–), MZ B (CD19+CD21hiCD23lo/–), FO B (CD19+CD21intCD23hi), GC B (CD19+PNA+Fas+), B-1 cells (CD19+B220lo/–), plasma cells (Lin–CD138hiCD28+), total B cells (CD19+B220+), and granulocytes (Gr1+Mac1+). Lineage-positive (Lin+) cells were MACS depleted, and then electronically gated away with the following lineage marker antibodies: B220, CD19, CD4, CD8a, TCRβ, DX5, Gr1, Ly6C, Mac1, CD11c, and Ter119 (for MPPs, CLPs, ALPs, and BLPs); and CD4, CD8a, CD21, and F4/80 (for plasma cells). WT mature B cells were purified for ChIP-seq experiments from lymph nodes by MACS depletion of non–B cells with anti-PE beads after staining with PE-labeled TCRβ, CD4, CD8a, Mac1, Gr1, DX5, and Ly6C antibodies. For RNA sequencing, FO B cells were FACS sorted as B220+CD19+CD21lo/–CD23+IgMloIgDhi cells and B-1a cells as CD19+B220lo/–CD5+ cells from the spleen (Fig. S5).
Competitive bone marrow transplantation.
Bone marrow cells from Cd23-Cre mice (Ly5.2) and Cd23-Cre Ebf1fl/fl mice (Ly5.2) were twice depleted of T cells by MACS depletion with TCRβ, CD4, and CD8a antibodies, and then each mixed at a 1:1 ratio with bone marrow cells from WT mice (C57BL/6-Ly5.1) before intravenous injection into C57BL/6-Ly5.1 mice, which were γ-irradiated with a single dose of 12 Gy 24 h before. Peripheral blood was analyzed by flow cytometry 10 wk after bone marrow transplantation.
Calcium fluorometry.
FO B cells (2 × 106) were isolated from lymph nodes by MACS depletion of non–B cells with anti-PE beads after staining with PE-labeled TCRβ, CD4, CD8a, Mac1, and DX5 antibodies. Purified B cells (2 × 106) were loaded with indo-1 acetoxymethylester (Invitrogen) at a final concentration of 1 µM in 1 ml of RPMI-1640 medium containing 10% fetal calf serum. After incubation at 37°C for 45 min, the cells were washed twice and reincubated at 37°C for an additional 30 min. Propidium iodide was added, and the fluorescence ratio of Indo-1 emission at 405/485 nm was measured in live cells on a LSR flow cytometer (BD). The acquisition of the data was initiated 30 s before the addition of the monoclonal anti-IgM antibody M41.42 (at 10 µg/ml). The data were collected for 180 s and analyzed using FlowJo software (Tree Star).
In vitro B cell stimulation experiments.
Mature B cells were isolated from the lymph nodes by depletion of non–B cells or enrichment of B220+ B cells by MACS sorting. The purified B cells were plated at 0.5 × 106 cells/ml in IMDM medium supplemented with 10% fetal calf serum, and were subsequently treated for up to 4 d with 10 µg/ml anti-IgM (M41.42) antibody alone (Fig. 6 B), with 10 µg/ml anti-IgM antibody and 20 ng/ml rIL-4 (R&D Systems; Fig. 6 C), or with 1 µg/ml anti-CD40 antibody (HM-40-3; eBioscience) and 20 ng/ml rIL-4 (Fig. 6, C and D). For cell proliferation analysis, the purified B cells were first stained with 5 µM CellTrace Violet reagent (Invitrogen) before stimulation, as described above.
Immunizations and plasma cell analysis.
SRBCs were washed in PBS and resuspended at 109 cells/ml, followed by intraperitoneal injection of 100 µl into an adult mouse. The immune response to a specific antigen was studied by intraperitoneal injection of 100 µg of 4-hydroxy-3-nitrophenylacetyl–conjugated keyhole limpet hemocyanin (in Alum). The frequencies of anti–NP-IgG1 ASCs were determined in the spleen and bone marrow by ELISpot assay as previously described (Smith et al., 1997). NP4-BSA– and NP23-BSA–coated plates were used for capturing high-affinity or total anti–NP-IgG1 antibodies secreted by individual cells, respectively. Spots were visualized with goat anti–mouse IgG1 antibodies conjugated to alkaline phosphatase (SouthernBiotech), and color was developed by the addition of BCIP/NBT Plus solution (SouthernBiotech). After extensive washing, the spots were counted with an AID ELISpot reader system (AID Diagnostika).
The serum titer of NP-specific IgG1 antibodies was determined by ELISA (Smith et al., 1997) by using plates that were coated with 25 µg/ml of NP4-BSA or NP23-BSA to capture high-affinity or total NP-specific IgG1 antibodies, respectively. The serum concentration of NP-specific IgG1 was determined relative to that of a standard anti-NP IgG1 antibody (hybridoma SSX2.1).
Histological analysis.
Cryosections of the spleen from immunized mice were stained with a FITC-anti-IgD antibody (11-26c.2a; BD) and biotinylated PNA (B-1075; Vector Laboratories). FITC-anti-IgD was detected with an alkaline phosphatase–coupled anti-FITC antibody (Roche) that was visualized by incubation with Fast Blue (Sigma-Aldrich). Biotinylated PNA was detected with horseradish peroxidase–conjugated streptavidin (BD), followed by incubation with DAB (Sigma-Aldrich).
Antibodies.
A rabbit polyclonal EBF1 antibody was raised against a bacterially expressed His-tagged EBF1 polypeptide, which contained N-terminal sequences from amino acid residue 15 (Met) to residue 259 (His) of mouse EBF1 (Hagman et al., 1993), and was affinity-purified with immobilized antigen. A rabbit polyclonal Pax5 antibody (directed against the Pax5 residues 17–145) and a mouse monoclonal TATA-binding protein (TBP) antibody (clone TBP-3G3; obtained from L. Tora) were used as controls for immunoblot analysis.
ChIP analysis of EBF1 binding.
Pro–B cells isolated from the bone marrow of Rag2−/− mice were expanded in vitro for 4–5 d on OP9 cells in the presence of IL-7, and mature FO B cell were purified from lymph nodes of WT C57BL/6 mice as described above. The Rag2−/− pro–B cells and purified FO B cells were subjected to ChIP with an EBF1 antibody (see above) as described (Schebesta et al., 2007). The ChIP efficiency was controlled by real-time PCR analysis (Decker et al., 2009) before Solexa sequencing (Illumina) of the precipitated DNA.
cDNA preparation for RNA sequencing.
RNA was isolated from ex vivo–sorted cells using the RNeasy Plus Mini kit (QIAGEN), and mRNA was obtained by two rounds of poly(A) selection using the Dynabeads mRNA purification kit (Invitrogen) followed by fragmentation by heating at 94°C for 3 min. The fragmented mRNA was used as template for first-strand cDNA synthesis with random hexamers using the SuperScript VILO cDNA Synthesis kit (Invitrogen). dNTPs were removed on a Mini Quick Spin Column (Roche) before the second-strand cDNA synthesis with 100 mM dATP, dCTP, dGTP, and dUTP in the presence of RNase H, E. coli DNA polymerase I and DNA ligase (Invitrogen). The incorporation of dUTP allowed elimination of the second strand during library preparation (see below), thereby preserving strand specificity (Parkhomchuk et al., 2009).
Solexa deep sequencing.
Approximately 5 ng of cDNA or ChIP-precipitated DNA were used as starting material for the generation of single-end sequencing libraries as described by Illumina’s ChIP Sequencing sample preparation protocol. DNA fragments of 200–350 bp and 150–700 bp were selected for ChIP-seq and RNA-seq experiments, respectively. For strand-specific RNA-sequencing, the uridines present in one cDNA strand were digested with uracil-N-glycosylase (New England Biolabs) as previously described (Parkhomchuk et al., 2009), followed by PCR amplification. Completed libraries were quantified with the Agilent Bioanalyzer dsDNA 1000 assay kit and Agilent QPCR NGS library quantification kit. Cluster generation and sequencing was performed by using the Illumina/Solexa Genome Analyzer II and IIx systems according to the manufacturer’s guidelines.
Peak calling of ChIP-seq data.
Peaks were called using the MACS program version 1.3.6.1 (Zhang et al., 2008) with default parameters, a read length of 36, a genome size of 2,654,911,517 bp (mm9) and the appropriate input control sample. Peaks were filtered for p-values of <10−10. This stringent cutoff efficiently removed false positive (unique) peaks of technical replicas.
Analysis of RNA-seq data.
For analysis of differential gene expression, the RNA-seq samples were cut down to a common read length (30 bp) and aligned to the mouse transcriptome using the TopHat version 1.3.1 (Trapnell et al., 2009). The number of reads per gene was counted using the HTseq version 0.5.3 (http://www-huber.embl.de/users/anders/HTSeq) with the overlap resolution mode option set to “union.” As two independent RNA-seq experiments were available for each cell type shown in Fig. 9 (A–C), the corresponding RNA-seq data were compared by using the R package DESeq version 1.6.1 (Anders and Huber, 2010) to calculate the significance level of differential expression. To this end, the samples were normalized, and the dispersions were estimated using the default DESeq settings. Genes with an adjusted p-value of <0.1 were called as differentially expressed.
Accession nos.
The ChIP-seq and RNA-seq data discussed in this paper are available at the Gene Expression Omnibus (GEO) database under the accession no. GSE35857.
Online supplemental material.
Fig. S1 describes the generation of the Ebf1ihCd2 allele. Fig. S2 describes the generation of the Ebf1fl allele. Fig. S3 describes the generation of the R26CAEbf1 allele. Fig. S4 deals with the binding of EBF1 to it target genes in mature B cells. Fig. S5 documents the FACS sorting and reanalysis of FO and B-1a B cells used for RNA-sequencing. Table S1 shows the sequence coordinates of EBF1 peaks in FO B cells. Table S2 is a description of Solexa sequencing experiments generated for this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20112422/DC1.
Acknowledgments
We thank G. Schmauss and T. Lendl for FACS sorting, C. Theussl and J. Wojciechowski for blastocyst injection, P. Eckerstorfer for help with calcium flux measurements, L. Nitschke for advice on B-1 cell biology, R. Grosschedl for providing the Ebf1+/2 mouse, H. Tagoh for generating the DNase I hypersensitivity data, M. Novatchkova for VH and Vκ sequence analysis and A. Sommer, and his team at the CSF Next Generation Sequencing Unit for Solexa sequencing.
This research was supported by Boehringer Ingelheim, the Austrian GEN-AU initiative (financed by the Bundesminsterium für Bildung und Wissenschaft), the European Union Sixth Framework Programme FP6 (funding the EuTRACC project), and the Austrian Science Fund (grant P21604-B13).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ALP
- all-lymphoid progenitor
- ASC
- antibody-secreting cell
- BAC
- bacterial artificial chromosome
- BCR
- B cell receptor
- BLP
- B-cell–biased lymphoid progenitor
- ChIP
- chromatin immunoprecipitation
- CLP
- common lymphoid progenitor
- EBF1
- early B cell factor 1
- FO
- follicular
- GC
- germinal center
- HSC
- hematopoietic stem cell
- IRES
- internal ribosome entry sequence
- Lin
- lineage
- MPP
- multipotent progenitor
- MZ
- marginal zone
- PI3K
- phosphoinositide 3-kinase
- PNA
- peanut agglutinin
- RPKM
- reads per kilobase of exon per million mapped sequence reads
- RPM
- reads per gene per million mapped sequence reads
- SRBC
- sheep red blood cell
- TBP
- TATA-binding protein
References
- Ahearn J.M., Fischer M.B., Croix D., Goerg S., Ma M., Xia J., Zhou X., Howard R.G., Rothstein T.L., Carroll M.C. 1996. Disruption of the Cr2 locus results in a reduction in B-1a cells and in an impaired B cell response to T-dependent antigen. Immunity. 4:251–262 10.1016/S1074-7613(00)80433-1 [DOI] [PubMed] [Google Scholar]
- Allman D., Pillai S. 2008. Peripheral B cell subsets. Curr. Opin. Immunol. 20:149–157 10.1016/j.coi.2008.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G., Maandag E.C.R., Izon D.J., Amsen D., Kruisbeek A.M., Weintraub B.C., Krop I., Schlissel M.S., Feeney A.J., van Roon M., et al. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 79:885–892 10.1016/0092-8674(94)90077-9 [DOI] [PubMed] [Google Scholar]
- Baumgarth N. 2011. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nat. Rev. Immunol. 11:34–46 10.1038/nri2901 [DOI] [PubMed] [Google Scholar]
- Berland R., Wortis H.H. 2002. Origins and functions of B-1 cells with notes on the role of CD5. Annu. Rev. Immunol. 20:253–300 10.1146/annurev.immunol.20.100301.064833 [DOI] [PubMed] [Google Scholar]
- Blasi F., Carmeliet P. 2002. uPAR: a versatile signalling orchestrator. Nat. Rev. Mol. Cell Biol. 3:932–943 10.1038/nrm977 [DOI] [PubMed] [Google Scholar]
- Brekke K.M., Garrard W.T. 2004. Assembly and analysis of the mouse immunoglobulin kappa gene sequence. Immunogenetics. 56:490–505 10.1007/s00251-004-0659-0 [DOI] [PubMed] [Google Scholar]
- Brooks S.R., Li X., Volanakis E.J., Carter R.H. 2000. Systematic analysis of the role of CD19 cytoplasmic tyrosines in enhancement of activation in Daudi human B cells: clustering of phospholipase C and Vav and of Grb2 and Sos with different CD19 tyrosines. J. Immunol. 164:3123–3131 [DOI] [PubMed] [Google Scholar]
- Brown K.S., Blair D., Reid S.D., Nicholson E.K., Harnett M.M. 2004. FcgammaRIIb-mediated negative regulation of BCR signalling is associated with the recruitment of the MAPkinase-phosphatase, Pac-1, and the 3′-inositol phosphatase, PTEN. Cell. Signal. 16:71–80 10.1016/S0898-6568(03)00113-X [DOI] [PubMed] [Google Scholar]
- Buhl A.M., Cambier J.C. 1999. Phosphorylation of CD19 Y484 and Y515, and linked activation of phosphatidylinositol 3-kinase, are required for B cell antigen receptor-mediated activation of Bruton’s tyrosine kinase. J. Immunol. 162:4438–4446 [PubMed] [Google Scholar]
- Carter J.H., Lefebvre J.M., Wiest D.L., Tourtellotte W.G. 2007. Redundant role for early growth response transcriptional regulators in thymocyte differentiation and survival. J. Immunol. 178:6796–6805 [DOI] [PubMed] [Google Scholar]
- Casola S., Otipoby K.L., Alimzhanov M., Humme S., Uyttersprot N., Kutok J.L., Carroll M.C., Rajewsky K. 2004. B cell receptor signal strength determines B cell fate. Nat. Immunol. 5:317–327 10.1038/ni1036 [DOI] [PubMed] [Google Scholar]
- de Boer J., Williams A., Skavdis G., Harker N., Coles M., Tolaini M., Norton T., Williams K., Roderick K., Potocnik A.J., Kioussis D. 2003. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33:314–325 10.1002/immu.200310005 [DOI] [PubMed] [Google Scholar]
- Decker T., Pasca di Magliano M., McManus S., Sun Q., Bonifer C., Tagoh H., Busslinger M. 2009. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 30:508–520 10.1016/j.immuni.2009.01.012 [DOI] [PubMed] [Google Scholar]
- Fearon D.T., Carroll M.C. 2000. Regulation of B lymphocyte responses to foreign and self-antigens by the CD19/CD21 complex. Annu. Rev. Immunol. 18:393–422 10.1146/annurev.immunol.18.1.393 [DOI] [PubMed] [Google Scholar]
- Fischer M.B., Goerg S., Shen L., Prodeus A.P., Goodnow C.C., Kelsoe G., Carroll M.C. 1998. Dependence of germinal center B cells on expression of CD21/CD35 for survival. Science. 280:582–585 10.1126/science.280.5363.582 [DOI] [PubMed] [Google Scholar]
- Fuxa M., Busslinger M. 2007. Reporter gene insertions reveal a strictly B lymphoid-specific expression pattern of Pax5 in support of its B cell identity function. J. Immunol. 178:8222–8228 [DOI] [PubMed] [Google Scholar]
- Fuxa M., Skok J., Souabni A., Salvagiotto G., Roldán E., Busslinger M. 2004. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 18:411–422 10.1101/gad.291504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman J., Belanger C., Travis A., Turck C.W., Grosschedl R. 1993. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 7:760–773 10.1101/gad.7.5.760 [DOI] [PubMed] [Google Scholar]
- Hao Z., Rajewsky K. 2001. Homeostasis of peripheral B cells in the absence of B cell influx from the bone marrow. J. Exp. Med. 194:1151–1164 10.1084/jem.194.8.1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R.R., Carmack C.E., Shinton S.A., Riblet R.J., Hayakawa K. 1989. A single VH gene is utilized predominantly in anti-BrMRBC hybridomas derived from purified Ly-1 B cells. Definition of the VH11 family. J. Immunol. 142:3643–3651 [PubMed] [Google Scholar]
- Hardy R.R., Kincade P.W., Dorshkind K. 2007. The protean nature of cells in the B lymphocyte lineage. Immunity. 26:703–714 10.1016/j.immuni.2007.05.013 [DOI] [PubMed] [Google Scholar]
- Hobeika E., Thiemann S., Storch B., Jumaa H., Nielsen P.J., Pelanda R., Reth M. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. USA. 103:13789–13794 10.1073/pnas.0605944103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horcher M., Souabni A., Busslinger M. 2001. Pax5/BSAP maintains the identity of B cells in late B lymphopoiesis. Immunity. 14:779–790 10.1016/S1074-7613(01)00153-4 [DOI] [PubMed] [Google Scholar]
- Huntington N.D., Xu Y., Puthalakath H., Light A., Willis S.N., Strasser A., Tarlinton D.M. 2006. CD45 links the B cell receptor with cell survival and is required for the persistence of germinal centers. Nat. Immunol. 7:190–198 10.1038/ni1292 [DOI] [PubMed] [Google Scholar]
- Inlay M.A., Bhattacharya D., Sahoo D., Serwold T., Seita J., Karsunky H., Plevritis S.K., Dill D.L., Weissman I.L. 2009. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 23:2376–2381 10.1101/gad.1836009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida D., Su L., Tamura A., Katayama Y., Kawai Y., Wang S.F., Taniwaki M., Hamazaki Y., Hattori M., Minato N. 2006. Rap1 signal controls B cell receptor repertoire and generation of self-reactive B1a cells. Immunity. 24:417–427 10.1016/j.immuni.2006.02.007 [DOI] [PubMed] [Google Scholar]
- Johnston C.M., Wood A.L., Bolland D.J., Corcoran A.E. 2006. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J. Immunol. 176:4221–4234 [DOI] [PubMed] [Google Scholar]
- Katagiri K., Ohnishi N., Kabashima K., Iyoda T., Takeda N., Shinkai Y., Inaba K., Kinashi T. 2004. Crucial functions of the Rap1 effector molecule RAPL in lymphocyte and dendritic cell trafficking. Nat. Immunol. 5:1045–1051 10.1038/ni1111 [DOI] [PubMed] [Google Scholar]
- Katso R.M., Pardo O.E., Palamidessi A., Franz C.M., Marinov M., De Laurentiis A., Downward J., Scita G., Ridley A.J., Waterfield M.D., Arcaro A. 2006. Phosphoinositide 3-Kinase C2beta regulates cytoskeletal organization and cell migration via Rac-dependent mechanisms. Mol. Biol. Cell. 17:3729–3744 10.1091/mbc.E05-11-1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti T., Ruppelt A., Wang J.H., Liss M., Wagner R., Taskén K., Saksela K. 2007. Reciprocal regulation of SH3 and SH2 domain binding via tyrosine phosphorylation of a common site in CD3epsilon. J. Immunol. 179:878–885 [DOI] [PubMed] [Google Scholar]
- Kwon K., Hutter C., Sun Q., Bilic I., Cobaleda C., Malin S., Busslinger M. 2008. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 28:751–762 10.1016/j.immuni.2008.04.014 [DOI] [PubMed] [Google Scholar]
- Lin H., Grosschedl R. 1995. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 376:263–267 10.1038/376263a0 [DOI] [PubMed] [Google Scholar]
- Lin Y.C., Jhunjhunwala S., Benner C., Heinz S., Welinder E., Mansson R., Sigvardsson M., Hagman J., Espinoza C.A., Dutkowski J., et al. 2010. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 11:635–643 10.1038/ni.1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffucci T., Cooke F.T., Foster F.M., Traer C.J., Fry M.J., Falasca M. 2005. Class II phosphoinositide 3-kinase defines a novel signaling pathway in cell migration. J. Cell Biol. 169:789–799 10.1083/jcb.200408005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H., Ostraat R., Gao H., Fields S., Shinton S.A., Medina K.L., Ikawa T., Murre C., Singh H., Hardy R.R., Hagman J. 2004. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat. Immunol. 5:1069–1077 10.1038/ni1119 [DOI] [PubMed] [Google Scholar]
- Martin F., Kearney J.F. 2002. Marginal-zone B cells. Nat. Rev. Immunol. 2:323–335 10.1038/nri799 [DOI] [PubMed] [Google Scholar]
- Medina K.L., Pongubala J.M., Reddy K.L., Lancki D.W., Dekoter R., Kieslinger M., Grosschedl R., Singh H. 2004. Assembling a gene regulatory network for specification of the B cell fate. Dev. Cell. 7:607–617 10.1016/j.devcel.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Medvedovic J., Ebert A., Tagoh H., Busslinger M. 2011. Pax5: a master regulator of B cell development and leukemogenesis. Adv. Immunol. 111:179–206 10.1016/B978-0-12-385991-4.00005-2 [DOI] [PubMed] [Google Scholar]
- Mikhalap S.V., Shlapatska L.M., Yurchenko O.V., Yurchenko M.Y., Berdova G.G., Nichols K.E., Clark E.A., Sidorenko S.P. 2004. The adaptor protein SH2D1A regulates signaling through CD150 (SLAM) in B cells. Blood. 104:4063–4070 10.1182/blood-2004-04-1273 [DOI] [PubMed] [Google Scholar]
- Miller T., Williams K., Johnstone R.W., Shilatifard A. 2000. Identification, cloning, expression, and biochemical characterization of the testis-specific RNA polymerase II elongation factor ELL3. J. Biol. Chem. 275:32052–32056 10.1074/jbc.M005175200 [DOI] [PubMed] [Google Scholar]
- Molina H., Holers V.M., Li B., Fung Y., Mariathasan S., Goellner J., Strauss-Schoenberger J., Karr R.W., Chaplin D.D. 1996. Markedly impaired humoral immune response in mice deficient in complement receptors 1 and 2. Proc. Natl. Acad. Sci. USA. 93:3357–3361 10.1073/pnas.93.8.3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecino-Rodriguez E., Dorshkind K. 2012. B-1 B cell development in the fetus and adult. Immunity. 36:13–21 10.1016/j.immuni.2011.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecino-Rodriguez E., Leathers H., Dorshkind K. 2006. Identification of a B-1 B cell-specified progenitor. Nat. Immunol. 7:293–301 10.1038/ni1301 [DOI] [PubMed] [Google Scholar]
- Nutt S.L., Kee B.L. 2007. The transcriptional regulation of B cell lineage commitment. Immunity. 26:715–725 10.1016/j.immuni.2007.05.010 [DOI] [PubMed] [Google Scholar]
- Nutt S.L., Urbánek P., Rolink A., Busslinger M. 1997. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 11:476–491 10.1101/gad.11.4.476 [DOI] [PubMed] [Google Scholar]
- O’Riordan M., Grosschedl R. 1999. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 11:21–31 10.1016/S1074-7613(00)80078-3 [DOI] [PubMed] [Google Scholar]
- O’Rourke L.M., Tooze R., Turner M., Sandoval D.M., Carter R.H., Tybulewicz V.L., Fearon D.T. 1998. CD19 as a membrane-anchored adaptor protein of B lymphocytes: costimulation of lipid and protein kinases by recruitment of Vav. Immunity. 8:635–645 10.1016/S1074-7613(00)80568-3 [DOI] [PubMed] [Google Scholar]
- Offenhäuser N., Borgonovo A., Disanza A., Romano P., Ponzanelli I., Iannolo G., Di Fiore P.P., Scita G. 2004. The eps8 family of proteins links growth factor stimulation to actin reorganization generating functional redundancy in the Ras/Rac pathway. Mol. Biol. Cell. 15:91–98 10.1091/mbc.E03-06-0427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhomchuk D., Borodina T., Amstislavskiy V., Banaru M., Hallen L., Krobitsch S., Lehrach H., Soldatov A. 2009. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 37:e123 10.1093/nar/gkp596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar K., Mostoslavsky G., Sattler M., Tai Y.T., Hayashi T., Catley L.P., Hideshima T., Mulligan R.C., Chauhan D., Anderson K.C. 2004. Critical role for hematopoietic cell kinase (Hck)-mediated phosphorylation of Gab1 and Gab2 docking proteins in interleukin 6-induced proliferation and survival of multiple myeloma cells. J. Biol. Chem. 279:21658–21665 10.1074/jbc.M305783200 [DOI] [PubMed] [Google Scholar]
- Pongubala J.M.R., Northrup D.L., Lancki D.W., Medina K.L., Treiber T., Bertolino E., Thomas M., Grosschedl R., Allman D., Singh H. 2008. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat. Immunol. 9:203–215 10.1038/ni1555 [DOI] [PubMed] [Google Scholar]
- Punnonen J., Cocks B.G., Carballido J.M., Bennett B., Peterson D., Aversa G., de Vries J.E. 1997. Soluble and membrane-bound forms of signaling lymphocytic activation molecule (SLAM) induce proliferation and Ig synthesis by activated human B lymphocytes. J. Exp. Med. 185:993–1004 10.1084/jem.185.6.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reininger L., Ollier P., Poncet P., Kaushik A., Jaton J.-C. 1987. Novel V genes encode virtually identical variable regions of six murine monoclonal anti-bromelain-treated red blood cell autoantibodies. J. Immunol. 138:316–323 [PubMed] [Google Scholar]
- Roberts T., Snow E.C. 1999. Cutting edge: recruitment of the CD19/CD21 coreceptor to B cell antigen receptor is required for antigen-mediated expression of Bcl-2 by resting and cycling hen egg lysozyme transgenic B cells. J. Immunol. 162:4377–4380 [PubMed] [Google Scholar]
- Rodríguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25:139–140 10.1038/75973 [DOI] [PubMed] [Google Scholar]
- Roessler S., Györy I., Imhof S., Spivakov M., Williams R.R., Busslinger M., Fisher A.G., Grosschedl R. 2007. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol. Cell. Biol. 27:579–594 10.1128/MCB.01192-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safford M., Collins S., Lutz M.A., Allen A., Huang C.T., Kowalski J., Blackford A., Horton M.R., Drake C., Schwartz R.H., Powell J.D. 2005. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat. Immunol. 6:472–480 10.1038/ni1193 [DOI] [PubMed] [Google Scholar]
- Scharenberg A.M., Humphries L.A., Rawlings D.J. 2007. Calcium signalling and cell-fate choice in B cells. Nat. Rev. Immunol. 7:778–789 10.1038/nri2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schebesta A., McManus S., Salvagiotto G., Delogu A., Busslinger G.A., Busslinger M. 2007. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 27:49–63 10.1016/j.immuni.2007.05.019 [DOI] [PubMed] [Google Scholar]
- Sebzda E., Bracke M., Tugal T., Hogg N., Cantrell D.A. 2002. Rap1A positively regulates T cells via integrin activation rather than inhibiting lymphocyte signaling. Nat. Immunol. 3:251–258 10.1038/ni765 [DOI] [PubMed] [Google Scholar]
- Seet C.S., Brumbaugh R.L., Kee B.L. 2004. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J. Exp. Med. 199:1689–1700 10.1084/jem.20032202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl K.J., MacKenzie J.D., Wang D., Kantor A.B., Kabat E.A., Herzenberg L.A., Herzenberg L.A. 1997. Frequent occurrence of identical heavy and light chain Ig rearrangements. Int. Immunol. 9:689–702 10.1093/intimm/9.5.689 [DOI] [PubMed] [Google Scholar]
- Sigvardsson M., O’Riordan M., Grosschedl R. 1997. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 7:25–36 10.1016/S1074-7613(00)80507-5 [DOI] [PubMed] [Google Scholar]
- Smith K.G., Light A., Nossal G.J., Tarlinton D.M. 1997. The extent of affinity maturation differs between the memory and antibody-forming cell compartments in the primary immune response. EMBO J. 16:2996–3006 10.1093/emboj/16.11.2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.M., Gisler R., Sigvardsson M. 2002. Cloning and characterization of a promoter flanking the early B cell factor (EBF) gene indicates roles for E-proteins and autoregulation in the control of EBF expression. J. Immunol. 169:261–270 [DOI] [PubMed] [Google Scholar]
- Smith E., Lin C., Shilatifard A. 2011. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 25:661–672 10.1101/gad.2015411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souabni A., Cobaleda C., Schebesta M., Busslinger M. 2002. Pax5 promotes B lymphopoiesis and blocks T cell development by repressing Notch1. Immunity. 17:781–793 10.1016/S1074-7613(02)00472-7 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Okuno T., Yamamoto M., Pasterkamp R.J., Takegahara N., Takamatsu H., Kitao T., Takagi J., Rennert P.D., Kolodkin A.L., et al. 2007. Semaphorin 7A initiates T-cell-mediated inflammatory responses through alpha1beta1 integrin. Nature. 446:680–684 10.1038/nature05652 [DOI] [PubMed] [Google Scholar]
- Tallquist M.D., Soriano P. 2000. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 26:113–115 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25:1105–1111 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber T., Mandel E.M., Pott S., Györy I., Firner S., Liu E.T., Grosschedl R. 2010. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 32:714–725 10.1016/j.immuni.2010.04.013 [DOI] [PubMed] [Google Scholar]
- Tuveson D.A., Carter R.H., Soltoff S.P., Fearon D.T. 1993. CD19 of B cells as a surrogate kinase insert region to bind phosphatidylinositol 3-kinase. Science. 260:986–989 10.1126/science.7684160 [DOI] [PubMed] [Google Scholar]
- Xi H., Kersh G.J. 2004. Early growth response gene 3 regulates thymocyte proliferation during the transition from CD4-CD8- to CD4+CD8+. J. Immunol. 172:964–971 [DOI] [PubMed] [Google Scholar]
- Yurchenko M.Y., Kovalevska L.M., Shlapatska L.M., Berdova G.G., Clark E.A., Sidorenko S.P. 2010. CD150 regulates JNK1/2 activation in normal and Hodgkin’s lymphoma B cells. Immunol. Cell Biol. 88:565–574 10.1038/icb.2010.14 [DOI] [PubMed] [Google Scholar]
- Zandi S., Mansson R., Tsapogas P., Zetterblad J., Bryder D., Sigvardsson M. 2008. EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J. Immunol. 181:3364–3372 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Cotta C.V., Stephan R.P., deGuzman C.G., Klug C.A. 2003. Enforced expression of EBF in hematopoietic stem cells restricts lymphopoiesis to the B cell lineage. EMBO J. 22:4759–4769 10.1093/emboj/cdg464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Muller M., Chen C.H., Zeng L., Farooq A., Zhou M.M. 2005. New insights into the catalytic activation of the MAPK phosphatase PAC-1 induced by its substrate MAPK ERK2 binding. J. Mol. Biol. 354:777–788 10.1016/j.jmb.2005.10.006 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]