Loss of the telomere component TRF1 and the DNA repair mediator 53BP1 enhances the ATR-mediated DNA damage response, promotes the homology-directed repair of dysfunctional telomeres, and exacerbates DNA damage.
Abstract
TRF1 protects mammalian telomeres from fusion and fragility. Depletion of TRF1 leads to telomere fusions as well as accumulation of γ-H2AX foci and activation of both the ataxia telangiectasia mutated (ATM)– and the ataxia telangiectasia and Rad3 related (ATR)–mediated deoxyribonucleic acid (DNA) damage response (DDR) pathways. 53BP1, which is also present at dysfunctional telomeres, is a target of ATM that accumulates at DNA double-strand breaks and favors nonhomologous end-joining (NHEJ) repair over ATM-dependent resection and homology-directed repair (homologous recombination [HR]). To address the role of 53BP1 at dysfunctional telomeres, we generated mice lacking TRF1 and 53BP1. 53BP1 deficiency significantly rescued telomere fusions in mouse embryonic fibroblasts (MEFs) lacking TRF1, but they showed evidence of a switch from the NHEJ- to HR-mediated repair of uncapped telomeres. Concomitantly, double-mutant MEFs showed evidence of hyperactivation of the ATR-dependent DDR. In intact mice, combined 53BP1/TRF1 deficiency in stratified epithelia resulted in earlier onset of DNA damage and increased CHK1 phosphorylation during embryonic development, leading to aggravation of skin phenotypes.
Introduction
The ends of linear chromosomes are formed by a special heterochromatic structure, known as telomeres, which protect chromosome ends from degradation and repair activities and, therefore, are essential to ensure chromosome stability (van Steensel et al., 1998; Chin et al., 1999; Karlseder et al., 1999; d’Adda di Fagagna et al., 2003; Takai et al., 2003; Celli and de Lange, 2005; de Lange, 2005; Blasco, 2007; Palm and de Lange, 2008). Mammalian telomeres are formed by tandem repeats of the TTAGGG sequence bound by a specialized six-protein complex, known as shelterin, which has roles in telomere capping and the regulation of telomerase activity at chromosome ends. The shelterin complex is composed of six core proteins, TRF1 and TRF2 (the telomeric repeat binding factors 1 and 2, respectively), the TIN2 (TRF1-interacting protein 2), POT1 (protection of telomeres protein 1), TPP1, and RAP1 (Smogorzewska and de Lange, 2004; de Lange, 2005; Palm and de Lange, 2008; Martínez and Blasco, 2010, 2011). Complete abrogation of most these components results in early embryonic lethality in mice with the exception of Rap1, which is also dispensable for telomere protection (Karlseder et al., 2003; Chiang et al., 2004; Celli and de Lange, 2005; Hockemeyer et al., 2006; Lazzerini Denchi et al., 2006; Wu et al., 2006; Kibe et al., 2010; Sfeir et al., 2010; Martínez and Blasco, 2011). When telomeres become critically short or unprotected because of shelterin deficiencies, they trigger an ataxia telangiectasia mutated (ATM)– or ataxia telangiectasia and Rad3 related (ATR)–dependent DNA damage response (DDR) at chromosome ends, which are then recognized as double-strand breaks (DSBs; de Lange, 2009; Martínez and Blasco, 2010; Tejera et al., 2010).
Mammalian DSBs are repaired by homologous recombination (HR) or nonhomologous end joining (NHEJ). HR initiates error-free repair between homologous repeated sequences. It requires the MRN complex to tether DNA ends and CtIP to generate the 3′ single-stranded DNA (ssDNA) substrate necessary for HR (Sartori et al., 2007; Mimitou and Symington, 2008; Williams et al., 2008). NHEJ is an error-prone repair that also requires the MRN complex and is composed of two pathways, the classical NHEJ (C-NHEJ) and the alternative NHEJ (A-NHEJ; Corneo et al., 2007; Nussenzweig and Nussenzweig, 2007; Soulas-Sprauel et al., 2007; Yan et al., 2007; Lieber, 2008; Mahaney et al., 2009). The C-NHEJ requires Ku70/86 and the DNA ligase IV (Lig4)–XRCC4 complex. The A-NHEJ fuses DNA ends that display microhomology and is dependent on PARP1 (poly (ADP-ribose) polymerase-1) and the XRCC1–DNA ligase III (Lig3) complex (Audebert et al., 2004; Wang et al., 2005; Haber, 2008). As in the case of DSBs, uncapped telomeres are repaired by activation of the HR and NHEJ pathways, leading to telomere length changes and end to end fusions, respectively (de Lange, 2009; Denchi, 2009). End to end fusions can arise from the activation of either the C-NHEJ or the A-NHEJ pathways, depending on how telomeres are rendered dysfunctional (Rai et al., 2010). For example, fusions arising upon TRF2 depletion are mediated by the C-NHEJ, whereas fusions induced by TPP1-POT1 depletion are mediated by the A-NHEJ pathway (Rai et al., 2010).
53BP1 is a C-NHEJ component and an ATM target that accumulates at DSBs and at uncapped telomeres (Rappold et al., 2001; Fernandez-Capetillo et al., 2002; Wang et al., 2002; Takai et al., 2003). The interaction of 53BP1 with chromatin involves the binding of its tudor domains to H4K20me2 and an MDC1-dependent interaction with γ-H2AX (Ward et al., 2003a; Bekker-Jensen et al., 2005; Botuyan et al., 2006). Although 53BP1 is dispensable for DNA damage signaling, the NHEJ of DSBs is severely affected by 53BP1 deficiency (Manis et al., 2004; Ward et al., 2004). Deletion of 53BP1 in cells with severe telomere uncapping as a result of TRF2 deficiency established that NHEJ of dysfunctional telomeres is strongly dependent on the binding of 53BP1 to damaged chromosome ends (Dimitrova et al., 2008; Rai et al., 2010). Binding of 53BP1 close to DNA breaks impacts the dynamic behavior of the local chromatin and facilitates the NHEJ repair reactions that involve distant sites (Dimitrova et al., 2008). Depletion of 53BP1 has been recently shown to restore HR defects in BRCA1-deficient cells and to abrogate the ATM-dependent checkpoint response and G2 cell cycle arrest triggered by the accumulation of DNA breaks (Bouwman et al., 2010; Bunting et al., 2010). In particular, removal of 53BP1 from DSBs is crucial for the ATM-dependent resection to produce recombinogenic ssDNA that precedes the homology-directed repair of the break (Bunting et al., 2010). Thus, the current notion is that removal of end-joining proteins, such as 53BP1, from breaks enables resection by CtIP, RPA loading onto ssDNA regions, and further RPA displacement by RAD51 promoting strand invasion at the homologous region of the sister chromatid (Bunting et al., 2010).
TRF1 deletion in mice produces embryonic lethality at the blastocyst stage, without apparent defects in telomere length or telomere capping (Karlseder et al., 2003). The recent development of conditional TRF1 alleles demonstrated that TRF1 deletion leads to rapid induction of senescence in differentiated cells, which is concomitant with the formation of telomeric γ-H2AX foci and phosphorylation of ATM and the ATM/ATR downstream checkpoint kinases CHK1 and CHK2 (Martínez et al., 2009; Sfeir et al., 2009). Abrogation of the p53 and retinoblastoma (Rb) pathways in these cells bypasses senescence but leads to chromosome instability, including sister chromatid fusions, chromosome concatenation, and occurrence of multitelomeric signals (MTSs) associated with replication fork stalling at telomeres (Martínez et al., 2009; Sfeir et al., 2009) in the absence of changes in telomere length (Martínez et al., 2009). Mice conditionally deleted for TRF1 in stratified epithelia, TRF1Δ/Δ K5-cre mice, show perinatal lethality and severe skin morphogenesis defects, which are concomitant with increased telomere damage, activation of the p53–p21 and p16 pathways, and cell cycle arrest in vivo (Martínez et al., 2009). Interestingly, stratified epithelia from TRF1-deficient mice also develop preneoplastic lesions (dysplasia and hyperkeratosis), which progress to invasive and genetically unstable squamous cell carcinomas in combination with p53 deficiency (Martínez et al., 2009). In turn, p53 deficiency rescues hair follicle stem cell defects, skin hyperpigmentation, and mouse survival, indicating that proliferative defects associated with TRF1 abrogation are mediated by p53.
Here, we set out to dissect the role of 53BP1 at telomeres rendered dysfunctional by TRF1 deficiency. Given that TRF1-deleted cells showed abundant end to end fusions and that 53BP1 is required for the C-NHEJ–mediated ligation of DNA ends, we set out to address the contributions of aberrant chromosome rearrangements to the deleterious effects of TRF1 deletion in vivo. Chromosome-type end to end fusions were significantly rescued in TRF1- and 53BP1-deficient mouse embryonic fibroblasts (MEFs), but this was accompanied by an increase in HR at telomeres. TRF1Δ/Δ 53BP1−/− MEFs showed a stronger CHK1 activation and increased RPA recruitment to telomeres, suggesting presence of recombinogenic ssDNA at telomeres and hyperactivation of the ATR-dependent DDR. Indeed, in marked contrast to the complete rescue of phenotypes induced by 53BP1 deficiency in the context of the BRCA1 knockout mice, TRF1Δ/Δ K5-Cre 53BP1−/− mice showed aggravated phenotypes compared with the TRF1-deficient controls. The TRF1Δ/Δ K5-Cre 53BP1−/− mice showed earlier onset of telomeric DNA damage and of ATR-mediated DDR activation than the TRF1Δ/Δ K5-Cre 53BP1+/+ littermates during embryonic development. Keratinocytes derived from TRF1Δ/Δ K5-Cre 53BP1−/− newborns also showed a stronger G2 arrest and increased polyploidy compared with TRF1Δ/Δ K5-Cre 53BP1+/+ keratinocytes. These results are the first to reveal that the in vivo loss of a telomeric protein in combination with a DNA repair component enhances early demise.
Results
53BP1 deficiency does not rescue growth defects of TRF1-deleted MEFs
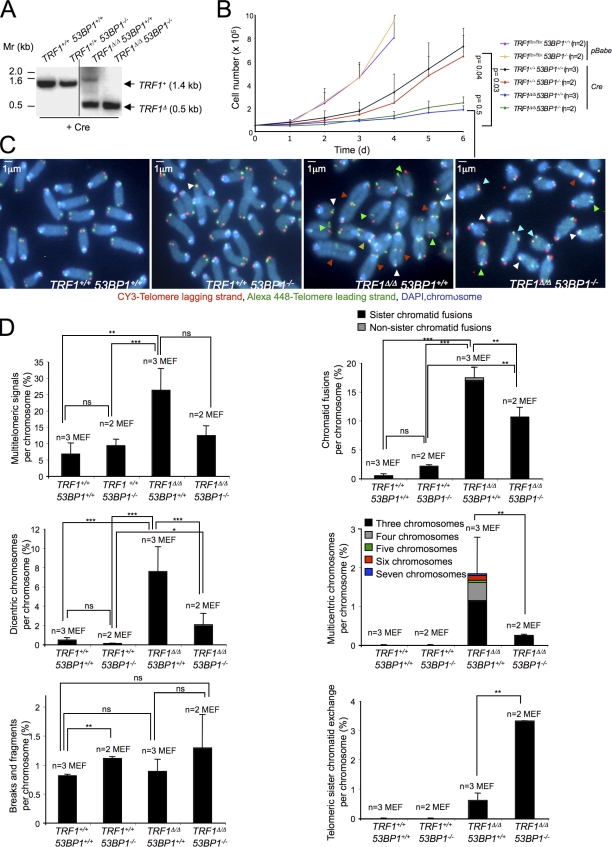
To study the impact of 53BP1 deficiency in cells deleted for TRF1, we generated MEFs of the TRF1+/+ 53BP1+/+, TRF1+/+ 53BP1−/−, TRF1flox/flox 53BP1+/+, and TRF1flox/flox 53BP1−/− genotypes by crossing 53BP1−/− (Ward et al., 2003b) with TRF1flox/flox mice (Martínez et al., 2009). To prevent induction of p53/Rb-dependent senescence as a consequence of TRF1 deletion (Martínez et al., 2009), we first immortalized MEFs with SV40 large T antigen (LT), which abrogates the p53 and Rb pathways, and then transduced them with a pBabe-Cre retrovirus to generate the TRF1Δ allele (Fig. 1 A). TRF1Δ/Δ 53BP1+/+ and TRF1Δ/Δ 53BP1−/− Cre-LT MEFs grew at similar rates, and both failed to proliferate further than two-cell divisions (Fig. 1 B). As expected, no differences in growth rates were detected between wild-type and 53BP1−/− cells. No significant differences were detected in the cell cycle profile between TRF1Δ/Δ 53BP1+/+ and TRF1Δ/Δ 53BP1−/− Cre-LT MEFs (Fig. S1).
Figure 1.
MEFs doubly deficient for TRF1 and 53BP1 show decreased chromosome-type end to end fusions but increased telomere recombination. (A) TRF1 deletion in TRF1flox/flox SV40 LT-immortalized MEFs upon retroviral infection with Cre recombinase was confirmed by PCR amplification of TRF1 alleles. The black line indicates that intervening lanes have been spliced out. (B) Growth curves of the indicated independent MEF lines. n, number of independent MEF cultures of the indicated genotypes. (C) Representative images of metaphase spreads of the indicated genotypes. Green arrowheads indicate MTS, white arrowheads indicate sister chromatid fusions, red arrowheads indicate dicentric chromosomes, the gold arrowhead indicates multicentric chromosomes, and blue arrowheads indicate T-SCE (yellow signals). (D) Frequency of the indicated aberrations in metaphase spreads of the indicated genotypes. n, independent MEFs used per genotype. From 6 to 11 metaphases were taken per independent MEF, making a total of ≥20 metaphases analyzed per genotype. Error bars indicate standard deviation. Statistical differences were calculated using the Student’s t test, and p-values are indicated. Mr, molecular marker. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
53BP1 deficiency rescues chromosome-type end to end fusions but results in increased telomere recombination associated with TRF1 abrogation
To evaluate the contribution of 53BP1 in the generation of chromosomal aberrations arising upon TRF1 deletion, we performed metaphase chromosome orientation FISH (CO-FISH) on TRF1Δ/Δ 53BP1+/+ and TRF1Δ/Δ 53BP1−/− Cre-LT MEFs (Fig. 1 C), which allows the visualization of both the leading and lagging telomere strands and can be used to determine the frequency of telomeric sister chromatid exchange (T-SCE) events (see Materials and methods). TRF1 deficiency has been previously shown to lead to telomeric DNA breakage associated with stalled replication forks, which results in chromosome ends with more than one telomere signal, the so-called MTS (Martínez et al., 2009; Sfeir et al., 2009). In agreement with this, TRF1-deleted MEFs showed a very high incidence of MTS (>20% of chromosomes; Fig. 2 D). Simultaneous deficiency in 53BP1, however, did not significantly affect MTS frequency as compared with TRF1Δ/Δ controls, indicating that 53BP1 does not impact telomere fragility associated with TRF1 depletion. Interestingly, TRF1Δ/Δ 53BP1−/− cells showed significantly less chromosome-type fusions, dicentric and multicentric chromosomes, compared with TRF1Δ/Δ 53BP1+/+ MEFs (Fig. 1 D). These findings are in agreement with a role for 53BP1 in the repair of dysfunctional telomeres through the C-NHEJ pathway. Nevertheless, the fact that chromosome fusions are still observed in the absence of 53BP1 indicates that end to end fusions associated with TRF1 deficiency can also be generated by the A-NHEJ pathway, although to a much lesser extent. The frequency of sister chromatid-type fusions, although significantly lower in MEFs lacking both TRF1 and 53BP1 compared with MEFs lacking only TRF1, was remarkably high, reaching values >10% of total chromosomes (Fig. 1 D). Therefore, 53BP1 deficiency merely suppresses sister chromatid-type fusion events. These results indicate that chromatid-type fusion events are either also mediated by the A-NHEJ pathway and/or originate from sister chromatid bridges at fragile sites. The frequency of chromosome breaks and fragments was not affected by 53BP1 deficiency (Fig. 1 D).
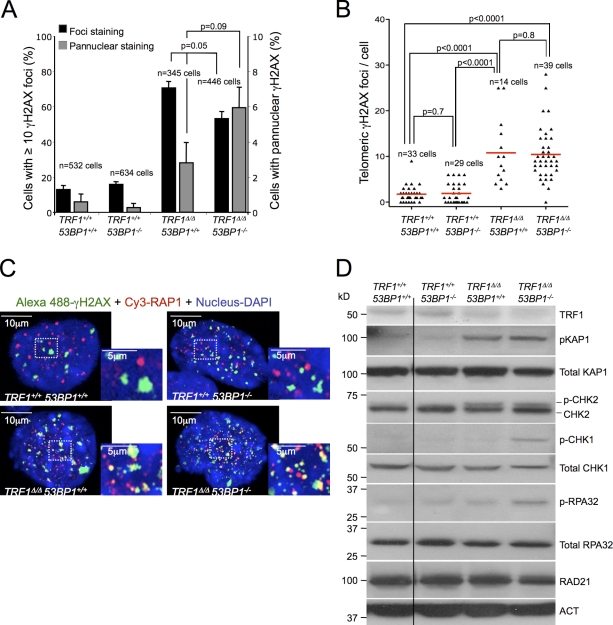
Figure 2.
53BP1 deficiency in TRF1Δ/Δ-immortalized MEFs leads to stronger CHK1 activation. (A) Percentage of nuclei exhibiting ≥10 γ-H2AX foci and pannuclear γ-H2AX staining in Cre-infected LT-immortalized MEFs 6 d after selection. Error bars indicate standard deviation. (B) Number of TIF per nuclei determined by immunofluorescence detection of γ-H2AX and RAP1. The number of cells analyzed in each case is indicated. Statistical differences were calculated using the Student’s t test, and p-values are indicated. Red lines correspond to mean values. (C) Representative combined images of γ-H2AX foci, RAP1, and DAPI of the indicated genotypes. Insets represent larger magnification of the squared areas, and TIF are visualized as yellow spots. (D) Western blot detection of DDR factors in Cre-infected LT-immortalized MEFs 4 d after selection of the indicated genotypes. RAD21 and actin were used as loading controls. The black line indicates that intervening lanes have been spliced out. ACT, actin.
Strikingly, 53BP1 deficiency increased HR events between sister chromatids (T-SCE) by fivefold in TRF1Δ/Δ 53BP1−/− compared with TRF1Δ/Δ 53BP1+/+ MEFs (Fig. 1 D). These results indicate that, in the absence of 53BP1, repair of dysfunctional telomeres caused by TRF1 deficiency is unbalanced from NHEJ toward HR. Finally, we did not observe any differences in the length of the G-strand overhang between TRF1Δ/Δ 53BP1+/+ and TRF1Δ/Δ 53BP1−/− MEFs as determined by terminal restriction fragment (TRF) analysis (Fig. S2, A–C).
53BP1 deficiency in TRF1-null MEFs leads to stronger CHK1 activation and telomeric RPA foci
A switch from the NHEJ to the HR pathway as a consequence of 53BP1 abrogation in TRF1-deficient MEFs may also affect the DDR. To explore this, we first evaluated the impact of 53BP1 deletion on the presence of uncapped telomeres associated with TRF1 deficiency as determined by γ-H2AX foci (Modesti and Kanaar, 2001; d’Adda di Fagagna et al., 2003; Takai et al., 2003; Martínez et al., 2009; Sfeir et al., 2009). 53BP1 deficiency did not affect the number of cells showing >10 γH2AX foci but had an impact on the number of cells showing γ-H2AX pannuclear staining (Fig. 2 A). Similarly, we did not see an impact of 53BP1 deficiency in the number of γ-H2AX foci colocalizing to telomeres, the so-called telomeric-induced damage foci (TIF; Fig. 2, B and C). These results indicate that the massive uncapping of telomeres that occurs as a consequence of TRF1 deletion is not affected by 53BP1 deficiency in MEFs.
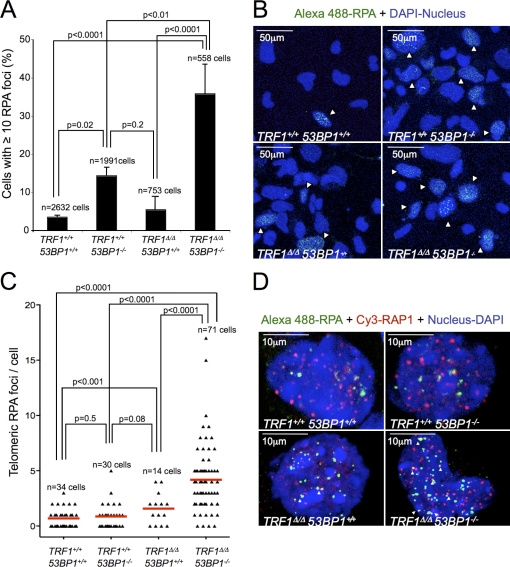
TIF induction after TRF1 deletion is accompanied by phosphorylation of ATM and the ATM/ATR downstream checkpoint kinases CHK2 and CHK1 (Martínez et al., 2009; Sfeir et al., 2009). To address the impact of 53BP1 deficiency on DDR activation as a consequence of TRF1 deletion, we performed Western blotting of different components of the ATM and ATR axis (Fig. 2, D and E). No differences in the ATM signaling pathway were observed between TRF1Δ/Δ 53BP1−/− and TRF1Δ/Δ 53BP1+/+ MEFs, as seen by similar levels of phospho-KAP1 and phospho-CHK2 in both settings (Fig. 2 D). In contrast, TRF1Δ/Δ 53BP1−/− cells showed a much stronger signal of the ATR downstream checkpoint kinase phospho-CHK1 and higher levels of phopho-RPA32 compared with TRF1Δ/Δ 53BP1+/+ cells (Fig. 2 D and Fig. S3, A and B). These results are consistent with previous findings showing that abrogation of 53BP1 in BRCA1−/− cells affects the levels of phosphorylated RPA upon DNA damage but does not affect the ATM–CHK2–p53 signaling pathway (Cao et al., 2009; Bunting et al., 2010). Increased levels of phosphorylated RPA could indicate the presence of ssDNA overhangs at telomeres resulting from DSB resection before their repair by HR (Jazayeri et al., 2006; Sartori et al., 2007; Mimitou and Symington, 2009). Furthermore, RPA bound to ssDNA is required for the recruitment of ATR to sites of DNA damage and for ATR-mediated CHK1 activation for checkpoint signaling (Zou and Elledge, 2003). To address whether RPA was specifically increased at TRF1-depleted telomeres in the absence of 53BP1, we performed double immunofluorescence with RPA and the telomeric protein RAP1 (Fig. 3, A–D). The number of cells containing RPA foci (>10 foci per cell) was fivefold higher in TRF1Δ/Δ 53BP1−/− compared with TRF1Δ/Δ 53BP+/+ MEFs (Fig. 3, A and B). Moreover, these foci were significantly increased at telomeres in TRF1Δ/Δ 53BP1−/− compared with TRF1Δ/Δ 53BP+/+ MEFs (Fig. 3, C and D), suggesting the presence of ssDNA at telomeric regions and consistent with the observed increase in telomeric recombination frequencies.
Figure 3.
53BP1 deficiency in TRF1Δ/Δ-immortalized MEFs leads to increased RPA foci at telomeres. (A) The percentage of nuclei exhibiting ≥10 RPA foci determined in Cre-infected LT-immortalized MEFs 6 d after selection. Error bars indicate standard deviation. (B) Representative immunofluorescence images of RPA staining of the indicated genotypes. (C) Number of telomeric RPA foci (TIF) per nuclei determined by immunofluorescence detection of RPA and RAP1. Two to three independent MEF lines were assayed per genotype, and the total number of cells analyzed in each case is indicated. Statistical differences were calculated using the Student’s t test, and p-values are indicated. Red lines correspond to mean values. (D) Representative combined images of RPA foci, RAP1, and DAPI of the indicated genotypes. RPA and RAP1 colocalization events are indicated by arrowheads.
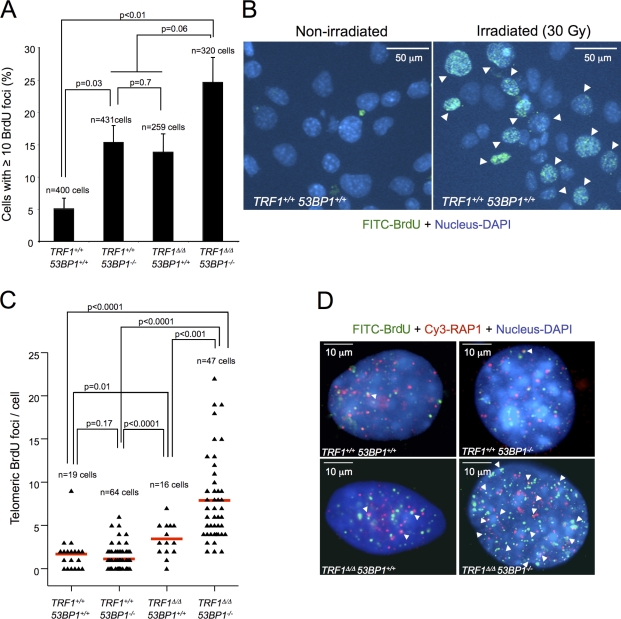
To confirm telomeric ssDNA, we performed a BrdU staining under nondenaturing conditions. A higher number of cells presenting >10 BrdU foci was detected in MEFS lacking 53BP1 or TRF1 as compared with wild-type cells, and this number was further increased in cells lacking both 53BP1 and TRF1 (Fig. 4 A). As the positive control, BrdU staining was increased in γ-irradiated cells as compared with nonirradiated cells (Fig. 4 B). We observed an increased colocalization of telomeres detected by RAP1 staining with BrdU-visualized ssDNA in TRF1Δ/Δ 53BP1−/− cells, strongly supporting the presence of ssDNA at telomeres (Fig. 4, C and D). In summary, these results suggest that 53BP1 deficiency in TRF1-deleted cells results in a switch from the NHEJ to the HR repair mode, which is accompanied by accumulation of ssDNA at telomeres and a hyperactivation of the ATR signaling pathway, including higher levels of CHK1 and phosphorylated RPA.
Figure 4.
53BP1 deficiency in TRF1Δ/Δ MEFs results in increased telomeric ssDNA. (A) The percentage of nuclei exhibiting ≥10 BrdU foci determined in Cre-infected LT-immortalized MEFs 4 d after selection. Error bars indicate standard deviation. (B) Representative immunofluorescence images of BrdU staining of wild-type cells nonirradiated and after 30 Gy treatment of ionizing radiation. As a positive control for ssDNA-BrdU staining, all the MEF lines used for this study were irradiated (30 Gy) and incubated for 90 min to let DNA resection to proceed. (C) Number of telomeric ssDNA foci per nuclei determined by immunofluorescence detection of BrdU and RAP1. Two independent MEF lines were assayed per genotype, and the total number of cells analyzed in each case is indicated. Statistical differences were calculated using the Student’s t test, and p-values are indicated. Red lines correspond to mean values. (D) Representative combined images of BrdU foci, RAP1, and DAPI of the indicated genotypes. BrdU and RAP1 colocalization events are indicated by arrowheads.
Aggravation of TRF1 loss–mediated in vivo pathologies by 53BP1 deficiency
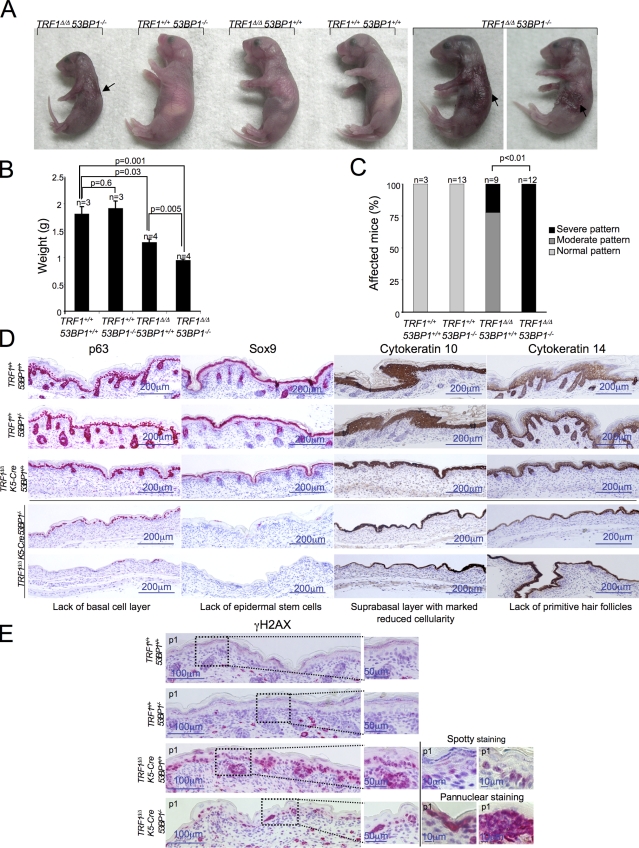
The impact of activation of the ATR versus the ATM pathway in the context of the organism is dramatically different. Although deficiencies in components of the ATM pathway do not lead to embryonic lethality in mice, mice deleted for the components of the ATR pathway are embryonically lethal, most likely reflecting an essential role of the HR pathway in mammals (Elson et al., 1996; de Klein et al., 2000). The results described here suggest that 53BP1 is a key component in the balance between these two modes of repair, favoring the NHEJ pathway over the HR pathway. To study the in vivo impact of 53BP1 abrogation in the context of TRF1-deficient mice, we crossed conditional TRF1flox/+ K5-Cre (Martínez et al., 2009) with TRF1+/+ 53BP1−/− mice (Ward et al., 2003b) to obtain TRF1Δ/Δ K5-Cre 53BP1−/− mice. Doubly deficient TRF1Δ/Δ K5-Cre 53BP1−/− mice were born at the expected Mendelian ratios (Table 1), but none survived past the first day postpartum (P1) in contrast to single-mutant TRF1Δ/Δ K5-Cre mice, in which 8% of the newborns reached day 3 postpartum (P3; Martínez et al., 2009). These results indicate that abrogation of 53BP1 enhances early demise associated with TRF1 deficiency. At the macroscopic level, the skin of TRF1Δ/Δ K5-Cre 53BP1−/− newborns showed severe epithelial necrosis, resulting in ulcers and hemorrhages, which were not observed in the 53BP1+/+ littermates (Fig. 5 A). TRF1Δ/Δ K5-Cre 53BP1−/− mice also showed significantly lower weight at birth than the TRF1Δ/Δ K5-Cre newborns (Fig. 5 B). Histopathological analysis of the different stratified epithelia in the organism revealed that 100% of double knockout mice present a severe pattern of epithelial atrophy, whereas 75% of the single TRF1-deleted skin present a moderate pattern (Fig. 5 C and Fig. S4).
Table 1.
Offspring from TRF1flox/flox 53BP1−/− and TRF1flox/+ K5-Cre+/T 53BP1+/− and TRF1flox/flox 53BP1−/− and TRF1flox/+ K5-Cre+/T 53BP1−/− mouse intercrosses
| Crosses | Total pups | Genotype | Pups (expected), p-value |
| TRF1flox/flox 53BP1−/− and TRF1flox/+ K5-Cre+/T 53BP1+/− | 131 | TRF1flox/flox K5-Cre+/T 53BP1+/− and TRF1flox/flox K5-Cre+/T 53BP1−/− | 12 (16), P = 0.4 and 14 (16), P = 0.85 |
| TRF1flox/flox 53BP1−/− and TRF1flox/+ K5-Cre+/T 53BP1−/− | 67 | TRF1flox/flox K5-Cre+/T 53BP1−/− and TRF1flox/flox 53BP1−/− | 10 (17), P = 0.2 and14 (17), P = 0.7 |
Figure 5.
In vivo deleterious genetic interactions between TRF1 and 53BP1. (A) Representative images of newborn mice of the indicated genotype at birth (P1). (right) Images of two independent TRF1Δ/Δ K5-Cre 53BP1−/− mice (P1). Arrows indicate areas of epithelial necrosis, ulcers, and hemorrhages. Note the smaller size and more aggravated phenotype of the TRF1Δ/Δ K5-Cre 53BP1−/− mice compared with an age-matched TRF1Δ/Δ K5-Cre mouse. (B) Mean weight at birth of the indicated genotypes. Error bars represent standard errors, and the number of mice is indicated in each case. Statistical differences were calculated using the Student’s t test, and p-values are indicated. (C) Quantification of histopathological findings at birth in different stratified epithelia of the indicated genotypes. Severe pattern refers to a high-grade dysplasia in the skin, tongue, palate, esophagus, nonglandular stomach, nails, and urinary bladder, with marked cytological atypia with disturbed polarity of cells, irregular epithelia stratification, loss of nuclear polarity, prominent nucleoli, and nuclear polymorphism. All tissues present focal hyperplasia and hyperkeratosis. The back skin shows epithelial necrosis and complete absence of hair follicles. Moderate pattern is characterized by a lower grade of dysplasia in skin, tongue, nonglandular stomach, and esophagus, with cells larger than normal, often with irregular stratification, enlarged nuclei, and in some cases, loss of nuclear polarity. The back skin shows primordial hair follicles, and the nails are unaffected. The number of mice analyzed in each case is indicated. Statistical differences are calculated using the χ2 test, and p-values are indicated. (D and E) Representative images of back skin sections stained for p63, Sox9, cytokeratin 10, cytokeratin 14 (D), and γ-H2AX (E) at birth of mice from the indicated genotypes. (E, right) Larger magnifications in which different γ-H2AX staining patterns are shown. Note that, in the TRF1Δ/Δ K5-Cre, the γ-H2AX–positive cells display a foci staining in contrast to TRF1Δ/Δ K5-Cre 53BP1−/− epidermal γ-H2AX–positive cells that present a pannuclear staining. Note the absence of p63 and Sox9 expression in TRF1Δ/Δ K5-Cre 53BP1−/− skin.
Aggravation of skin phenotypes in TRF1Δ/Δ K5-Cre 53BP1−/− newborns with respect to TRF1Δ/Δ K5-Cre newborns was further confirmed by various markers of skin morphogenesis. In particular, TRF1Δ/Δ K5-Cre 53BP1−/− skin showed almost complete absence of p63-positive cells, a marker of the basal cell layer, and no staining for SOX9, a marker for epidermal stem cells (Fig. 5 D). Staining with the differentiation markers cytokeratins K10 and K14 also showed an aggravation of the skin phenotypes associated with single TRF1 deficiency, such as a marked reduction in the cellularity of the suprabasal skin layer as well as the complete absence of primitive hair follicles (Fig. 5 D).
TRF1Δ/Δ K5-Cre 53BP1−/− mice show earlier onset of DNA damage and of DDR activation
To address whether the aggravation of the skin phenotypes produced by 53BP1 deficiency was associated with increased DNA damage at birth, we first determined presence of γ-H2AX in newborn skin. As expected, γ-H2AX–positive cells were completely absent from TRF1 wild-type newborn skin independently of the 53BP1 status. In contrast, 100% of the skin basal layer cells were positive for γ-H2AX in both TRF1Δ/Δ and TRF1Δ/Δ 53BP1−/− newborns (Fig. 5 E). Strikingly, TRF1Δ/Δ 53BP1−/− skin cells showed a different γ-H2AX staining pattern compared with TRF1Δ/Δ K5-Cre skin. Although TRF1Δ/Δ 53BP1+/+ cells showed a spotted pattern of γ-H2AX foci, characteristic of DSBs and uncapped telomeres, TRF1Δ/Δ 53BP1−/− cells showed a nonapoptotic-related pannuclear γ-H2AX staining (Fig. 5 E and Fig. S5). Pannuclear γ-H2AX staining has been associated with accumulation of ssDNA and activation of the ATR pathway as a consequence of replicative stress or replication fork stalling (Murga et al., 2009). These results suggest that 53BP1 abrogation switches the type of DDR induced by TRF1 deletion.
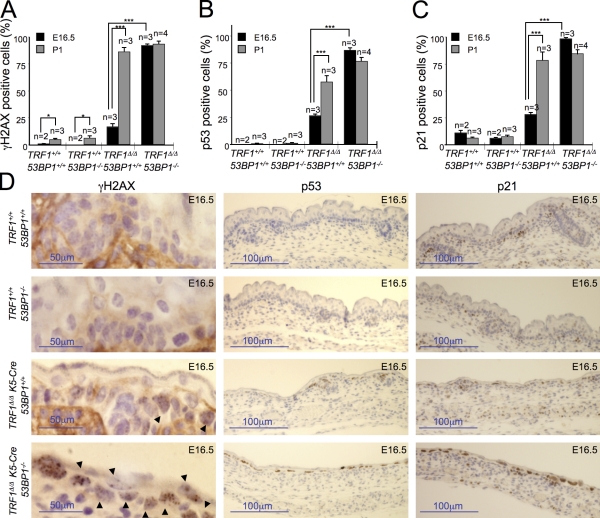
The fact that TRF1Δ/Δ K5-Cre 53BP1−/− newborn mice showed aggravated skin phenotypes compared with the single TRF1Δ/Δ K5-Cre controls prompted us to analyze DDR activation before birth. To this end, we analyzed the levels of γ-H2AX, p53, and p21 in the developing skin of embryonic day 16.5 (E16.5) embryos (Fig. 6, A–D). TRF1Δ/Δ K5-Cre 53BP1−/− embryos showed a fourfold increase in γ-H2AX–positive cells in the developing skin compared with TRF1Δ/Δ 53BP1+/+ embryos (Fig. 6, A and D). Similarly, p53- and p21-positive cells were elevated more than twofold in TRF1Δ/Δ K5-Cre 53BP1−/− embryos compared with single-mutant TRF1Δ/Δ K5-Cre controls (Fig. 6, B–D). Together, these results indicate an earlier onset of DNA damage and subsequent DDR activation associated with 53BP1 deficiency, which is in agreement with the more severe skin morphogenesis defects at birth in TRF1Δ/Δ K5-Cre 53BP1−/− newborns compared with the TRF1 single-mutant controls.
Figure 6.
53BP1 deficiency leads to an earlier DDR onset during embryonic development. (A–C) Percentage of γ-H2AX (A)–, p53 (B)-, and p21 (C)-positive cells in the back skin of embryos of the indicated genotypes at either day 16.5 postcoitum (E16.5) or in 1-d-old newborns (P1). n, number of embryos or mice analyzed. Error bars represent standard error. Statistical differences were calculated using the Student’s t test, and p-values are indicated. *, P < 0.05; ***, P < 0.001. (D) Representative images of back skin sections stained for γ-H2AX, p53, and p21 from embryos (E16.5) of the indicated genotypes. Arrowheads mark γ-H2AX–positive cells.
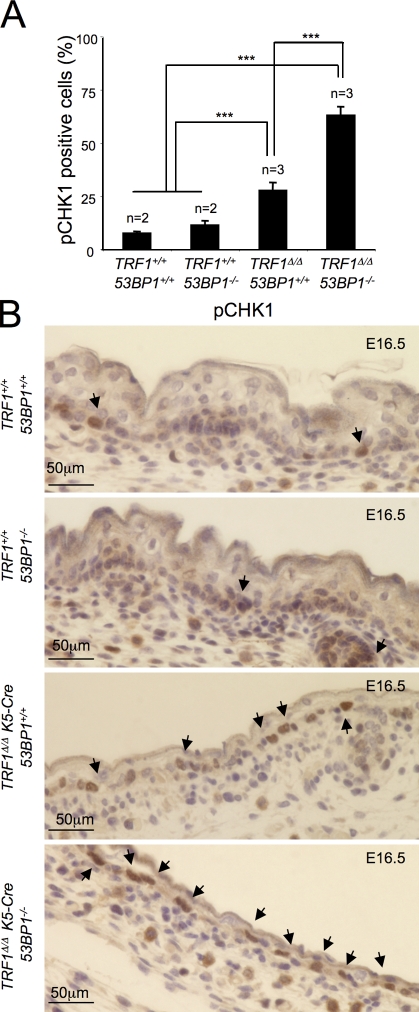
Based on the stronger CHK1 activation observed in TRF1Δ/Δ 53BP1−/− as compared with TRF1Δ/Δ 53BP1+/+ MEFs (Fig. 2), we tested phosphorylated CHK1 (p-CHK1) levels in the epidermis of E16.5 embryos (Fig. 7, A and B). In the developing epidermis of TRF1Δ/Δ K5-Cre 53BP1−/− embryos, a threefold increase of p-CHK1–positive cells was readily observed as compared with the levels detected in TRF1Δ/Δ K5-Cre embryos. These results are consistent with the higher percentage of p53–p21-positive cells observed in E16.5 TRF1Δ/Δ K5-Cre 53BP1−/− embryos (Fig. 6). Interestingly, no p-CHK1–positive cells were detected in newborn skin of any of the aforementioned genotypes under study (unpublished data). It has been reported that massive ATR–CHK1 activation takes places especially during embryonic development when tissues are highly proliferative (Murga et al., 2009).
Figure 7.
53BP1 deficiency leads to a hyperactivation of the ATR-dependent DDR during embryonic development. (A) Percentage of p-CHK1–positive cells in the back skin of embryos of the indicated genotypes at day 16.5 postcoitum (E16.5). n, number of analyzed embryos. Error bars represent standard error. Statistical differences were calculated using the Student’s t test, and p-values are indicated. ***, P < 0.001. (B) Representative images of back skin sections stained for p-CHK1 (Ser345) from embryos (E16.5) of the indicated genotypes. Arrows mark p-CHK1–positive cells.
Massive arrest of TRF1Δ/Δ K5-Cre 53BP1−/− keratinocytes in G2
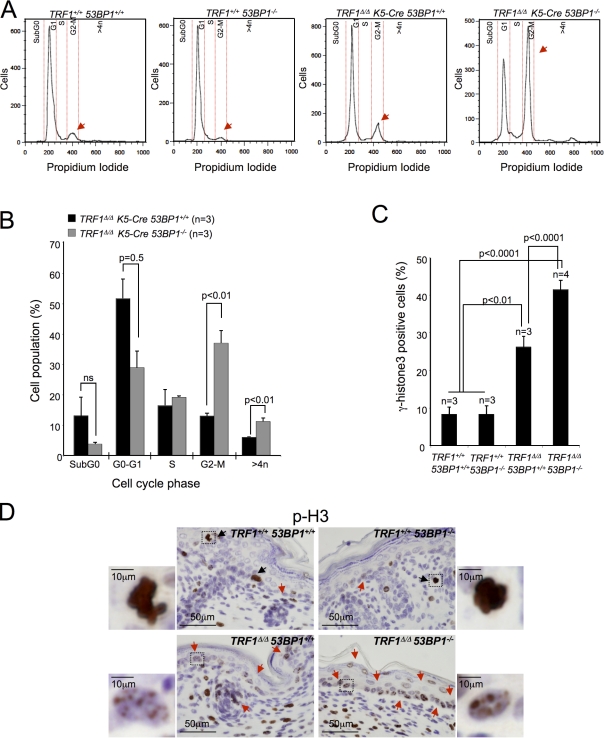
To further confirm aggravation of TRF1-associated cellular phenotypes as a consequence of 53BP1 deletion, we performed flow cytometry analysis of isolated newborn skin keratinocytes to determine cell cycle profile and polyploidy. TRF1Δ/Δ K5-Cre 53BP1−/− keratinocytes showed a threefold increase in the percentage of cells with 4n DNA content and a twofold increase in polyploid cells (>4n) compared with TRF1Δ/Δ K5-Cre keratinocytes (Fig. 8, A and B). The accumulation of cells with a 4n DNA content might correspond to either cells arrested in G2/M or to polyploid cells arrested in G1. To distinguish between these two possibilities, we analyzed the number as well as the staining pattern of phospho–Histone H3 (Ser10)-positive cells in newborn mice. Mitotic phosphorylation of H3 initiates nonrandomly in pericentromeric heterochromatin in late G2 interphase cells, in which staining is visualized as discrete foci. After initiation, H3 phosphorylation spreads throughout the condensing chromatin, and the staining increases progressively until complete nuclear staining of the mitotic chromatin is achieved (Hendzel et al., 1997). In contrast to wild-type and 53BP1−/− skin in which <10% of the cells were p-H3 positive, TRF1Δ/Δ K5-Cre and TRF1Δ/Δ K5-Cre 53BP1−/− epidermis showed a three- and fivefold increase in p-H3–positive cells, respectively (Fig. 8, C and D). The H3 staining pattern between TRF1-proficient and TRF1-deficient cells was also remarkably different. In particular, although p-H3–positive cells in TRF1-proficient skin showed both foci (typical of G2) and pannuclear (typical of mitosis) staining, all p-H3–positive cells from the TRF1-deficient epidermis presented foci and, therefore, corresponded to G2 (Fig. 8 D). These findings demonstrate that a large proportion of TRF1-deleted cells are arrested in G2 and fail to proceed to mitosis and that this defect is exacerbated by 53BP1 deficiency.
Figure 8.
Keratinocytes doubly deficient for TRF1 and 53BP1 are arrested in G2. (A) Representative FACS profiles of isolated newborn keratinocytes of the indicated genotypes. The different cell cycle phases are marked between the red dotted lines. The arrows indicate the peaks corresponding to G2–M phase of the cell cycle. (B) Quantification of the percentage of cells at different phases of the cell cycle by FACS analysis of newborn TRF1Δ/Δ K5-Cre and TRF1Δ/Δ K5-Cre 53BP1−/− keratinocytes. Cells were stained with propidium iodide. n, number of independent newborn mice used per genotype. (C) Percentage of phospho–Histone 3 (S10)-positive cells in the back skin of mice of the indicated genotypes at birth (P1). n, number of mice analyzed. (B and C) Error bars represent standard error. Statistical differences were calculated using the Student’s t test, and p-values are indicated. (D) Representative images of back skin sections stained for p-H3. Insets represent larger magnifications of the cell within the squares. Black arrows indicate cells in mitosis (intense and whole nuclear staining), and red arrows show cells in G2 (foci staining).
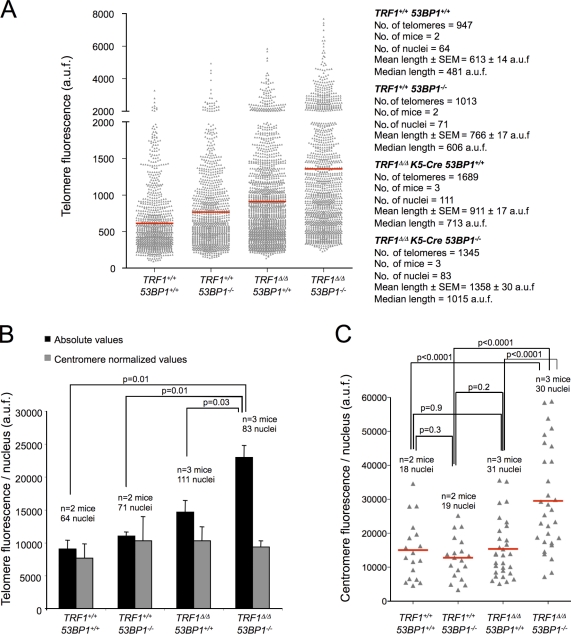
Longer telomeres in TRF1Δ/Δ K5-Cre 53BP1−/− mouse epidermis
To address whether the severe developmental defects of the TRF1Δ/Δ K5-Cre 53BP1−/− epidermis were caused by telomere length defects associated with 53BP1 deficiency, we performed telomere quantitative FISH (Q-FISH) directly on skin sections (Fig. 9, A–C). Centromere fluorescence was also performed to correct for differences in ploidy. Interestingly, the skin of TRF1Δ/Δ K5-Cre 53BP1−/− mice showed higher telomere fluorescence than that of TRF1Δ/Δ K5-Cre 53BP1+/+ controls, both when measuring individual telomere dots and when considering mean telomere fluorescence per nuclei (Fig. 9, A and B). In particular, TRF1Δ/Δ K5-Cre 53BP1−/− skin showed an increased number of telomeres with high fluorescence (>2,000 arbitrary units of fluorescence) and a decrease in the amount of telomeres showing low fluorescence (<250 arbitrary units of fluorescence) compared with TRF1Δ/Δ K5-Cre 53BP1+/+ littermates (Fig. 9 A). Because 53BP1 deficiency also results in increased telomeric recombination (Fig. 1, C and D), we speculate that the presence of individual telomere signals showing high fluorescence in TRF1Δ/Δ K5-Cre 53BP1−/− skin (Fig. 9 A) may be the result of increased HR repair at telomeres. Increased total nuclear telomere fluorescence in TRF1Δ/Δ K5-Cre 53BP1−/− skin is associated with increased ploidy compared with TRF1Δ/Δ K5-Cre 53BP1+/+ skin, as indicated by increased centromeric fluorescence in TRF1Δ/Δ K5-Cre 53BP1−/− skin compared with both wild-type and TRF1Δ/Δ K5-Cre controls (Fig. 9 C). Indeed, when mean total nuclear telomere fluorescence was normalized to mean total nuclear centromere fluorescence, no significant differences could be detected between the different genotypes (Fig. 9 B).
Figure 9.
Keratinocytes doubly deficient for TRF1 and 53BP1 show increased telomere and centromere fluorescence. (A) Telomere fluorescence distribution of individual telomere dots in back skin sections of mice of the indicated genotype as determined by Q-FISH analysis. n, number of mice used for the analysis. The total number of telomeres analyzed. The mean and median fluorescence as well as standard error values are shown for each genotype. (B) Quantification of total telomere fluorescence per nucleus in back skin sections from mice of the indicated genotypes. Absolute fluorescence values and centromere normalized values are depicted. The number of mice used for the analysis and the total number of nuclei analyzed for each genotype are indicated. Bars represent standard errors. (C) Total centromere fluorescence per nucleus distribution in back skin sections from mice of the indicated genotypes. The number of mice used for the analysis and the total number of nuclei analyzed for each genotype are indicated. Red lines correspond to mean values. (B and C) The Student’s t test was used for statistical analysis, and p-values are indicated. a.u.f., arbitrary units of fluorescence.
Discussion
53BP1 is required for the C-NHEJ–mediated ligation of DNA ends (Wang et al., 2002; Manis et al., 2004; Ward et al., 2004; Dimitrova et al., 2008; Martínez et al., 2009; Rai et al., 2010). Here, we address the role of 53BP1 at dysfunctional telomeres caused by TRF1 deficiency. In particular, we study the impact of 53BP1 deletion on both the chromosomal defects as well as the severe cellular and organismal phenotypes associated with TRF1 deletion in vivo. We initially reasoned that 53BP1 deficiency could significantly rescue end to end fusions in TRF1-depleted cells by blocking the C-NHEJ at dysfunctional telomeres, and this, in turn, might impact favorably on the survival of TRF1Δ/Δ K5-Cre mice. As expected, at the chromosomal level, 53BP1 deficiency in TRF1-deleted MEFs led to a significant rescue of chromosome-type end to end fusions but also to an unexpected simultaneous increase in telomeric recombination events, suggesting a switch from C-NHEJ– to HR-mediated repair. This was accompanied by a hyperactivation of the ATR-dependent DDR as indicated by increased RPA at telomeres and augmented phosphorylation of the ATR downstream checkpoint kinase CHK1. At the organismal level, these molecular events resulted in an aggravation of the skin phenotypes associated with TRF1 deletion, which was associated with an earlier onset of DNA damage and DDR activation during embryonic development, leading to massive G2 arrest in the skin of TRF1Δ/Δ K5-Cre 53BP1−/− newborn mice. This phenotypic aggravation resulting from combined TRF1 and 53BP1 deficiencies is in contrast to the recently described impact of 53BP1 deletion in rescuing phenotypes associated with BRCA1 deletion. However, in that scenario, the phenotypes observed were attributed to defects in HR (Cao et al., 2009; Bouwman et al., 2010; Bunting et al., 2010).
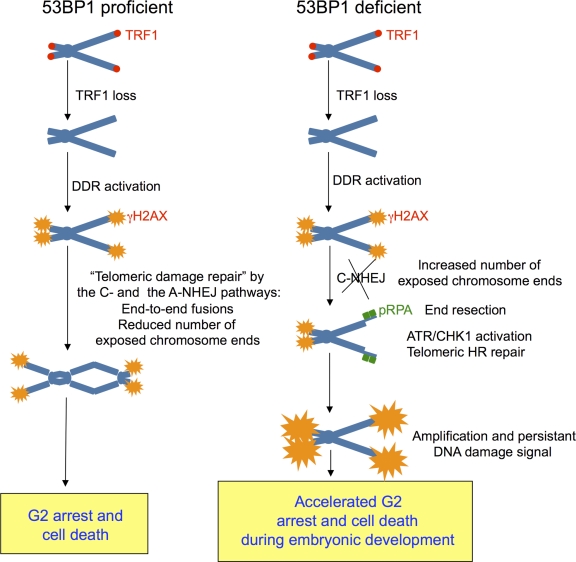
In summary, our data support a model in which deletion of 53BP1 in TRF1-deficient cells impairs the C-NHEJ repair pathway, decreasing the occurrence of chromosome-type end to end telomere fusions while inducing a persistent DNA damage signal that activates the ATR-dependent DDR and the HR repair pathway (Fig. 10). In particular, in the absence of 53BP1, dysfunctional telomeres accumulate higher levels of ssDNA and RPA indicative of increased resection (Fig. 3 and Fig. 4). This, in turn, triggers a hyperactivation of the ATR–CHK1 pathway that further amplifies DDR signaling (Fig. 2 D and Fig. 7) and that may lead to the G2 arrest observed in vivo in TRF1Δ/Δ K5-Cre 53BP1−/− mice (Fig. 8). Finally, resected telomeres may be recognized as substrates by the HR machinery, which may explain the increased frequency of telomeric sister chromatid exchange triggered by 53BP1 abrogation in TRF1-deficient cells (Fig. 1). Our results are in agreement with the model proposed by Bunting et al. (2010) in which removal of 53BP1 from the DSB is crucial for the ATM-dependent resection to produce HR-competent ssDNA. 53BP1 binding to dysfunctional TRF1-null telomeres might function by blocking telomere resection, thereby interfering with HR. Processing of exposed telomeres by HR-dependent repair generates interchromosomal recombination events that lead to elongation of telomeric tracts, a mechanism known as alternative lengthening of telomeres (Dunham et al., 2000; Varley et al., 2002). In support of this notion, 53BP1 loss also leads to a higher proportion of long telomeres in TRF1-deficient keratinocytes (Fig. 9). Unexpectedly, neither the decreased frequency of end to end fusions nor the activation of the HR repair pathway at uncapped telomeres upon 53BP1 loss alleviated the deleterious effects of TRF1 deficiency in vivo; instead, they were aggravated. These observations suggest that fusion of dysfunctional telomeres may represent a mechanism to temporally heal dysfunctional telomeres and, in this manner, reduce DDR signaling. These results also suggest that severe phenotypes present in the skin of TRF1Δ/Δ k5-Cre mice are not solely caused by chromosome fusions but rather by increased DNA damage signaling at telomeres (Martínez et al., 2009; Sfeir et al., 2009). Although an activated HR could be expected to rescue fork stalling at telomeres, the fact that neither the incidence of MTS nor a rescue in lethality was detected upon 53BP1 abrogation supports a model in which TRF1 has a more specific function in telomere replication. In fact, TRF1 has been proposed to facilitate telomeric DNA replication by recruiting/activating the BLM and RTEL helicases involved in the removal of G4 DNA structures formed in single-stranded TTAGG repeats (Sfeir et al., 2009).
Figure 10.
Model used in this study. TRF1 loss yields dysfunctional/uncapped telomeres that are marked with γ-H2AX foci. Dysfunctional telomeres elicit a DNA damage response (DDR) by activation of upstream kinases, DNA protein kinase, ATM, and ataxia telangiectasia and Rad3 related (ATR) that leads to a p53–p21-mediated cell cycle arrest until the damage is repaired. Activation of either the classical or alternative nonhomologous end-joining (C-NHEJ or A-NHEJ, respectively) repair pathways results in end to end fusions. The joining of dysfunctional telomeres results in the abolishment of the DNA damage signal by reducing the number of exposed chromosome ends and, thereby, can be considered as a temporal healing of the damage at least until the following mitosis, in which fused chromosomes will lead to further DSBs. In 53BP1-deficient cells with an impaired C-NHEJ pathway, the DNA damage signal associated with dysfunctional telomeres persists. Absence of 53BP1 at dysfunctional telomeres facilitates resection of telomeric DNA that gets coated by RPA enhancing ATR–CHK1 activation that further amplifies the DDR signaling, leading to an accelerated G2 arrest and eventual cell death. Moreover, resected telomeres act as substrates for the HR machinery, resulting in a higher incidence of inter- and intrachromosomal telomeric recombination events that impact telomere length changes. As a consequence, mice doubly deficient for TRF1 and 53BP1 in skin accumulate telomeric DNA damage more rapidly than TRF1 single knockout, severely blocking skin morphogenesis early during embryonic development.
The aggravation of phenotypes associated with TRF1 depletion by 53BP1 deficiency is a consequence of an increased ATR signaling caused by either aberrant processing of dysfunctional telomeres and/or by an increased number of exposed chromosome ends as a result of reduced end to end fusion frequency. Previous studies have suggested that proteins of the NHEJ pathway can have suppressive effects on HR (Pierce et al., 2001; Xie et al., 2007). More specifically, the frequency of HR is enhanced in the absence of the NHEJ proteins Ku70, XRCC4, and DNA-PKcs (Pierce et al., 2001) as well as by overexpression of a dominant-negative 53BP1 fragment (Xie et al., 2007). However, rescue of genomic instability in BRCA1−/− cells is solely achieved by loss of 53BP1 and not by the loss of any other component of the NHEJ pathway (Bunting et al., 2010). At telomeres, the Ku70–Ku80 complex has also been shown to repress exchange between sister telomeres upon TRF2 loss (Celli et al., 2006). In accordance with our findings, loss of 53BP1 in TRF2-deleted cells also led to an increase in T-SCE (Rai et al., 2010). However, to our knowledge, the in vivo effects of either Ku70 or 53BP1 abrogation in combination with a deficiency in a shelterin component has not been reported until now. A similar synthetic genetic interaction has been previously described for DNA-PKcs deficiency in late generation Terc−/−-deficient mice presenting very short telomeres (Maser et al., 2007; Wong et al., 2007). Thus, the G4-Terc−/− mice in combination with DNA-PKc deletion experienced an accelerated loss of organismal viability and reduction in median lifespan (Espejel et al., 2004; Wong et al., 2007). Future work in which lack of a shelterin component is combined with a deficiency in any other component of the classical NHEJ, such as Ku80 and Lig4, will provide insights on whether the observed in vivo phenotypes are solely caused by the persistence of exposed chromosome ends or, on the contrary, is specific of 53BP1 deficiency.
Finally, it has recently been reported that p53 not only regulates cell cycle arrest at the G1/S in response to telomere dysfunction but also at the G2/M transition (Karlseder et al., 1999; Thanasoula et al., 2010). Telomere uncapping caused by TRF1 deletion delays mitotic entry in a p53–p21-dependent manner (Martínez et al., 2009; Thanasoula et al., 2010). Our results strongly reinforce this notion. The earlier onset of telomeric damage during embryonic development in TRF1Δ/Δ K5-Cre 53BP1−/− compared with TRF1Δ/Δ K5-Cre 53BP1+/+ triggers a G2 cell cycle arrest severely impairing morphological development of the skin. Our work provides the first evidence of deleterious genetic interaction between a shelterin component and a DNA damage repair component, TRF1 and 53BP1, respectively.
Materials and methods
Generation of conditional double TRF1/53BP1 knockout mice
To generate TRF1Δ/Δ K5-Cre 53BP1−/− mice, TRF1+/Δ K5-Cre 53BP1+/+ mice (Martínez et al., 2009) were first crossed with TRF1+/+ 53BP1−/− mice (Ward et al., 2003b). The crosses between double-heterozygote TRF1+/flox 53BP1+/− and TRF1+/Δ K5-Cre 53BP1+/− mice generated double-homozygote TRF1Δ/Δ K5-Cre 53BP1−/− mice. Both parental mouse strains used to generate the TRF1Δ/Δ K5-Cre 53BP1−/− mouse model have a C57BL/6 genetic background. All mice were generated and maintained at the Spanish National Cancer Centre under specific pathogen-free conditions in accordance with the recommendation of the Federation of European Laboratory Animal Science Associations.
Immunoblotting
Whole-cell extracts from LT-Cre–infected MEFs were prepared by resuspending the cells in lysis buffer (50 mM Tris, pH 7.5, 200 mM NaCl, 0.2% NP-40, 0.5% Tween, and 50 mM glycerophosphate) containing PMSF and protease inhibitors. 50 µg of total protein was analyzed by gel electrophoresis followed by Western blotting. The blocking reagent (PhosphoBLOCKER; Cell Biolabs, Inc.) was used to block the membranes. The following antibodies were used for immunoblotting: anti-TRF1 mouse monoclonal antibody (Abcam), antiphospho-KAP1 (S824) rabbit polyclonal, antiphospho-KAP1 (S473) rabbit polyclonal (BioLegend), antiphospho–anti-CHK2 mouse monoclonal (Millipore), antiphospho-CHK1 (Ser345) rabbit polyclonal (Cell Signaling Technology), anti-CHK1 mouse monoclonal (Novocastra), antiphospho-RPA32 (S4/S8) rabbit polyclonal (Bethyl Laboratories, Inc.), anti-RPA32 rat monoclonal (Cell Signaling Technology), and anti-RAD21 rabbit polyclonal (gift from A. Losada, Spanish National Cancer Centre, Madrid, Spain). Antibody binding was detected after incubation with a secondary antibody coupled to horseradish peroxidase using enhanced chemiluminescence.
Retroviral infections
Retroviral infections were performed as previously described (Martínez et al., 2009). The excision of TRF1 exon I by Cre-mediated recombination was confirmed by PCR analysis using primers forward E1 popout, 5′-ATAGTGATCAAAATGTGGTCCTGGG-3′, and reverse SA1, 5′-GCTTGCCAAATTGGGTTGG-3′. Amplification of the wild-type, flox, and knockout alleles renders 1.4-, 1.5-, and 0.48-kb fragments, respectively.
Immunofluorescence staining techniques
Cre-infected MEFs were treated for 5 min with Triton X-100 buffer (Muñoz et al., 2009) for nuclear extraction, fixed 10 min in 4% buffered formaldehyde, permeabilized with 0.1% PBS–Triton X-100 for 10 min, and blocked with 2% fetal bovine serum in PBS for 1 h. Sections were incubated overnight at 4°C with the primary antibody at a 1:200 dilution. TIF were detected using a rabbit polyclonal to RAP1 (Bethyl Laboratories, Inc.) and a mouse monoclonal antibody raised against phospho–Histone 2 H2AX-Ser139 (Millipore) and further incubated with Cy3 goat anti–rabbit and 488 Alexa Fluor goat anti–mouse antibody (1:400; Jackson ImmunoResearch Laboratories, Inc.). Telomeric RPA foci were detected using a rabbit polyclonal to RAP1 (Bethyl Laboratories, Inc.) and a rat polyclonal antibody raised against RPA32 (4E4; Cell Signaling Technology) and further incubated with Cy3 goat anti–rabbit, 488 Alexa Fluor goat anti–rat, and 488 Alexa Fluor goat anti–mouse antibody (1:400; Jackson ImmunoResearch Laboratories, Inc.). For ssDNA immunofluorescence, cells were grown with 10 µM BrdU for 48 h and then treated with either 0 or 30 Gy of ionizing radiation and left for 90 min at 37°C. Cells were fixed with ice-cold methanol for 1 h at −20°C. BrdU was detected using a FITC-conjugated mouse monoclonal anti-BrdU (RPN20AB; GE Healthcare). Slides were mounted in Vectashield (Vector Laboratories) with DAPI. Confocal microscopy was performed at room temperature with a laser-scanning microscope (TCS SP5; Leica) using a Plan Apochromat 63× NA oil immersion objective (HCX; Leica). The pictures show the maximal projection of z stacks generated using advance fluorescence software (LAS; Leica).
Measurement of sister telomere recombination events using CO-FISH
Exponentially growing primary MEFs were subcultured in the presence of BrdU (Sigma-Aldrich) at a final concentration of 10−5 M and then allowed to replicate their DNA once at 37°C for 24 h. Colcemid was added at a concentration of 0.1 µg/ml during the last 4 h. Cells were then recovered, subjected to hypotonic shock, and fixed in methanol/acetic acid (3:1) as previously described (Samper et al., 2000). CO-FISH was performed as previously described (Bailey et al., 2004) using first a telomeric (CCCTAA)7 peptide nucleic acid (PNA) probe labeled with Cy3 and then a second telomeric (TTAGGG)7 PNA probe labeled with Alexa Fluor 488. Metaphase spreads were captured on a fluorescence microscope (DMRB; Leica). For analysis of chromosomal aberrations, metaphases were analyzed by superimposing the telomere image on the DAPI image using the TFL-Telo software (provided by P. Lansdorp, British Columbia Cancer Research Center, Vancouver, British Columbia, Canada).
TRF and G-strand overhang analyses
One million Cre-infected LT-immortalized MEFs were included in agarose plugs, and TRF analysis was performed as previously described (Samper et al., 2000). After overnight digestion in LDS buffer (10 mM Tris, pH 8, 100 mM EDTA, and 1% lithium dodecyl sulphate), the plugs were digested with 0 or 100 U of mung bean nuclease. Then, the plugs were digested with MboI overnight and run on pulse-field electrophoresis gels. Then, sequential in-gel hybridizations in native and denaturing conditions were performed. Values of the G-strand overhang signals were corrected by the telomere signal in denaturing gel conditions.
Histopathological analyses and immunohistochemistry staining techniques
Skin samples and other mouse tissues were fixed in 10% buffered formalin, dehydrated, and embedded in paraffin. For histopathological analysis, 4-µm sections were deparaffinized and stained with hematoxylin and eosin according to standard procedures.
Immunohistochemistry was performed on deparaffinized skin sections processed with 10 mM sodium citrate, pH 6.5, and cooked under pressure for 2 min. Slides were washed in water and then in buffer TBS with 0.5% Tween 20, blocked with peroxidase, washed with TBS with 0.5% Tween 20 again, and blocked with fetal bovine serum followed by another wash. The slides were incubated with the following primary antibodies: rabbit polyclonal to p53 (CM5; Novocastra), goat polyclonal to p21 (C-19-G; Santa Cruz Biotechnology, Inc.), mouse monoclonal to phospho–Histone H2AX (Ser139), rabbit monoclonal to p-CHK1 (Ser345), rabbit polyclonal to SOX9 (Millipore), mouse monoclonal to p63 (p63-4A4; NeoMarkers), rabbit polyclonal to cytokeratin 14 (AF64; Covance), rabbit polyclonal to cytokeratin 10 (Covance), rabbit polyclonal to caspase 3 active (R&D Systems), and rabbit polyclonal to phospho–Histone 3 H3 (Ser10; Millipore). Slides were then incubated with secondary antibodies conjugated with peroxidase obtained from Dako. For signal development, DAB (Dako) was used as a substrate. Sections were lightly counterstained with hematoxylin and analyzed by light microscopy.
The pathologies observed were classified as severe or moderate. The severe pattern is characterized as a high-grade dysplasia in the skin, tongue, palate, esophagus, nonglandular stomach, nails, and urinary bladder, with marked cytological atypia with disturbed polarity of cells, irregular epithelia stratification, loss of nuclear polarity, prominent nucleoli, and nuclear polymorphism. All tissues present focal hyperplasia and hyperkeratosis. The back skin shows epithelial necrosis and complete absence of hair follicles. Moderate pattern is characterized by a lower grade of dysplasia in skin, tongue, nonglandular stomach, and esophagus, with cells larger than normal, often with irregular stratification, enlarged nuclei, and in some cases, loss of nuclear polarity. The back skin shows that primordial hair follicles and the nails are unaffected.
Flow cytometric analysis (FACS)
Primary keratinocyte from newborn mice (P1) were obtained after floating the skin overnight with dermis side down on trypsin without EDTA at 4°C to separate the epidermis from the dermis previously described (Muñoz et al., 2005). The epidermis was then minced with a razor blade, resuspended in culture medium (CnT-02; CELLnTEC) supplemented with 0.2 mM CaCl, and stirred for 30 min at room temperature. Cell suspension was filtered through a 70-µm mesh nylon gauze, and keratinocytes were washed twice with PBS and fixed/permeabilized with ice-cold 70% ethanol. Fixed cells were washed with PBS and resuspended in 1 ml PBS containing 0.2 µg propidium iodide and 100 µg RNase. The samples were incubated for 30 min at 37°C, and the FACS analysis was performed in a flow cytometry system (FACSCanto II; BD). The data were analyzed with the software FACSDiva v5.1.1 (BD).
Telomere and centromere fluorescence analyses on skin sections
Telomere and centromere Q-FISH directly on skin sections was performed as previously described (Martínez et al., 2009) using a telomeric (CCCTAA)7 and a centromeric major satellite (GGGTTA)7 PNA probes labeled with Cy3, respectively. The images were capture at room temperature using a charge-coupled device camera (FK512; Cohu Electronics) on a fluorescence microscope (DMRB) with a Plan Apochromat 100× 1.40 NA oil immersion objective (HCX) using Q-FISH software (Leica) in a linear acquisition mode to prevent the saturation of fluorescence intensity. The TFL-Telo program was used to quantify the telomere fluorescence intensity of individual telomeres.
Online supplemental material
Fig. S1 shows the cell cycle profile of Cre-infected LT-immortalized MEFs. Fig. S2 shows normal telomere length and G-strand overhangs in TRF1Δ/Δ 53BP1−/− LT-immortalized MEFs. Fig. S3 shows that 53BP1 deficiency in TRF1Δ/Δ-immortalized MEFs leads to increased levels of phosphorylated CHK1 and phosphorylated RPA. Fig. S4 shows histopathological findings in different stratified epithelia of TRF1Δ/Δ K5-Cre 53BP1−/− mice at birth. Fig. S5 shows that no apoptotic cells are detected in TRF1Δ/Δ K5-Cre 53BP1−/− neonate skin. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201110124/DC1.
Acknowledgments
We thank R.M. Serrano for expert mouse care, M. Stagno for carefully reading the manuscript, and the Comparative Pathology Unit at Centro Nacional de Investigaciones Oncológicas for technical assistance.
P. Martínez is a Ramon y Cajal Program senior scientist. Work in the Blasco laboratory was funded by Spanish Ministry of Science and Innovation projects SAF2008-05384 and CSD2007-00017, European Union FP7 projects 2007-A-201630 (GENICA) and 2007-A-200950 (TELOMARKER), European Research Council Advanced grant GA#232854, the Körber Foundation, Fundación Botín, and Fundación Lilly.
Author contributions: M.A. Blasco and P. Martínez designed experiments and wrote the manuscript. P. Martínez performed the experiments, and J.M. Flores performed the histopathology analyses described in Fig. S4.
Footnotes
Abbreviations used in this paper:
- A-NHEJ
- alternative NHEJ
- ATM
- ataxia telangiectasia mutated
- ATR
- ataxia telangiectasia and Rad3 related
- C-NHEJ
- classical NHEJ
- CO-FISH
- chromosome orientation FISH
- DDR
- DNA damage response
- DSB
- double-strand break
- HR
- homologous recombination
- LT
- large T antigen
- MEF
- mouse embryonic fibroblast
- MTS
- multitelomeric signal
- NHEJ
- nonhomologous end joining
- PNA
- peptide nucleic acid
- Q-FISH
- quantitative FISH
- Rb
- retinoblastoma
- ssDNA
- single-stranded DNA
- TIF
- telomeric-induced damage foci
- TRF
- terminal restriction fragment
- T-SCE
- telomeric sister chromatid exchange
References
- Audebert M., Salles B., Calsou P. 2004. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 279:55117–55126 10.1074/jbc.M404524200 [DOI] [PubMed] [Google Scholar]
- Bailey S.M., Brenneman M.A., Goodwin E.H. 2004. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 32:3743–3751 10.1093/nar/gkh691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S., Lukas C., Melander F., Bartek J., Lukas J. 2005. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J. Cell Biol. 170:201–211 10.1083/jcb.200503043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco M.A. 2007. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 8:299–309 10.1038/nrg2047 [DOI] [PubMed] [Google Scholar]
- Botuyan M.V., Lee J., Ward I.M., Kim J.E., Thompson J.R., Chen J., Mer G. 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 127:1361–1373 10.1016/j.cell.2006.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P., Aly A., Escandell J.M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q., et al. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17:688–695 10.1038/nsmb.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting S.F., Callén E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 141:243–254 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Xu X., Bunting S.F., Liu J., Wang R.H., Cao L.L., Wu J.J., Peng T.N., Chen J., Nussenzweig A., et al. 2009. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol. Cell. 35:534–541 10.1016/j.molcel.2009.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli G.B., de Lange T. 2005. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7:712–718 10.1038/ncb1275 [DOI] [PubMed] [Google Scholar]
- Celli G.B., Denchi E.L., de Lange T. 2006. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 8:885–890 10.1038/ncb1444 [DOI] [PubMed] [Google Scholar]
- Chiang Y.J., Kim S.H., Tessarollo L., Campisi J., Hodes R.J. 2004. Telomere-associated protein TIN2 is essential for early embryonic development through a telomerase-independent pathway. Mol. Cell. Biol. 24:6631–6634 10.1128/MCB.24.15.6631-6634.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L., Artandi S.E., Shen Q., Tam A., Lee S.L., Gottlieb G.J., Greider C.W., DePinho R.A. 1999. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 97:527–538 10.1016/S0092-8674(00)80762-X [DOI] [PubMed] [Google Scholar]
- Corneo B., Wendland R.L., Deriano L., Cui X., Klein I.A., Wong S.Y., Arnal S., Holub A.J., Weller G.R., Pancake B.A., et al. 2007. Rag mutations reveal robust alternative end joining. Nature. 449:483–486 10.1038/nature06168 [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 426:194–198 10.1038/nature02118 [DOI] [PubMed] [Google Scholar]
- de Klein A., Muijtjens M., van Os R., Verhoeven Y., Smit B., Carr A.M., Lehmann A.R., Hoeijmakers J.H. 2000. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr. Biol. 10:479–482 10.1016/S0960-9822(00)00447-4 [DOI] [PubMed] [Google Scholar]
- de Lange T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19:2100–2110 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- de Lange T. 2009. How telomeres solve the end-protection problem. Science. 326:948–952 10.1126/science.1170633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi E.L. 2009. Give me a break: how telomeres suppress the DNA damage response. DNA Repair (Amst.). 8:1118–1126 10.1016/j.dnarep.2009.04.013 [DOI] [PubMed] [Google Scholar]
- Dimitrova N., Chen Y.C., Spector D.L., de Lange T. 2008. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 456:524–528 10.1038/nature07433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham M.A., Neumann A.A., Fasching C.L., Reddel R.R. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26:447–450 10.1038/82586 [DOI] [PubMed] [Google Scholar]
- Elson A., Wang Y., Daugherty C.J., Morton C.C., Zhou F., Campos-Torres J., Leder P. 1996. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl. Acad. Sci. USA. 93:13084–13089 10.1073/pnas.93.23.13084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejel S., Klatt P., Ménissier-de Murcia J., Martín-Caballero J., Flores J.M., Taccioli G., de Murcia G., Blasco M.A. 2004. Impact of telomerase ablation on organismal viability, aging, and tumorigenesis in mice lacking the DNA repair proteins PARP-1, Ku86, or DNA-PKcs. J. Cell Biol. 167:627–638 10.1083/jcb.200407178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O., Chen H.T., Celeste A., Ward I., Romanienko P.J., Morales J.C., Naka K., Xia Z., Camerini-Otero R.D., Motoyama N., et al. 2002. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat. Cell Biol. 4:993–997 10.1038/ncb884 [DOI] [PubMed] [Google Scholar]
- Haber J.E. 2008. Alternative endings. Proc. Natl. Acad. Sci. USA. 105:405–406 10.1073/pnas.0711334105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M.J., Wei Y., Mancini M.A., Van Hooser A., Ranalli T., Brinkley B.R., Bazett-Jones D.P., Allis C.D. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 106:348–360 10.1007/s004120050256 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D., Daniels J.P., Takai H., de Lange T. 2006. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 126:63–77 10.1016/j.cell.2006.04.044 [DOI] [PubMed] [Google Scholar]
- Jazayeri A., Falck J., Lukas C., Bartek J., Smith G.C., Lukas J., Jackson S.P. 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 8:37–45 10.1038/ncb1337 [DOI] [PubMed] [Google Scholar]
- Karlseder J., Broccoli D., Dai Y., Hardy S., de Lange T. 1999. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 283:1321–1325 10.1126/science.283.5406.1321 [DOI] [PubMed] [Google Scholar]
- Karlseder J., Kachatrian L., Takai H., Mercer K., Hingorani S., Jacks T., de Lange T. 2003. Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol. Cell. Biol. 23:6533–6541 10.1128/MCB.23.18.6533-6541.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibe T., Osawa G.A., Keegan C.E., de Lange T. 2010. Telomere protection by TPP1 is mediated by POT1a and POT1b. Mol. Cell. Biol. 30:1059–1066 10.1128/MCB.01498-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini Denchi E., Celli G., de Lange T. 2006. Hepatocytes with extensive telomere deprotection and fusion remain viable and regenerate liver mass through endoreduplication. Genes Dev. 20:2648–2653 10.1101/gad.1453606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M.R. 2008. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 283:1–5 10.1074/jbc.R700039200 [DOI] [PubMed] [Google Scholar]
- Mahaney B.L., Meek K., Lees-Miller S.P. 2009. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 417:639–650 10.1042/BJ20080413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis J.P., Morales J.C., Xia Z., Kutok J.L., Alt F.W., Carpenter P.B. 2004. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 5:481–487 10.1038/ni1067 [DOI] [PubMed] [Google Scholar]
- Martínez P., Blasco M.A. 2010. Role of shelterin in cancer and aging. Aging Cell. 9:653–666 10.1111/j.1474-9726.2010.00596.x [DOI] [PubMed] [Google Scholar]
- Martínez P., Blasco M.A. 2011. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat. Rev. Cancer. 11:161–176 10.1038/nrc3025 [DOI] [PubMed] [Google Scholar]
- Martínez P., Thanasoula M., Muñoz P., Liao C., Tejera A., McNees C., Flores J.M., Fernández-Capetillo O., Tarsounas M., Blasco M.A. 2009. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 23:2060–2075 10.1101/gad.543509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser R.S., Wong K.K., Sahin E., Xia H., Naylor M., Hedberg H.M., Artandi S.E., DePinho R.A. 2007. DNA-dependent protein kinase catalytic subunit is not required for dysfunctional telomere fusion and checkpoint response in the telomerase-deficient mouse. Mol. Cell. Biol. 27:2253–2265 10.1128/MCB.01354-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 455:770–774 10.1038/nature07312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. 2009. DNA end resection: many nucleases make light work. DNA Repair (Amst.). 8:983–995 10.1016/j.dnarep.2009.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesti M., Kanaar R. 2001. DNA repair: spot(light)s on chromatin. Curr. Biol. 11:R229–R232 10.1016/S0960-9822(01)00112-9 [DOI] [PubMed] [Google Scholar]
- Muñoz P., Blanco R., Flores J.M., Blasco M.A. 2005. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 37:1063–1071 10.1038/ng1633 [DOI] [PubMed] [Google Scholar]
- Muñoz P., Blanco R., de Carcer G., Schoeftner S., Benetti R., Flores J.M., Malumbres M., Blasco M.A. 2009. TRF1 controls telomere length and mitotic fidelity in epithelial homeostasis. Mol. Cell. Biol. 29:1608–1625 10.1128/MCB.01339-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga M., Bunting S., Montaña M.F., Soria R., Mulero F., Cañamero M., Lee Y., McKinnon P.J., Nussenzweig A., Fernandez-Capetillo O. 2009. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat. Genet. 41:891–898 10.1038/ng.420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig A., Nussenzweig M.C. 2007. A backup DNA repair pathway moves to the forefront. Cell. 131:223–225 10.1016/j.cell.2007.10.005 [DOI] [PubMed] [Google Scholar]
- Palm W., de Lange T. 2008. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42:301–334 10.1146/annurev.genet.41.110306.130350 [DOI] [PubMed] [Google Scholar]
- Pierce A.J., Hu P., Han M., Ellis N., Jasin M. 2001. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 15:3237–3242 10.1101/gad.946401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Zheng H., He H., Luo Y., Multani A., Carpenter P.B., Chang S. 2010. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J. 29:2598–2610 10.1038/emboj.2010.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappold I., Iwabuchi K., Date T., Chen J. 2001. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage–signaling pathways. J. Cell Biol. 153:613–620 10.1083/jcb.153.3.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper E., Goytisolo F.A., Slijepcevic P., van Buul P.P., Blasco M.A. 2000. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 1:244–252 10.1093/embo-reports/kvd051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. 2007. Human CtIP promotes DNA end resection. Nature. 450:509–514 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de Lange T. 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 138:90–103 10.1016/j.cell.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A., Kabir S., van Overbeek M., Celli G.B., de Lange T. 2010. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 327:1657–1661 10.1126/science.1185100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A., de Lange T. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73:177–208 10.1146/annurev.biochem.73.071403.160049 [DOI] [PubMed] [Google Scholar]
- Soulas-Sprauel P., Le Guyader G., Rivera-Munoz P., Abramowski V., Olivier-Martin C., Goujet-Zalc C., Charneau P., de Villartay J.P. 2007. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J. Exp. Med. 204:1717–1727 10.1084/jem.20070255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H., Smogorzewska A., de Lange T. 2003. DNA damage foci at dysfunctional telomeres. Curr. Biol. 13:1549–1556 10.1016/S0960-9822(03)00542-6 [DOI] [PubMed] [Google Scholar]
- Tejera A.M., Stagno d’Alcontres M., Thanasoula M., Marion R.M., Martinez P., Liao C., Flores J.M., Tarsounas M., Blasco M.A. 2010. TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev. Cell. 18:775–789 10.1016/j.devcel.2010.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanasoula M., Escandell J.M., Martinez P., Badie S., Muñoz P., Blasco M.A., Tarsounas M. 2010. p53 prevents entry into mitosis with uncapped telomeres. Curr. Biol. 20:521–526 10.1016/j.cub.2010.01.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska A., de Lange T. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell. 92:401–413 10.1016/S0092-8674(00)80932-0 [DOI] [PubMed] [Google Scholar]
- Varley H., Pickett H.A., Foxon J.L., Reddel R.R., Royle N.J. 2002. Molecular characterization of inter-telomere and intra-telomere mutations in human ALT cells. Nat. Genet. 30:301–305 10.1038/ng834 [DOI] [PubMed] [Google Scholar]
- Wang B., Matsuoka S., Carpenter P.B., Elledge S.J. 2002. 53BP1, a mediator of the DNA damage checkpoint. Science. 298:1435–1438 10.1126/science.1076182 [DOI] [PubMed] [Google Scholar]
- Wang H., Rosidi B., Perrault R., Wang M., Zhang L., Windhofer F., Iliakis G. 2005. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 65:4020–4030 10.1158/0008-5472.CAN-04-3055 [DOI] [PubMed] [Google Scholar]
- Ward I.M., Minn K., Jorda K.G., Chen J. 2003a. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 278:19579–19582 10.1074/jbc.C300117200 [DOI] [PubMed] [Google Scholar]
- Ward I.M., Minn K., van Deursen J., Chen J. 2003b. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23:2556–2563 10.1128/MCB.23.7.2556-2563.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I.M., Reina-San-Martin B., Olaru A., Minn K., Tamada K., Lau J.S., Cascalho M., Chen L., Nussenzweig A., Livak F., et al. 2004. 53BP1 is required for class switch recombination. J. Cell Biol. 165:459–464 10.1083/jcb.200403021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.S., Moncalian G., Williams J.S., Yamada Y., Limbo O., Shin D.S., Groocock L.M., Cahill D., Hitomi C., Guenther G., et al. 2008. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 135:97–109 10.1016/j.cell.2008.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K.K., Maser R.S., Sahin E., Bailey S.T., Xia H., Ji H., McNamara K., Naylor M., Bronson R.T., Ghosh S., et al. 2007. Diminished lifespan and acute stress-induced death in DNA-PKcs-deficient mice with limiting telomeres. Oncogene. 26:2815–2821 10.1038/sj.onc.1210099 [DOI] [PubMed] [Google Scholar]
- Wu L., Multani A.S., He H., Cosme-Blanco W., Deng Y., Deng J.M., Bachilo O., Pathak S., Tahara H., Bailey S.M., et al. 2006. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 126:49–62 10.1016/j.cell.2006.05.037 [DOI] [PubMed] [Google Scholar]
- Xie A., Hartlerode A., Stucki M., Odate S., Puget N., Kwok A., Nagaraju G., Yan C., Alt F.W., Chen J., et al. 2007. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell. 28:1045–1057 10.1016/j.molcel.2007.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C.T., Boboila C., Souza E.K., Franco S., Hickernell T.R., Murphy M., Gumaste S., Geyer M., Zarrin A.A., Manis J.P., et al. 2007. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 449:478–482 10.1038/nature06020 [DOI] [PubMed] [Google Scholar]
- Zou L., Elledge S.J. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 300:1542–1548 10.1126/science.1083430 [DOI] [PubMed] [Google Scholar]