Human telomere replication initiates either from within telomere repeats or from within the subtelomere using a chromosome-specific replication program that appears conserved between different cell types.
Abstract
Telomeric and adjacent subtelomeric heterochromatin pose significant challenges to the DNA replication machinery. Little is known about how replication progresses through these regions in human cells. Using single molecule analysis of replicated DNA (SMARD), we delineate the replication programs—i.e., origin distribution, termination site location, and fork rate and direction—of specific telomeres/subtelomeres of individual human chromosomes in two embryonic stem (ES) cell lines and two primary somatic cell types. We observe that replication can initiate within human telomere repeats but was most frequently accomplished by replisomes originating in the subtelomere. No major delay or pausing in fork progression was detected that might lead to telomere/subtelomere fragility. In addition, telomeres from different chromosomes from the same cell type displayed chromosome-specific replication programs rather than a universal program. Importantly, although there was some variation in the replication program of the same telomere in different cell types, the basic features of the program of a specific chromosome end appear to be conserved.
Introduction
Mammalian telomeres are specialized structures that cap chromosome termini, consisting of hundreds to thousands of tandem TTAGGG repeats complexed with several proteins including telomere-specific shelterins. Telomere ends are organized into protective structures termed t-loops (Griffith et al., 1999), which prevent telomeres from being mistaken as broken or damaged chromosomes by the DNA repair machinery. Formation of t-loops protects chromosome ends against inappropriate repair activities that could lead to fusions and deleterious recombination-mediated events. Maintenance of telomere structure and function requires efficient replication of telomeric DNA. It is known that the majority of telomere DNA is duplicated by conventional semiconservative DNA replication (for review see Gilson and Géli, 2007). However, the features of telomere replication programs (i.e., origin distribution, the efficiency of origin firing, termination site location, fork rate and direction, and timing) and how these programs influence replication efficiency are largely unknown.
Telomeres challenge replication machinery because of the combination of their repetitive G-rich sequence and extensive heterochromatization. Structural elements of telomeres, including secondary structures such as G-quadruplexes (Paeschke et al., 2005; Lipps and Rhodes, 2009; Smith et al., 2011) and more complex structures such as t-loops, present potential impediments to replication fork passage. Several studies in yeast and human cells suggest that telomeric DNA has an inherent ability to pause or stall replication forks (Ivessa et al., 2002; Makovets et al., 2004; Miller et al., 2006; Verdun and Karlseder, 2006; Anand et al., 2012). We and others have shown that telomeric DNA is difficult to replicate, leading to telomere fragility under replication stress (Miller et al., 2006; Sfeir et al., 2009). Replication of G-rich sequences by cellular DNA polymerases appears to require assistance from other proteins. For example, the Saccharomyces cerevisiae Pif1 DNA helicase has been shown to play a key role in replication fork progression through quadruplex motifs in G-rich regions at nontelomeric sites in the genome (Paeschke et al., 2011). With specific regard to telomeres, the studies of Sfeir et al. (2009) have revealed that efficient replication of mammalian telomeres requires the involvement of the shelterin protein TRF1. A similar requirement for yeast telomere replication has been demonstrated for the TRF1/TRF2 homologue TAZ1 (Miller et al., 2006).
Cytological examination of fluorescently labeled replicated telomeres in metaphase spreads has provided valuable information on telomere replication (for review see Williams et al., 2011). However, this approach cannot be used to determine the specific characteristics of a replication program. More detailed analysis of telomere replication has been performed using 2D gel electrophoresis (Ivessa et al., 2002; Makovets et al., 2004; Miller et al., 2006; Anand et al., 2012). Although 2D gel methodology can map origins and termination regions, as well as provide information on fork progression, in specific chromosomal segments, it is limited to analysis of small (2–10 kb) segments. Moreover, the data obtained from 2D analysis comes from a population of molecules; therefore events within individual molecules cannot be discriminated. Recently, we applied an individual molecule approach termed single molecule analysis of replicated DNA (SMARD) to examine mouse telomere replication (Sfeir et al., 2009). Although this initial study was performed on a population of total genomic telomeric molecules, the design of SMARD allows for all features of replication programs to be mapped over large genomic regions, spanning as many as 500 kb, in specific individual molecules (Norio and Schildkraut, 2001, 2004).
The replication of telomeres had been assumed to begin at initiation sites (origins) within the subtelomere, with telomeres being replicated by forks progressing from subtelomere to telomere (Oganesian and Karlseder, 2009). However, the evidence for lack of initiation within telomeric DNA came primarily from yeast, where initiation occurs at well-defined autonomously replicating sequence (ARS) sequences. Origin-dependent initiation within telomeric DNA has been demonstrated in vitro in a Xenopus laevis cell-free system (Kurth and Gautier, 2010). In addition, studies linking the shelterin protein TRF2 to the recruitment of origin recognition complex (ORC) proteins to telomeres raised the possibility of initiation within human telomere repeats (Atanasiu et al., 2006; Deng et al., 2007; Tatsumi et al., 2008). Our recent SMARD studies on the replication of mouse telomeres demonstrated that initiation can occur within telomeres (Sfeir et al., 2009). These studies suggested that initiation events in mouse telomeres are not common, implying that replication of telomeric DNA initiates predominantly from origins within adjacent subtelomeric regions or further upstream. With regard to the overall program of replication, it is currently unknown whether mammalian telomeres replicate using telomere-specific or universal replication programs. It is also not known if telomere replication programs are unique to, and thus defining features of, specific cell types. We have demonstrated that replication programs for several gene loci such as OCT4 and the immunoglobulin heavy chain (Igh) region vary between cell types (Norio et al., 2005; Guan et al., 2009; Schultz et al., 2010). In our previous study, replication of total genomic telomeric DNA was analyzed, which provided only limited information regarding replication programs (Sfeir et al., 2009). In the present study we have used SMARD to examine the replication of specific human telomeres and determine how replication progresses through human telomeres and their flanking subtelomeres. We demonstrate, for the first time, that the replication of human telomeres is most frequently accomplished by replisomes originating in the subtelomere. In addition, we find that telomere/subtelomeres from different chromosomes from the same cell type exhibit chromosome-specific programs rather than a universal program. Importantly, we observe that, although some variation was seen in the replication program of the same telomere in the different cell types examined here, the basic features of the replication program appear to be conserved. Finally, we also have evidence that replication can initiate within the telomere repeats in certain human chromosomes.
Results
Analysis of telomere replication programs
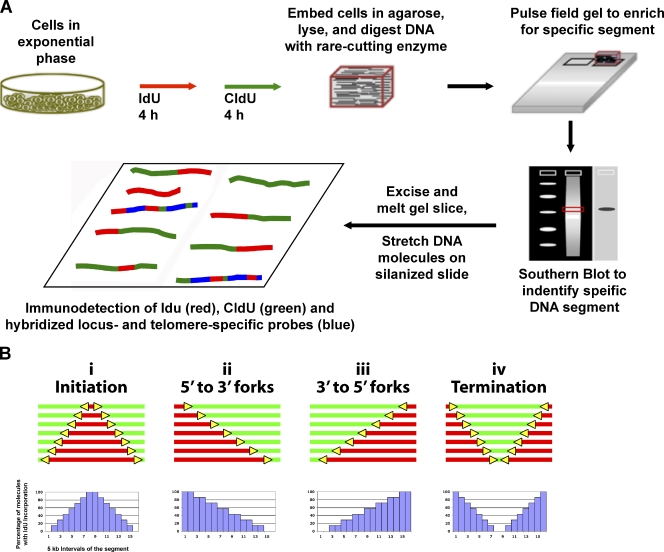
The extensive studies on yeast telomere replication by 2D gel electrophoresis represent the most detailed analysis of telomere replication program features to date. However, because we wanted to obtain detailed information on individual molecules over an extensive length and 2D analysis does not discriminate events in individual molecules, we used SMARD to analyze the replication of specific human telomeres in single DNA molecules. SMARD is a direct approach to capture images of individual replicated DNA molecules (Fig. 1). In brief, exponentially growing cells are sequentially pulsed with two different halogenated nucleosides, iododeoxyuridine (IdU) followed by chlorodeoxyuridine (CldU), to label replicating DNA. The replicated DNA is isolated and stretched on silanized glass slides, and the halogenated nucleosides incorporated in the replicated DNA were detected by immunostaining. Portions of molecules replicated during the IdU (first) pulse stain red, whereas areas of CldU incorporation (second pulse) stain green. Asymmetrically placed biotinylated FISH probes (blue) create a “bar code” used to identify molecules of interest and to align the images of individual molecules, producing a composite profile of replication. Replication program events that can be detected by SMARD include: initiation sites (Fig. 1 B, i), replication forks (yellow arrows) moving in the 5′-to-3′ (Fig. 1 b, ii) and 3′-to-5′ (Fig. 1 b, iii) directions, and terminations (Fig. 1 b, iv). Fork orientation designation is based on the observed transitions from IdU to CldU (red to green), corresponding to forks that were active during the IdU-to-CldU transition. Molecules of each replication event class are arranged in order of increasing amounts of IdU incorporation. This was done in a nonsubjective manner by first identifying the specific stretched DNA strands of interest by FISH, and then arranging the photographic images of these molecules from the lowest to highest amount of incorporated IdU (red stain) as determined by visual inspection.
Figure 1.
SMARD. (A) Exponentially growing cells were labeled with the halogenated nucleosides IdU for 4 h followed by CldU for 4 h. Genomic DNA was then isolated. To avoid the breakage of large molecules, the cells were embedded in agarose. We used rare cutting restriction endonucleases to obtain unique large molecules of genomic DNA, which were separated by pulse field gel electrophoresis to enrich by size for the particular subtelomere segments of interest. Southern blotting was performed to identify the target segment within the gel, which was excised. The gel slice containing the segment of interest was melted and the enriched DNA in the melted gel solution was stretched on silanized glass slides. Fluorescent antibodies were used to identify regions where IdU (red) or CldU (green) were incorporated into the DNA. Immunodetected biotinylated FISH probes (blue) were used to identify the target molecules and to align the images of individual molecules to produce a composite profile of replication. (B) Specific replication events are identified by characteristic patterns in the aligned molecule images. DNA molecules with increasing red stain from one end indicate replication forks progressing in a single direction through the aligned region (ii and iii). Initiation events are indicated by a red tract flanked on both sides by green (i), whereas a green tract flanked on both sides by red (iv) indicates a termination event. Molecule alignments can be graphically depicted as histograms of the percentage of IdU incorporation along each 5 kb of a segment. Specific replication events produce characteristic features in these replication profiles. Positions of the centers of initiation zones are indicated by peaks (i), whereas termination events are indicated by valleys (iv). Replication progressing through the segment primarily in one direction (5′ to 3′ or 3′ to 5′) from an external origin is observed as a progressive decrease in the percentage of IdU incorporation from one end to the other across the segment (x axis; ii and iii).
In addition to the alignments of micrographic images of the molecules, replication events can be graphically represented as replication profiles, i.e., histograms of the percentage of IdU incorporation along each 5 kb of a segment (Fig. 1 b). Specific replication events produce characteristic features in replication profiles. Peaks represent early replicating regions, with sharp peaks indicating origins that are used frequently while broad peaks indicate initiation zones. Valleys represent later replicating regions and sharp valleys indicate a termination site or zone. The progressive decrease in the percentage of IdU incorporation from one end of the segment to the other is indicative of replication progressing through the segment primarily in one direction from an external origin. In many cases, the replication profile represents a mixture of the above situations and reflects the predominant event in a population of DNA molecules.
We characterized the replication programs of several human telomere loci and their associated subtelomeric regions (140–270 kb) by SMARD to establish the extent of program variability. Four different human telomere/subtelomere loci, located at the ends of chromosome arms 5p, 7q, 10q, and 11q, were examined. These loci were selected based on the availability of restriction endonuclease sites that permit excision of the end of a chromosome in a segment of suitable length (140–300 kb) for SMARD analysis. SMARD was performed on unsynchronized, exponentially growing, early passage cultures of cells grown under physiological conditions. Telomere replication programs were determined for several cell types including embryonic stem (ES) cells (lines H1 and H9), HeLa cervical carcinoma cells, and primary somatic cells: microvascular endothelial cells (MECs) and lung fibroblasts (IMR-90). Utilization of the SMARD approach required generation of specific FISH probes for identification of these segments, an important step not used in our studies on mouse telomeres. Because all telomeres consist of the identical sequence, the biotinylated locus–specific FISH probes used to identify particular telomeric/subtelomeric chromosomal segments had to be based on specific subtelomeric sequences. Telomeres within the segments were identified with a telomere-specific biotinylated peptide nucleic acid (PNA) probe. It should be noted that the biotinylated telomeric PNA probe, which hybridizes only to the G-rich strand, interferes with the detection of IdU and CldU in the telomere repeats in that strand. In principle, therefore, IdU and CldU incorporation in the telomere cannot be detected in half of the replicated molecules examined, the segments where the G-rich strand is the nascent (substituted) strand of the duplex. Consequently, labeled molecules whose telomeres are stained with PNA only (blue without any red or green) are observed, which presumably contain IdU and/or CldU substitutions in the G-rich strand.
Initiation events occur throughout the human chromosome 5p telomere-containing region, and replication forks move across the segment from both directions
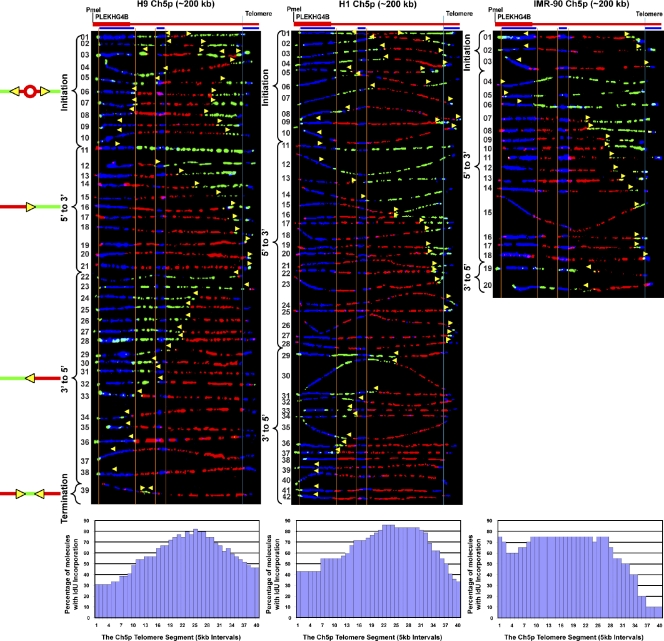
Genomic regions are replicated by a variety of programs. For example, a region can be replicated by several forks from outside of the region or many initiation sites within the region. These initiation sites are often present in a zone where many origins can be activated in an apparently stochastic manner. To determine how telomere-containing regions are replicated, we analyzed the replication of an ∼200-kb PmeI restriction fragment containing the chromosome 5p telomere and its associated subtelomeric region in three different cell lines by SMARD. This analysis revealed common features in the replication of this telomere in the cell types examined. We observed frequent initiation events (a red patch surrounded by green) in the subtelomere, which occurred in a zone centered 65–115 kb away from the telomere, depending on the cell type (Fig. 2). Initiation events within the 5p telomere were rare, as few molecules had patterns consistent with such events. Only in the H9 ES line were these molecules, i.e., molecules containing 3′-to-5′ forks with short (<10 kb) red tracts at the 3′ end, detected (Fig. 2, H9 molecules 22 and 23). These results indicated that the 5p telomere is replicated primarily by forks progressing from subtelomere to telomere. In some cases forks progressing toward the telomere (5′ to 3′) were detected >120 kb away from the telomere (Fig. 2, H9 molecules 11–13, H1 molecules 11–14, IMR-90 molecules 4–6), which suggests that in these molecules the telomere was replicated from fairly distant origins. In addition, there was only one molecule containing more than one initiation event, and only one molecule contained a termination event (Fig. 2, H9 molecules 3 and 39, respectively) which suggests that the segment is replicated primarily from a single origin per molecule per cell cycle. Notably, there was no evidence of major fork pausing or stalling observed in the molecules examined. Specifically, we did not detect multiple molecules with IdU to CldU transitions (replication forks) in the same location, which is indicative of a major pause or stall site, either in the telomere or subtelomere.
Figure 2.
Initiation events occur throughout the subtelomere in the human 5p segment in three cell lines, and replication forks progress through the segment in both directions. SMARD analysis of three different cell lines indicated that the 5p telomere is replicated primarily by forks progressing from the subtelomere to the telomere. Alignments of replicated molecules fully labeled with both IdU (red) and CldU (green) are shown. A map of the 5p locus is depicted above each alignment, with the positions of the FISH probes (blue bars below) used for identifying and orienting the molecules indicated. Vertical orange lines indicate the positions of the ends of the subtelomeric FISH signals used to align the molecules. The boundary between the subtelomere and telomere is delineated by a vertical blue line. Yellow arrows mark sites of transition from IdU incorporation to CldU incorporation and indicate the direction of fork progression at the moment of transition during the replication of the molecule. Replication profiles, histograms of the percentage of molecules containing IdU per 5-kb interval along the segment, are shown below each alignment. Initiation events (red tracts surrounded by green) occur at multiple locations. The origins appear to be clustered around a 40–90-kb region centered 65–115-kb from the telomere, seen as a peak in the replication profiles. Aside from a leftward broadening of the initiation zone in IMR-90 cells, there was not much variation in the basic features of the replication program of the 5p telomere in the two ES cell lines (H1 and H9) and the primary fibroblasts (IMR-90).
Some program variation was observed in the replication program of the 5p telomere in different cells, such as in the width of the initiation zone (Fig. 2) and the percentages of 5′-to-3′ and 3′-to-5′ single fork molecules (Table 1). Specifically, in H9 ES cells the subtelomeric initiation zone spanned ∼40 kb, whereas the zone was larger in H1 cells, spanning 60 kb, and larger still in IMR-90 primary fibroblast cells (∼90 kb). In the H1 and H9 ES cells, there were roughly equal numbers of 5′-to-3′ and 3′-to-5′ forks (Fig. 2 and Table 1). However, in the IMR-90 cells, molecules with single forks were almost exclusively 5′-to-3′ forks. This may be caused by a larger number of origins and/or more proximal origins outside of the segment in IMR-90 cells. Most importantly, although some program variation was seen in the replication program of the 5p telomere region in different cells, the basic features of the replication program were conserved.
Table 1.
Replication events in telomeric chromosomal segments analyzed by SMARD
| Telomere segment of chromosome | Approximate size of the segment | Cell type | Replication events | |||
| Initiation | 5′-to-3′ fork | 3′-to-5′ fork | Termination | |||
| 5p | 200 kb | ES (H1) | 10 (24%) | 18 (43%) | 14 (33%) | 0 (0%) |
| 5p | 200 kb | ES (H9) | 10 (26%) | 11 (28%) | 17 (44%) | 1 (2%) |
| 5p | 200 kb | Lung fibroblast (IMR-90) | 3 (15%) | 15 (75%) | 2 (10%) | 0 (0%) |
| 7q | 200 kb | ES (H9) | 3 (8%) | 18 (49%) | 12 (32%) | 4 (11%) |
| 10q | 210 kb | ES (H1) | 0 (0%) | 28 (97%) | 0 (0%) | 1 (3%) |
| 10q | 210 kb | ES (H9) | 2 (8%) | 17 (68%) | 2 (8%) | 4 (16%) |
| 10q | 270 kb | Lung fibroblast (IMR-90) | 0 (0%) | 19 (100%) | 0 (0%) | 0 (0%) |
| 10q | 210 kb | MEC | 2 (6%) | 22 (65%) | 2 (6%) | 8 (23%) |
| 10q | 240 kb | HeLa 1.3 | 3 (13%) | 13 (54%) | 2 (8%) | 6 (25%) |
| 11q | 140 kb | ES (H1) | 19 (32%) | 22 (37%) | 16 (27%) | 2 (4%) |
| 11q | 140 kb | ES (H9) | 17 (36%) | 21 (45%) | 7 (15%) | 2 (4%) |
| 11q | 140 kb | MEC | 6 (25%) | 12 (50%) | 3 (12.5%) | 3 (12.5%) |
A summary of all replication events detected in the SMARD analysis of telomere/subtelomere segments (Figs. 2–6) from the various cell types studied is shown. The number of fully IdU- and CldU-substituted (red-green) molecules containing initiations, terminations, or a single visible fork (5′ to 3′ or 3′ to 5′) is indicated. The percentage of a given event of the total events observed is indicated in parentheses.
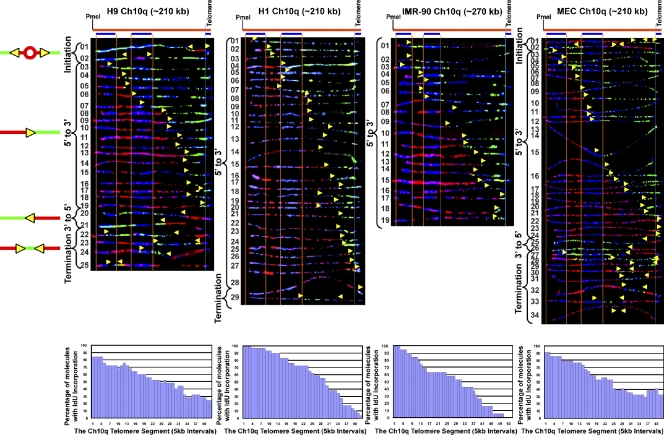
Replication progresses from origins located >100 kb from the human chromosome 10q telomere-containing region, predominantly from subtelomere to telomere
Our findings for the 5p telomere-containing segment indicated that the telomere was replicated primarily by forks progressing from subtelomere to telomere. This result was the first direct evidence in support of the idea that human telomere replication mainly initiates in the subtelomere. To determine if this was generally the case for human telomeres and whether the program exhibited by the 5p telomere was common to other telomeres, we next examined the replication program of the chromosome 10q locus. Unlike the 5p segment, initiation events were infrequently observed in the10q telomere/subtelomere segment for any of the cell types examined. In H1 and IMR-90 cells, no initiations were seen, while only two were detected (out of 25 molecules analyzed) in H9 cells and two (out of 34 molecules analyzed) in MECs (Fig. 3, H9 molecules 1 and 2, and MEC molecules 1 and 2). Although only a small number of molecules (four) with clear initiations sites (red patches surrounded by green) were detected, some of the few molecules that contained forks moving away from the telomere (Fig. 3, H9 molecule 23, H1 molecule 29, and MEC molecules 28 and 29) did have patterns (forks with short, <10-kb, red tracts at the 3′ end), which strongly suggest initiation within the telomere.
Figure 3.
Replication forks progress toward the telomere from origins in the subtelomere of chromosome 10q in three cell types. SMARD analysis of the 10q telomere segment in ES lines H9 and H1, primary lung fibroblast, IMR-90 cells, and primary microvascular endothelial MECs indicated that the four cell lines replicate this segment using very similar programs. In these lines, the 10q telomere is replicated primarily by forks progressing from the subtelomere to the telomere. This is reflected in the replication profile histograms, which show a progressive decrease in the percentage of IdU incorporation from 5′ to 3′ across the segment, which is indicative of replication progressing through the segment primarily in one direction from an external origin. These distal subtelomeric origins are located predominantly >200 kb from the telomere in all four lines. A region of mixed staining (blue-green or blue-red) is seen in the right half of many molecules resulting from nonspecific hybridization of the FISH probes. The yellow arrows indicate the direction of replication fork progression, and the vertical orange and blue lines demarcate the boundaries of sequences where fish probes bind, as described in the legend to Fig. 2.
Importantly, in the majority of molecules with single forks, replication progressed in one direction, from subtelomere to telomere (5′ to 3′; Fig. 3 and Table 1), and these forks initiated from origins >100 kb from the telomere. Thus, in contrast to the 5p telomere, the 10q telomere appeared to be predominantly passively replicated by forks arising from distant origins (Fig. 3). Furthermore, as in the case of the 5p telomere, there was some variation in the 10q programs between the cell types studied. Replication progressed solely from subtelomere to telomere in IMR-90 10q and almost exclusively from subtelomere to telomere in H1 10q. In MEC 10q, replication progressed mainly from subtelomere to telomere, and more frequently from subtelomere to telomere than from telomere to subtelomere in H9. Accordingly, termination events were more frequently detected in H9 (molecules 22–25) and MEC (molecules 27–34), only occasionally seen in H1 cells (molecule 29), and not detected in IMR-90 cells (Table 1). In addition, as in the 5p telomere, fork pausing or stalling was not detected.
Thus, most importantly, our comparison of the replication of different telomeres (Ch 5p and 10q) from the same cell types demonstrated that different individual telomeres in the same cell are replicated by chromosome arm–specific, rather than universal, programs.
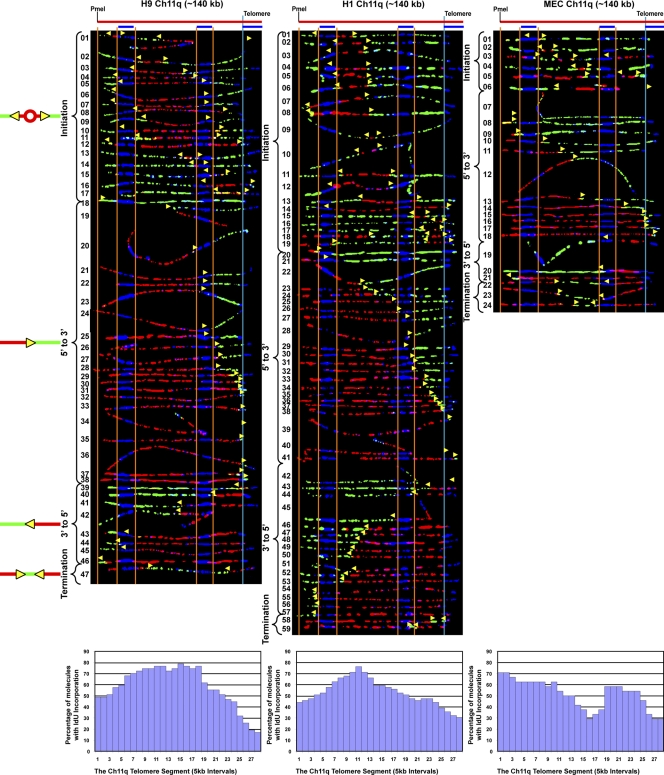
Initiation events occur at many positions in the human chromosome 11q telomere-containing region, and forks move across the segment in both directions
Our SMARD results indicated that the 5p and 10q loci displayed different replication programs, particularly with respect to origin site selection. One factor we considered that may influence initiation site selection is the timing of telomere replication. It has been shown that although human telomere replication occurs throughout S phase, specific telomeres replicate early and others replicate late (Arnoult et al., 2010). Furthermore, it has been shown that the telomeres of the 5p and 10q segments, which exhibited differences in origin site selection, are replicated predominantly in mid and late S phase, respectively. Therefore, to determine if origin selection could be dependent on timing, we next examined the replication program of the chromosome 11q telomere, another late replicating telomere, and its associated subtelomeric region. Our analysis of the 11q segment revealed that in the cell types studied there were similar program features. We observed frequent initiation events in the segment, which occurred at many positions in 140-kb segment (Fig. 4). This differed from the 10q segment, where initiations were infrequently seen. It also differed from the program of the 5p segment, another locus that exhibited frequent initiations, but where initiations were clustered in a smaller 40–90-kb region rather than throughout the segment. However, within the 11q segment there did appear to be preferred sites of initiation within a zone in the ES cell lines, which were located in similar locations in these cells (Fig. 4 and replication profiles). Initiations were more frequently detected than terminations in 11q, particularly in the ES cell lines, which suggests that replication of the segment initiates predominantly from either one origin or two or more closely spaced origins. Importantly, although both the 11q and 10q telomeres replicate late in S phase, they have distinctly different replication programs, which suggests that timing is not major determinant of program features. Furthermore, only one molecule (Fig. 4, H9 molecule 39) exhibited a pattern that was consistent with initiation within the 11q telomere. Collectively, these findings indicated that the 11q telomere is replicated primarily by forks progressing from subtelomere to telomere. In addition, the proportion of particular types of replication events (initiations, terminations, and forks moving 5′-to-3′ and 3′-to-5′) of the total replication events observed was similar in the cell lines examined (Table 1). Thus, as with the 5p and 10q telomeres, our SMARD analysis of the 11q telomere indicates that the replication program of a specific telomere appears similar in different cell types.
Figure 4.
Initiation events occur throughout the human 11q segment and forks move across the segment in both directions in three cell lines.SMARD analysis of the Ch11q telomere segment in human ES H9, H1, and IMR-90 fibroblast cells revealed similar program features. Frequent initiation events occurred throughout the full 140-kb length of the segment. Preferred sites of initiation, seen as peaks in the replication profile histograms, could be detected. In all three lines, the 11q telomere is replicated primarily by forks progressing from the subtelomere to the telomere. The yellow arrows indicate the direction of replication fork progression, and the vertical orange and blue lines demarcate the boundaries of sequences where fish probes bind, as described in the legend to Fig. 2.
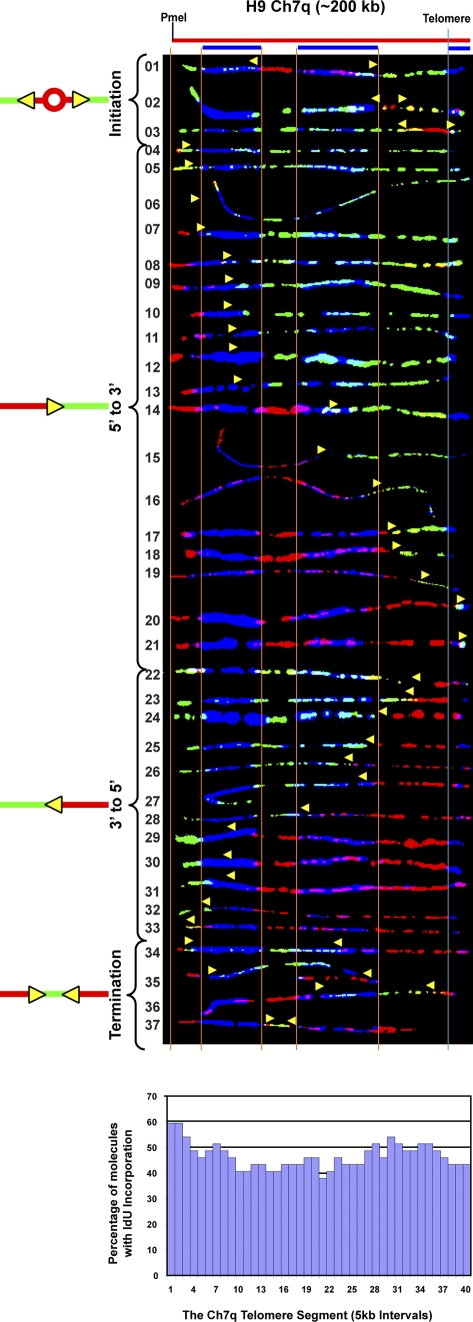
The human 7q telomere segment in H9 ES cells is replicated by forks originating either within or near to the telomere, or from outside of the segment
Our findings from the 5p, 10q, and 11q segments indicated that telomere replication was most frequently accomplished by forks that initiated in the subtelomere. This suggested that subtelomere sequence organization could affect initiation site location. Human subtelomeres often contain large (>1 kb) segments (duplicons), which are duplicated in other locations in the genome (for review see Riethman, 2008). All of the segments examined (5p, 10q, and 11q) contained duplicons (Fig. S1). To determine whether the absence of duplicons would influence subtelomeric origin use, we examined the replication of the chromosome 7q telomere, whose subtelomere is devoid of duplicons (Fig. S1). Our examination of the replication program of a 200-kb PmeI segment of chromosome 7 in H9 ES cells containing the 7q telomere and adjacent subtelomeric region revealed distinct differences from the other telomere programs. In contrast to the H9 5p and 11q segments (Figs. 2 and 4), initiation in the subtelomeric portion of the H9 7q segment was relatively rare (only three initiations detected in 37 molecules with forks), which indicates that no initiation zone was present (Fig. 5) in the most telomere-proximal 200 kb of the subtelomere. In addition, unlike the 10q segment where molecules with 3′-to-5′ forks were rarely detected (Fig. 3), similar percentages of molecules containing 5′-to-3′ and 3′-to-5′ forks were detected for the 7q segment (Fig. 5 and Table 1). Notably, the comparable percentages of 5′-to-3′ and 3′-to-5′ single fork molecules, along with the low number of molecules detected with initiation events (three), indicated that the 7q segment is replicated with similar frequency by forks originating either within (or near to) the telomere or from outside of the segment. This conclusion was further supported by the observed low frequency of termination events (Table 1), which occur when two simultaneously active forks collide.
Figure 5.
Initiation sites were rarely detected in the subtelomere of Ch 7q. SMARD analysis of the Ch 7 telomere/subtelomere segment of human ES H9 cells is shown. Few initiation or termination events were detected in the molecules examined. The lack of distinct peaks or valleys in the replication profile histogram indicates the absence of preferred initiation or termination sites. Similar numbers of replication forks proceed in the telomere-to-subtelomere (3′-to-5′) and subtelomere-to-telomere (5′-to-3′) directions. Many of the 5′-to-3′ forks appear to initiate from origins at least 150 kb from the telomere. The yellow arrows indicate the direction of replication fork progression, and the vertical orange and blue lines demarcate the boundaries of sequences where fish probes bind, as described in the legend to Fig. 2.
Importantly, the program observed for the 7q telomere differed from the programs of the other telomeres (5p, 10q, and 11q) from the same cell line (H9), further demonstrating that individual telomeres in the same cell are replicated by telomere-specific, rather than universal, programs.
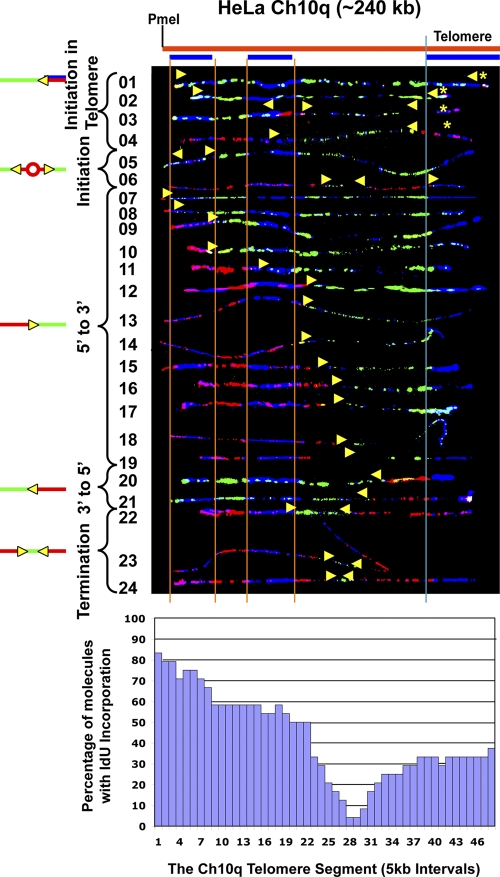
Initiation events are more frequently detected in 10q telomeres with an increased length
Our results indicated that telomeres were most frequently replicated by forks originating in the subtelomere. However, in almost all the telomeric regions studied here, there is at least one and often many forks proceeding from the region of the telomere repeats, which is suggestive of initiation within or near the telomere. Initiation events (a significant red tract surrounded by green) are difficult to detect within the human telomere repeats because the repeats are only 10 kb in average size. To more firmly establish whether initiation occurs within the telomere repeats, we analyzed telomere replication in HeLa 1.3 cells, a line with a mean telomere length of 23 kb, more than twice as long as most human cells (Takai et al., 2010). We chose two telomere loci to examine: 7q and 10q. The 7q locus was chosen because it showed the most potential for telomeric initiation of the segments studied (see the preceding paragraphs); therefore, it should be the most likely segment to observe initiation within the telomere. The 10q locus was chosen because it showed few potential telomeric initiations, allowing us to determine if increased telomere length could promote initiation in the telomere. Our initial SMARD examination of the 7q segment revealed that its telomeres were of similar size as the other cells examined; thus, 7q was not studied further. However, we found that the telomeres of the 10q segment were considerably larger, averaging 25 kb. The results for the 10q segment are shown in Fig. 6. Replication forks proceeding preferentially but not predominantly from 5′ to 3′ were observed, differing somewhat from the other cell lines examined (Fig. 3). In addition, termination events were frequently detected (Table 1) in a termination zone, seen as a valley in the replication profile histogram, centered ∼60 kb from the telomere/subtelomere border (Fig. 6). The presence of this zone indicates that the segment is frequently replicated by simultaneously active forks originating from separate regions of the chromosome. Importantly, we observed clear evidence for initiation in the telomere repeats in the HeLa cells. Telomeric initiation events were detected in molecules containing terminations where the red tract of the 3′-to-5′ fork does not extend out from the telomere or extends only a few kilobases out from the telomere (Fig. 6, molecules 1–4). In these cases, the origins that gave rise to these forks must have been located within the telomere. The small number of initiation events within the subtelomere further indicates that forks that progress in the 3′-to-5′ direction initiate in the telomere. Thus, similar to our observations in mouse cells (Sfeir et al., 2009; unpublished data), we find that initiation does occur within human telomere repeats. Moreover, because the 10q termination zone, which is associated with initiation in the telomere, was seen in HeLa but not in the other cell lines studied (Fig. 3), this implies that replication initiates more frequently in the longer 10q telomeres of the HeLa cells.
Figure 6.
Replication forks initiate within the telomere repeats in chromosome 10q in a HeLa cell line with long telomeres. SMARD analysis of the Ch10q telomere segment in HeLa 1.3, a line with a mean telomere length of 23 kb, more than twice as long as in most human cells (Takai et al., 2010). Based on the length of the signals for the FISH probes, we estimate that the mean length of the telomeres in Ch10q is ∼40 kb. Telomeric initiation events are detected in molecules containing terminations where the red tract of the 3′-to-5′ fork does not extend out of the telomere or extends only a few kilobases out from the telomere (molecules 1–4). The centers of telomeric initiation events are indicated by yellow asterisks. A termination zone, seen as a valley in the replication profile histogram centered ∼60 kb from the telomere/subtelomere border, indicates the frequent use of two origins separated by at least 120 kb, which fire at similar times to replicate the segment. The yellow arrows indicate the direction of replication fork progression, and the vertical orange and blue lines demarcate the boundaries of sequences where fish probes bind, as described in the legend to Fig. 2.
Discussion
Uniqueness of replication programs
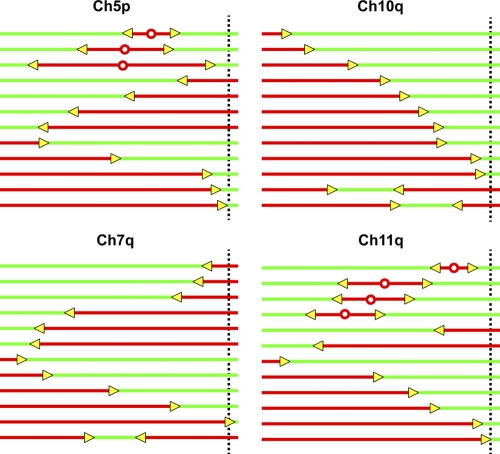
Our studies reveal a common, fundamental feature of the replication programs of human telomeres. The replication of human telomeres is most frequently accomplished by replisomes originating in the subtelomere. However, the distances that subtelomeric replisomes travel to reach the telomere can differ significantly from one particular chromosome to another, reflecting variation in initiation profiles (replication programs; Fig. 7). In some subtelomeres (Ch 11q), zones where initiation events are frequently supported occur within relatively close proximity to the telomere. In contrast, initiations are uncommon near the telomere in other subtelomeres (Ch 10q), and forks from distant origins replicate the telomere. Thus, rather than using a universal program, it appears that different telomeres from the same cell type use telomere-specific replication programs. With telomere replication primarily originating from the subtelomere, it is not unexpected that telomere replication programs would be chromosome arm–specific, given the high degree of interchromosomal subtelomere variation (Fig. S1; for review see Riethman, 2008). Surprisingly, although a particular subtelomere can exhibit sequence polymorphism (for review see Riethman, 2008), the basic features of the replication program of a specific telomere appear to be conserved from one cell type to another in the cell types examine here.
Figure 7.
Schematic representation of the replication program of four different telomere/subtelomere segments of the ES and primary somatic cells studied here. This diagrammatic summary of representative molecules indicates that human telomeres do not exhibit a universal replication program. The dotted vertical line indicates the boundary between the telomere and subtelomere.
Significance of program features
The sequence orientation of telomeres is such that when replication forks proceed from the subtelomere into the telomere, the G-rich strand of the duplex serves as the template strand for lagging strand synthesis. When single-stranded, the G-rich strand has the potential to form G-quadruplexes or other structures that can challenge fork progression (Lipps and Rhodes, 2009). Because lagging strand templates have more extensive single-stranded portions than leading strand templates, these structures should have a higher likelihood of forming in lagging strand (G-rich) templates. Therefore, it could be potentially more difficult to replicate telomeres by forks moving from the subtelomere to the telomere than from the telomere toward the subtelomere (Fig. S2). However, although our findings indicate that initiation does occur in certain telomeres, and origins within the telomere (preferentially near the end) would be advantageous, human telomere replication initiates predominantly in the subtelomere.
Compensatory mechanisms appear to exist for dealing with difficulties in replicating G-rich strands by lagging strand synthesis, including the use of duplex-unwinding helicases. Efficient telomere lagging strand synthesis has been shown to be reliant on the Rec Q helicase WRN, which could act to resolve replication-challenging structures (Crabbe et al., 2004). Studies in yeast have suggested, however, that both lagging and leading strand telomere replication challenges the replication machinery. Telomeric sequences that were ectopically inserted into an internal, nonterminal position in a yeast chromosome were found to be equally capable of stalling replication forks (in the absence of Taz1), regardless of their orientation with respect to the direction of replication (Miller et al., 2006). Furthermore, it was recently shown that the up-frameshift 1 (UPF1) helicase supports leading strand synthesis of human telomeres (Chawla et al., 2011). Thus, it appears that cells may be equally able to replicate telomeres from either direction.
Replication forks can stall at particular sequences and result in double-strand breaks, leading to genome instability (Torres et al., 2004; Mirkin and Mirkin, 2007; Tourrière and Pasero, 2007; Branzei and Foiani, 2010). The potential for replication forks to pause or stall within human telomeres has been suggested (Verdun and Karlseder, 2006). The heterochromatic nature of subtelomeres may also present a potential hindrance to replication fork passage. It has been shown that replication forks move slowly through S. cerevisiae telomeres (Makovets et al., 2004). However, the rate of fork movement in the last 100–200 kb of the human chromosomes examined here (Table 1) was very similar to that measured for internal genomic regions such as the Igh, OCT4, and other loci (Schultz et al., 2010). In addition, we did not observe evidence of fork pausing for an extended length of time. Specifically, we did not detect multiple molecules with IdU-to-CldU transitions (replication forks) in the same location, either in the telomere or subtelomere, as has been observed at pause sites in the family of repeated sequences in the Epstein–Barr virus genome (Norio and Schildkraut, 2004). Pausing and stalling of forks in S. cerevisiae telomeric sequences has been well documented using 2D gels (Ivessa et al., 2002; Makovets et al., 2004; Anand et al., 2012). Replication intermediates formed by very brief pausing at a specific site, such as the subtelomere/telomere boundary, would represent a relatively small number of molecules containing forks at the same position along the molecule. The small number of molecules containing these pauses would not be detected by SMARD but can be detected by the 2D gels used in the yeast studies. In addition, differences in observed replication fork pausing may, in part, be caused by differences in structure-forming abilities of yeast and human telomeric sequences. Moreover, a recent study has strongly suggested that stalling in yeast telomeric sequences is protein-mediated rather than DNA structure–mediated (Anand et al., 2012). Thus, the differences in detected pausing/stalling between human and yeast telomeres may also reflect differences in telomere-bound protein contributions in addition to DNA secondary structure.
Determinants of telomere replication programs
Because the majority of human telomere replication initiates in the subtelomere, the principal determinants of telomere replication programs are likely subtelomeric rather than telomeric in nature. Along with subtelomere sequence composition (Fig. S1), other potential factors that may contribute to the chromosome arm–specific replication programs include epigenetic modifications, local transcription, and timing of replication. Studies suggest that replication initiation can be associated with transcription start sites and nucleosome-free sites (Berbenetz et al., 2010; Karnani et al., 2010; Ding and MacAlpine, 2011; Lubelsky et al., 2011; Mesner et al., 2011; Valenzuela et al., 2011; for review see Aladjem, 2007; Méchali, 2010; Schepers and Papior, 2010). However, human subtelomeric DNA has been shown to be heavily methylated (Cross et al., 1990; de Lange et al., 1990; Brock et al., 1999), and mammalian subtelomeric chromatin has been shown to be hypoacetylated and to contain repressive trimethyl marks (Benetti et al., 2007), which are features of compact, silenced DNA.
Nevertheless, human subtelomeres can be transcriptionally active (Riethman, 2008). Interestingly, of the telomere/subtelomere segments studied here, subtelomeric initiation events occur much more frequently within the Ch 11q segment, which has only a single transcript mapped to it, compared with the Ch10q segment, which has over a dozen transcripts mapped to it. In addition to subtelomere transcription, the transcription of telomeres into telomeric repeat-containing RNA (TERRA), driven by CpG-island promoters located in the directly adjacent subtelomere, has recently been demonstrated (Azzalin et al., 2007; Nergadze et al., 2009). However, we find that the presence of these TERRA CpG promoter sites (Nergadze et al., 2009) does not appear to be associated with an increase of initiation at these loci. Initiation events very near or at the telomere/subtelomere boundary in segments containing TERRA CpG promoters (Nergadze et al., 2009) are no more frequent than in segments lacking these promoters. It should be noted that it has also been shown that TERRA transcription appears to occur regardless of the presence of 61-29-37 CpG repeats. (Nergadze et al., 2009). For example, TERRA transcripts originating from 11q and Xp/Yp chromosome ends have been detected (Azzalin et al., 2007), although the same chromosome ends are apparently devoid of 61-29-37 promoters (Nergadze et al., 2009). Importantly, a recent genome-wide study found that although moderately transcribed regions were generally associated with high initiation frequency in mammalian cells, few initiation events were detected in highly transcribed regions (Martin et al., 2011). Thus, not only the presence of transcriptional activity, but also the level of activity appear to be key in determining whether local transcription promotes or represses initiation. In addition, TERRA transcripts may also have effects on origin site selection beyond those attributed to transcription-mediated local changes to chromatin. It has been recently proposed, based on in vitro observations, that TERRA and heterogeneous nuclear protein A1 regulate the timely exchange at telomeres between replication protein A (RPA) and POT1, both single-stranded DNA (ssDNA)-binding proteins, during S phase (Flynn et al., 2011). In this light, TERRA molecules might mediate a molecular cross-talk between subtelomeric and telomeric replication factors, thereby contributing to initiation site selection.
With regard to replication timing during the S phase, it has been shown that specific human telomeres replicate early and others later in S phase (Arnoult et al., 2010). Three of the telomeres of the segments we examined (7q, 10q, and 11q) were found to similarly replicate late in S phase, whereas the fourth (5p) replicated in early to mid S phase (Arnoult et al., 2010). Given the distinctly different programs we observe, it appears that replication timing does not play a major role in shaping telomere replication programs. Interestingly, the timing of telomere replication has also been shown to be chromosome-arm specific (Arnoult et al., 2010). From these studies, it has been proposed that subtelomeric sequences that direct nuclear localization act as determinants of replication timing. However, because we did not observe a link between timing and other replication program features, it appears that sequence determinants of replication timing do not influence other features of telomere programs. Thus additional factors, possibly including ones telomeric in nature, contribute to subtelomeric initiation site selection and other program features. Moreover, telomere replication programs are likely the result of combined contributions by many or all of the above factors rather than determined by a single factor.
Importance of telomere replication programs
Improper replication can lead to dysfunctional telomeres that can result in genomic instability, cellular senescence, and apoptosis. Certain replication programs may render telomeres more susceptible to dysfunction than other programs. Along these lines, the ability to initiate replication within the telomere may safeguard against dysfunction by providing for rescue of stalled telomere replication. The present characterization of human telomere replication programs is an essential step in understanding their role in telomere function.
Materials and methods
Cell culture
Human ES cell lines and H1 (WA01) and H9 (WA09) were cultured in HES medium (DME/F12 supplemented with 20% knockout serum replacement [Invitrogen], l-glutamine, penicillin-streptomycin and nonessential amino acids, 0.1 mM β-mercaptoethanol, and 4 ng/ml FGF2 [R&D Systems] on mouse embryonic fibroblasts as described previously (Schultz et al., 2010). Cells were transferred (after Dispase [Worthington] treatment) onto Matrigel (BD) to eliminate the MEF population before nucleoside labeling. Matrigel-plated ES cells were also analyzed for the pluripotency markers Oct4, Nanog, SSEA-4, TRA-1-60, and TRA-1-81 by immunostaining and FACS (Schultz et al., 2010). In brief, cells were fixed in 4% paraformaldehyde and 0.015% picric acid (20 min at room temperature), washed in PBS, then blocked in PBS, 0.5% BSA, and 0.3% Triton X-100 (50 min at room temperature). The cells were then washed in PBS and then incubated overnight at 4°C with primary antibodies diluted in PBS–1% BSA (mouse anti-Oct3/4 [Santa Cruz Biotechnology, Inc.], goat anti-Nanog [R&D Systems], mouse anti-SSEA-4, [Developmental Studies Hybridoma Bank], mouse anti–TRA1-60, and mouse anti–TRA1-81 [Millipore]). For cell surface marker (SSEA-4, TRA1-60, and TRA1-81) detection, Triton X-100 was omitted from the PBS-BSA blocking solution. After overnight incubation, cells were washed with PBS then incubated with secondary antibodies (Alexa Fluor 488 anti–mouse antibody for SSEA-4, TRA1-60, and TRA1-81; Alexa Fluor 555 anti–mouse antibody for Oct3/4, and Alexa Fluor 488 anti–goat antibody for Nanog (all from Invitrogen/Molecular Probes) diluted in PBS–1% BSA for 1 h at room temperature. The cells were then counterstained with DAPI (100 ng/ml for 5 min at room temperature) followed by a PBS wash. The same protocol was used for FACS analysis except the cells were maintained in suspension and on ice.
IMR-90 fetal lung fibroblasts (Coriell) cells were cultured in MEM/EBSS (Hyclone) supplemented with 15% FBS, l-glutamine, penicillin-streptomycin, and nonessential amino acids. Primary human cortex MECs (Cell Systems) were cultured on gelatin-coated tissue culture dishes in M199 media supplemented with 20% newborn calf serum (Invitrogen), 5% human serum (Biocell), l-glutamine, 0.1 g/ml heparin, 0.05 g/ml ascorbic acid (Sigma-Aldrich), endothelial cell growth factor (Sigma-Aldrich), bovine brain extract (Clonetics), and penicillin-streptomycin as previously described (Schultz et al., 2010).
HeLa 1.3 cervical carcinoma cells, a HeLa subclone with a mean telomere length of 23 kb (Takai et al., 2010), were a generous gift of T. de Lange (The Rockefeller University, New York, NY). The cells were cultured in DME (Hyclone) supplemented with FBS, l-glutamine, penicillin-streptomycin, and nonessential amino acids.
SMARD
SMARD was performed essentially as described previously (Fig. 1; Norio and Schildkraut, 2001, 2004; Schultz et al., 2010). Exponentially growing cells were sequentially pulse labeled with 30 µM IdU (for 4 h) followed by 30 µM CldU (4 h). Labeled cells were embedded in 0.5% low melting agarose (InCert; FMC) at 106 cells per 80-µl agarose cell plug and lysed overnight at 50°C in 1% n-lauroylsarcosine, and 0.5 M EDTA, pH 8, containing 20 mg/ml proteinase K. The treated agarose cell plugs were then washed several times (1 h at 50°C) with TE (10 mM Tris, pH 8, and 1 mM EDTA), once with 200 µM phenylmethanesulfonyl fluoride in TE (1 h at 50°C), then several more times in TE (1 h at room temperature). The plugs were then equilibrated in restriction enzyme digestion buffer (New England Biolabs) for 2 h at room temperature and the DNA in the plugs was digested in situ with PmeI (50 U/plug). The treated plugs were cast into 0.7% gels (SeaPlaque GTG; Lonza), and the digested DNA was separated by pulse field gel electrophoresis using a CHEF-DRII system (Bio-Rad Laboratories). Specific telomere/subtelomere chromosome segments within the gel were then located by performing Southern blotting on portion of the gel using subtelomere-specific probes. After Southern blotting, pulse field gel slices containing the telomere/subtelomere segments of interest were excised and melted (20 min at 70°C), and the DNA in the gel solution was stretched on microscope slides coated with 3-aminopropyltriethoxysilane (Sigma-Aldrich). The stretched DNA was denatured in alkali buffer (0.1 M NaOH in 70% ethanol and 0.1% β-mercaptoethanol) and fixed in alkali buffer containing 0.5% glutaraldehyde. The denatured, fixed DNA was hybridized overnight with biotinylated probes at 37°C in humidified chamber. Biotinylated DNA FISH probes based on the following subtelomeric sequences were prepared by nick translation in the presence of biotin-16-dUTP (Roche) and used to identify specific telomeric/subtelomeric segments. Ch 5p: nucleotides 100,065–105,023 (probe 1), 104,858–108,992 (probe 2), and 136,605–177,232 (probe 3); Ch 7q: nucleotides 158,980,894–159,014,530 (probe 1), and 159,037,147–159,082,858 (probe 2); Ch 10q: nucleotides 135,365,825–135,402,175 (probe 1) and 135,429,047–135,466,998 (probe 2); and Ch 11q: nucleotides 134,841,259–134,846,641 (probe1), 134,846,750–134,850,820 (probe2), 134,850,242–134,854,333 (probe 3), 134,907,552–134,909,757 (probe 4), and 134,912,615–134,917,753 (probe 5; all map coordinates were based on National Center for Biotechnology Information Human Genome Map Build 37.3). A biotin-OO-(CCCTAA)4 PNA probe (50 nM; BioSythesis) was used to identify the telomeric portion of the segment. After hybridization, the slides were blocked with 1% BSA for a minimum of 20 min and then incubated with an Alexa Fluor 350–conjugated NeutrAvidin antibody (Invitrogen) followed two rounds of incubation first with a biotinylated anti-avidin antibody (Vector) and then the Alexa Fluor 350–conjugated NeutrAvidin antibody to detect the hybridized biotinylated probes. Incorporated halogenated nucleotides were detected by incubating the slides with a mouse anti-IdU monoclonal antibody (BD) and a rat anti-CldU monoclonal antibody (Accurate) followed by Alexa Fluor 568–conjugated goat anti–mouse (Invitrogen) and Alexa Fluor 488–conjugated goat anti–rat secondary antibodies (Invitrogen).
Microphotographic image acquisition
Images of immunostained molecules were acquired at room temperature using a fluorescence microscope (Axioskop 2; Carl Zeiss) equipped with a Plan Apochromatic 100× 1.4 NA oil objective lens and a charge-coupled device camera (CoolSNAP HQ; Photometrics) using IPLab software (BD). Images were processed with Photoshop (Adobe) and aligned according to the FISH probe pattern using Illustrator software (Adobe).
Online supplemental material
Fig. S1 depicts the sequence organization of human telomere/subtelomere segments examined in this study. Fig. S2 illustrates telomere replication by forks progressing from telomeric versus subtelomeric origins. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201112083/DC1.
Acknowledgments
We thank Drs. Sherri Schultz for characterizing the human ES and endothelial cells and additional assistance; Harold Reithman and Carol Elias for advice; and James Borowiec, Jeannine Gerhardt, Paolo Norio, and Joel Huberman for valuable suggestions in writing the manuscript.
S.G. Calderano was supported by a fellowship from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP). This work was supported by National Institute of General Medicine Sciences grant 5R01-GM045751 (to C.L. Schildkraut) and the Empire State Stem Cell Fund through New York State contract C024348 (to C.L. Schildkraut). The authors have no commercial affiliations or conflicts of interest.
Footnotes
Abbreviations used in this paper:
- CldU
- chlorodeoxyuridine
- ES
- embryonic stem
- IdU
- iododeoxyuridine
- MEC
- microvascular endothelial cell
- PNA
- peptide nucleic acid
- SMARD
- single molecule analysis of replicated DNA
- TERRA
- telomeric repeat-containing RNA
References
- Aladjem M.I. 2007. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat. Rev. Genet. 8:588–600 10.1038/nrg2143 [DOI] [PubMed] [Google Scholar]
- Anand R.P., Shah K.A., Niu H., Sung P., Mirkin S.M., Freudenreich C.H. 2012. Overcoming natural replication barriers: differential helicase requirements. Nucleic Acids Res. 40:1091–1105 10.1093/nar/gkr836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult N., Schluth-Bolard C., Letessier A., Drascovic I., Bouarich-Bourimi R., Campisi J., Kim S.H., Boussouar A., Ottaviani A., Magdinier F., et al. 2010. Replication timing of human telomeres is chromosome arm-specific, influenced by subtelomeric structures and connected to nuclear localization. PLoS Genet. 6:e1000920 10.1371/journal.pgen.1000920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu C., Deng Z., Wiedmer A., Norseen J., Lieberman P.M. 2006. ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 7:716–721 10.1038/sj.embor.7400730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J. 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 318:798–801 10.1126/science.1147182 [DOI] [PubMed] [Google Scholar]
- Benetti R., García-Cao M., Blasco M.A. 2007. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat. Genet. 39:243–250 10.1038/ng1952 [DOI] [PubMed] [Google Scholar]
- Berbenetz N.M., Nislow C., Brown G.W. 2010. Diversity of eukaryotic DNA replication origins revealed by genome-wide analysis of chromatin structure. PLoS Genet. 6:e1001092 10.1371/journal.pgen.1001092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M. 2010. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11:208–219 10.1038/nrm2852 [DOI] [PubMed] [Google Scholar]
- Brock G.J., Charlton J., Bird A. 1999. Densely methylated sequences that are preferentially localized at telomere-proximal regions of human chromosomes. Gene. 240:269–277 10.1016/S0378-1119(99)00442-4 [DOI] [PubMed] [Google Scholar]
- Chawla R., Redon S., Raftopoulou C., Wischnewski H., Gagos S., Azzalin C.M. 2011. Human UPF1 interacts with TPP1 and telomerase and sustains telomere leading-strand replication. EMBO J. 30:4047–4058 10.1038/emboj.2011.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe L., Verdun R.E., Haggblom C.I., Karlseder J. 2004. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 306:1951–1953 10.1126/science.1103619 [DOI] [PubMed] [Google Scholar]
- Cross S., Lindsey J., Fantes J., McKay S., McGill N., Cooke H. 1990. The structure of a subterminal repeated sequence present on many human chromosomes. Nucleic Acids Res. 18:6649–6657 10.1093/nar/18.22.6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T., Shiue L., Myers R.M., Cox D.R., Naylor S.L., Killery A.M., Varmus H.E. 1990. Structure and variability of human chromosome ends. Mol. Cell. Biol. 10:518–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z., Dheekollu J., Broccoli D., Dutta A., Lieberman P.M. 2007. The origin recognition complex localizes to telomere repeats and prevents telomere-circle formation. Curr. Biol. 17:1989–1995 10.1016/j.cub.2007.10.054 [DOI] [PubMed] [Google Scholar]
- Ding Q., MacAlpine D.M. 2011. Defining the replication program through the chromatin landscape. Crit. Rev. Biochem. Mol. Biol. 46:165–179 10.3109/10409238.2011.560139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn R.L., Centore R.C., O’Sullivan R.J., Rai R., Tse A., Songyang Z., Chang S., Karlseder J., Zou L. 2011. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 471:532–536 10.1038/nature09772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Géli V. 2007. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 8:825–838 10.1038/nrm2259 [DOI] [PubMed] [Google Scholar]
- Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T. 1999. Mammalian telomeres end in a large duplex loop. Cell. 97:503–514 10.1016/S0092-8674(00)80760-6 [DOI] [PubMed] [Google Scholar]
- Guan Z., Hughes C.M., Kosiyatrakul S., Norio P., Sen R., Fiering S., Allis C.D., Bouhassira E.E., Schildkraut C.L. 2009. Decreased replication origin activity in temporal transition regions. J. Cell Biol. 187:623–635 10.1083/jcb.200905144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa A.S., Zhou J.Q., Schulz V.P., Monson E.K., Zakian V.A. 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 16:1383–1396 10.1101/gad.982902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N., Taylor C.M., Malhotra A., Dutta A. 2010. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol. Biol. Cell. 21:393–404 10.1091/mbc.E09-08-0707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth I., Gautier J. 2010. Origin-dependent initiation of DNA replication within telomeric sequences. Nucleic Acids Res. 38:467–476 10.1093/nar/gkp929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipps H.J., Rhodes D. 2009. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 19:414–422 10.1016/j.tcb.2009.05.002 [DOI] [PubMed] [Google Scholar]
- Lubelsky Y., Sasaki T., Kuipers M.A., Lucas I., Le Beau M.M., Carignon S., Debatisse M., Prinz J.A., Dennis J.H., Gilbert D.M. 2011. Pre-replication complex proteins assemble at regions of low nucleosome occupancy within the Chinese hamster dihydrofolate reductase initiation zone. Nucleic Acids Res. 39:3141–3155 10.1093/nar/gkq1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovets S., Herskowitz I., Blackburn E.H. 2004. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol. Cell. Biol. 24:4019–4031 10.1128/MCB.24.9.4019-4031.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M.M., Ryan M., Kim R., Zakas A.L., Fu H., Lin C.M., Reinhold W.C., Davis S.R., Bilke S., Liu H., et al. 2011. Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res. 21:1822–1832 10.1101/gr.124644.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méchali M. 2010. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat. Rev. Mol. Cell Biol. 11:728–738 10.1038/nrm2976 [DOI] [PubMed] [Google Scholar]
- Mesner L.D., Valsakumar V., Karnani N., Dutta A., Hamlin J.L., Bekiranov S. 2011. Bubble-chip analysis of human origin distributions demonstrates on a genomic scale significant clustering into zones and significant association with transcription. Genome Res. 21:377–389 10.1101/gr.111328.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K.M., Rog O., Cooper J.P. 2006. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 440:824–828 10.1038/nature04638 [DOI] [PubMed] [Google Scholar]
- Mirkin E.V., Mirkin S.M. 2007. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 71:13–35 10.1128/MMBR.00030-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nergadze S.G., Farnung B.O., Wischnewski H., Khoriauli L., Vitelli V., Chawla R., Giulotto E., Azzalin C.M. 2009. CpG-island promoters drive transcription of human telomeres. RNA. 15:2186–2194 10.1261/rna.1748309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P., Schildkraut C.L. 2001. Visualization of DNA replication on individual Epstein-Barr virus episomes. Science. 294:2361–2364 10.1126/science.1064603 [DOI] [PubMed] [Google Scholar]
- Norio P., Schildkraut C.L. 2004. Plasticity of DNA replication initiation in Epstein-Barr virus episomes. PLoS Biol. 2:e152 10.1371/journal.pbio.0020152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P., Kosiyatrakul S., Yang Q., Guan Z., Brown N.M., Thomas S., Riblet R., Schildkraut C.L. 2005. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol. Cell. 20:575–587 10.1016/j.molcel.2005.10.029 [DOI] [PubMed] [Google Scholar]
- Oganesian L., Karlseder J. 2009. Telomeric armor: the layers of end protection. J. Cell Sci. 122:4013–4025 10.1242/jcs.050567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K., Simonsson T., Postberg J., Rhodes D., Lipps H.J. 2005. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 12:847–854 10.1038/nsmb982 [DOI] [PubMed] [Google Scholar]
- Paeschke K., Capra J.A., Zakian V.A. 2011. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 145:678–691 10.1016/j.cell.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethman H. 2008. Human telomere structure and biology. Annu. Rev. Genomics Hum. Genet. 9:1–19 10.1146/annurev.genom.8.021506.172017 [DOI] [PubMed] [Google Scholar]
- Schepers A., Papior P. 2010. Why are we where we are? Understanding replication origins and initiation sites in eukaryotes using ChIP-approaches. Chromosome Res. 18:63–77 10.1007/s10577-009-9087-1 [DOI] [PubMed] [Google Scholar]
- Schultz S.S., Desbordes S.C., Du Z., Kosiyatrakul S., Lipchina I., Studer L., Schildkraut C.L. 2010. Single-molecule analysis reveals changes in the DNA replication program for the POU5F1 locus upon human embryonic stem cell differentiation. Mol. Cell. Biol. 30:4521–4534 10.1128/MCB.00380-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de Lange T. 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 138:90–103 10.1016/j.cell.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.S., Chen Q., Yatsunyk L.A., Nicoludis J.M., Garcia M.S., Kranaster R., Balasubramanian S., Monchaud D., Teulade-Fichou M.P., Abramowitz L., et al. 2011. Rudimentary G-quadruplex-based telomere capping in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 18:478–485 10.1038/nsmb.2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K.K., Hooper S., Blackwood S., Gandhi R., de Lange T. 2010. In vivo stoichiometry of shelterin components. J. Biol. Chem. 285:1457–1467 10.1074/jbc.M109.038026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi Y., Ezura K., Yoshida K., Yugawa T., Narisawa-Saito M., Kiyono T., Ohta S., Obuse C., Fujita M. 2008. Involvement of human ORC and TRF2 in pre-replication complex assembly at telomeres. Genes Cells. 13:1045–1059 10.1111/j.1365-2443.2008.01224.x [DOI] [PubMed] [Google Scholar]
- Torres J.Z., Schnakenberg S.L., Zakian V.A. 2004. Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol. Cell. Biol. 24:3198–3212 10.1128/MCB.24.8.3198-3212.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H., Pasero P. 2007. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst.). 6:900–913 10.1016/j.dnarep.2007.02.004 [DOI] [PubMed] [Google Scholar]
- Valenzuela M.S., Chen Y., Davis S., Yang F., Walker R.L., Bilke S., Lueders J., Martin M.M., Aladjem M.I., Massion P.P., Meltzer P.S. 2011. Preferential localization of human origins of DNA replication at the 5′-ends of expressed genes and at evolutionarily conserved DNA sequences. PLoS ONE. 6:e17308 10.1371/journal.pone.0017308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun R.E., Karlseder J. 2006. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 127:709–720 10.1016/j.cell.2006.09.034 [DOI] [PubMed] [Google Scholar]
- Williams E.S., Cornforth M.N., Goodwin E.H., Bailey S.M. 2011. CO-FISH, COD-FISH, ReD-FISH, SKY-FISH. Methods Mol. Biol. 735:113–124 10.1007/978-1-61779-092-8_11 [DOI] [PubMed] [Google Scholar]