Triple-negative breast cancers with elevated MYC are sensitized to CDK inhibition.
Abstract
Estrogen, progesterone, and HER2 receptor-negative triple-negative breast cancers encompass the most clinically challenging subtype for which targeted therapeutics are lacking. We find that triple-negative tumors exhibit elevated MYC expression, as well as altered expression of MYC regulatory genes, resulting in increased activity of the MYC pathway. In primary breast tumors, MYC signaling did not predict response to neoadjuvant chemotherapy but was associated with poor prognosis. We exploit the increased MYC expression found in triple-negative breast cancers by using a synthetic-lethal approach dependent on cyclin-dependent kinase (CDK) inhibition. CDK inhibition effectively induced tumor regression in triple-negative tumor xenografts. The proapoptotic BCL-2 family member BIM is up-regulated after CDK inhibition and contributes to this synthetic-lethal mechanism. These results indicate that aggressive breast tumors with elevated MYC are uniquely sensitive to CDK inhibitors.
Clinically relevant biomarkers used to guide the use of targeted therapeutics for breast cancer include the overexpression of HER2 (human epidermal growth factor receptor 2) and the expression of the estrogen and progesterone receptors. For breast tumors that are positive for these receptors (receptor positive), several targeted therapeutic strategies have been successfully developed in the past decades. These include the use of small molecule kinase inhibitors, treatment with inhibitory monoclonal antibodies, and antihormonal therapies. Unfortunately, no such biomarker to predict response to selective therapeutics has been established for the most challenging receptor triple-negative subtype of breast cancer (Carey et al., 2006; Bauer et al., 2007; Liedtke et al., 2008). Clearly, further investigation of the biology of triple-negative breast cancer is required if effective therapies are to be developed (Irvin and Carey, 2008; Schneider et al., 2008).
Gene expression profiling of human primary breast tumors has identified several distinct molecular subtypes including luminal A and B, HER2+, basal-like, and normal-like (Perou et al., 2000; Sørlie et al., 2001). Approximately 70% of triple-negative tumors belong to the basal subtype (Bertucci et al., 2008), which often exhibits aggressive characteristics such as poor differentiation, a higher rate of proliferation, and increased metastatic capability (Livasy et al., 2006; Sarrió et al., 2008). In clinical studies, patients with triple-negative tumors have been found to respond to neoadjuvant chemotherapy with equal or better efficacy than those with receptor-positive tumors (Carey et al., 2007; Liedtke et al., 2008), presumably as a result of the higher mitotic index observed in triple-negative tumors. However, a complete pathological response is rarely achieved in patients with triple-negative tumors, who have a tendency to experience early relapse and a diminished 5-yr disease-free survival (Bauer et al., 2007; Dent et al., 2007). The molecular events that occur in triple-negative breast cancer have not been elucidated and, therefore, the mechanism for the poor prognosis of this subtype remains unclear. Thus, there is significant interest in identifying signaling pathways that distinguish triple-negative breast cancer from other breast cancer subtypes.
Several converging studies have suggested that the MYC proto-oncogene may play an important function in aggressive breast cancers. MYC is a basic helix-loop-helix zipper (bHLHZ) motif–containing transcription factor whose activity is tightly regulated by its direct binding to another bHLHZ protein MAX. MYC activation can lead to transcriptional activation or repression of specific genes (Eilers and Eisenman, 2008). The global transcriptional influence of MYC is also mediated through a MYC regulatory network whereby MYC activity is precisely controlled by the activity of multiple competing repressive MAX binding partners (i.e., MAD, MGA, MXD4, and MNT; Grandori et al., 2000; Cowling and Cole, 2006). MYC plays roles in multiple signaling pathways including those involved in cell growth, cell proliferation, metabolism, microRNA regulation, cell death, and cell survival (Dang, 1999; Eilers and Eisenman, 2008; Meyer and Penn, 2008). Furthermore, MYC signaling has recently been shown to be up-regulated in high-grade mammary tumors with presumptive cancer stem cell properties (Ben-Porath et al., 2008; Wong et al., 2008).
The genomic locus, 8q24, which harbors the MYC oncogene, is among the most frequently amplified region in breast cancers of various subtypes (Jain et al., 2001). The amplified region, however, contains numerous transcripts and, therefore, amplification is not strictly correlated with elevated MYC expression. More recent studies have identified a MYC transcriptional gene signature associated with the basal molecular subtype (Alles et al., 2009; Chandriani et al., 2009; Gatza et al., 2010). Other studies have examined staining of primary breast tumor tissues for MYC protein expression and did not find a clear connection between MYC overexpression and patient outcome (Bland et al., 1995; Naidu et al., 2002). Thus, evaluating the contribution of MYC signaling to triple-negative breast cancer would open up new mechanistic insights and potentially new therapeutic approaches to treat this aggressive tumor type.
Several outstanding questions about the relevance of MYC signaling in triple-negative breast cancer remain unresolved. Is MYC expression alone altered in triple-negative tumors or is the expression of other MYC signaling components (i.e., competing MAX binding proteins) also changed? Furthermore, does MYC signaling alter response to conventional chemotherapeutics that are routinely used to treat triple-negative breast cancer? Finally, if MYC signaling is up-regulated in triple-negative tumors, can one take advantage of this feature to develop new targeted therapeutics? In this study, we use several complementary approaches to address these questions.
Synthetic lethality has been proposed as a treatment strategy against cancers that lack obvious druggable targets (Reinhardt et al., 2009). As an example, small molecule inhibition of poly (ADP-ribose) polymerase (PARP) in patients with BRCA1/2 mutations has provided a proof of concept for exploiting synthetic lethality in the clinic (Fong et al., 2009; Tutt et al., 2010). Our previous work found a synthetic-lethal interaction between MYC overexpression and inhibition of the mitotic cyclin-dependent kinase (CDK) 1 by using transgenic MYC-driven models of lymphoma and liver cancer (Goga et al., 2007). However, whether this synthetic-lethal approach has utility in treating triple-negative breast cancers remains unknown.
In this study, we investigated whether exploiting a synthetic-lethal approach dependent on elevated MYC signaling is effective in treating triple-negative breast cancer. To gain a comprehensive understanding for the role of MYC in triple-negative breast cancer, in the current study we combine gene expression and protein analysis from patient tumors with functional studies in cell lines and tumor models. Here, we examine the efficacy of a CDK inhibitor currently in clinical development against triple-negative breast cancer and investigate the mechanism of synthetic lethality.
RESULTS
MYC expression is disproportionally elevated in triple-negative breast cancer
We sought to understand whether MYC signaling is an important oncogenic event in triple-negative breast tumors. The I-SPY TRIAL (Investigation of Serial Studies to Predict Your Therapeutic Response with Imaging and Molecular Analysis) is an NCI-sponsored multi-institutional study of Stage II and III breast cancer patients to identify diagnostic markers, validate hypothesis, and develop new treatment strategies against breast cancer (Barker et al., 2009; Jones, 2010; Esserman et al., 2012). This clinical trial collects extensive clinical annotation and global mRNA expression from primary tumor samples. Using mRNA expression data from 146 primary patient tumor samples for which hormone receptor and HER2 expression were available, we found that MYC mRNA levels are significantly elevated in triple-negative tumors (Fig. 1 A). We next examined MYC protein and phospho-MYC (T58/S62) expression in 208 independent primary breast tumor samples from a separate cohort using reverse-phase quantitative protein arrays. The 66 triple-negative tumors expressed significantly elevated MYC protein and phospho-MYC (Fig. 1 B) compared with the 142 tumors that were receptor positive. MYC phosphorylation at these sites has been shown to enhance MYC transcriptional activity as well as alter protein stability (Chang et al., 2000; Sears et al., 2000; Seo et al., 2008). Finally, using previously published mRNA expression profiling data (Neve et al., 2006), we examined whether MYC expression in 50 established human breast cell lines correlated with the receptor status. These 50 human breast cell lines have been shown to accurately model aspects of breast cancer biology (Neve et al., 2006) and have been extensively used for mechanistic studies. Compared with cell lines that are not triple negative, triple-negative cell lines had substantially elevated MYC mRNA (Fig. 1 C). Collectively, these three independent datasets confirm the association between elevated MYC expression and triple-negative tumors.
Figure 1.
Elevated MYC expression in human triple-negative cancers. (A) MYC mRNA expression in triple-negative versus receptor-positive primary breast tumors collected through the I-SPY TRIAL (P < 0.0001). (B) MYC and phospho-MYC (T58/S62) protein expression in triple-negative primary tumors. Shown is an independent cohort of 208 patients for which quantitative reverse-phase protein arrays were performed. (C) Relative expression of MYC mRNA in a panel of established human breast cell lines (Neve et al., 2006). The error bars represent means ± SEM. P-values were calculated by two-tailed Student’s t test.
MYC signaling is up-regulated in triple-negative tumors
To evaluate MYC activity in primary tumors, we applied a previously validated MYC-regulated transcriptional signature (Chandriani et al., 2009), which is comprised of 352 genes, to the I-SPY dataset. We ordered the patient tumor samples by the Pearson correlation to the MYC gene expression signature (Fig. 2 A), and tumors were divided into high, intermediate, and low correlation groups (see Materials and methods). Triple-negative breast tumors were significantly enriched in the high MYC gene expression group (P < 0.005; Fig. 2 A). These results indicate that a disproportionate number of primary triple-negative breast tumors exhibit elevated MYC function. A MYC gene expression signature has previously been correlated with a basal molecular subtype of breast cancer (Alles et al., 2009; Chandriani et al., 2009; Gatza et al., 2010), which encompasses ∼70% of triple-negative cancers. Whether MYC signaling is also increased in other molecular subtypes of human breast cancer remains unclear. We therefore examined the MYC gene expression signature across different molecular subtypes of breast cancer in the I-SPY dataset. Consistent with prior studies, we found that a high MYC gene expression signature correlated most strongly with the basal subtype; however, we also observed that the luminal B molecular subtype had significantly higher MYC signature than luminal A or HER2 subtypes (Fig. 2 B). Although most luminal B tumors express hormone receptors, this subtype is often associated with increased tumor proliferation and worse outcome than luminal A (Voduc et al., 2010). Our results indicate that along with basal, the luminal B molecular subtype is also associated with increased MYC signaling, which may, at least in part, explain the worse outcome for this tumor type (Cheang et al., 2009).
Figure 2.
Elevated MYC signaling in human triple-negative breast cancers. (A) 149 primary tumors ordered by each tumor’s Pearson correlation to a 352 gene MYC signature centroid (P < 0.005; Fisher’s exact test). Triple-negative tumor samples are indicated with a red dot, whereas molecular subtypes are indicated with colored bars. (B) Correlation between MYC gene expression signature and breast cancer molecular subtypes. Relative MYC gene expression was based on each tumor’s Pearson correlation to the MYC gene signature. The error bars represent means ± SEM. P-values were calculated by two-tailed Student’s t test for each comparison. (C) Expression of multiple genes within the MYC signaling pathway is altered in triple-negative breast cancers. Schema shows various MAX-interacting genes that are deregulated and can positively or negatively modulate MYC transcriptional activity. Genes are shaded green if expression is suppressed in triple-negative versus receptor-positive tumors, and red if expression is increased in triple-negative tumors. MAX, P = 0.02; MYC, P < 0.0001; MYCN, P = 0.06; MXD4, P = 0.03 (two-tailed Student’s t test).
We next sought to understand the molecular basis for the increased MYC activity in triple-negative breast cancers. Activation or repression of MYC target genes involves multiple signaling complexes comprised of MAX bound to an activator, such as MYC or MYCN, or bound to a repressor such as MXD1-4 or MAX (Grandori et al., 2000; Cowling and Cole, 2006). Thus, MYC/MAX heteromeric complexes can activate transcription, whereas MXD/MAX or MAX/MAX complexes can suppress activation of MYC target genes (Eilers and Eisenman, 2008). Changes in MYC pathway activity in breast cancer are therefore caused by alterations in the architecture of the MYC regulatory network through changing the relative abundance of MAX interacting partners. To our knowledge, the topology of the MYC regulatory network in primary breast tumors has not been previously described. Therefore, we sought to determine if, besides MYC, the mRNA expression of other MAX-interacting genes is also significantly altered in triple-negative primary tumors. Analyzing the mRNA array data available from the I-SPY dataset, we found that multiple MAX-interacting genes are altered in triple-negative tumors (Fig. 2 C). In addition to MYC, we found that the activator MYCN is also up-regulated in triple-negative tumors, whereas the repressor molecules MXD4 and MAX were significantly down-regulated (Fig. 2 C). Thus, the enhanced MYC activity in triple-negative tumors may be a result not only of MYC up-regulation but also of alterations in the expression of other MYC regulatory genes.
Up-regulation of MYC signaling is associated with poor prognosis
We next examined the clinical outcome of patients based on their MYC gene signature in the neoadjuvant I-SPY TRIAL. We found that an increased MYC gene signature was associated with significantly shortened disease-free survival with a median follow-up of 3.9 yr (P = 0.005; Fig. 3 A). Surprisingly, we found that the response of primary tumors at the time of surgery, immediately after the completion of conventional neoadjuvant chemotherapy, did not differ significantly based on MYC signature scores (Fig. 3 B). To further examine the association between MYC signaling and response to neoadjuvant chemotherapy, we divided the total patient population into two categories. One group was composed of those patients who exhibited either complete response or minimal residual cancer burden (RCB 0/I; Symmans et al., 2007) after conventional chemotherapy. The other group was composed of those with substantial residual disease (RCB II/III). For patients who had a dramatic response to neoadjuvant chemotherapy (RCB 0/I), increased MYC signaling did not significantly alter prognosis (Fig. 3 C). In contrast, for those patients whose tumors had only minimal response to neoadjuvant chemotherapy (RCB II/III), an increased MYC signature was associated with early disease recurrence (Fig. 3 D; P = 0.001).
Figure 3.
Elevated MYC signaling is associated with poor outcome. (A) Overall risk of breast cancer recurrence in patients with varying levels of MYC pathway activation. I-SPY TRIAL patients (n = 149) were divided into tertiles based on the relative expression of the genes in the MYC signature in their pretreatment biopsy samples. Differences in risk of disease recurrence between these groups were assessed using a Cox proportional hazards model and Wald’s test. (B) Correlation of MYC pathway activation with tumor burden after neoadjuvant chemotherapy. The MYC pathway activation tertile for the I-SPY TRIAL patients with gene expression data, and for which RCB (0-III; Symmans et al., 2007) was determined at the time of surgery (n = 133), was examined. The association between residual tumor burden and MYC pathway activation tertile was assessed using Fisher’s exact test. (C) Disease recurrence by MYC signature in RCB 0/I patients. (D) Disease recurrence by MYC signature in RCB II/III patients. (E) Multivariate analysis considering receptor status and MYC pathway activation as a continuous variable.
Although there was a strong association between elevated MYC expression and triple-negative breast cancer (Figs. 1 and 2), there also appeared to be considerable heterogeneity with regard to receptor status and MYC pathway activation (Fig. 2 A), leading us to explore whether the poor outcome associated with MYC pathway activation (univariate Cox proportional hazards ratio = 19.0 for MYC pathway activation as a continuous measure; 95% confidence interval [CI]: 4.4–82.3; P < 0.001; n = 146) reflects the aggressiveness of triple-negative disease (univariate Cox proportional hazards ratio = 2.2 for triple-negative tumors relative to receptor-positive tumors; 95% CI: 1.2–4.0; P = 0.015; n = 146) or whether MYC pathway activation might be an independent predictor of poor prognosis. In multivariate models that consider both receptor status and MYC pathway activation, the association between receptor status and outcome is considerably diminished (hazard ratio [HR] = 1.6; 95% CI: 0.8–3.0; P = 0.176; n = 146), whereas MYC pathway activation and outcome remain significantly associated (HR = 14.2; 95% CI: 3.1–64.4; P < 0.001; n = 146; Fig. 3 E). These results suggest that MYC pathway activation contains additional information about risk of recurrence that is not reflected in receptor status alone.
Triple-negative breast cells with elevated MYC expression are sensitive to CDK inhibition
Given the poor outcome of patients that have tumors with elevated MYC activity, we sought to identify a therapeutic strategy that could target these tumors. Synthetic lethality between MYC overexpression and the inhibition of the mitotic kinase CDK1 has previously been observed in engineered cells and transgenic mouse models (Goga et al., 2007). However, this synthetic lethality has not been examined in breast cancer. Purvalanol A, an experimental small molecule CDK inhibitor that has higher specificity toward CDK1 (Gray et al., 1998), induced apoptosis in lymphoma cells and hepatocytes engineered to overexpress MYC (Goga et al., 2007). Importantly, CDK1 inhibition has been found to have little effect on the viability of control cells or normal mouse tissues (Goga et al., 2007).
We reasoned that triple-negative breast cancer cells that have arisen to overexpress MYC might also be sensitive to a synthetic-lethal interaction with CDK inhibition. To model our observations from the I-SPY clinical samples, we tested a panel of triple-negative cell lines with elevated MYC expression and a panel of receptor-positive lines that were expected to have lower MYC protein expression based on previously published mRNA data (Neve et al., 2006) for their sensitivity to CDK inhibitors (Fig. 4 A). In these experiments, we tested two CDK inhibitors: purvalanol A, and another CDK inhibitor dinaciclib (Parry et al., 2010) that is currently in phase II clinical trials against various tumor types (Dickson and Schwartz, 2009). Dinaciclib inhibits CDK1, 2, 5, and 9 at concentrations of 1–5 nM, which can be readily achieved in vivo, and exhibits improved pharmacokinetic (PK) and pharmacodynamic (PD) properties, compared with other CDK inhibitors previously evaluated in clinical trials.
Figure 4.
Elevated MYC expression sensitizes triple-negative cancers to CDK inhibition. (A) A panel of triple-negative as well as receptor-positive breast cells, together with a matched pair of nontumorigenic model epithelial cells (RPE cells) engineered to overexpress MYC, was treated with CDK inhibitors purvalanol A (10 µM) or dinaciclib (10 nM) for 72 h and subjected to viability assay. The dashed line indicates the relative starting cell number at the time of adding CDK inhibitors (time 0). Positive numbers indicate cell growth and negative numbers indicate cell death. The experiment was independently repeated five times. The error bars represent means ± SEM. P-values were calculated by two-tailed Student’s t test for comparisons of cell lines treated with each of the two inhibitors. Western blots showing MYC and actin protein expression from the indicated cell lines are shown. (B) Cell cycle profiles of three triple-negative cell lines and three receptor-positive cell lines after treatment with 10 µM purvalanol A or 10 nM dinaciclib for 72 h. The percentage of cells in G1 and G2-M phases of the cell cycle, as determined by DNA content based on propidium iodide staining, is indicated. (C) A panel of triple-negative, as well as receptor-positive, breast cancer cells was treated with siRNA against CDK1 or CDK2 for 72 h and assessed for cell viability. The experiment was independently repeated three times. The error bars represent means ± SEM. P-values were calculated by two-tailed Student’s t test. Western blots showing CDK1, CDK2, and actin protein expression are shown.
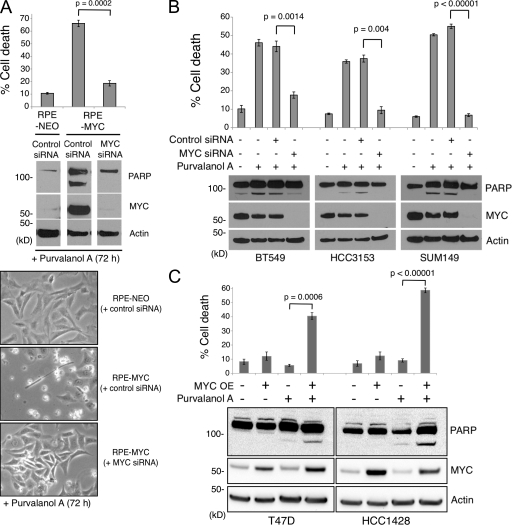
Breast cell lines were treated with 10 µM purvalanol A or 10 nM dinaciclib, respectively, and were subjected to a cell viability assay. These chosen concentrations induce cell cycle arrest without causing cell death in normal human epithelial cells (Fig. 4 A; Goga et al., 2007). Human epithelial cells engineered to overexpress MYC (RPE-MYC) undergo cell death when treated with purvalanol A, whereas those with endogenous levels of MYC expression (RPE-NEO) do not, serving as positive and negative controls, respectively (Fig. 4 A; Goga et al., 2007). Dinaciclib exhibited ∼1,000-fold higher potency compared with purvalanol A in inducing apoptosis in epithelial cells engineered to overexpress MYC but not in control RPE-NEO cells (Fig. 4 A). In breast cell lines, CDK inhibitor treatment induced substantial cell death in each of the five triple-negative cell lines tested (range 25–65%), whereas most of the receptor-positive lines showed resistance to such treatment (Fig. 4 A). One receptor-positive line, SKBR3, was sensitive to these inhibitors (eliciting >25% cell death); however, this may be the result of increased MYC expression found in these cells (Fig. 4 A), which was not predicted from previously published mRNA profiling data (Neve et al., 2006). Cell cycle analysis by flow cytometry showed that treatment of both triple-negative and receptor-positive breast cancer cell lines with purvalanol A induced an increased accumulation of G2-M arrested cells, whereas treatment with dinaciclib resulted in cells arrested in both G1-S and G2-M (Fig. 4 B). This is consistent with results showing that purvalanol A is highly selective for CDK1 (Gray et al., 1998), whereas dinaciclib targets both CDK1 and CDK2 with similar potency (Parry et al., 2010).
Although these small molecule CDK inhibitors are selective for either CDK1 (purvalanol A) or for a few CDKs (dinaciclib), it is possible that other non-CDK kinases may also be inhibited and thus may contribute to the overall cell death phenotypes. To address this issue of specificity and further investigate synthetic lethality between MYC overexpression and CDK inhibition, we performed siRNA experiments to knock down specific CDKs in a panel of triple-negative as well as receptor-positive cell lines (Fig. 4 C). We found that the treatment of all three triple-negative cell lines with CDK1 siRNA for 72 h resulted in a significant amount of cell death, whereas the viability of receptor-positive lines was only modestly affected (Fig. 4 C). Interestingly, we found that CDK2 siRNA could also induce cell death, but to a lesser extent in two of the triple-negative lines and in one of the receptor-positive lines, which is in agreement with recent observations that CDK2 function may be essential for the viability of certain cancer types (Molenaar et al., 2009). The higher potency of cell killing induced by dinaciclib may be, in part, a result of its ability to inhibit both CDK1 and CDK2 (Parry et al., 2010). Unexpectedly, we found that CDK1 siRNA treatment dramatically increased CDK2 protein expression in all of the three triple-negative lines and one of the receptor-positive cell lines, whereas CDK2 siRNA did not alter CDK1 protein expression (Fig. 4 C). It remains to be determined whether this CDK2 up-regulation indicates a compensatory mechanism within the cell cycle encountering the loss of the only mitotic CDK.
One possible explanation for the sensitivity of breast tumor cells with high MYC to CDK inhibition is that they are more proliferative. One would expect these cells to therefore also be more sensitive to other chemotherapeutics that target cell proliferation mechanisms. To investigate this possibility, we tested the sensitivity of the breast lines used in this study to the chemotherapeutic agents paclitaxel and doxorubicin (Table 1) that are commonly used in clinical practice to treat breast cancer, including in the I-SPY TRIAL. The cell lines used in this study did not show any MYC expression-specific sensitivity to the chemotherapeutic agents. These cell-based studies are consistent with the observations from the I-SPY TRIAL that a MYC signature was not associated with improved response of primary tumors to conventional chemotherapeutic agents. It is therefore likely that other functions of MYC, in addition to its role in cell proliferation, contribute to the mechanism of synthetic lethality observed.
Table 1.
Sensitivity (LC50) of human breast cell lines to chemotherapeutic drugs
| Cell line | Receptor status | Elevated MYC | Paclitaxel | Doxorubicin |
| µM | µM | |||
| MDA-MB-231 | TN | Yes | 0.82 | 0.73 |
| HCC3153 | TN | Yes | 0.15 | >5.0 |
| BT549 | TN | Yes | 0.14 | 0.61 |
| MCF10AMYC | TN | Yes | 0.21 | 0.77 |
| SUM149 | TN | Yes | 0.67 | 4.6 |
| HCC1428 | RP | No | >5.0 | 0.72 |
| UACC812 | RP | No | 1.3 | 0.85 |
| T47D | RP | No | 0.06 | 0.37 |
| LY2 | RP | No | >5.0 | 0.55 |
| SKBR3 | RP | Intermediate | 0.05 | 0.91 |
MYC protein expression by quantitative Western blotting in triple-negative (TN) and receptor-positive (RP) cell lines is indicated. No correlation between elevated MYC expression and sensitivity to paclitaxel or doxorubicin was observed (two-tailed Student’s t test).
We next asked whether the observed CDK inhibition-induced cell death in cells with elevated MYC expression is dependent on their MYC status. To address this question, we first used an RNAi approach to knock down MYC protein expression. We found that pretreatment of RPE-MYC cells with MYC-specific siRNA significantly reduced the extent of CDK inhibition-dependent cell death (Fig. 5 A). Similarly, the sensitivities of three triple-negative cell lines treated with MYC siRNA before the addition of purvalanol A were greatly reduced (Fig. 5 B). We next used a lentiviral transduction method to overexpress MYC in receptor-positive cells to examine whether MYC overexpression alone is sufficient to render these otherwise resistant cells sensitive to CDK inhibition. We found that purvalanol A treatment of T47D and HCC1428 cells engineered to overexpress MYC resulted in significantly increased cell death (Fig. 5 C). Thus, these results suggest that elevated MYC expression is necessary and it increases the sensitivity of breast cancer cells to CDK inhibitors. Interestingly, whereas MYC RNAi demonstrated a dramatic decrease in MYC protein expression (Fig. 5 B), it did not appreciably alter cell viability (not depicted).
Figure 5.
CDK inhibitor induced cell death is MYC dependent. (A) MYC dependency of cell death induced by CDK inhibition in RPE cells. RPE-MYC cells were first treated with either MYC siRNA or control nonspecific siRNA for 24 h, and then treated with purvalanol A for 72 h. The collected cells were analyzed for cell viability by Guava ViaCount assay and for PARP activation by Western blotting. The experiments were repeated three times in triplicate. The error bars represent means ± SEM. P-values were calculated by two-tailed Student’s t test. (B) siRNA-mediated MYC knockdown in triple-negative cells undergoing purvalanol A treatment. The experiment was independently repeated three times. The error bars represent means ± SEM. P-values were calculated by two-tailed Student’s t test. (C) Receptor-positive cell lines T47D and HCC1428, engineered to overexpress MYC and exposed for 72 h to purvalanol A treatment. The experiments were repeated three times in triplicate. The error bars represent means ± SEM. P-values were calculated by two-tailed Student’s t test.
CDK inhibition induces cell death in triple-negative cells in a three-dimensional (3D) culture system
Although we saw sensitivity of triple-negative cells to CDK inhibition in monolayer cultures (Fig. 4), there is evidence that tumor cells can differ in their response to anti-cancer agents between in vitro and in vivo conditions (Weaver et al., 2002). As a step toward addressing this question, we initially examined the effect of CDK inhibition on cell viability in a 3D Matrigel culture system. 3D epithelial cultures have been used extensively as an in vitro system to more closely approximate the physiological microenvironment of the epithelial cells (Lee et al., 2007). Indeed, it has been shown that a 3D gene signature can more accurately predict outcome in several independent datasets (Fournier et al., 2006; Martin et al., 2008). Two receptor-positive cell lines (T47D and HCC1428) and two triple-negative cell lines (SUM149 and BT549) were seeded onto a solid Matrigel matrix, allowed to form colonies, and subsequently treated with 10 µM purvalanol or 10 nM dinaciclib and allowed to grow for 6 d. In this assay, if the cells undergo cell death, the mean colony size decreases between the initial day and the last day of the treatment incubation time (day 0 and day 6). However, if the cells are insensitive to CDK inhibition-induced cell death, the size of the colonies remains the same in the case of cell cycle arrest or may increase. We found that the mean colony size in the two triple-negative cell lines significantly decreased over a 6-d period after purvalanol A or dinaciclib treatment (Fig. 6, A and B), indicating that the cells in these cultures underwent extensive cell death. In contrast, the colonies formed by the two receptor-positive cell lines did not significantly change (Fig. 6, A and B), suggesting the induction of cell cycle arrest. As positive controls, DMSO-treated cultures for all the four cell lines showed robust growth at day 6 (Fig. 6, A and B). Thus, consistent with what we observed in 2D cultures, receptor-positive cells expressing low MYC appear to be resistant to cell death, whereas triple-negative cells with elevated MYC expression are particularly sensitive to CDK inhibition in 3D cultures.
Figure 6.
CDK inhibition decreases colony size in triple-negative cancer cells in 3D Matrigel matrices. (A) Low- and high-magnification images of 3D Matrigel cultures of T47D, HCC1428, BT549, and SUM149 cell lines at day 0 of treatment and day 6 of DMSO-treated, purvalanol A–treated, and dinaciclib-treated cultures. Nonrefractile dark particles indicated the presence of cell death. Bars, 10 µm. (B) Quantification of colony size of 3D Matrigel cultures. Low-magnification images of Matrigel cultures were obtained, and the mean size of the colonies in each culture was determined both at the initial day of treatment (day 0) and at the final day of treatment (day 6). The mean colony size at day 0 was set to 100% for each cell line and the colony size at day 6 was compared with the day 0 size. The experiment was independently repeated three times. The error bars represent means ± SEM. P-values were calculated by two-tailed Student’s t test.
CDK inhibition induces in vivo tumor regression in mouse xenograft models of triple-negative breast cancer
A stringent test of any therapeutic strategy is the ability to inhibit or regress in vivo tumor growth. Using xenograft transplant models of breast cancer, we examined the in vivo efficacy of inhibiting CDKs in human triple-negative tumors with elevated MYC expression. We focused on dinaciclib because of its higher potency and improved PK properties compared with purvalanol A. We attempted to generate tumor xenografts of three triple-negative and three receptor-positive human breast cancer cell lines. Tumor cells were transplanted into BALB/c Nu/Nu mice and were allowed to form measurable tumors (200–250 mm3 in volume). Two triple-negative cell lines with elevated MYC expression, MDA-MB-231 and HCC3153, formed tumors with high penetrance and were used for subsequent studies. Our attempts to grow tumors of receptor-positive cells were not successful either because of the inability to form tumors or because only small sporadic tumors formed, despite our efforts to increase tumor engraftment by using Matrigel.
The triple-negative tumor-bearing mice were then treated twice weekly for 2 wk with dinaciclib (50 mg/kg/dose i.p.) or vehicle control. The mice treated with diluent experienced a ∼120–150% increase in growth of tumors during the experimental period (Fig. 7, A and B). In contrast, dinaciclib-treated mice had a dramatic response with ∼50% tumor regression (Fig. 7, A and B). In independent experiments, we also examined the effects of dinaciclib on the established MDA-MB-231 and HCC3153 xenografts 24 h after treatment. We found a dramatic decrease in the overall phosphorylation levels of presumptive CDK substrates that have the consensus CDK motif, as well as in the phosphorylation of a validated CDK1 substrate PP1-α (protein phosphatase 1–α; T320; Dohadwala et al., 1994; Blethrow et al., 2008) in primary tumors (Fig. 7 C). This was accompanied by the induction of apoptosis as demonstrated by PARP cleavage in tumor tissues (Fig. 7 C). Collectively, these results demonstrate that small molecule inhibition of CDKs represents a novel and feasible treatment strategy against human triple-negative breast cancers.
Figure 7.
CDK inhibition is effective in treating xenografted triple-negative tumors in mice. (A) Tumor growth in mouse xenograft models of triple-negative breast cancer. Representative photos of tumors after 2 wk of treatment with either vehicle alone or with dinaciclib (50 mg/kg i.p. twice weekly) are shown. (B) Growth of triple-negative tumors in nude mice treated with the CDK inhibitor dinaciclib (50 mg/kg i.p. twice weekly) for 2 wk. Each treatment group per tumor cell type contained the indicated number of mice. The error bars represent means ± SEM. P-values were calculated by two-tailed Student’s t test. (C) PARP cleavage and serine phosphorylation of presumptive CDK substrates occurs in tumors within 24 h of dinaciclib administration. This antibody recognizes amino acid sequences that contain phosphorylated CDK consensus epitope, which is (K/R)(phosphorylated-S)(PX)(K/R) where X can be any amino acid. Two independent tumor samples for each treatment (vehicle or dinaciclib) are shown per cell line.
BIM up-regulation mediates CDK inhibition-dependent cell death
We next sought to understand the mechanism that underlies the synthetic-lethal interaction between elevated MYC expression and CDK inhibition in epithelial cells. We previously reported that CDK inhibition–induced apoptosis was independent of p53 but required activation of the mitochondrial intrinsic apoptotic pathway (Goga et al., 2007). We therefore reasoned that inhibition of CDKs might either increase the activity of proapoptotic BCL-2 family members or decrease the activity of prosurvival factors. To test this hypothesis, we examined the protein expression of components of the mitochondrial intrinsic pathway in matched RPE cells engineered to overexpress MYC (RPE-MYC) or control cells (RPE-NEO). RPE cells have modest levels of endogenous MYC expression (Fig. 4 A), which makes them a suitable model to study their response to MYC overexpression. We found that a proapoptotic BH3-only member BIM was substantially up-regulated in RPE-MYC cells (Fig. 8 A) as has been previously observed in other cell types engineered to overexpress MYC (Egle et al., 2004; Hemann et al., 2005). Surprisingly, BIM was dramatically further up-regulated after RPE-MYC cells were treated with purvalanol A, whereas BIM protein levels remained undetectable in RPE-NEO cells throughout the time course (Fig. 8 A). BIM up-regulation in RPE-MYC cells coincided with the cleavage of PARP, an event which indicated induction of apoptosis (Fig. 8 A). Protein quantification revealed that the extent of BIM protein up-regulation in this particular cell line was 2.2-fold (Fig. 8 B). This level of BIM up-regulation is likely to be significant because BIM up-regulation induces apoptosis unless its activity is concomitantly suppressed by overexpression of antiapoptotic BCL-2 family members (O’Connor et al., 1998). Quantitative PCR (TaqMan) analysis showed up-regulation of BIM mRNA (∼2.5-fold) after treatment of RPE-MYC cells with purvalanol A (Fig. 8 C). These results suggest that BIM up-regulation after CDK inhibition is, in part, a result of increased BIM mRNA levels. BIM mRNA also increased in RPE-NEO cells after purvalanol A treatment but remained substantially lower than the levels observed in RPE-MYC cells (Fig. 8 C). Interestingly, we found that dinaciclib treatment of RPE-MYC cells resulted in the up-regulation of not only the BIM-EL isoform found with purvalanol A treatment but also the shorter isoforms BIM-L and BIM-S (Fig. 8 D), which have been shown to be significantly more potent in inducing apoptosis (O’Connor et al., 1998). This may, at least in part, explain the higher potency associated with dinaciclib compared with that of purvalanol A (Fig. 4 A).
Figure 8.
BIM contributes to CDK inhibition-induced cell death in cells with elevated MYC expression. (A) RPE cells, with or without constitutive MYC overexpression, were treated with 10 µM purvalanol A for 72 h and were tested for protein expression of multiple pro- and antiapoptotic BCL2 family members, as well as cleaved PARP and loading control β-actin. The results shown are representatives of at least five independent experiments. (B) Relative fold change in BIM protein expression in RPE-MYC cells treated with purvalanol A for 72 h. The purvalanol A time course experiments were repeated at least five times and BIM protein was quantified as described in Materials and methods. The error bars represent means ± SEM. (C) BIM mRNA expression after purvalanol A treatment. BIM mRNA levels were quantified by qPCR in RPE cells with or without constitutive MYC overexpression. Mean of three experiments ± SEM are shown. (D) BIM expression in RPE-MYC cells treated with dinaciclib for 36 h. (E) RPE-MYC cells were transfected with either a pool of specific BIM siRNAs or a pool of control nontargeting siRNAs and cell viability was determined after treatment with 10 µM purvalanol A for 72 h. The experiment was repeated three times. The error bars represent ± SEM. (F) BIM protein expression in a panel of triple-negative cells. Cell lines were tested for BIM protein expression after treatment with 10 µM purvalanol A for 72 h. The results shown are the representatives of at least three independent experiments. (G) Quantification of BIM protein expression. Shown are the means from at least three independent experiments that yielded the following SEMs, respectively: ±0.2 (HCC3153), ±0.88 (BT549), ±0.48 (MCF10AMYC), and ±0.57 (SUM149PT). (H) shRNA-mediated BIM knockdown in two triple-negative cell lines, MCF10AMYC and BT549. shRNA against GFP was used as negative control. (I) BIM knockdown effects on cell death of MCF10AMYC and BT549 cells after purvalanol A treatment. The experiments were repeated at least three times in triplicate. Error bars represent means ± SEM. P-values were calculated by two-tailed Student’s t test.
We postulated that the cell death observed in MYC-overexpressing cells after CDK inhibition is dependent on a threshold level of BIM, which is not reached in RPE-NEO cells. To test this hypothesis, we asked whether BIM is necessary for cell death induced by CDK inhibition in the context of MYC overexpression. Pretreatment of RPE-MYC cells with BIM-specific siRNAs protected the cells from purvalanol A–induced cell death (Fig. 8 E). These findings establish BIM as a major contributor to CDK inhibitor-induced apoptosis. We next tested triple-negative breast cell lines with endogenously elevated MYC expression and observed similar BIM up-regulation upon CDK inhibitor treatment (Figs. 8, F–G). To determine if BIM was required for the induction of cell death in these cells after CDK inhibitor treatment, we generated stable cell lines expressing a control or BIM shRNA (Fig. 8 H). We found that BIM depletion attenuates cell death after purvalanol A treatment (Fig. 8 I). These results indicate that the mechanism of CDK inhibitor–induced apoptosis in triple-negative cells with elevated MYC expression involves BIM up-regulation.
In addition to the proapoptotic BCL-2 family member BIM, we found that all of the three prosurvival BCL-2 family members, BCL-2, BCL-xL, and MCL-1, were also up-regulated in RPE-MYC cells compared with RPE-NEO cells in the absence of purvalanol A (Fig. 8 A). This is consistent with the hypothesis that cancers that have evolved to sustain high MYC expression have reached a balance between pro- and antiapoptotic factors to limit spontaneous apoptosis (Lowe et al., 2004; Hemann et al., 2005). This is also consistent with previous observation that co-overexpression of MYC and BCL-2 occurs in high-grade human breast tumors (Sierra et al., 1999). In our study, however, the expression levels of these prosurvival members did not change appreciably upon CDK inhibition (Fig. 8 A). Thus, treatment of tumors with elevated MYC expression, such as triple-negative breast cancers, with CDK inhibitors may tip the balance in favor of apoptosis by increasing BIM expression.
DISCUSSION
There is an evident and urgent need to develop targeted therapeutic strategies against triple-negative breast cancer. To identify these therapeutic targets, a better understanding of the biology of triple-negative breast cancer is therefore needed. In this study, we investigated the biology of triple-negative cancers and identified that MYC signaling is elevated in these tumors. Prior studies have examined MYC expression in breast cancers; however, we find, for the first time, that the expression of multiple MAX binding partners, which can regulate MYC activity, is altered in triple-negative tumors and may therefore contribute to MYC pathway activity.
Several studies have shown that the basal breast cancer subtype exhibits enrichment for a MYC transcriptional gene signature (Alles et al., 2009; Chandriani et al., 2009; Gatza et al., 2010). However, basal breast tumors account for only ∼70% of triple-negative tumors, and the importance of MYC signaling in the remaining 30% has previously been unknown. In clinical practice, tumors are routinely evaluated for estrogen receptor, progesterone receptor, and HER2 receptor status and, thus, the pathological determination of the triple-negative subtype is more clinically relevant for deciding on a course of treatment. We found that 33 of 36 triple-negative tumors (92%) in the I-SPY TRIAL had a high or intermediate MYC gene signature score (Fig. 2 A) that correlated with worse outcome (Fig. 3, A and D). The present study also demonstrates, for the first time, that MYC signaling is associated with diminished disease-free survival in patients whose tumors exhibited poor response to neoadjuvant chemotherapy (Fig. 3 D).
To determine if elevated MYC signaling can be exploited to treat triple-negative breast cancer, we assessed the utility of a synthetic-lethal approach between MYC up-regulation and CDK inhibition. Small molecule inhibition of CDK activity was effective at inducing significant cell death in triple-negative cell lines with elevated MYC expression as well as in mouse xenograft models. We found that the mechanism of such cell death included the up-regulation of the proapoptotic BCL-2 family member BIM. Thus, this study represents a significant step forward in identifying apoptotic mechanisms for treating triple-negative breast cancers.
Although prior studies have focused on the many biological functions of MYC, how this information could be translated to develop novel ways to treat breast cancer remains unclear. In an emerging era of personalized medicine, it is crucial to identify what specific patient populations would most benefit from a given treatment strategy. We found that, for patients who experienced a limited tumor response to conventional neoadjuvant chemotherapy, disease-free survival was strongly influenced by MYC expression. Insight into components of MYC signaling, therefore, will provide potential targets that could be exploited as new therapies for this subset of breast cancer patients. However, despite up-regulation of MYC signaling in a variety of human cancers, the potential utility of direct MYC inhibition remains unclear. For instance, MYC knockdown via RNAi has not been found to diminish the viability of cultured tumor-derived cells (Guan et al., 2007), consistent with our observations in this study with breast cancer cells. In contrast, inhibition of MYC-MAX dimerization using an experimental compound, 10058-F4, led to the induction of apoptosis in cultured human leukemia cells (Huang et al., 2006). Furthermore, in vivo inhibition of MYC transcriptional activity using a conditional dominant-negative mutant induced cell death and caused tumor regression in a KRAS-initiated mouse lung tumor model (Soucek et al., 2008). Whether the differences among these observations are a result of the methods used (i.e., knocking down MYC protein expression versus inhibition of its transcriptional activity) or to the extent that each cancer type depends on deregulated MYC activity requires further investigation.
An alternative approach to directly inhibiting MYC is to use a MYC-dependent synthetic-lethal strategy. We previously identified a form of synthetic-lethal interaction between MYC overexpression and CDK1 inhibition using engineered cell lines as well as in vivo model systems (Goga et al., 2007). More recent studies have shown that RNAi-mediated or small molecule inhibition of two additional cell cycle kinases, CDK2 and aurora kinase B, respectively, has synthetic-lethal interactions with MYC overexpression in certain cancer cell types (Molenaar et al., 2009; Yang et al., 2010). Another study has also reported that agonist-mediated activation of a TRAIL (tumor necrosis factor–related apoptosis-inducing ligand) receptor (DR5) induces MYC-dependent synthetic lethality (Wang et al., 2004). These distinct forms of synthetic lethality appear to require fundamentally different cellular mechanisms of cell death, including different requirements for an intact p53 tumor suppressor pathway (Wang et al., 2004; Goga et al., 2007; Molenaar et al., 2009; Yang et al., 2010). For example, 44–82% of primary basal breast tumors either lack p53 or harbor p53 mutant alleles (Sørlie et al., 2001; Carey et al., 2006). Therefore, CDK1-MYC synthetic lethality, which is p53 independent (Goga et al., 2007), may be particularly useful in treating these tumors.
The idea of targeting cell cycle kinases to selectively kill tumor cells, which often exhibit higher proliferation rates than nontumorigenic cells, is appealing and has indeed led to the clinical development of several small-molecule CDK inhibitors (Shapiro, 2006; Malumbres et al., 2008). However, there have been several issues associated with their clinical development. Previous generations of CDK inhibitors generally suffered from low in vivo potency and poor PK/PD properties. More recent third-generation CDK inhibitors, including dinaciclib, exhibit significantly improved PK/PD properties and offer greater promise for in vivo use. Indeed, dinaciclib was well tolerated in phase I trials and is currently being evaluated in phase II trials against various tumor types (Dickson and Schwartz, 2009; Parry et al., 2010). Prior clinical studies have also suffered from a lack of understanding of which tumor types are the most likely to be responsive to CDK inhibitors (Malumbres et al., 2008). Therefore, selection of patient cohorts based on molecular targets, such as MYC overexpression, may improve the therapeutic potential of CDK inhibitors in clinical trials.
In the present study, we found that CDK inhibition increases BIM protein levels not only in the model cells engineered to overexpress MYC but also in a panel of patient-derived triple-negative breast cancer cell lines with elevated MYC expression. Elevated BIM plays a direct role in CDK inhibition–induced cell death. Prior studies have found that BIM isoforms can be regulated at both the transcriptional and posttranslational levels. Protein expression of the shortest isoform Bim-S is sufficient to potently induce apoptosis (O’Connor et al., 1998). In contrast, the activity of the longest isoform BIM-EL can also be modulated by distinct phosphorylation mechanisms that can either stabilize the protein or induce its degradation via the proteasome pathway (Hübner et al., 2008). In our studies, we found that both isoforms can be up-regulated in epithelial cells after CDK inhibitor treatment.
Previous studies have shown that BIM expression is elevated upon MYC overexpression (Egle et al., 2004; Hemann et al., 2005). Having uncovered that CDK inhibition triggers induction of BIM expression, we became interested in studying the relationship between the protein expression levels of MYC and BIM in a panel of untreated breast cancer cell lines. We did not find a correlation between MYC and BIM expression in these cells regardless of their molecular subtypes or receptor status (unpublished data). Therefore, in a given cellular context, it is likely that the relative increase in BIM activity in response to CDK inhibition, not the absolute basal level of BIM expression, determines whether or not apoptosis can be initiated. Thus, the protein stoichiometry among BIM and antiapoptotic BCL-2 family members such as BCL-2, BCL-xL, and MCL-1 is likely to dictate if apoptosis is triggered. In this respect, a combinatorial approach of CDK inhibition with inhibition of antiapoptotic BCL-2 family members would be predicted to have a synergistic effect in inducing cell death in MYC overexpressing triple-negative tumors. Indeed, several BH3 mimetics (Chonghaile and Letai, 2008), namely ABT-737/263 (Oltersdorf et al., 2005) and obatoclax (Nguyen et al., 2007), are currently under development for clinical use.
In conclusion, we have shown utility for small molecule CDK inhibitors in the treatment of triple-negative breast tumors with elevated MYC expression. It is likely that CDK inhibitors will be effective not only against MYC-overexpressing triple-negative tumors but also for aggressive receptor-positive tumors with elevated MYC expression (i.e., luminal B). Methods for immunohistochemical analysis on paraffin-embedded primary tumor biopsies for MYC protein expression have been previously described (Gurel et al., 2008; Ruzinova et al., 2010). In addition, gene expression profiling methods on primary breast tumors are becoming available for use in the clinic (van ’t Veer et al., 2002). Such technologies could be applied to assess whether MYC signaling is elevated in patient tumor samples (Alles et al., 2009; Chandriani et al., 2009; Gatza et al., 2010). Thus, these detection methods for MYC activity have the potential to be translated for routine use in the clinic. Considering the lack of established targeted therapeutics against triple-negative tumors, we propose that MYC status will prove useful as a predictive biomarker of response to CDK inhibitors for the treatment of triple-negative breast cancers.
MATERIALS AMD METHODS
I-SPY Trial.
Expression microarray results were available for 149 tumors before treatment. Three tumors could not be designated as triple negative or receptor positive, owing to a lack of staining for one or more receptor types, leaving 146 available for analysis. Details of the I-SPY Trial protocol, patient array data, and characteristics of enrolled patients are described elsewhere (Esserman et al., 2012) and are also available at the NCI I-SPY Data Portal (http://ispy.nci.nih.gov/). All patients included in our analysis had pretreatment core biopsies for which gene expression measurements had been determined using 44K microarrays (Agilent Technologies). Microarray hybridization was performed as per the manufacturer, the background was subtracted, and Lowess-normalized log2 ratio of Cy3 and Cy5 intensity values were calculated. For MYC mRNA levels, a MYC-specific probe (A_32_P60687) was compared across the 146 samples for which receptor status was known. Patients enrolled in the trial received conventional neoadjuvant chemotherapy that included doxorubicin and cyclophosphamide, and/or paclitaxel per the I-SPY protocol before definitive surgical resection. The extent of residual disease was quantified using RCB and reported by RCB class (RCB 0-III; Symmans et al., 2007).
Bioinformatics and analysis of the I-SPY data.
Expression of the MYC signature (Chandriani et al., 2009) in the I-SPY dataset was examined by first converting the platform-specific probe identifiers in both studies to UNIGENE cluster identifiers (Unigene Build 219 Homo sapiens). Data from multiple probes mapping to the same UNIGENE cluster IDs were averaged. The MYC pathway activity refers to the MYC gene signature score centroid that was calculated as previously described (Chandriani et al., 2009). The Pearson correlation of each tumor’s expression profile of these genes to the MYC signature centroid was determined. The tumors were then ordered, from high to low, by the Pearson correlation value. Tumors that lacked conclusive receptor status information were excluded from analysis. The remaining tumors were divided into tertiles representing three groups with high, intermediate, or low correlation to the MYC signature centroid. Recurrence-free survival was analyzed using Cox Proportional Hazards models that were evaluated using Wald’s Test. This analysis was performed using the survival package in the R Environment for Statistical Computing (http://www.r-project.org/). Associations between categorical variables were evaluated using Fisher’s Exact Test. This analysis was performed using the stats package in the R Environment for Statistical Computing (http://www.r-project.org/).
Reverse-phase protein array analysis of MYC and phospho-MYC expression.
Protein was extracted from the human tumors, and reverse-phase protein lysate microarray was done as described previously (Tibes et al., 2006; Hennessy et al., 2007; Hu et al., 2007; Liang et al., 2007). Briefly, lysis buffer was used to lyse frozen tumors by homogenization. Tumor lysates were normalized to 1 µg/µl concentration with the use of bicinchoninic acid assay and were boiled with 1% SDS, and the supernatants were manually diluted in six or eight twofold serial dilutions with lysis buffer. An arrayer (2470; Aushon Biosystems) created 1,056 sample arrays on nitrocellulose-coated FAST slides (Schleicher & Schuell BioScience) from the serial dilutions. A slide was then probed with a validated primary MYC and phospho-MYC T58/S62 antibody (Cell Signaling Technology), and the signals were amplified with a catalyzed system (Dako). A secondary antibody was used as a starting point for amplification. The slides were scanned, analyzed, and quantitated with the use of MicroVigene software (VigeneTech Inc.) to generate serial dilution signal intensity curves for each sample with the logistic fit model: ln(y) = a + (b − a)/(1 + expc*[d−ln(x)]). A representative natural logarithmic value of each sample curve on the slide (curve mean) was then used as a relative quantification of the amount of each protein in each sample. The levels of unphosphorylated and phosphorylated-MYC (T58/S62) in each sample was expressed as a log-mean centered value after correction for protein loading with the use of the mean expression levels of >150 proteins as previously described (Tibes et al., 2006; Hennessy et al., 2007; Hu et al., 2007; Liang et al., 2007; Stemke-Hale et al., 2008; Gonzalez-Angulo et al., 2009).
Cell culture.
The propagation of human breast cell lines used in this study and their global mRNA expression profiling has been previously described (Neve et al., 2006). Engineered human epithelial cell lines RPE-NEO and RPE-MYC cells were previously described (Goga et al., 2007). The MYC-overexpressing versions of the receptor-positive cell lines T47D and HCC1428 were established by infecting the cells with the lentivirus prepared using pLVX-AcGFP-N1 plasmid with a full-length human MYC cDNA cloned into the EcoRI–XhoI sites or with recombinant retrovirus expressing MYC to generate the MCF10A-MYC cells..
Cell viability assays.
CellTiter (Promega) cell viability assay shown in Fig. 4 A was performed in 96-well plates, using a Safire2 plate reader (TECAN) that runs Magellan software, according to the manufacturer’s instruction. Each cell line was plated onto 10 wells per experiment and the assay was repeated at least five times. For the rest of the cell viability experiments described throughout this manuscript, a flow cytometry–based Guava ViaCount viability assay (Millipore) was performed according to the manufacturer’s instruction.
Cell cycle analysis.
Cell lines were treated with DMSO, 10 µM purvalanol A, or 10 nM dinaciclib (provided by the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute [Bethesda, MD] and MERCK) for 72 h. After treatment, cells were fixed in 70% ethanol and stained with propidium iodide to measure DNA content. Samples were analyzed on a LSRII flow cytometer. Cell populations were gated to exclude cell debris and doublets and cell cycle distribution was determined using FlowJo analysis software (Tree Star).
Protein lysate preparation and Western blotting analysis.
Cultured cells were washed with ice-cold PBS and harvested directly into radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, 150 mM NaCl, 0.5% sodium-deoxycholate, 1% NP-40, 0.1% SDS, 2 mM EDTA, pH 7.5) containing COMPLETE protease inhibitor cocktail (Roche) and phosphatase inhibitors (Santa Cruz Biotechnology, Inc.). Isolated tumor tissues were first washed in ice-cold PBS and homogenized on ice using Tissue Tearor (Biospec Products, Inc.) in RIPA buffer containing protease inhibitors and phosphatase inhibitors. Protein concentrations were determined by performing DC Protein Assay (Bio-Rad Laboratories) using BSA as standard. The following antibodies were used for western analyses: MYC (Epitomics), β-actin (Sigma-Aldrich), PARP (Cell Signaling Technology), BIM (Assay Designs), Puma (Cell Signaling Technology), Bid (R&D Systems), Bax (Cell Signaling Technology), Bak (Santa Cruz Biotechnology, Inc.), BCL-2 (Cell Signaling Technology), BCL-xl (Santa Cruz Biotechnology, Inc.), MCL-1 (Abcam), Phospho-(Ser) CDK substrates (Cell Signaling Technology), PP1-α (Epitomics), PP1-α (pT320; Epitomics), CDK1 (Santa Cruz Biotechnology, Inc.), and CDK2 (Santa Cruz Biotechnology, Inc.).
Determination of relative protein expression levels.
VersaDoc Imaging System (4000 MP; Bio-Rad Laboratories) was used to quantify protein expression. BIM up-regulation at the protein level was determined by normalizing presaturated BIM signals to those of β-actin. To determine the relative MYC protein expression levels across a panel of breast cell lines, the MYC signals acquired through anti-MYC Western blotting were normalized against total fluorescence from the Ponceau S–stained bands as previously described (Nijjar et al., 2005). This method was necessary as a result of significantly different protein expression levels of major house keeping proteins across breast cell lines used in this study.
Small molecule CDK inhibitors.
Purvalanol A (Sigma-Aldrich) was reconstituted in 100% DMSO. Unless otherwise indicated, it was used at a concentration (10 µM) previously shown to induce cell cycle arrest in various mammalian cell lines (Gray et al., 1998; Goga et al., 2007). Dinaciclib was reconstituted either in 100% DMSO for cell culture use or in 20% HPBCD (hydroxypropyl β cyclodextrin) for mouse studies.
Matrigel 3D cultures.
The establishment of 3D cultures was performed as previously described (Lee et al., 2007). Briefly, cells were seeded on top of the solidified Matrigel (BD) layer at 2,000 cells per well in 96-well plates and incubated for 6 d to allow for multicellular colonies to form. Phase contrast images were obtained at this point and designated as day 0. Cultures were treated with DMSO, 10 µM purvalanol A, or 10 nM dinaciclib and incubated for an additional 6 d before obtaining images of treated and control cultures and designated as day 6. Images were analyzed using ImageJ image analysis software (National Institutes of Health). The background was subtracted to remove differences in intensity as a result of shadow effects within the culture images, and the mean area of the colonies within the field was determined using the particle analysis algorithm within ImageJ. To decrease the chance of debris being included in the analysis, particles with an area of 5,000 pixels (3.1 µm) or less (the mean size of noncellular debris within the images as determined empirically) were excluded. The mean surface area of the day 0 cultures for each cell line was set to 100%, and the mean surface area of day 6–treated cultures were determined relative to day 0. The Student’s t test was used to determine statistical significance.
Mouse xenograft studies.
For all the breast cancer cell lines used in this study, 107 cells in 200 µl PBS were subcutaneously injected into immunodeficient female mice (BALB/c nude/nude) aged ∼6–8 wk. The tumors were allowed to grow for 3–4 wk (with the tumors reaching ∼200–250 mm3 in volume) before the animals were treated with either dinaciclib at 50 mg/kg or vehicle alone (20% HPBCD) via i.p. injection. All animal experiments were approved by the University of California San Francisco institutional animal care and use committee.
siRNA/shRNA experiments.
Unless otherwise noted, siRNA against human CDK1, CDK2, MYC, and BIM (BCL2L11), respectively, and a pool of nontargeting control siRNA (siGENOME SMART pool siRNA; Dharmacon) were used according to the manufacturer’s protocol. For the MYC knockdown experiments described in Fig. 5 A, the liposomal siRNA preparations against human MYC and luciferase (negative control), respectively, were provided by Alnylam Pharmaceuticals, Inc. The retroviral shRNA constructs used in this study are pMKO shRNA Bim (plasmid 17235; Addgene; Schmelzle et al., 2007) and pMKO shRNA GFP (plasmid 10675; Addgene; Masutomi et al., 2005).
Analysis of BIM mRNA levels using quantitative PCR.
RPE cells, with or without constitutive MYC overexpression, were treated with purvalanol A for 0, 24, 48, and 72 h. Total RNA was isolated from cells using mirVana (Ambion) and digested with DNaseI to remove contaminating DNA (Ambion). cDNA was prepared from 1 µg of total RNA using Superscript II reverse transcription kit (Invitrogen). Real-time PCR was performed using probes specific for human BIM and β-ACTIN (ABI), according to the manufacturer’s instructions. Samples were run in triplicate on a Real-Time Thermal Cycler (Bio-Rad Laboratories). Variation of BIM expression was calculated using the ΔΔCT method (Livak and Schmittgen, 2001) with β-ACTIN mRNA as an internal control.
Online supplemental material.
A list of 352 genes that comprise the MYC gene signature used for the analysis in this study is provided in Table S1. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20111512/DC1.
Acknowledgments
We thank the patients, investigators, and institutions that participated in the I-SPY TRIAL. We are grateful to Drs. Luika Timmerman, Koei Chin, Wen-Lin Kuo, Adi Gazdar, Stephen Ethier, and Joe Gray for providing cell lines. We thank Drs. Clifford Hudis and J. Michael Bishop, and Ms. Brittany Anderton for comments and critical reading of the manuscript.
We acknowledge the following support: California Breast Cancer Research Program post-doctoral (D. Horiuchi and L. Kusdra) and pre-doctoral (N.E. Huskey) fellowships, NCI 2T32CA108462 post-doctoral training grant (L. Kusdra), the Howard Hughes Medical Institute (S. Chandriani), the UC Cancer Coordinating Committee (N.E. Huskey), AHA Scientist Development Grant SDG3420042 (J.W. Smyth), NCI 1K23CA121994 (A.M. Gonzalez-Angulo), the Susan G. Komen Foundation (A.M.Gonzalez-Angulo, G.B. Mills, and A. Goga), NCI 1K08CA104032, 1R01CA136717 (A. Goga), a UCSF SPORE 5P50CA058207 Developmental Project (A. Goga), and V-Foundation Scholar Award (A. Goga). We also acknowledge the I-SPY program for additional statistical support: NCI SPORE, CA58207; ACRIN, U01 CA079778 and CA080098; and CALGB, CA31964 and CA3360.
The authors do not declare any competing financial interests.
Footnotes
Abbreviations used:
- CDK
- cyclin-dependent kinase
- PD
- pharmacodynamic
- PK
- pharmacokinetic
- RCB
- residual cancer burden
References
- Alles M.C., Gardiner-Garden M., Nott D.J., Wang Y., Foekens J.A., Sutherland R.L., Musgrove E.A., Ormandy C.J. 2009. Meta-analysis and gene set enrichment relative to er status reveal elevated activity of MYC and E2F in the “basal” breast cancer subgroup. PLoS ONE. 4:e4710 10.1371/journal.pone.0004710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker A.D., Sigman C.C., Kelloff G.J., Hylton N.M., Berry D.A., Esserman L.J. 2009. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin. Pharmacol. Ther. 86:97–100. 10.1038/clpt.2009.68 [DOI] [PubMed] [Google Scholar]
- Bauer K.R., Brown M., Cress R.D., Parise C.A., Caggiano V. 2007. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 109:1721–1728. 10.1002/cncr.22618 [DOI] [PubMed] [Google Scholar]
- Ben-Porath I., Thomson M.W., Carey V.J., Ge R., Bell G.W., Regev A., Weinberg R.A. 2008. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40:499–507. 10.1038/ng.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci F., Finetti P., Cervera N., Esterni B., Hermitte F., Viens P., Birnbaum D. 2008. How basal are triple-negative breast cancers? Int. J. Cancer. 123:236–240. 10.1002/ijc.23518 [DOI] [PubMed] [Google Scholar]
- Bland K.I., Konstadoulakis M.M., Vezeridis M.P., Wanebo H.J. 1995. Oncogene protein co-expression. Value of Ha-ras, c-myc, c-fos, and p53 as prognostic discriminants for breast carcinoma. Ann. Surg. 221:706–718. 10.1097/00000658-199506000-00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blethrow J.D., Glavy J.S., Morgan D.O., Shokat K.M. 2008. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc. Natl. Acad. Sci. USA. 105:1442–1447. 10.1073/pnas.0708966105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey L.A., Perou C.M., Livasy C.A., Dressler L.G., Cowan D., Conway K., Karaca G., Troester M.A., Tse C.K., Edmiston S., et al. 2006. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 295:2492–2502. 10.1001/jama.295.21.2492 [DOI] [PubMed] [Google Scholar]
- Carey L.A., Dees E.C., Sawyer L., Gatti L., Moore D.T., Collichio F., Ollila D.W., Sartor C.I., Graham M.L., Perou C.M. 2007. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin. Cancer Res. 13:2329–2334. 10.1158/1078-0432.CCR-06-1109 [DOI] [PubMed] [Google Scholar]
- Chandriani S., Frengen E., Cowling V.H., Pendergrass S.A., Perou C.M., Whitfield M.L., Cole M.D. 2009. A core MYC gene expression signature is prominent in basal-like breast cancer but only partially overlaps the core serum response. PLoS ONE. 4:e6693 10.1371/journal.pone.0006693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.W., Claassen G.F., Hann S.R., Cole M.D. 2000. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol. Cell. Biol. 20:4309–4319. 10.1128/MCB.20.12.4309-4319.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheang M.C., Chia S.K., Voduc D., Gao D., Leung S., Snider J., Watson M., Davies S., Bernard P.S., Parker J.S., et al. 2009. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J. Natl. Cancer Inst. 101:736–750. 10.1093/jnci/djp082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chonghaile T.N., Letai A. 2008. Mimicking the BH3 domain to kill cancer cells. Oncogene. 27:S149–S157. 10.1038/onc.2009.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling V.H., Cole M.D. 2006. Mechanism of transcriptional activation by the Myc oncoproteins. Semin. Cancer Biol. 16:242–252. 10.1016/j.semcancer.2006.08.001 [DOI] [PubMed] [Google Scholar]
- Dang C.V. 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent R., Trudeau M., Pritchard K.I., Hanna W.M., Kahn H.K., Sawka C.A., Lickley L.A., Rawlinson E., Sun P., Narod S.A. 2007. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. 13:4429–4434. 10.1158/1078-0432.CCR-06-3045 [DOI] [PubMed] [Google Scholar]
- Dickson M.A., Schwartz G.K. 2009. Development of cell-cycle inhibitors for cancer therapy. Curr. Oncol. 16:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohadwala M., da Cruz e Silva E.F., Hall F.L., Williams R.T., Carbonaro-Hall D.A., Nairn A.C., Greengard P., Berndt N. 1994. Phosphorylation and inactivation of protein phosphatase 1 by cyclin-dependent kinases. Proc. Natl. Acad. Sci. USA. 91:6408–6412. 10.1073/pnas.91.14.6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egle A., Harris A.W., Bouillet P., Cory S. 2004. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc. Natl. Acad. Sci. USA. 101:6164–6169. 10.1073/pnas.0401471101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M., Eisenman R.N. 2008. Myc’s broad reach. Genes Dev. 22:2755–2766. 10.1101/gad.1712408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esserman L.J., Berry D.A., DeMichele A., Carey L.A., Davis S.E., Buxton M., Hudis C., Gray J.W., Perou C., Yau C., et al. 2012. Pathologic complete response predicts recurrence-free survival more effectively by cancer subset: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657). J. Clin. Oncol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O’Connor M.J., et al. 2009. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361:123–134. 10.1056/NEJMoa0900212 [DOI] [PubMed] [Google Scholar]
- Fournier M.V., Martin K.J., Kenny P.A., Xhaja K., Bosch I., Yaswen P., Bissell M.J. 2006. Gene expression signature in organized and growth-arrested mammary acini predicts good outcome in breast cancer. Cancer Res. 66:7095–7102. 10.1158/0008-5472.CAN-06-0515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatza M.L., Lucas J.E., Barry W.T., Kim J.W., Wang Q., Crawford M.D., Datto M.B., Kelley M., Mathey-Prevot B., Potti A., Nevins J.R. 2010. A pathway-based classification of human breast cancer. Proc. Natl. Acad. Sci. USA. 107:6994–6999. 10.1073/pnas.0912708107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goga A., Yang D., Tward A.D., Morgan D.O., Bishop J.M. 2007. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat. Med. 13:820–827. 10.1038/nm1606 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Angulo A.M., Stemke-Hale K., Palla S.L., Carey M., Agarwal R., Meric-Berstam F., Traina T.A., Hudis C., Hortobagyi G.N., Gerald W.L., et al. 2009. Androgen receptor levels and association with PIK3CA mutations and prognosis in breast cancer. Clin. Cancer Res. 15:2472–2478. 10.1158/1078-0432.CCR-08-1763 [DOI] [PubMed] [Google Scholar]
- Grandori C., Cowley S.M., James L.P., Eisenman R.N. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16:653–699. 10.1146/annurev.cellbio.16.1.653 [DOI] [PubMed] [Google Scholar]
- Gray N.S., Wodicka L., Thunnissen A.M., Norman T.C., Kwon S., Espinoza F.H., Morgan D.O., Barnes G., LeClerc S., Meijer L., et al. 1998. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science. 281:533–538. 10.1126/science.281.5376.533 [DOI] [PubMed] [Google Scholar]
- Guan Y., Kuo W.L., Stilwell J.L., Takano H., Lapuk A.V., Fridlyand J., Mao J.H., Yu M., Miller M.A., Santos J.L., et al. 2007. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin. Cancer Res. 13:5745–5755. 10.1158/1078-0432.CCR-06-2882 [DOI] [PubMed] [Google Scholar]
- Gurel B., Iwata T., Koh C.M., Jenkins R.B., Lan F., Van Dang C., Hicks J.L., Morgan J., Cornish T.C., Sutcliffe S., et al. 2008. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod. Pathol. 21:1156–1167. 10.1038/modpathol.2008.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann M.T., Bric A., Teruya-Feldstein J., Herbst A., Nilsson J.A., Cordon-Cardo C., Cleveland J.L., Tansey W.P., Lowe S.W. 2005. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 436:807–811. 10.1038/nature03845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy B.T., Lu Y., Poradosu E., Yu Q., Yu S., Hall H., Carey M.S., Ravoori M., Gonzalez-Angulo A.M., Birch R., et al. 2007. Pharmacodynamic markers of perifosine efficacy. Clin. Cancer Res. 13:7421–7431. 10.1158/1078-0432.CCR-07-0760 [DOI] [PubMed] [Google Scholar]
- Hu J., He X., Baggerly K.A., Coombes K.R., Hennessy B.T., Mills G.B. 2007. Non-parametric quantification of protein lysate arrays. Bioinformatics. 23:1986–1994. 10.1093/bioinformatics/btm283 [DOI] [PubMed] [Google Scholar]
- Huang M.J., Cheng Y.C., Liu C.R., Lin S., Liu H.E. 2006. A small-molecule c-Myc inhibitor, 10058-F4, induces cell-cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp. Hematol. 34:1480–1489. 10.1016/j.exphem.2006.06.019 [DOI] [PubMed] [Google Scholar]
- Hübner A., Barrett T., Flavell R.A., Davis R.J. 2008. Multisite phosphorylation regulates Bim stability and apoptotic activity. Mol. Cell. 30:415–425. 10.1016/j.molcel.2008.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin W.J., Jr, Carey L.A. 2008. What is triple-negative breast cancer? Eur. J. Cancer. 44:2799–2805. 10.1016/j.ejca.2008.09.034 [DOI] [PubMed] [Google Scholar]
- Jain A.N., Chin K., Børresen-Dale A.L., Erikstein B.K., Eynstein Lonning P., Kaaresen R., Gray J.W. 2001. Quantitative analysis of chromosomal CGH in human breast tumors associates copy number abnormalities with p53 status and patient survival. Proc. Natl. Acad. Sci. USA. 98:7952–7957. 10.1073/pnas.151241198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. 2010. Adaptive trials receive boost. Nat. Rev. Drug Discov. 9:345–348. 10.1038/nrd3174 [DOI] [PubMed] [Google Scholar]
- Lee G.Y., Kenny P.A., Lee E.H., Bissell M.J. 2007. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods. 4:359–365. 10.1038/nmeth1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Shao S.H., Xu Z.X., Hennessy B., Ding Z., Larrea M., Kondo S., Dumont D.J., Gutterman J.U., Walker C.L., et al. 2007. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat. Cell Biol. 9:218–224. 10.1038/ncb1537 [DOI] [PubMed] [Google Scholar]
- Liedtke C., Mazouni C., Hess K.R., André F., Tordai A., Mejia J.A., Symmans W.F., Gonzalez-Angulo A.M., Hennessy B., Green M., et al. 2008. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 26:1275–1281. 10.1200/JCO.2007.14.4147 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Livasy C.A., Karaca G., Nanda R., Tretiakova M.S., Olopade O.I., Moore D.T., Perou C.M. 2006. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod. Pathol. 19:264–271. 10.1038/modpathol.3800528 [DOI] [PubMed] [Google Scholar]
- Lowe S.W., Cepero E., Evan G. 2004. Intrinsic tumour suppression. Nature. 432:307–315. 10.1038/nature03098 [DOI] [PubMed] [Google Scholar]
- Malumbres M., Pevarello P., Barbacid M., Bischoff J.R. 2008. CDK inhibitors in cancer therapy: what is next? Trends Pharmacol. Sci. 29:16–21. 10.1016/j.tips.2007.10.012 [DOI] [PubMed] [Google Scholar]
- Martin K.J., Patrick D.R., Bissell M.J., Fournier M.V. 2008. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS ONE. 3:e2994 10.1371/journal.pone.0002994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutomi K., Possemato R., Wong J.M., Currier J.L., Tothova Z., Manola J.B., Ganesan S., Lansdorp P.M., Collins K., Hahn W.C. 2005. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl. Acad. Sci. USA. 102:8222–8227. 10.1073/pnas.0503095102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N., Penn L.Z. 2008. Reflecting on 25 years with MYC. Nat. Rev. Cancer. 8:976–990. 10.1038/nrc2231 [DOI] [PubMed] [Google Scholar]
- Molenaar J.J., Ebus M.E., Geerts D., Koster J., Lamers F., Valentijn L.J., Westerhout E.M., Versteeg R., Caron H.N. 2009. Inactivation of CDK2 is synthetically lethal to MYCN over-expressing cancer cells. Proc. Natl. Acad. Sci. USA. 106:12968–12973. 10.1073/pnas.0901418106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidu R., Wahab N.A., Yadav M., Kutty M.K. 2002. Protein expression and molecular analysis of c-myc gene in primary breast carcinomas using immunohistochemistry and differential polymerase chain reaction. Int. J. Mol. Med. 9:189–196. [PubMed] [Google Scholar]
- Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.P., Tong F., et al. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 10:515–527. 10.1016/j.ccr.2006.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M., Marcellus R.C., Roulston A., Watson M., Serfass L., Murthy Madiraju S.R., Goulet D., Viallet J., Bélec L., Billot X., et al. 2007. Small molecule obatoclax (GX15-070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proc. Natl. Acad. Sci. USA. 104:19512–19517. 10.1073/pnas.0709443104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijjar T., Bassett E., Garbe J., Takenaka Y., Stampfer M.R., Gilley D., Yaswen P. 2005. Accumulation and altered localization of telomere-associated protein TRF2 in immortally transformed and tumor-derived human breast cells. Oncogene. 24:3369–3376. 10.1038/sj.onc.1208482 [DOI] [PubMed] [Google Scholar]
- O’Connor L., Strasser A., O’Reilly L.A., Hausmann G., Adams J.M., Cory S., Huang D.C. 1998. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 17:384–395. 10.1093/emboj/17.2.384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltersdorf T., Elmore S.W., Shoemaker A.R., Armstrong R.C., Augeri D.J., Belli B.A., Bruncko M., Deckwerth T.L., Dinges J., Hajduk P.J., et al. 2005. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 435:677–681. 10.1038/nature03579 [DOI] [PubMed] [Google Scholar]
- Parry D., Guzi T., Shanahan F., Davis N., Prabhavalkar D., Wiswell D., Seghezzi W., Paruch K., Dwyer M.P., Doll R., et al. 2010. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol. Cancer Ther. 9:2344–2353. 10.1158/1535-7163.MCT-10-0324 [DOI] [PubMed] [Google Scholar]
- Perou C.M., Sørlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., et al. 2000. Molecular portraits of human breast tumours. Nature. 406:747–752. 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- Reinhardt H.C., Jiang H., Hemann M.T., Yaffe M.B. 2009. Exploiting synthetic lethal interactions for targeted cancer therapy. Cell Cycle. 8:3112–3119. 10.4161/cc.8.19.9626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzinova M.B., Caron T., Rodig S.J. 2010. Altered subcellular localization of c-Myc protein identifies aggressive B-cell lymphomas harboring a c-MYC translocation. Am. J. Surg. Pathol. 34:882–891. 10.1097/PAS.0b013e3181db83af [DOI] [PubMed] [Google Scholar]
- Sarrió D., Rodriguez-Pinilla S.M., Hardisson D., Cano A., Moreno-Bueno G., Palacios J. 2008. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 68:989–997. 10.1158/0008-5472.CAN-07-2017 [DOI] [PubMed] [Google Scholar]
- Schmelzle T., Mailleux A.A., Overholtzer M., Carroll J.S., Solimini N.L., Lightcap E.S., Veiby O.P., Brugge J.S. 2007. Functional role and oncogene-regulated expression of the BH3-only factor Bmf in mammary epithelial anoikis and morphogenesis. Proc. Natl. Acad. Sci. USA. 104:3787–3792. 10.1073/pnas.0700115104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider B.P., Winer E.P., Foulkes W.D., Garber J., Perou C.M., Richardson A., Sledge G.W., Carey L.A. 2008. Triple-negative breast cancer: risk factors to potential targets. Clin. Cancer Res. 14:8010–8018. 10.1158/1078-0432.CCR-08-1208 [DOI] [PubMed] [Google Scholar]
- Sears R., Nuckolls F., Haura E., Taya Y., Tamai K., Nevins J.R. 2000. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 14:2501–2514. 10.1101/gad.836800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.R., Kim J., Bae S., Soh J.W., Lee Y.S. 2008. Cdk5-mediated phosphorylation of c-Myc on Ser-62 is essential in transcriptional activation of cyclin B1 by cyclin G1. J. Biol. Chem. 283:15601–15610. 10.1074/jbc.M800987200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro G.I. 2006. Cyclin-dependent kinase pathways as targets for cancer treatment. J. Clin. Oncol. 24:1770–1783. 10.1200/JCO.2005.03.7689 [DOI] [PubMed] [Google Scholar]
- Sierra A., Castellsague X., Escobedo A., Moreno A., Drudis T., Fabra A. 1999. Synergistic cooperation between c-Myc and Bcl-2 in lymph node progression of T1 human breast carcinomas. Breast Cancer Res. Treat. 54:39–45. 10.1023/A:1006120006471 [DOI] [PubMed] [Google Scholar]
- Sørlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.B., van de Rijn M., Jeffrey S.S., et al. 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA. 98:10869–10874. 10.1073/pnas.191367098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L., Whitfield J., Martins C.P., Finch A.J., Murphy D.J., Sodir N.M., Karnezis A.N., Swigart L.B., Nasi S., Evan G.I. 2008. Modelling Myc inhibition as a cancer therapy. Nature. 455:679–683. 10.1038/nature07260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemke-Hale K., Gonzalez-Angulo A.M., Lluch A., Neve R.M., Kuo W.L., Davies M., Carey M., Hu Z., Guan Y., Sahin A., et al. 2008. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 68:6084–6091. 10.1158/0008-5472.CAN-07-6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmans W.F., Peintinger F., Hatzis C., Rajan R., Kuerer H., Valero V., Assad L., Poniecka A., Hennessy B., Green M., et al. 2007. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J. Clin. Oncol. 25:4414–4422. 10.1200/JCO.2007.10.6823 [DOI] [PubMed] [Google Scholar]
- Tibes R., Qiu Y., Lu Y., Hennessy B., Andreeff M., Mills G.B., Kornblau S.M. 2006. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol. Cancer Ther. 5:2512–2521. 10.1158/1535-7163.MCT-06-0334 [DOI] [PubMed] [Google Scholar]
- Tutt A., Robson M., Garber J.E., Domchek S.M., Audeh M.W., Weitzel J.N., Friedlander M., Arun B., Loman N., Schmutzler R.K., et al. 2010. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 376:235–244. 10.1016/S0140-6736(10)60892-6 [DOI] [PubMed] [Google Scholar]
- van ’t Veer L.J., Dai H., van de Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., et al. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 415:530–536. 10.1038/415530a [DOI] [PubMed] [Google Scholar]
- Voduc K.D., Cheang M.C., Tyldesley S., Gelmon K., Nielsen T.O., Kennecke H. 2010. Breast cancer subtypes and the risk of local and regional relapse. J. Clin. Oncol. 28:1684–1691. 10.1200/JCO.2009.24.9284 [DOI] [PubMed] [Google Scholar]
- Wang Y., Engels I.H., Knee D.A., Nasoff M., Deveraux Q.L., Quon K.C. 2004. Synthetic lethal targeting of MYC by activation of the DR5 death receptor pathway. Cancer Cell. 5:501–512. 10.1016/S1535-6108(04)00113-8 [DOI] [PubMed] [Google Scholar]
- Weaver V.M., Lelièvre S., Lakins J.N., Chrenek M.A., Jones J.C., Giancotti F., Werb Z., Bissell M.J. 2002. beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2:205–216. 10.1016/S1535-6108(02)00125-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D.J., Liu H., Ridky T.W., Cassarino D., Segal E., Chang H.Y. 2008. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2:333–344. 10.1016/j.stem.2008.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Liu H., Goga A., Kim S., Yuneva M., Bishop J.M. 2010. Therapeutic potential of a synthetic lethal interaction between the MYC proto-oncogene and inhibition of aurora-B kinase. Proc. Natl. Acad. Sci. USA. 107:13836–13841. 10.1073/pnas.1008366107 [DOI] [PMC free article] [PubMed] [Google Scholar]