Crystal structure and mutagenesis analyses suggest a MAIT TCR–MR1 docking mode distinct from the NKT TCR-CD1d docking mode.
Abstract
Mucosal-associated invariant T (MAIT) cells express a semiinvariant αβ T cell receptor (TCR) that binds MHC class I–like molecule (MR1). However, the molecular basis for MAIT TCR recognition by MR1 is unknown. In this study, we present the crystal structure of a human Vα7.2Jα33-Vβ2 MAIT TCR. Mutagenesis revealed highly conserved requirements for the MAIT TCR–MR1 interaction across different human MAIT TCRs stimulated by distinct microbial sources. Individual residues within the MAIT TCR β chain were dispensable for the interaction with MR1, whereas the invariant MAIT TCR α chain controlled specificity through a small number of residues, which are conserved across species and located within the Vα-Jα regions. Mutagenesis of MR1 showed that only two residues, which were centrally positioned and on opposing sides of the antigen-binding cleft of MR1, were essential for MAIT cell activation. The mutagenesis data are consistent with a centrally located MAIT TCR–MR1 docking that was dominated by the α chain of the MAIT TCR. This candidate docking mode contrasts with that of the NKT TCR–CD1d-antigen interaction, in which both the α and β chain of the NKT TCR is required for ligation above the F′-pocket of CD1d.
Antigen (Ag)-specific immunity is orchestrated by T cells that determine the specificity of immune responses via their clonally distributed αβ TCRs. A wide array of Ags is presented to TCRs, and to counteract this diversity, the host possesses a vast T cell repertoire. TCR diversity is generated via gene rearrangement within the variable (V) domains of the TCR; namely, the V and junction (J) gene segments define the Vα chain, whereas the Vβ chain is composed of rearranged V, D (diversity), and J gene segments, in addition to non–nucleotide-encoded diversity (N) at these gene segment boundaries. This diversity is manifested in the complementarity determining regions (CDR) of the TCR, providing key sites that interact with the Ag-presenting molecule (Clements et al., 2006). Despite this TCR diversity and its importance in immunity, there are numerous examples of TCR bias in protective immunity and autoimmunity in which restricted V gene usage and/or sequence conservation within CDR3 loops defines the immune response to specific Ags (Turner et al., 2006; Godfrey et al., 2008; Gras et al., 2008). Why this should occur, and the factors that shape biased TCR usage, remain unclear.
TCRs recognize peptides complexed to the MHC (pMHC), and the insight gleaned from structures of TCR–pMHC complexes has been extremely informative in understanding how the TCR simultaneously, and specifically, focuses with host MHC and fragments of foreign peptide Ag (Rudolph et al., 2006; Marrack et al., 2008; Burrows et al., 2010). However, there are other Ag-presenting molecules of the immune system that the TCR specifically interacts with. For example, the CD1 family presents lipid-based Ags to T cells (Godfrey et al., 2008). The most extensively studied group of T cells that interact with lipid-based Ags are the natural killer T (NKT) cells, which express an NKT TCR that specifically recognizes CD1d-Ag in mice and humans (Bendelac et al., 2007; Godfrey et al., 2010a). Similar to some MHC-restricted responses, NKT cells use a limited range of TCR genes, such that human NKT cells typically express an invariant Vα24-Jα18 rearranged TCR α chain and most express a Vβ11 TCR β chain (Godfrey et al., 2010a). The most widely studied glycolipid Ag for activating NKT cells is a synthetic α-glycolipid, α-galactosylceramide. The structures of human and mouse NKT TCRs in complex with CD1d-α-galactosylceramide and other lipid Ags have recently been determined (Godfrey and Rossjohn, 2011). These NKT TCR–CD1d-Ag complexes demonstrate a conserved docking strategy that differs from all known TCR-pMHC interactions, whereupon the NKT TCR adopted a tilted and parallel docking mode in relation to the CD1d Ag-binding cleft (Godfrey et al., 2008). The invariant NKT TCR α chain dominated this interaction, with the CDR1α loop predominantly interacting with the lipid Ag, whereas the CDR3α loop played a central role, contacting CD1d and the lipid Ag. The roles of the human Vβ11 and the homologous mouse Vβ8.2 chain were essentially restricted to the CDR2β loop that interacted with CD1d (Godfrey et al., 2010a; Joyce et al., 2011). Nevertheless, the CDR3β loop can play an important role in determining CD1d autoreactivity (Matulis et al., 2010; Mallevaey et al., 2011). Thus, the structural studies have been very informative in understanding how a TCR can recognize a lipid-laden Ag-presenting molecule, but it remains unclear how TCRs can interact with other specialized Ag-presenting molecules of the immune system.
Mucosal-associated invariant T (MAIT) cells are a subpopulation of T cells that are restricted by the monomorphic MHC class 1–like molecule (MR1; Treiner et al., 2003; Gapin, 2009; Le Bourhis et al., 2011). The role of MAIT cells in immunity is emerging and is supported by their conservation across species such as humans, cattle, and mice, as well as recent data implying protective function in autoimmunity and certain infections (Gold et al., 2010; Le Bourhis et al., 2010; Le Bourhis et al., 2011; Miyazaki et al., 2011; Chiba et al., 2012). In some regards, MAIT cells are reminiscent of CD1d-restricted NKT cells, although in humans, MAIT cells are much more abundant, comprising 1–10% of peripheral blood T cells when compared with their NKT cell counterparts (typically <0.1%), and MAIT cells can constitute up to ∼50% of liver T cells (Godfrey et al., 2010b; Dusseaux et al., 2011). Indeed, MAIT cells are readily detected in human blood, the gastrointestinal mucosa, and mesenteric lymph nodes. Furthermore, MAIT cells, like NKT cells, rapidly produce a broad range of cytokines upon activation (Kawachi et al., 2006; Martin et al., 2009). There are further parallels between MR1-restricted MAIT cells and CD1d-restricted NKT cells in that, like NKT cells, MAIT cells express a semiinvariant TCR comprised of an invariant TCR α chain (Vα19Jα33 in mice, or the homologous Vα7.2Jα33 in humans) in combination with TCR-Vβ6 or Vβ8 in mice, TCR-Vβ2 or Vβ13 in humans (Tilloy et al., 1999). The semiinvariant and evolutionarily conserved nature of the MAIT TCR suggests that MAIT cells may potentially be specific for an important, albeit limited and atypical, class of Ags presented by the MR1 molecule. Nevertheless, despite the limited TCR repertoire of MAIT cells, they respond to a surprisingly broad range of microorganisms, including diverse strains of bacteria and yeast, suggesting the existence of a conserved Ag (or family of Ags), which is common to these cellular organisms, presented to MAIT cells in an MR1-dependent manner (Gold et al., 2010; Le Bourhis et al., 2010). Indeed, endogenous cell surface expression levels of MR1 are very low, suggesting that a bacterial Ag is required to stabilize MR1 and/or increase its MR1 presentation (Chua et al., 2011). Alternatively, it is possible that bacterial infection may indirectly result in MAIT cell activation through activation of Toll-like receptors or other innate pathways, as occurs for NKT cells (Brigl et al., 2011). However, this seems unlikely for MAIT cells, as previous studies indicate that their activation can occur independently of My88D, TRIF, Nod1,-2, N1rp3, Asc, Ips TLR2, and TLR4 (Gold et al., 2010; Le Bourhis et al., 2010). Furthermore, MAIT cells can be activated by fixed APCs cultured in the presence of bacteria in an MR1-dependent manner (Le Bourhis et al., 2010). Hence, it is a simpler proposition that MAIT cell activation results from TCR recognition of MR1 complexed with a bacterial Ag, although the nature of a candidate bacterial Ag has yet to be identified.
Because MAIT cells are largely defined by their unique, semiinvariant TCR, their Ag-restricting element (MR1), and, potentially, an unusual Ag specificity (Huang et al., 2008, 2009), a critical step in understanding the biology of these cells will be to understand their TCR-specificity.
RESULTS
MAIT TCR structure
In humans, the MAIT TCR comprises the TRAV1-2 α chain (Vα7.2-Jα33) and is generally assembled with either the TRBV6 (Vβ13) or TRBV20 (Vβ2) β chains, with the CDR3β loop being hypervariable (Tilloy et al., 1999). In mouse MAIT cells, Vα19-Jα33 assembles with either TRBV-19 (Vβ6) or TRBV13 (Vβ8). We expanded and cloned MAIT cells from the PBMCs of a healthy donor and isolated the TCR α and β chain cDNAs for further investigation. Three clones were selected, each expressing the Vα7.2-Jα33 assembled with either a Vβ13.3 (clone BV6-1), Vβ13.5 (clone BV6-4), or Vβ2 β chain (clone BV20). TRBV6-1 (Vβ13.3) and TRBV20 (Vβ2) sequences were derived from published sequence data (Tilloy et al., 1999). The TRBV6-4 (Vβ13.5) sequence was derived from FACS single-cell sorting and RT-PCR analysis. A sequence alignment of the CDR loops of the three MAIT TCRs is shown (Fig. S1 and Fig. S2).
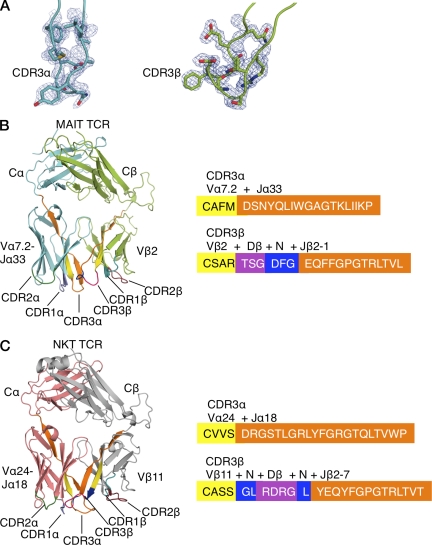
To begin to understand MAIT TCR function, the extracellular domains of the two Vβ13 and single Vβ2 MAIT TCRs were expressed in Escherichia coli, refolded into a native conformation, and purified. The functional integrity of the refolded TCRs was assessed by gel filtration chromatography and ELISA reactivity with the TCR-specific conformation-dependent mAb 12H8 (unpublished data). Although the two Vβ13 MAIT TCRs failed to crystallize (not depicted), the Vβ2 MAIT TCR crystallized, and its structure was subsequently determined to 1.7 Å resolution (Table 1). Given the resolution, the quality of the electron density for the Vβ2 MAIT TCR, and in particular the Ag-binding site, was excellent (Fig. 1 A).
Table 1.
Data collection and refinement statistics
| MAIT TCR | |
| Data collection | |
| Temperature (K) | 100 |
| Resolution limits (Å) | 50-1.7 (1.79-1.70) |
| Space group | P212121 |
| Cell dimensions (Å) | a = 41.96; b = 64.51; c = 155.12; α = γ = β = 90.00° |
| Total no. observations | 351,526 |
| No. unique observations | 45,957 |
| Multiplicitya | 7.6 (7.4) |
| Data completenessa | 97.1 (92.6) |
| I/σ1a | 13.7 (2.5) |
| Rp.i.mab (%) | 3.5 (28.5) |
| Refinement statistics | |
| Rfactorc (%) | 19.1 |
| Rfreed (%) | 23.1 |
| Nonhydrogen atoms: protein/ water | 3,472/271 |
| Ramachandran plot: most favored/allowed region (%) | 89.1/10.9 |
| B-factors: average main chain/average side chain/water (Å2) | 17.3/20.4/26.7 |
| rmsd bonds (Å) | 0.014 |
| rmsd angles (°) | 1.500 |
Values in parentheses refer to the highest resolution bin.
Rp.i.m = Σhkl [1/(N − 1)]1/2 Σi | Ihkl, i− <Ihkl> | / Σhkl <Ihkl>
Rfactor = (Σ | |Fo|− |Fc| |) / (Σ |Fo|), for all data except as indicated in footnote d.
5% of data were used for the Rfree calculation
Figure 1.
Structures of MAIT and NKT TCRs. (A) 2Fo-Fc electron density map shown as a blue mesh and contoured at 1 sigma for the CDR3α and CDR3β of the MAIT TCR. (B) Structure of MAIT TCR and amino acid composition of CDR3 loops. TCRα chain, cyan; TCRβ chain, light green; CDR1α, purple; CDR2α, dark green; CDR1β, teal, CDR2β, ruby; CDR3 loops, color coded according to their genetic origin (right). (C) Structure of NKT TCR and amino acid composition of CDR3 loops. TCRα chain, salmon; TCRβ chain, gray; CDR loops, color coded as in B.
The overall structure of the MAIT Vβ2 TCR resembles that of other MHC- and CD1d-restricted αβ TCRs, namely it comprises four immunoglobulin-like domains with a constant (C) and variable (V) domain in each chain, with their respective domains packing against each other (Fig. 1 B; Kjer-Nielsen et al., 2002; Rudolph et al., 2006). The V domain of each chain is composed of three CDRs and together these six hypervariable loops form a ligand-binding site for the Vβ2 MAIT TCR (Fig. 1 B). All six CDR loops were well-ordered in the electron density, suggesting little mobility of these CDR loops, in contrast to the flexibility of the CDR loops typical of several MHC-restricted TCRs (Garcia et al., 1998; Reiser et al., 2002; Godfrey et al., 2008), implying that the MAIT TCR possesses a rigid, preformed MR1-binding interface.
Unless explicitly stated, comparative structural analyses have been restricted to the semiinvariant human Vα24-Vβ11 NKT TCR (NKT15; Kjer-Nielsen et al., 2006; Fig. 1 C). The root mean square deviation for the pairwise superposition between the Vβ2 MAIT TCR and the Vβ11 NKT TCR was 0.90 Å (347 residues), highlighting that these semiinvariant TCRs adopt very similar topologies. Although the constant domains of the Vβ2 MAIT and Vβ11 NKT TCR superpose closely with each other (root mean square deviation [rmsd], 0.33 Å; 160 residues), differences in juxtaposition between the Vα and Vβ domains were observed, reflecting differences in the interchain pairing of the NKT TCR and the MAIT TCR. Moreover, comparative analyses revealed structural divergences within the respective Vα and Vβ domains. For example, pairwise superposition between MAIT and NKT Vα domains was 0.45 Å (75 residues), whereas the pairwise superposition between the Vβ domains was 0.70 Å (75 residues), showing that the Vβ domain was more divergent than the Vα domain, as expected from the markedly differing nature of the Vβ chains. Further, although the CDR1α loop adopts a similar conformation to the corresponding loop in the NKT TCR (Kjer-Nielsen et al., 2006), the remaining CDR loop conformations are markedly different, presumably reflecting the fact that the MAIT and NKT TCRs engage MR1 and CD1d, respectively (Fig. 1 and Fig. 2).
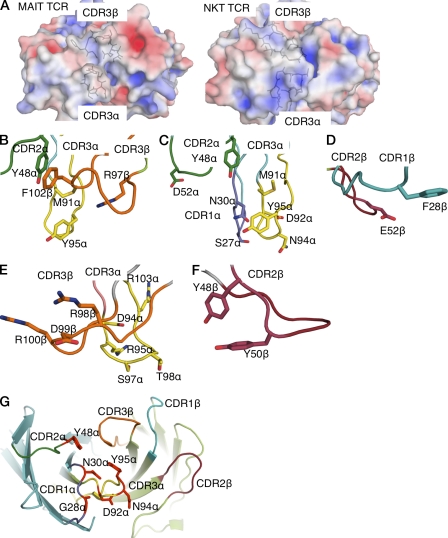
Figure 2.
Electrostatic surface and close-up view of CDR loops. (A) Electrostatic calculations were performed using the MAIT TCR and NKT TCR (Protein Data Bank accession code 2EYS). Coordinate preparation was performed using the PDB2PQR server (v1.7; Dolinsky et al., 2007). Electrostatic calculations were subsequently performed using the APBS plug-in in PyMOL (DeLano, 2002; v1.1.0 and v1.2.x, respectively) with 0.15 M concentration for the +1 and −1 ion species. (B) Central binding pocket of the MAIT TCR. CDR2α, dark green; CDR3α, yellow; CDR3β, orange. (C) Residues at the tip of CDRα loops of MAIT TCR. CDR1α, purple; CDR2α and CDR3α, color coded as in B. (D) CDR1β (teal) and CDR2β (ruby) loops of the MAIT TCR. (E) Central binding pocket of the NKT TCR. CDR loops, color coded as in B. (F) Tyrosines of the CDR2β loop (ruby) of the NKT TCR. (G) Energetically important residues at the binding interface of the MAIT TCR. TCR α chain, cyan; TCRβ chain, light green; CDR1α, purple; CDR2α, dark green; CDR3α, yellow; CDR1β, teal, CDR2β, ruby; CDR3β, orange. Energetically important residues are shown in red.
The MAIT TCR Vα-Vβ interface uses both the Vα7.2 and Jα33 gene segment of the invariant chain. The sequences encoded by the invariant Jα and hypervariable Dβ-N-Jβ region create the CDR3 loops that are centrally located at the Ag-binding interface (Fig. 1 B). Namely, the residues lining this region are mostly apolar in nature, in stark contrast to the electropositive surface of NKT TCR (Fig. 2 A). The central binding pocket of the MAIT TCR is comprised of Met91α and Arg97β, which is flanked by Tyr95α from the CDR3α loop and Phe102β from the CDR3β loop, the latter of which packs against framework residue Tyr48α from the CDR2α loop (Fig. 2 B). The tips of the CDR1α, CDR2α, and CDR3α loops are polar (Ser27α, Asn30α, Asp52α, Asp92α, and Asn94α), if not featureless (Fig. 2 C). The residues from the germline-encoded regions of the Vβ chain are mostly flat and featureless, with Glu52β from the CDR2β loop, and Phe28β from the CDR1β loop representing prominent side chains, nevertheless Phe28β lies parallel with, and away from the Ag-binding interface (Fig. 2 D). Thus, in marked contrast to that of the NKT TCR, which possesses a large number of charged residues within the CDR3β loop, the Jα18-encoded CDR3α loop, and two tyrosine residues encoded within the CDR2β loop (Fig. 2, E and F), a defining feature of the MAIT TCR is the cluster of aromatic residues within the central region of its combining site (Fig. 2 B).
The MAIT TCR Vα chain is highly conserved across species (Tilloy et al., 1999), with the surface exposed residues from the CDR1α loop (Ser27α, Gly28α, and Asn30α), the CDR2α loop (Tyr48α, Val50α, Leu51α, Asp52α, and Gly53α), and the CDR3α loop (Asp92α, Asn94α, Tyr95α, and Ile98α) conserved across four species (Goldfinch et al., 2010). However, there is less conservation in the germline-encoded CDR loops of the Vβ2 and Vβ13 chains, with a conserved Asn-His-Asp motif present in the CDR1β loop of mouse Vβ6 and Vβ8.1 (Fig. S2). However, human Vβ2 and Vβ13 do not contain this motif, and there is little conservation of CDR2β loops across, or within, human and mouse sequences apart from a Tyr/Thr-Ser motif at the junction of this loop (Fig. S2). Accordingly, the structural data suggests that although the MAIT TCR resembles the overall architecture of MHC- and CD1d-restricted TCRs, MAIT TCR recognition is more dependent on the invariant encoded α chain, with a less obvious contribution from the Vβ residues.
MAIT–MR1 recognition
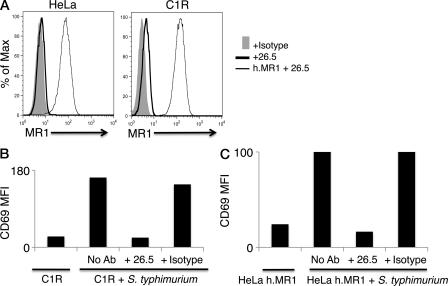
It is thought that MAIT cells may be able to potentially recognize unknown microbial Ags presented by MR1 (Gold et al., 2010; Le Bourhis et al., 2010), although definitive identification of a bacterially encoded Ag is lacking at present. To explore the activation of human MAIT TCRs by MR1, we transduced genes for MAIT TCRs using TRBV20 (Vβ2), TRBV6-1 (Vβ13.3), or TRBV6-4 (Vβ13.5) β chains into either of the two human leukemia T cell lines SKW3 (Hundhausen et al., 1992; Gras et al., 2010) and Jurkat (Gillis and Watson, 1980), thus creating clonal cellular reagents for T cell recognition (SKW3.TRBV20, SKW3.TRBV6-1, and SKW3.TRBV6-4; Jurkat.BV20, Jurkat.BV6-1, and Jurkat.BV6-4). To generate Ag-presenting cells, HeLa cells and the MHC-I–deficient human lymphoblastoid cell line C1R were transduced with the gene for human MR1. C1R cells express low levels of endogenous MR1, and thus were also used as untransduced APCs (Fig. 3 A). T cell activation was assayed by CD69 up-regulation that was determined by flow cytometry. Mindful that MAIT cells can be activated by various strains of bacteria (Gold et al., 2010; Le Bourhis et al., 2011), we first determined that SKW3 T cells transduced with MAIT TCRs were activated by Ag-presenting cells infected with Salmonella enterica serovar Typhimurium. SKW3.TRBV20 MAIT cells were activated by C1R cells (expressing endogenous MR1) only when infected with S. typhimurium, and this activation was blocked by addition of the MR1-specific mAb 26.5 (Huang et al., 2005) but not by the MHC-I-specific mAb W6/32 (Parham et al., 1979; Fig. 3 B). Similar results were obtained when SKW3.TRBV6-1 and SKW3.TRBV6-4 cells were used as responder cells (unpublished data). Similarly, HeLa cells transduced with MR1 could activate Jurkat.BV6-1 cells (Fig. 3 C), as well as Jurkat.BV20 and Jurkat.BV6-4 cells (not depicted). Activation of MAIT TCR–transduced SKW3 and Jurkat cells was dependent on the multiplicity of infection (MOI) of S. typhimurium infection, and this activation was blocked by the MR1-specific mAb 26.5. Thus, both the SKW3 and Jurkat cell lines transduced with MAIT TCRs were activated in an MR1-restricted manner.
Figure 3.
Characterization of APC lines and MAIT TCR-transduced T cell lines. (A) Staining of HeLa and C1R cells with the MR1-reactive mAb 26.5 by indirect immunofluorescence. FACS histograms compare intensity of staining with either an isotype control antibody 8A5 (+ Isotype), or with anti-MR1 antibody (+26.5). HeLa and C1R cells transduced with MR1 were also stained with 26.5 (h.MR1 + 26.5). (B) SKW3.TRBV20 cells were incubated with C1R cells infected with Salmonella typhimurium, either in the absence (No Ab) or presence of either an MR1-reactive mAb (+ 26.5) or the HLA class I–reactive mAb W6/32 (+ Isotype). SKW3.TRBV20 cells were subsequently stained for CD69 cell surface expression (mean fluorescence intensity [MFI] values shown) and analyzed by flow cytometry. (C) Jurkat.TRBV6-1 cells were incubated with MR1-transduced HeLa cells infected with S. typhimurium, either in the absence (No Ab) or presence of either an MR1-reactive mAb (+ 26.5) or the HLA class I–reactive mAb W6/32 (+ Isotype). Jurkat.TRBV6-1 cells were subsequently stained for CD69 cell surface expression (MFI values shown) and analyzed by flow cytometry. Experiments in A–C were performed three times, with similar results.
Specificity determinants of the MAIT-MR1 interaction
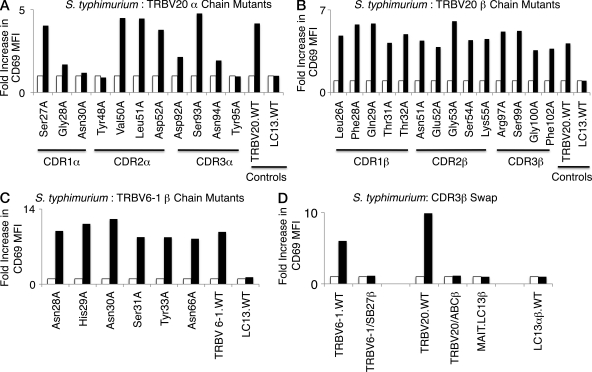
To further probe the underlying basis of the MR1-induced MAIT TCR activation, we generated a panel of MAIT TCR mutants based on the nonliganded Vβ2 (TRBV20) MAIT TCR structure. In total, 25 single-site alanine-scanning mutations were made in the TRBV20 MAIT TCR, 11 in the Vα chain (CDR1α: Ser27, Gly28, and Asn30; CDR2α: Tyr48, Val50, Leu51, and Asp52; CDR3α: Asp92, Ser93, Asn94, Tyr95), and 14 in the Vβ chain (CDR1β: Leu26, Phe28, Gln29, Thr31, and Thr32; CDR2β: Asn51, Glu52, Gly53, Ser54, and Lys55; CDR3β: Arg97, Ser99, Gly100, and Phe102). Leu26βAla was remote from the likely MAIT TCR–MR1-Ag–binding site and was selected as a negative control, whereas all the other mutants were at surface exposed positions within the CDR loops. As these residues were surface exposed, they were unlikely to affect the conformation of the CDR loop per se, and thus their mutation was predicted to not indirectly impact on MR1-induced activation. Thus, any effect observed by these alanine mutations could be attributable to impacting directly on MR1 binding.
Constructs containing MAIT TCR alanine mutations were transduced into SKW3 and Jurkat cells, and the cell surface expression of MAIT TCR was closely matched as determined by flow cytometry using staining for cell surface CD3 expression and coexpression of GFP (unpublished data). We then tested the impact of these mutants using C1R APCs infected with S. typhimurium (Fig. 4). We observed that mutations at six residues markedly impaired responses (Gly28α, Asn30α, Tyr48α, Asp92α, Asn94α, and Tyr95α; Fig. 4 A). Surprisingly, none of the β chain mutants showed decreased activation responses to S. typhimurium, suggesting that no individual residue within the β chain was essential for the MAIT TCR–MR1 recognition (Fig. 4 B). All 25 of the mutant SKW3 cells expressing TRBV20 Ala-substitutions were activated by beads coated with anti-CD3 and anti-CD28 mAbs indicating intact signaling capacity was retained by the mutant TCRs (unpublished data). Similar results were obtained using transduced Jurkat cells as T cells responders and we observed the Jurkat cells were slightly less sensitive than the transduced SKW3 cells (unpublished data). Thus, the mutational data suggest a limited number of Vα residues of the MAIT TCR are critical for the MR1-induced activation.
Figure 4.
Activation of mutant SKW3.BV20 cells by S. typhimurium. 11 mutant α chain (A) and 14 mutant β chain (B) SKW3.TRBV20 cell lines were incubated with C1R cells infected with S. typhimurium at a MOI of 100. Shaded bars show the fold increase in CD69 surface expression (fold increase in MFI) of mutant SKW3.TRBV20 cells co-incubated with C1R cells infected with S. typhimurium compared with SKW3.BV20 cells co-incubated with uninfected C1R cells (open bars). A positive control, wild-type SKW3.TRBV20 (TRBV20.WT), and a negative control, SKW3.LC13 (LC13.WT), were included. The mutant CDR1β Leu26A cell line was included as an internal control where activation was expected to remain intact. (C) MAIT.TRBV6-1 TCRs with six separate solvent-exposed residues of the β chain mutated to alanine were transduced into SKW3 cells and tested in the same manner as the mutant SKW3.TRBV20 cell lines in A and B. (D) SKW3 cells transduced with MAIT TCRs containing CDR3β regions of the MAIT.TRBV6-1 TCR or the MAIT.TRBV20 TCR exchanged with the CDR3β regions of known functional TCRs using TRBV6-1 or TRBV20, respectively, were then tested in the same manner as the mutant SKW3.TRBV20 cell lines in A and B. The following SKW3-transduced cell lines were tested: wild-type SKW3.TRBV6-1 cells (TRBV6-1.WT); SKW3.TRBV6-1 cells with a TCR containing the SB27 TRBV6-1 CDR3β region (TRBV6-1/SB27β); wild-type SKW3.TRBV20 cells (TRBV20.WT); SKW3.TRBV20 cells with a TCR containing the ABC TRBV20 CDR3β region (TRBV20/ABCβ; to be described elsewhere); SKW3 cells with a TCR containing the MAIT invariant α chain paired with the LC13 TCR β chain (MAIT.LC13β); and SKW3 cells with the wild-type LC13 TCR (LC13αβ.WT). The experiments shown in A–D were also performed at an MOI of 1 and 10 and yielded similar results (not depicted). The experiments shown in A and B and C and D were done three times and twice, respectively, with similar results.
Role of the Vβ interactions in MAIT specificity
To further examine the role of Vβ residues in MAIT TCR activation we examined the impact of alanine mutations in five solvent exposed CDR1β residues (Asn28, His29, Asn30, Ser31, and Tyr33; as well as a control residue, Asn66, of the MAIT TRBV6-1 TCR. The structure of the SB27 TCR (Tynan et al., 2005b) was used to determine solvent exposed residues in the CDR1β loop of the TRBV6-1 β chain from the MAIT TRBV6-1 TCR. Constructs containing these alanine mutations were transduced into SKW3 cells that were matched for TCR expression as described above. Testing the impact of these mutants using wild-type C1R APCs infected with S. typhimurium revealed that none of the mutants individually had an adverse impact on MR1-restricted activation by the mutated MAIT TCRs (Fig. 4 C). These findings underscore the observation that no individual germline-encoded residue within the Vβ chain is essential in mediating MR1-restricted activation of the MAIT TCR.
Next, given that none of the individual CDR3β mutations in TRBV20 had any apparent impact on MAIT cell activation, we created T cells expressing TCRs where the whole CDR3β of a MAIT TCR was swapped for that of another with non–MAIT TCR specificity. For this purpose, we selected two unrelated T cell clones with Vβ usage similar to MAIT clone TRBV6-1 and clone TRBV20. The first, SB27, recognizes HLA-B*3501 complexed to a peptide from an Epstein–Barr virus Ag and uses a TRBV6-1 β chain similar to the β chain of the MAIT clone BV6-1(Tynan et al., 2005a,b). Second, clone ABC recognizes HLA-B*5701 and the drug abacavir and expresses a TRBV20 β chain similar to the β chain of MAIT clone TRBV20. The CDR3β loop of MAIT clone 6–1 was replaced with the SB27 CDR3β loop, whereas the CDR3β loop of MAIT clone TRBV20 was replaced with the CDR3β loop of clone ABC, and these hybrid TCRs were expressed with Vα7.2Jα33 in SKW3 T cells (Fig. S3). When stimulated by C1R cells infected with S. typhimurium, neither the SKW3 cells expressing MAIT.TRBV6-1/SB27.CDR3β nor the cells expressing MAIT.TRBV20/ABC.CDR3β were activated (Fig. 4 D). Activation by anti-CD3 and anti-CD28 beads was identical in all transduced T cells (unpublished data), and the parental wild-type MAIT cells were activated normally by S. typhimurium–infected C1R cells. Accordingly, although single-residue mutants in the CDR3β of the MAIT-TRBV20 TCR had no impact on MAIT TCR activation, swapping the full CDR3β loop of both this MAIT-TRBV20 TCR and the MAIT-TRBV6-1 TCR completely abolished MAIT TCR activation in both of these clones. This indicates that redundancy in the important contacts from this loop or that the conformation of the CDR3β loop can impact on the MAIT TCR–MR1 interaction, possibly via steric hindrance mechanisms.
Identical TCR specificity pattern independent of bacterial stimulators
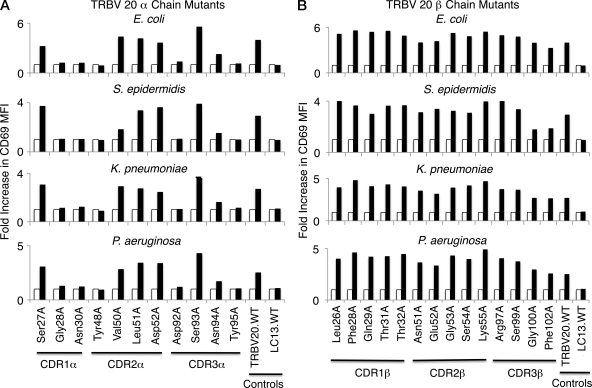
We next examined whether the specificity pattern observed in the TRBV20 SKW3 mutants stimulated by S. typhimurium was the same or different when the cells were stimulated by other bacteria. C1R APCs were independently infected with either the Gram-negative bacteria Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa or with the Gram-positive bacterium Staphylococcus epidermidis, and the infected APCs were used to stimulate the SKW3.TRBV20 mutant T cells (Fig. 5). The pattern of T cell activation was indistinguishable from that observed when the mutants were stimulated by C1R infected with S. typhimurium, such that Vα residues Gly28α, Asn30α, Tyr48α, Asp92α, Asn94α, and Tyr95α markedly reduced MAIT TCR activation. Again, there was no consistent influence of TCR β chain mutations on T cell activation by any of the bacteria, with the exception of assay variation in T cell responses of some of the Vβ mutants (e.g., S. epidermidis, Gly100βAla, and Phe102βAla). These results suggest that the MAIT TCR acts like a pattern recognition receptor, with a conserved MR1-binding mode, irrespective of the source of bacterial stimulation.
Figure 5.
Activation of mutant SKW3.TRBV20 cell lines by different bacterial species. 11 mutant α chain (A) and 14 mutant β chain (B) SKW3.TRBV20 cell lines were tested for activation by E. coli, S. epidermidis, K. pneumoniae, and P. aeruginosa. Shaded bars show the fold increase in CD69 surface expression (fold increase in MFI) of mutant SKW3.TRBV20 cells co-incubated with C1R cells infected with bacteria at an MOI of 100 compared with SKW3.TRBV20 cells co-incubated with uninfected C1R cells (open bars). The experiments shown in A and B were also performed at an MOI of 1 and 10, with similar results (not depicted). These experiments were performed three times, with similar results.
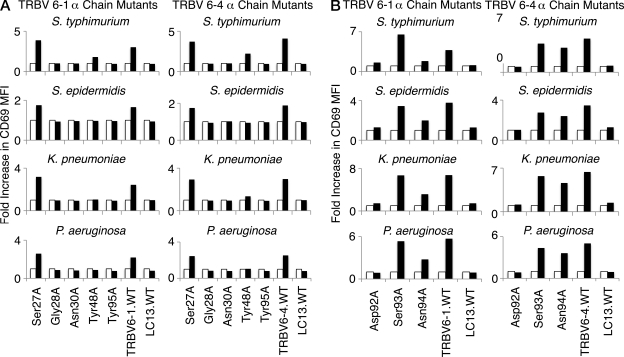
Conserved fine specificity by other MAIT TCRs
We next examined whether the same pattern of MAIT TCR specificity was evident when the mutated TRAV1-2 α chains (Vα7.2-Jα33; selected α chain mutants: Ser27, Gly28, Asn30, Tyr48, Asp92, Ser93, Asn94, and Tyr95) were paired with other MAIT Vβ families (Tilloy et al., 1999). Thus, we transduced SKW3 T cells with genes encoding the mutant TRAV1-2 (Vα7.2/Jα33) TCR α chain and either the TRBV6-1 or the TRBV6-4 β chains, representing the alternate Vβ13 family preferentially used by some human MAIT cells. The transduced cells were stimulated by C1R APCs infected with S. typhimurium, K. pneumoniae, P. aeruginosa, or S. epidermidis, and activation of SKW3 cells was determined by staining and flow cytometric analysis of surface CD69 expression (Fig. 6, A and B). Except for activation responses in SKW3.TRBV6-4 cells with the Asn94Ala mutation being only minimally impaired, the fine specificity pattern of reactivity of the TCR α chain mutants was otherwise very similar to that observed for the SKW3.TRBV20 cells, regardless of the source of the TCR β chain or the source of bacterial stimulation, again suggesting a passive role for the TCR β chain in direct Ag-specificity observed in our system. Specific responses were blocked by the anti-MR1 mAb 26.5 and were dose dependent (unpublished data). Accordingly, MR1-restricted MAIT TCR–mediated activation is not modulated appreciably by the TCR β chain, and is determined by the invariant α chain (Fig. 2 G).
Figure 6.
The α chain residues crucial to MAIT TCR recognition are conserved in two other MAIT TCRs. The mutant α chain residues observed to diminish bacterial activation in SKW3.TRBV20 cell lines (Gly28Ala, Asn30Ala, Tyr48Ala, Asp92Ala, Asn94Ala, and Tyr95Ala, as well as control Ser27Ala and Ser93Ala mutations) were introduced into the MAIT.TRBV6-1 and MAIT.TRBV6-4 TCRs before transduction of MAIT TCR genes into SKW3 cells. SKW3.TRBV6-1 and SKW3.TRBV6-4 cell lines transduced with mutants Ser27Ala, Gly28Ala, Asn30Ala, Tyr48Ala, or Tyr95Ala (A), or mutants Asp92Ala, Ser93Ala, or Asn94Ala (B) were then tested for activation by S. typhimurium, S. epidermidis, K. pneumoniae, and P. aeruginosa. Shaded bars show the fold increase in CD69 surface expression (fold increase in MFI) of mutant SKW3.TRBV6-1 or mutant SKW3.TRBV6-4 cells co-incubated with C1R cells infected with bacteria at an MOI of 100 compared with SKW3.TRBV6-1 or SKW3.TRBV6-4 cells co-incubated with uninfected C1R cells (open bars). These experiments were performed three times, with similar results.
A small energetic footprint on MR1
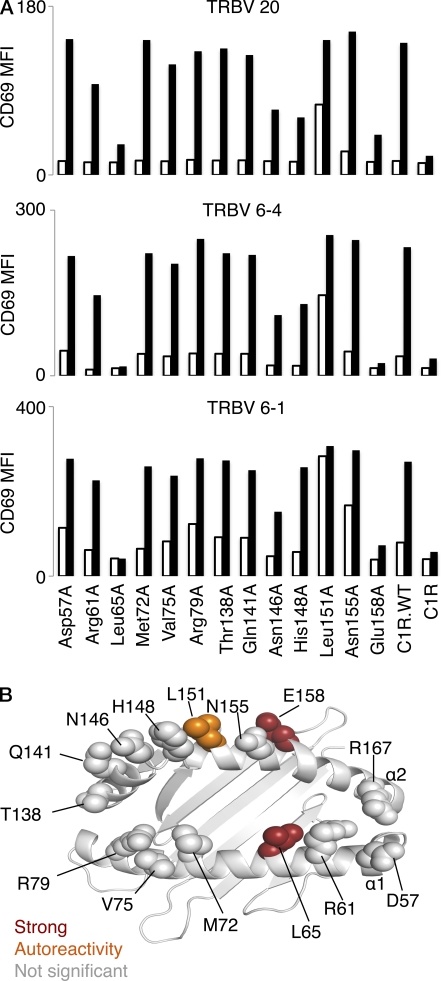
Having established that a small number of MAIT TCR residues were essential for MR1 recognition, we next explored whether the MAIT TCR–MR1 interaction was underpinned by a focused energetic “hot spot” on MR1. To establish this, we generated a homology model of human MR1 (see Materials and methods) and selected 14 residues, 6 on the α1-helix (Asp57, Arg61, Leu65, Met72, Val75, and Arg79) and 8 on the α2-helix (Thr138, Gln141, Asn146, His148, Leu151, Asn155, Glu158, and Arg167). The selected residues were exposed to solvent, and thus represented potential MAIT TCR contact points. Moreover, the residues targeted for mutagenesis included charged, polar, and hydrophobic residues, and ran along the length and breadth of the Ag-binding cleft of MR1. Accordingly, the MR1 mutagenesis approach enabled us to gain a broad perspective on the chemistry of the interaction, as well as the energetic requirement of the MR1 residues that are central to the interaction with the MAIT TCR. The 14 single-site alanine mutants were transfected into C1R cells, and cells were then sorted for high levels of MR1 expression by flow cytometry using coexpressed GFP (Fig. 7).
Figure 7.
The effect of mutation of MR1 residues on MAIT TCR activation. (A) 13 mutant MR1, as well as wild-type MR1 (C1R.WT) and parental C1R, cell lines were either not infected (open bars) or infected (shaded bars) with S. typhimurium at a multiplicity of infection (MOI) of 1. After infection, Jurkat.TRBV 20, Jurkat.TRBV 6–1, or Jurkat.TRBV 6–4 cell lines were then added for 16 h before measurement of increase in CD69 surface expression (MFI) by staining and flow cytometric analysis. This experiment was performed twice with similar results. Mutant Arg167Ala MR1 C1R cells activated Jurkat.MAIT cells similarly to wild-type MR1 C1R cells (not depicted). The experiment was also performed at an MOI of 10 with similar results (not depicted). (B) The effects of the MR1 mutants were mapped onto the human MR1 homology model. Mutations that had no impact on MAIT TCR activation is shown in gray; impact on autoreactivity is shown in orange; mutants that markedly reduced MAIT TCR activation are shown in red.
Jurkat cells expressing human MAIT TCRs (with either TRBV6-1, TRBV6-4, or TRBV20 pairing β chains) were incubated for 16–20 h, with C1R cells expressing either wild-type or each mutant MR1, in the absence or presence of S. typhimurium. Activation of MAIT TCR–expressing Jurkat cells was determined by flow cytometric analysis of up-regulation of cell surface CD69 expression. Of the 14 MR1 mutants tested, mutation of 9 residues (Asp57, Arg61, Met72, Val75, Arg79, Thr138, Gln141, Asn155, and Arg167) had no significant impact on MAIT activation; two residues (Asn146 and His148) had a minor effect on MAIT TCR activation; one mutation (Leu151Ala) resulted in enhanced autoreactivity in the absence of bacteria; and, most importantly, two mutants (Leu65 and Glu158) markedly reduced MAIT TCR activation (Fig. 7 A). The pattern of the effect of these mutants was the same, regardless of which MAIT TCR was used in the activation assay. Strikingly, these two key positions map centrally to the MR1-Ag–binding cleft, directly opposite each other (Fig. 7 B). Moreover, the “autoreactive” and less impacted positions are proximal to Glu158 on the α2-helix. The effect of these key positions (Leu65 and Glu158) indicates that there is a centrally located and extremely focused energetic hot spot on MR1 that underpins recognition by the MAIT TCR.
DISCUSSION
MAIT cells share many characteristics with NKT cells in that both cell types express invariant TCR-α chains paired preferentially with a restricted range of TCR-β chains (Godfrey et al., 2010b). Minor differences exist in that the NKT receptor α chain has little or no N-region variation at the V-J junction whereas MAIT α chains can tolerate some N-region variation at this junction (Tilloy et al., 1999; Bendelac et al., 2007). Nonetheless, both types of T cell have appropriated their largely innate receptors from the pool of αβ TCRs that are considered part of the adaptive immune repertoire. Both T cells are activated by MHC-I–like Ag-presenting molecules (CD1d and MR1 for NKT and MAIT cells, respectively). In contrast to NKT recognition, the underlying basis of the MAIT TCR–MR1 interaction is unclear, largely because the identity of the putative MR1-restricted ligand is unknown (Huang et al., 2008); indeed, it has not yet been definitively shown that MR1 presents self- or bacterially derived Ags. Nevertheless, we used a combined structural and mutagenesis approach (a proven approach used to understand biased TCR usage in pMHC and CD1d-Ag recognition; Borg et al., 2005; Scott-Browne et al., 2007; Wun et al., 2008) to provide the first insight into the specificity requirements that underscore MAIT cell activation by MR1. Initially, we determined the structure of the human MAIT TCR (Vα7.2Jα33-Vβ2), which showed that the CDR loops formed an ordered Ag-binding interface, thereby suggesting that there is limited conformational plasticity in the MAIT TCR–MR1 interaction. Further, the MAIT TCR structure showed that the central binding pocket, lined by the CDR3α and CDR3β loops, markedly contrasted to that of the human NKT TCR (Kjer-Nielsen et al., 2006), differences that presumably reflect that MAIT and NKT cells are activated by MR1 and CD1d, respectively.
Our MAIT TCR mutagenesis studies identified Vα residues Gly28 and Asn30 from CDR1α, Tyr48 from CDR2α, and Asp92, Asn94, and Tyr95 from CDR3α (Jα) as being important or crucial for MAIT TCR activation, regardless of whether the MAIT TCR expressed Vβ2 or Vβ13. Notably, the importance of these residues in MR1-restricted activation explains the invariant selection of the Vα7.2-Jα33 α chain segments that uniquely encode this cluster of residues. These residues are fully conserved in homologous Vα-Jα combinations selected across humans, mice, sheep, and cattle (Tilloy et al., 1999; Goldfinch et al., 2010; Fig. S4). This pattern of germline Vα and Jα residues defining cognate activation therefore has a similar basis to the invariant selection of Vα24-Jα18 by NKT cells in their CD1d-restricted recognition of glycolipids (Gadola et al., 2006; Kjer-Nielsen et al., 2006).
The small energetic footprint of the MAIT TCR was mirrored by the small energetic footprint on MR1, where mutation of only two residues, Leu65 and Glu158, markedly reduced MAIT TCR activation, regardless of the Vβ usage of the MAIT TCR. In addition, one mutation, Leu151Ala, promoted MAIT TCR–MR1 autoreactivity, and this position was previously shown to determine human and mouse MAIT TCR–MR1 cross-species reactivity (Huang et al., 2009). These three positions clustered closely, and were centrally located within the Ag-binding cleft of MR1, tentatively suggesting that the MAIT TCR docks centrally on MR1; it is also conceivable that these residues are involved in stabilization of Ag binding to MR1. A central MAIT TCR–MR1 docking mode would contrast with that of the NKT TCR–CD1d-Ag interaction, in which the NKT TCR is perched above the F′-pocket of CD1d, whereupon the CDR2β loop makes critical contacts with CD1d (Godfrey et al., 2010a).
In NKT cells, differential Vβ usage is observed, with some regions of sequence conservation observed between the different Vβ families. These conserved Vβ residues were important not only for NKT cognate interaction but also for the cross-species reactivity between mouse and human CD1d (Wun et al., 2008). Although MAIT cells also preferentially select a few Vβ families, there is limited sequence identity between the CDR1β and CDR2β regions of human Vβ2 and Vβ13 (Tilloy et al., 1999), suggesting that the reported cross reactivity of human MAIT cells on mouse MR1 may be largely underpinned by the Vα-Jα chain residues interacting with the MR1 “hotspot.” Indeed, our functional data suggests that differential Vβ usage in MAIT cells cannot be assigned to single-residue contributions. Importantly, using a variety of bacteria as sources of MAIT TCR stimulation, we demonstrated that individual residues within the Vβ chain did not play a controlling role in MAIT cell activation when analyzed by mutagenesis, which is perhaps consistent with the lack of β chain sequence conservation in humans. However, replacement of whole MAIT CDR3β loops with those from non–MAIT TCRs abolishes T cell activation indicating a more complex role for CDR3β residues in MAIT specificity. It is possible that the conformation of the CDR3β loop can impact on the conformation of the CDR3α loop, thereby impacting MR1 recognition in a manner similar to how Vβ-based changes impact the NKT TCR α chain conformation and resultant CD1d-Ag recognition (Kjer-Nielsen et al., 2006; Pellicci et al., 2009), or in a manner analogous to that recently observed in MHC-restricted immunity (Stadinski et al., 2011). It is also possible that the selective Vβ usage by MAIT cells is shaped by a putative positive selecting endogenous ligand, as suggested for NKT cells (Wei et al., 2006), and/or avoidance of self-MR1 reactivity with endogenous MR1-bound ligands.
In MHC-restricted immunity, the TCR can ligate the pMHC landscape through a range of docking modes to mediate viral immunity, alloreactivity, and autoimmunity (Burrows et al., 2010). This variability is attributable to the highly polymorphic nature of the MHC molecules, the variable peptide cargo, and the diversity of TCR Vα and Vβ usage. Moreover, MHC-restricted TCRs are inherently cross-reactive (Archbold et al., 2008), and this cross-reactivity is often manifested in conformational plasticity of the CDR loops (or the pMHC itself; Tynan et al., 2007; Archbold et al., 2009) upon TCR-pMHC ligation. In contrast, our data suggest that the MAIT TCR might be relatively rigid, which is characteristic of innate receptors styled to recognize monomorphic or only modestly variable Ags. Moreover, the crucial role for the same TCR Vα-Jα residues in recognizing different species of Gram-negative and -positive bacteria suggests a highly conserved MR1 docking mode that underpins MAIT cell activation by diverse microbes. Collectively, our studies have provided the first fundamental insight into the MAIT TCR–MR1 interaction and the basis for invariant TCRα chain selection.
MATERIALS AND METHODS
Generation of MR1-expressing APCs.
Full-length cDNA genes encoding either human or murine MR1 were cloned into a modified version of p-MSCV-IRES-eGFP (pMIG; D. Vignali, St. Jude Children’s Research Hospital, Memphis, TN; Liu et al., 1997; Pear et al., 1998). 293T packaging cells were then transfected with retroviral vectors using FuGENE 6 (Roche), and C1R or HeLa cells were transduced with retroviral supernatant. MR1 was detected on the surface of transduced cells with the MR1-reactive mAb 26.5 (Huang et al., 2005). A preTα-specific mAb 8A5 generated against recombinantly expressed preTCR (Pang et al., 2010) was used as an isotype control in staining experiments.
Generation of T cell lines expressing MAIT TCRs.
Full-length human cDNA TCR α and β chain genes were cloned into a self-cleaving 2A peptide-based pMIG vector (Szymczak et al., 2004) and transduced into parental hybridoma Jurkat or SKW3 T cells using 293T packaging cells. The human T cell line SKW3 lacks endogenous TCRαβ expression but possesses the CD3 and other components of the signaling apparatus. Jurkat lacks endogenous TCRβ but has an endogenous TCRα and CD3 components. Expression of heterologous TCRαβ genes in SKW3 and Jurkat restores parental TCR specificity that can be assayed by up-regulation of CD69 using flow cytometry (Fig. S5 and Fig. S6). SKW3 cells were also transduced with hybrid MAIT TCRs containing the CDR3β regions from either an abacavir-specific, TRBV20-positive T cell clone, ABC (Chessman et al., 2008); or from an Epstein–Barr-specific, TRBV6-1–positive T cell clone (SB27; Tynan et al., 2005b).
Activation studies with MAIT TCR–expressing T cell lines.
Jurkat or SKW3 T hybridoma cells expressing human MAIT TCRs were incubated for 16–20 h with APCs expressing human MR1 (or mutants thereof), in the absence or presence of S. typhimurium, P. aeruginosa, K. pneumoniae, E. coli, or S. epidermidis. Activation of MAIT TCR–expressing T cells was determined by flow cytometric analysis of up-regulation of cell surface CD69 expression. The 25 SKW3 cell lines expressing TRBV20 Ala substitutions were also incubated with beads coated with anti-CD3 and anti-CD28 mAbs titrated to produce activation that just achieved plateau levels of stimulation of SKW3 cells expressing a wild-type MAIT TCR.
Generation of MAIT TCR mutants.
MAIT TCR genes encoding single alanine-mutations were generated by the QuikChange Site-Directed Mutagenesis method (Stratagene). All mutant genes were fully sequenced before subcloning into the 2A peptide-based pMIG vector and subsequent retroviral transfer into Jurkat and SKW3 cells. Residues to be mutated were selected on the basis of the solved crystal structure of a MAIT TCR, and include the following CDR residues: CDR1α: Ser27, Gly28, and Asn30; CDR2α: Tyr48, Val50, Leu51, and Asp52; CDR3α: Asp92, Ser93, Asn94, and Tyr95; CDR1β: Phe28, Gln29, Thr31, Thr32, and Leu26 (Leu26 as a control); CDR2β: Asn51, Glu52, Gly53, Ser54, and Lys55; CDR3β: Arg97, Ser99, Gly100, and Phe102.
Protein expression and purification.
The MAIT TRBV20 and TRBV6-1 and TRBV6-4 TCRs were expressed, refolded, and purified using an engineered disulfide linkage in the constant domains between the TRAC and TRBC, essentially as described previously (Clements et al., 2002). In brief, the α and β chain of the MAIT TCR were expressed separately as inclusion bodies in BL21 E. coli strain. Inclusion bodies were resuspended in 8 M Urea, 20 mM Tris-HCl, pH 8.0, 0.5 mM Na-EDTA, and 1 mM DTT. TCRs were refolded by flash dilution in a solution containing 5 M Urea, 100 mM Tris, pH 8.0, 2 mM Na-EDTA, 400 mM l-arginine-HCl, 0.5 mM oxidized glutathione, 5 mM reduced glutathione, PMSF, and pepstatin A. The refolding solution was then dialyzed to eliminate the urea. The resulting refolded protein was then purified sequentially by DEAE anion exchange, gel filtration, and Mono-Q anion exchange chromatography.
Crystallization, structure determination, and refinement.
The MAIT TCR Vα7.2Jα33-Vβ2 (7–10 mg/ml in 10 mM Tris, pH 8.0, and 150 mM NaCl) crystallized at 20°C in 25% PEG 1500, 0.1 M MIB buffer (sodium malonate/Imidazole/boric acid in molar rations of 2:3:3), pH 5.0, via the hanging drop vapor diffusion technique. Equal ratio of the protein to mother liquor resulted in plate-like crystals after 1–3 d. The crystals were flash frozen before data collection in mother liquor containing 20% glycerol. The crystals diffracted to 1.7 Å at the MX2 beamline at the Australian Synchrotron facility in Melbourne, Australia (Table 1). The crystals belong to the space group P212121, with one molecule in the asymmetric unit. The crystal structure of the MAIT TCR was solved by molecular replacement method with the program Phaser from the CCP4 Suite (Collaborative Computational Project, Number 4, 1994), using ELS4 TCR (Protein Data Bank ID accession code 2NX5; Tynan et al., 2007) without the CDR loops and loops connecting the variable and constant domains as a model. ARP/wARP in the CCP4 suite (Collaborative Computational Project, Number 4, 1994) was used for automated model building, and Refmac was used for restrained refinement (Table 1). The quality of structure was confirmed at the Research Collaboratory for Structural Bioinformatics Protein Data Bank Data Validation and Deposition Services website (http://www.rcsb.org/pdb/home/home.do; Protein Data Bank accesion code 4DZB). All molecular graphics representations were created using PyMOL (DeLano, 2002).
Homology modeling of MR1.
The sequence of full-length human MR1 (UniProt accession no. Q95460) was used to generate a homology model of the protein via the PHYRE web server (Bennett-Lovsey et al., 2008). The sequence of human MR1 lacking the N-terminal signal peptide was also used as a query on the server. Homologue detection by the server against the human MR1 protein sequence was considered reliable, as sequence identity between the query sequence and retrieved homologous sequences was >40%. All generated homology models were inspected manually before selection of the final model for structural analysis. The homology model of MR1 used for final analysis was based on the high resolution crystal structure of a chicken MHC-I molecule YFCI*7.1 (Hee et al., 2010), to which it was found to have the highest sequence identity (42%) of all candidate homologues retrieved by the server, as well as a manual search for homologues using a BLAST search. The MR1 homology model contains residues 22–296 of human MR1. 14 residues on the α1 and α2 helices of the MR1 homology model, which did not appear to affect the local fold of the protein upon substitution by alanine and were predicted to be solvent exposed, were targeted for mutagenesis.
Generation of MR1 mutants.
14 MR1 genes encoding single alanine mutations were generated by the QuikChange Site directed mutagenesis method (Stratagene). All mutant genes were fully sequenced before subcloning into the pMIG vector and subsequent retroviral transfer into C1R cells. Residues to be mutated were selected on the basis of the human MR1 homology model, and include the following residues: Asp57, Arg61, Leu65, Met72, Val75, Arg79, Thr138, Gln141, Asn146, His148, Leu151, Asn155, Glu158, and Arg167. Expression of MR1 was confirmed by staining with the MR1-reactive mAb 26.5; as well as coexpression of GFP. High-expressing MR1-transduced C1R cells were sorted by flow cytometry using the coexpressed GFP gene, and sorted cells were subsequently used in activation assays with Jurkat cells expressing MAIT TCRs.
Online supplemental material.
Fig. S1 shows a comparison of the CDR3 amino acid sequences of MAIT TCRs using the β chains TRBV6-1, TRBV6-4, and TRBV20. Fig. S2 shows a comparison of the CDR1 and CDR2 regions of Vβ chains of human and mouse MAIT TCRs. Fig. S3 shows a comparison of the CDR3β amino acid sequences of four TCR β chains. Fig. S4 shows the alignment of MAIT Vα7.2/Jα33 amino acid sequences from four species. Fig. S5 shows the gating strategy for CD69 measurement. Fig. S6 shows the CD69 measurement of gated SKW3 cells. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20112095/DC1.
Acknowledgments
We thank the staff at the MX2 beamline of the Australian synchrotron for assistance in data collection and Daniel Pellicci for useful discussions.
This work is supported by program and project grants of the Australian National Health and Medical Research Council (NHMRC). D.I. Godfrey is an NHMRC Senior Principal Research Fellow; J. Rossjohn is an Australia fellow of the NHMRC. R. Reantragoon is a PhD scholar funded by the Faculty of Medicine, Chulalongkorn University (Bangkok, Thailand) and by the King Chulalongkorn Memorial Hospital, Thai Red Cross.
The authors have no competing financial interests.
Footnotes
Abbreviations used:
- CDR
- complementarity determining region
- D
- diversity
- J
- junction
- MAIT
- mucosal-associated invariant T
- MFI
- mean fluorescent intensity
- MOI
- multiplicity of infection
- MR1
- MHC class I–like molecule
- pMHC
- peptides complexed to MHC
- V
- variable
References
- Archbold J.K., Macdonald W.A., Burrows S.R., Rossjohn J., McCluskey J. 2008. T-cell allorecognition: a case of mistaken identity or déjà vu? Trends Immunol. 29:220–226 10.1016/j.it.2008.02.005 [DOI] [PubMed] [Google Scholar]
- Archbold J.K., Macdonald W.A., Gras S., Ely L.K., Miles J.J., Bell M.J., Brennan R.M., Beddoe T., Wilce M.C., Clements C.S., et al. 2009. Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition. J. Exp. Med. 206:209–219 10.1084/jem.20082136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A., Savage P.B., Teyton L. 2007. The biology of NKT cells. Annu. Rev. Immunol. 25:297–336 10.1146/annurev.immunol.25.022106.141711 [DOI] [PubMed] [Google Scholar]
- Bennett-Lovsey R.M., Herbert A.D., Sternberg M.J.E., Kelley L.A. 2008. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 70:611–625 10.1002/prot.21688 [DOI] [PubMed] [Google Scholar]
- Borg N.A., Ely L.K., Beddoe T., Macdonald W.A., Reid H.H., Clements C.S., Purcell A.W., Kjer-Nielsen L., Miles J.J., Burrows S.R., et al. 2005. The CDR3 regions of an immunodominant T cell receptor dictate the ‘energetic landscape’ of peptide-MHC recognition. Nat. Immunol. 6:171–180 10.1038/ni1155 [DOI] [PubMed] [Google Scholar]
- Brigl M., Tatituri R.V.V., Watts G.F.M., Bhowruth V., Leadbetter E.A., Barton N., Cohen N.R., Hsu F.-F., Besra G.S., Brenner M.B. 2011. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J. Exp. Med. 208:1163–1177 10.1084/jem.20102555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows S.R., Chen Z., Archbold J.K., Tynan F.E., Beddoe T., Kjer-Nielsen L., Miles J.J., Khanna R., Moss D.J., Liu Y.C., et al. 2010. Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability. Proc. Natl. Acad. Sci. USA. 107:10608–10613 10.1073/pnas.1004926107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chessman D., Kostenko L., Lethborg T., Purcell A.W., Williamson N.A., Chen Z., Kjer-Nielsen L., Mifsud N.A., Tait B.D., Holdsworth R., et al. 2008. Human leukocyte antigen class I-restricted activation of CD8+ T cells provides the immunogenetic basis of a systemic drug hypersensitivity. Immunity 28:822–832 [DOI] [PubMed] [Google Scholar]
- Chiba A., Tajima R., Tomi C., Miyazaki Y., Yamamura T., Miyake S. 2012. Mucosal-associated invariant T cells promote inflammation and exacerbate disease in murine models of arthritis. Arthritis Rheum. 64:153–161 10.1002/art.33314 [DOI] [PubMed] [Google Scholar]
- Chua W.-J., Kim S., Myers N., Huang S., Yu L., Fremont D.H., Diamond M.S., Hansen T.H. 2011. Endogenous MHC-related protein 1 is transiently expressed on the plasma membrane in a conformation that activates mucosal-associated invariant T cells. J. Immunol. 186:4744–4750 10.4049/jimmunol.1003254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements C.S., Kjer-Nielsen L., Brooks A.G., Purcell A.W., McCluskey J., Rossjohn J. 2002. The production, purification and crystallisation of a soluble, heterodimeric form of a highly selected T-cell receptor in its unliganded and liganded state. Acta Crystallogr. D Biol. Crystallogr. 58:2131–2134 10.1107/S0907444902015482 [DOI] [PubMed] [Google Scholar]
- Clements C.S., Dunstone M.A., Macdonald W.A., McCluskey J., Rossjohn J. 2006. Specificity on a knife-edge: the alphabeta T cell receptor. Curr. Opin. Struct. Biol. 16:787–795 10.1016/j.sbi.2006.09.004 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- DeLano W.L. 2002. The PyMOL Molecular Graphics System. http://www.pymol.org
- Dolinsky T.J., Czodrowski P., Li H., Nielsen J.E., Jensen J.H., Klebe G., Baker N.A. 2007. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35:W522-525 10.1093/nar/gkm276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusseaux M., Martin E., Serriari N., Péguillet I., Premel V., Louis D., Milder M., Le Bourhis L., Soudais C., Treiner E., Lantz O. 2011. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 117:1250–1259 10.1182/blood-2010-08-303339 [DOI] [PubMed] [Google Scholar]
- Gadola S.D., Koch M., Marles-Wright J., Lissin N.M., Shepherd D., Matulis G., Harlos K., Villiger P.M., Stuart D.I., Jakobsen B.K., et al. 2006. Structure and binding kinetics of three different human CD1d-alpha-galactosylceramide-specific T cell receptors. J. Exp. Med. 203:699–710 10.1084/jem.20052369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapin L. 2009. Where do MAIT cells fit in the family of unconventional T cells? PLoS Biol. 7:e70 10.1371/journal.pbio.1000070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia K.C., Degano M., Pease L.R., Huang M., Peterson P.A., Teyton L., Wilson I.A. 1998. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science. 279:1166–1172 10.1126/science.279.5354.1166 [DOI] [PubMed] [Google Scholar]
- Gillis S., Watson J. 1980. Biochemical and biological characterization of lymphocyte regulatory molecules. V. Identification of an interleukin 2-producing human leukemia T cell line. J. Exp. Med. 152:1709–1719 10.1084/jem.152.6.1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey D.I., Rossjohn J. 2011. New ways to turn on NKT cells. J. Exp. Med. 208:1121–1125 10.1084/jem.20110983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey D.I., Rossjohn J., McCluskey J. 2008. The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity. 28:304–314 10.1016/j.immuni.2008.02.004 [DOI] [PubMed] [Google Scholar]
- Godfrey D.I., Pellicci D.G., Patel O., Kjer-Nielsen L., McCluskey J., Rossjohn J. 2010a. Antigen recognition by CD1d-restricted NKT T cell receptors. Semin. Immunol. 22:61–67 10.1016/j.smim.2009.10.004 [DOI] [PubMed] [Google Scholar]
- Godfrey D.I., Rossjohn J., McCluskey J. 2010b. Fighting infection with your MAITs. Nat. Immunol. 11:693–695 10.1038/ni0810-693 [DOI] [PubMed] [Google Scholar]
- Gold M.C., Cerri S., Smyk-Pearson S., Cansler M.E., Vogt T.M., Delepine J., Winata E., Swarbrick G.M., Chua W.-J., Yu Y.Y.L., et al. 2010. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 8:e1000407 10.1371/journal.pbio.1000407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfinch N., Reinink P., Connelley T., Koets A., Morrison I., Van Rhijn I. 2010. Conservation of mucosal associated invariant T (MAIT) cells and the MR1 restriction element in ruminants, and abundance of MAIT cells in spleen. Vet. Res. 41:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gras S., Kjer-Nielsen L., Burrows S.R., McCluskey J., Rossjohn J. 2008. T-cell receptor bias and immunity. Curr. Opin. Immunol. 20:119–125 10.1016/j.coi.2007.12.001 [DOI] [PubMed] [Google Scholar]
- Gras S., Chen Z., Miles J.J., Liu Y.C., Bell M.J., Sullivan L.C., Kjer-Nielsen L., Brennan R.M., Burrows J.M., Neller M.A., et al. 2010. Allelic polymorphism in the T cell receptor and its impact on immune responses. J. Exp. Med. 207:1555–1567 10.1084/jem.20100603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hee C.S., Gao S., Loll B., Miller M.M., Uchanska-Ziegler B., Daumke O., Ziegler A. 2010. Structure of a classical MHC class I molecule that binds “non-classical” ligands. PLoS Biol. 8:e1000557 10.1371/journal.pbio.1000557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Gilfillan S., Cella M., Miley M.J., Lantz O., Lybarger L., Fremont D.H., Hansen T.H. 2005. Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. J. Biol. Chem. 280:21183–21193 10.1074/jbc.M501087200 [DOI] [PubMed] [Google Scholar]
- Huang S., Gilfillan S., Kim S., Thompson B., Wang X., Sant A.J., Fremont D.H., Lantz O., Hansen T.H. 2008. MR1 uses an endocytic pathway to activate mucosal-associated invariant T cells. J. Exp. Med. 205:1201–1211 10.1084/jem.20072579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Martin E., Kim S., Yu L., Soudais C., Fremont D.H., Lantz O., Hansen T.H. 2009. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc. Natl. Acad. Sci. USA. 106:8290–8295 10.1073/pnas.0903196106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundhausen T., Laus R., Müller-Ruchholtz W. 1992. New parental cell lines for generating human hybridomas. J. Immunol. Methods. 153:21–29 10.1016/0022-1759(92)90301-9 [DOI] [PubMed] [Google Scholar]
- Joyce S., Girardi E., Zajonc D.M. 2011. NKT cell ligand recognition logic: molecular basis for a synaptic duet and transmission of inflammatory effectors. J. Immunol. 187:1081–1089 10.4049/jimmunol.1001910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawachi I., Maldonado J., Strader C., Gilfillan S. 2006. MR1-restricted V alpha 19i mucosal-associated invariant T cells are innate T cells in the gut lamina propria that provide a rapid and diverse cytokine response. J. Immunol. 176:1618–1627 [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L., Clements C.S., Brooks A.G., Purcell A.W., McCluskey J., Rossjohn J. 2002. The 1.5 A crystal structure of a highly selected antiviral T cell receptor provides evidence for a structural basis of immunodominance. Structure. 10:1521–1532 10.1016/S0969-2126(02)00878-X [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L., Borg N.A., Pellicci D.G., Beddoe T., Kostenko L., Clements C.S., Williamson N.A., Smyth M.J., Besra G.S., Reid H.H., et al. 2006. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J. Exp. Med. 203:661–673 10.1084/jem.20051777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourhis L., Martin E., Péguillet I., Guihot A., Froux N., Coré M., Lévy E., Dusseaux M., Meyssonnier V., Premel V., et al. 2010. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 11:701–708 10.1038/ni.1890 [DOI] [PubMed] [Google Scholar]
- Le Bourhis L., Guerri L., Dusseaux M., Martin E., Soudais C., Lantz O. 2011. Mucosal-associated invariant T cells: unconventional development and function. Trends Immunol. 32:212–218 10.1016/j.it.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Liu X., Sun Y., Constantinescu S.N., Karam E., Weinberg R.A., Lodish H.F. 1997. Transforming growth factor beta-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc. Natl. Acad. Sci. USA. 94:10669–10674 10.1073/pnas.94.20.10669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallevaey T., Clarke A.J., Scott-Browne J.P., Young M.H., Roisman L.C., Pellicci D.G., Patel O., Vivian J.P., Matsuda J.L., McCluskey J., et al. 2011. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity. 34:315–326 10.1016/j.immuni.2011.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrack P., Scott-Browne J.P., Dai S., Gapin L., Kappler J.W. 2008. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu. Rev. Immunol. 26:171–203 10.1146/annurev.immunol.26.021607.090421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E., Treiner E., Duban L., Guerri L., Laude H., Toly C., Premel V., Devys A., Moura I.C., Tilloy F., et al. 2009. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 7:e54 10.1371/journal.pbio.1000054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulis G., Sanderson J.P., Lissin N.M., Asparuhova M.B., Bommineni G.R., Schümperli D., Schmidt R.R., Villiger P.M., Jakobsen B.K., Gadola S.D. 2010. Innate-like control of human iNKT cell autoreactivity via the hypervariable CDR3beta loop. PLoS Biol. 8:e1000402 10.1371/journal.pbio.1000402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki Y., Miyake S., Chiba A., Lantz O., Yamamura T. 2011. Mucosal-associated invariant T cells regulate Th1 response in multiple sclerosis. Int. Immunol. 23:529–535 10.1093/intimm/dxr047 [DOI] [PubMed] [Google Scholar]
- Pang S.S., Berry R., Chen Z., Kjer-Nielsen L., Perugini M.A., King G.F., Wang C., Chew S.H., La Gruta N.L., Williams N.K., et al. 2010. The structural basis for autonomous dimerization of the pre-T-cell antigen receptor. Nature. 467:844–848 10.1038/nature09448 [DOI] [PubMed] [Google Scholar]
- Parham P., Barnstable C.J., Bodmer W.F. 1979. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A,B,C, antigens. J. Immunol. 123:342–349 [PubMed] [Google Scholar]
- Pear W.S., Miller J.P., Xu L., Pui J.C., Soffer B., Quackenbush R.C., Pendergast A.M., Bronson R., Aster J.C., Scott M.L., Baltimore D. 1998. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 92:3780–3792 [PubMed] [Google Scholar]
- Pellicci D.G., Patel O., Kjer-Nielsen L., Pang S.S., Sullivan L.C., Kyparissoudis K., Brooks A.G., Reid H.H., Gras S., Lucet I.S., et al. 2009. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 31:47–59 10.1016/j.immuni.2009.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser J.B., Grégoire C., Darnault C., Mosser T., Guimezanes A., Schmitt-Verhulst A.M., Fontecilla-Camps J.C., Mazza G., Malissen B., Housset D. 2002. A T cell receptor CDR3beta loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex. Immunity. 16:345–354 10.1016/S1074-7613(02)00288-1 [DOI] [PubMed] [Google Scholar]
- Rudolph M.G., Stanfield R.L., Wilson I.A. 2006. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24:419–466 10.1146/annurev.immunol.23.021704.115658 [DOI] [PubMed] [Google Scholar]
- Scott-Browne J.P., Matsuda J.L., Mallevaey T., White J., Borg N.A., McCluskey J., Rossjohn J., Kappler J., Marrack P., Gapin L. 2007. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat. Immunol. 8:1105–1113 10.1038/ni1510 [DOI] [PubMed] [Google Scholar]
- Stadinski B.D., Trenh P., Smith R.L., Bautista B., Huseby P.G., Li G., Stern L.J., Huseby E.S. 2011. A role for differential variable gene pairing in creating T cell receptors specific for unique major histocompatibility ligands. Immunity. 35:694–704 10.1016/j.immuni.2011.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak A.L., Workman C.J., Wang Y., Vignali K.M., Dilioglou S., Vanin E.F., Vignali D.A.A. 2004. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol. 22:589–594 10.1038/nbt957 [DOI] [PubMed] [Google Scholar]
- Tilloy F., Treiner E., Park S.-H., Garcia C., Lemonnier F., de la Salle H., Bendelac A., Bonneville M., Lantz O. 1999. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J. Exp. Med. 189:1907–1921 10.1084/jem.189.12.1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiner E., Duban L., Bahram S., Radosavljevic M., Wanner V., Tilloy F., Affaticati P., Gilfillan S., Lantz O. 2003. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 422:164–169 10.1038/nature01433 [DOI] [PubMed] [Google Scholar]
- Turner S.J., Doherty P.C., McCluskey J., Rossjohn J. 2006. Structural determinants of T-cell receptor bias in immunity. Nat. Rev. Immunol. 6:883–894 10.1038/nri1977 [DOI] [PubMed] [Google Scholar]
- Tynan F.E., Borg N.A., Miles J.J., Beddoe T., El-Hassen D., Silins S.L., van Zuylen W.J., Purcell A.W., Kjer-Nielsen L., McCluskey J., et al. 2005a. High resolution structures of highly bulged viral epitopes bound to major histocompatibility complex class I. Implications for T-cell receptor engagement and T-cell immunodominance. J. Biol. Chem. 280:23900–23909 10.1074/jbc.M503060200 [DOI] [PubMed] [Google Scholar]
- Tynan F.E., Burrows S.R., Buckle A.M., Clements C.S., Borg N.A., Miles J.J., Beddoe T., Whisstock J.C., Wilce M.C., Silins S.L., et al. 2005b. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat. Immunol. 6:1114–1122 10.1038/ni1257 [DOI] [PubMed] [Google Scholar]
- Tynan F.E., Reid H.H., Kjer-Nielsen L., Miles J.J., Wilce M.C., Kostenko L., Borg N.A., Williamson N.A., Beddoe T., Purcell A.W., et al. 2007. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat. Immunol. 8:268–276 10.1038/ni1432 [DOI] [PubMed] [Google Scholar]
- Wei D.G., Curran S.A., Savage P.B., Teyton L., Bendelac A. 2006. Mechanisms imposing the Vbeta bias of Valpha14 natural killer T cells and consequences for microbial glycolipid recognition. J. Exp. Med. 203:1197–1207 10.1084/jem.20060418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wun K.S., Borg N.A., Kjer-Nielsen L., Beddoe T., Koh R., Richardson S.K., Thakur M., Howell A.R., Scott-Browne J.P., Gapin L., et al. 2008. A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. J. Exp. Med. 205:939–949 10.1084/jem.20072141 [DOI] [PMC free article] [PubMed] [Google Scholar]