SUMOylation of the ubiquitin ligase HERC2 promotes efficient chromatin licensing in the vicinity of DNA double-strand breaks.
Abstract
Nonproteolytic ubiquitylation of chromatin surrounding deoxyribonucleic acid (DNA) double-strand breaks (DSBs) by the RNF8/RNF168/HERC2 ubiquitin ligases facilitates restoration of genome integrity by licensing chromatin to concentrate genome caretaker proteins near the lesions. In parallel, SUMOylation of so-far elusive upstream DSB regulators is also required for execution of this ubiquitin-dependent chromatin response. We show that HERC2 and RNF168 are novel DNA damage–dependent SUMOylation targets in human cells. In response to DSBs, both HERC2 and RNF168 were specifically modified with SUMO1 at DSB sites in a manner dependent on the SUMO E3 ligase PIAS4. SUMOylation of HERC2 was required for its DSB-induced association with RNF8 and for stabilizing the RNF8–Ubc13 complex. We also demonstrate that the ZZ Zinc finger in HERC2 defined a novel SUMO-specific binding module, which together with its concomitant SUMOylation and T4827 phosphorylation promoted binding to RNF8. Our findings provide novel insight into the regulatory complexity of how ubiquitylation and SUMOylation cooperate to orchestrate protein interactions with DSB repair foci.
Introduction
DNA damage arises continuously from intracellular metabolism and replication errors and from exposure of cells to multiple exogenous genotoxic agents (Lindahl and Barnes, 2000). DNA double-strand breaks (DSBs) represent particularly cytotoxic lesions that pose a major threat to genome stability if not properly sensed and repaired (Wyman and Kanaar, 2006; Jackson and Bartek, 2009). To meet this challenge, cells have evolved a global DNA damage signaling response, which mounts a coordinated response to the lesion, impacting on processes such as cell cycle progression and DNA repair to facilitate reestablishment of genomic integrity (Jackson and Bartek, 2009; Ciccia and Elledge, 2010).
After DSB induction, multiple DNA damage signaling and repair factors become concentrated in DSB repair foci formed around the lesion (Misteli and Soutoglou, 2009; Bekker-Jensen and Mailand, 2010). Protein recruitment to such structures occurs in a highly dynamic and hierarchical manner, controlled by posttranslational modifications of the DSB-flanking chromatin (Bekker-Jensen and Mailand, 2010; Polo and Jackson, 2011). Phosphorylation of multiple DSB-signaling components by the ATM/ATR/DNA-PK kinases plays a central role in promoting this response. In addition, recent work demonstrated a key function of nonproteolytic ubiquitylation in orchestrating protein interactions with DSB sites (Morris and Solomon, 2004; Kim et al., 2007; Sobhian et al., 2007; Wang et al., 2007; Panier and Durocher, 2009; Bekker-Jensen and Mailand, 2010), triggered by the RNF8/RNF168 ubiquitin ligases, which catalyze nonproteolytic ubiquitylation of H2A-type histones and possibly other targets at DSB-modified chromatin to generate permissive conditions for recruitment of DNA repair factors such as 53BP1 and BRCA1. Through DSB-induced interaction with MDC1, RNF8 promotes initial Ubc13-dependent histone ubiquitylation at DSB sites (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007). The giant ubiquitin ligase HERC2 functions as an auxiliary factor for RNF8 in this process, enabling RNF8 to preferentially interact with Ubc13 among its cognate E2 enzymes (Bekker-Jensen et al., 2010). The local RNF8-dependent increase in histone polyubiquitylation facilitates recruitment of RNF168 via ubiquitin-binding MIU (motif interacting with ubiquitin) domains, allowing it to amplify K63-linked histone polyubiquitylation to levels sufficient of sustaining the retention of DSB repair factors (Doil et al., 2009; Stewart et al., 2009). Underscoring the physiological importance of this pathway, mutation of RNF168 underlies the RIDDLE syndrome, characterized by mental retardation, microcephaly, and other neurological defects (Stewart et al., 2007; Devgan et al., 2011).
Recent work uncovered a role for SUMO in promoting DSB-associated histone ubiquitylation, and SUMO1/2/3 conjugates accumulate at DSB sites (Galanty et al., 2009; Morris et al., 2009). The SUMO E3 ligases PIAS1 and PIAS4 were shown to be required for DSB-associated SUMOylation and recruitment of BRCA1 and 53BP1 to damaged chromatin (Galanty et al., 2009; Morris et al., 2009). Interestingly, these studies also suggested a more upstream role of SUMO in this response, as PIAS4- and SUMO1-dependent modification of as-yet elusive proteins were required for robust ubiquitylation of DSB-flanking chromatin and, consequently, for recruitment of DNA repair factors (Galanty et al., 2009; Morris et al., 2009). Specifically, PIAS4-depleted cells fail to recruit RNF168 but not RNF8 to sites of DNA damage, suggesting the involvement of PIAS4-mediated SUMOylation events in the RNF8-dependent process that promotes RNF168 accrual. Here, we provide evidence that both HERC2 and RNF168 are modified by DSB-inducible, PIAS4-dependent SUMOylation. SUMOylation of HERC2 is necessary for its stable interaction with RNF8, involving a novel SUMO-binding Zinc finger motif in HERC2. Our findings help to explain the requirement of PIAS4-mediated SUMOylation for efficient ubiquitylation of DSB-modified chromatin.
Results and discussion
SUMOylation of HERC2 and RNF168 in response to DSBs
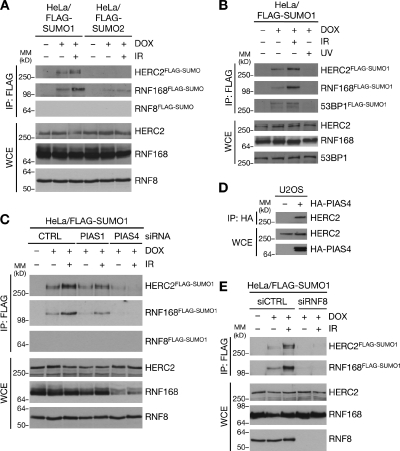
To investigate whether the DSB-responsive ubiquitin ligases RNF8, RNF168, and HERC2 are regulated by SUMOylation, we generated stable cell lines inducibly expressing FLAG-tagged SUMO1 or SUMO2 (Fig. S1 A) and monitored the presence of these E3 ligases in FLAG-SUMO immunoprecipitates (IPs). We noted that HERC2 and RNF168 could be modified with SUMO1 and, to a much lesser extent, SUMO2 (Fig. 1 A). SUMO1 modification of both HERC2 and RNF168 were markedly enhanced upon exposure of cells to DSB-inducing agents such as ionizing radiation (IR) but not other types of genotoxins (Fig. 1 [A and B] and not depicted), suggesting a regulatory function of such SUMOylation in cellular DSB responses. The HERC2 and RNF168 species isolated by FLAG IP from these cells showed retarded migration in SDS-PAGE (Fig. S1 B), and they were absent from cells that expressed a nonconjugatable FLAG-SUMO1 ΔGG allele or were depleted for the SUMO E2 enzyme Ubc9 (Fig. S1, C and D), demonstrating that HERC2 and RNF168 are direct targets of modification by SUMO1. On the other hand, we consistently failed to detect SUMOylation of endogenous RNF8 (Fig. 1 [A and C] and not depicted). These findings suggest that HERC2 and RNF168, but not RNF8, are novel SUMO-regulated E3 ubiquitin ligases and thus potentially important substrates in the SUMO-dependent pathway that orchestrates the DSB response (Galanty et al., 2009; Morris et al., 2009).
Figure 1.
PIAS4- and RNF8-dependent SUMOylation of HERC2 and RNF168 in response to DSBs. (A) HeLa/FLAG-SUMO cell lines left untreated or induced to express SUMO1 or SUMO2 by addition of doxycycline (DOX) for 24 h were subjected to IR or not and harvested 1 h later. Cells were lysed under denaturing conditions, and protein SUMOylation was analyzed by immunoblotting of FLAG IPs with the indicated antibodies. MM, molecular mass; WCE, whole-cell extract. (B) HeLa/FLAG-SUMO1 cells exposed to IR or UV were processed as in A. (C) HeLa/FLAG-SUMO1 cells transfected with control (CTRL), PIAS1, or PIAS4 siRNAs 24 h before addition of doxycycline were processed as in A. Knockdown efficiency of PIAS1 and PIAS4 siRNAs is shown in Fig. S1 E. (D) U2OS cells transfected or not with HA-tagged PIAS4 plasmid for 24 h were exposed to IR and harvested 1 h later. Cell extracts were subjected to HA IP followed by immunoblotting. (E) HeLa/FLAG-SUMO1 cells transfected with control or RNF8 siRNAs for 24 h were processed as in A.
PIAS4-dependent SUMOylation of HERC2 and RNF168 at DSB sites
Because HERC2 and RNF168 were preferentially modified with SUMO1, we reasoned that SUMOylation of these factors might underlie the requirement of PIAS4/SUMO1 for ubiquitin-dependent recruitment of DSB repair factors (Galanty et al., 2009; Morris et al., 2009). To test this, we asked whether SUMOylation of HERC2 and RNF168 required PIAS4 function. Indeed, the IR-induced SUMOylation of HERC2 and RNF168 were strongly suppressed in cells depleted for PIAS4 but not PIAS1, which regulates more downstream aspects of the DSB response (Figs. 1 C and S1 E; Galanty et al., 2009; Morris et al., 2009). Consistently, we found that HERC2 interacted with PIAS4 but not PIAS1 (Fig. 1 D and not depicted), suggesting that HERC2 is a direct target of PIAS4-mediated SUMOylation. We observed similar negative impact on HERC2 and RNF168 SUMOylation in RNF8-depleted cells (Fig. 1 E), which are defective for HERC2 and RNF168 retention at DSB-modified chromatin (Doil et al., 2009; Stewart et al., 2009; Bekker-Jensen et al., 2010), and RNF168 SUMOylation was also diminished upon HERC2 knockdown, which impairs RNF168 accumulation at DSB sites (Fig. S1 F; Bekker-Jensen et al., 2010). This suggests that their SUMOylation occurs predominantly at DSB sites, to which PIAS4 is recruited independently of the RNF8–RNF168 pathway (Galanty et al., 2009). Moreover, HERC2 SUMOylation was largely abolished in cells treated with PI3K-like kinase inhibitors, including caffeine (Fig. S1 G), which interfere with productive DSB-dependent signaling (Lavin, 2008). Collectively, these findings suggest that PIAS4 and HERC2/RNF168 are brought together at DSB sites to trigger SUMOylation of the latter proteins.
PIAS4 promotes DSB-induced association between RNF8 and HERC2
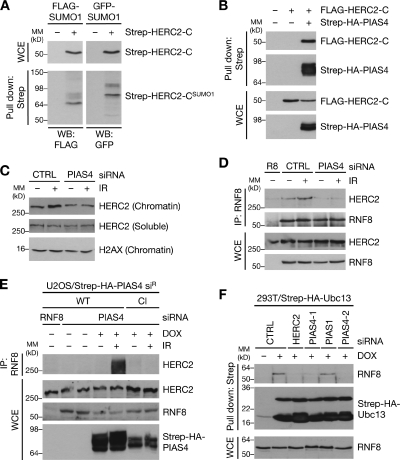
Having shown that HERC2 and RNF168 undergo DSB-inducible modification by SUMO1, we set out to delineate the regulatory significance of these events. We focused on dissecting the functional role of HERC2 SUMOylation. PIAS4 primarily SUMOylated and interacted with the HECT (homologous to E6AP C terminus) domain–containing C terminus of HERC2 (Figs. 2 [A and B] and S2 A). Despite the presence of many lysine residues in this region, none of these match known SUMO consensus motifs (Matic et al., 2010), and extensive mutagenesis of individual lysines in the HERC2 C terminus did not conclusively identify exact SUMOylation sites.
Figure 2.
SUMOylation of HERC2 promotes its interaction with RNF8. (A) SUMOylation of the C-terminal region of HERC2. U2OS cells were cotransfected with combinations of SUMO1- and HERC2-C (amino acids 4421–4834) expression constructs for 24 h, lysed, and processed for Strep-Tactin pulldowns. Bound complexes were analyzed by immunoblotting. Migration of molecular mass (MM) markers are indicated. WB, Western blot; WCE, whole-cell extract. (B) U2OS cells transfected with the indicated constructs were processed as in A. (C) U2OS cells transfected with siRNAs for 48 h were exposed to IR 1 h before lysis, separated into soluble and chromatin-enriched fractions, and immunoblotted with HERC2 and H2AX antibodies. CTRL, control. (D) U2OS cells transfected with indicated siRNAs for 48 h were exposed to IR and harvested 1 h later. Association between RNF8 and HERC2 was analyzed by immunoblotting of RNF8 IPs with RNF8 and HERC2 antibodies. R8, RNF8. (E) U2OS/Strep-HA-PIAS4 siR cell lines were transfected with siRNAs and induced or not with doxycycline (DOX) for 48 h, treated with IR, and harvested 1 h later. RNF8 IPs were analyzed by immunoblotting. (F) HEK293T/Strep-HA-Ubc13 cells transfected with the indicated siRNAs for 48 h were treated or not with doxycycline for an additional 24 h to induce expression of ectopic Strep-HA-Ubc13. Strep-Tactin pulldowns were immunoblotted with the indicated antibodies.
HERC2 promotes ubiquitin-dependent events at sites of DNA damage by facilitating RNF8–Ubc13 complex formation (Bekker-Jensen et al., 2010). Because ubiquitin-mediated DSB signaling downstream of RNF8 is defective in PIAS4-depleted cells (Galanty et al., 2009; Morris et al., 2009), we asked whether the RNF8-dependent accumulation of HERC2 at DSB-modified chromatin also requires functional PIAS4. As expected, siRNA-mediated depletion of PIAS4 suppressed IR-induced recruitment of RNF168 but not RNF8 to DSB sites (unpublished data). PIAS4 knockdown also prevented increased chromatin retention of HERC2 in response to IR (Fig. 2 C), suggesting that depletion of PIAS4 uncouples the DSB-responsive ubiquitin ligase cascade at the level of HERC2 accrual. Because HERC2 is attracted to DSB sites through direct interaction with RNF8 (Bekker-Jensen et al., 2010), we reasoned that PIAS4-mediated SUMOylation of HERC2 might be required for its DSB-induced interaction with RNF8. Indeed, the association between RNF8 and HERC2 was virtually abolished in cells treated with PIAS4 siRNAs (Fig. 2 D), and the DSB-induced RNF8–HERC2 interaction was reestablished by expression of wild-type (WT), but not catalytically inactive, PIAS4 in cells depleted of endogenous PIAS4 (Fig. 2 E). To further corroborate these findings, we tested whether PIAS4 was required to promote HERC2-dependent RNF8–Ubc13 complex formation. Similar to our previous findings for HERC2 (Bekker-Jensen et al., 2010), knockdown of PIAS4, but not PIAS1, suppressed the binding between RNF8 and Ubc13 (Fig. 2 F). These data argue for a regulatory role of HERC2 SUMOylation in promoting binding to RNF8, in turn facilitating RNF8–Ubc13 complex formation and RNF8-dependent protein recruitment to DSB repair foci.
HERC2 contains a novel SUMO-binding ZZ-type Zinc finger domain
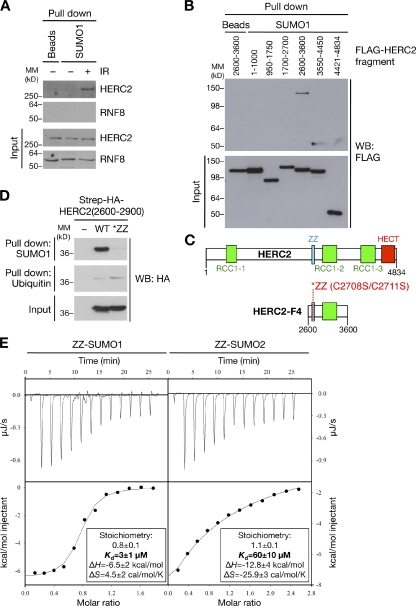
We sought to address the molecular mechanism underpinning the requirement of HERC2 SUMOylation for its interaction with RNF8. We previously showed that RNF8–HERC2 association involves direct recognition of phosphorylated T4827 on HERC2 by the forkhead-associated domain of RNF8 (Bekker-Jensen et al., 2010), and, hence, we tested whether the loss of RNF8–HERC2 interaction in PIAS4-depleted cells was a consequence of impaired HERC2 T4827 phosphorylation. We found that T4827 phosphorylation was unaffected by PIAS4 knockdown (Fig. S2 B), raising the possibility that RNF8 may harbor a domain that specifically recognizes SUMO-modified HERC2. To test this, we assessed the ability of agarose-immobilized SUMO to pull down RNF8 and HERC2. To our surprise, we found that HERC2 but not RNF8 is capable of binding to SUMO1 (Fig. 3 A). The SUMO-binding activity of endogenous HERC2 was strongly enhanced in cells exposed to IR, suggesting that DSB-induced phosphorylation of HERC2 may help to unmask its SUMO-binding region. To test the potential relevance of this SUMO-binding activity in HERC2 for its interaction with RNF8, we mapped the region responsible for SUMO binding. Using overlapping fragments spanning the entire HERC2 protein (Bekker-Jensen et al., 2010), we noted that the SUMO-binding activity localized specifically to amino acids 2600–3600 (HERC2-F4; Fig. 3, B and C). Inspection of this region did not reveal the presence of any obvious canonical SUMO-interacting motif (Hecker et al., 2006). Instead, HERC2-F4 harbors a Zinc finger motif of the ZZ type (Fig. 3 C), the biological function of which is unknown (Legge et al., 2004) and which is only found in HERC2 among the six mammalian HERC family members (Garcia-Gonzalo and Rosa, 2005). Strikingly, point mutations disrupting the Zinc-coordinating ability of the ZZ motif completely abrogated the SUMO-binding ability of HERC2 fragments containing this domain (Figs. 3 [C and D] and S3 A), suggesting that it confers the SUMO-binding ability of HERC2. Whereas an HERC2 fragment containing a WT but not mutated ZZ domain bound to both SUMO1 and SUMO2, it did not detectably interact with ubiquitin (Figs. 3 D and S3 A). To test whether the ZZ domain binds directly to SUMO, we performed isothermal titration calorimetry (ITC) experiments using purified and refolded recombinant HERC2 ZZ domain (Fig. S3, B and C). Indeed, the WT but not mutated ZZ domain showed high-affinity 1:1 binding to SUMO1, with a Kd of binding of ∼3 µM (Figs. 3 E and S3 D). The ZZ domain also bound SUMO2, albeit with a 20-fold lower affinity (Kd = ∼60 µM; Fig. 3 E). Importantly, consistent with pull-down experiments (Fig. 3 D), the HERC2 ZZ domain did not show detectable binding to ubiquitin under similar ITC conditions (Fig. S3 E), further underscoring the specificity of this motif for SUMO binding.
Figure 3.
The ZZ Zinc finger motif in HERC2 is a novel SUMO-binding domain. (A) Cell extracts from U2OS cells harvested 1 h after treatment or not with IR were incubated with SUMO1-immobilized agarose or unconjugated agarose beads. Bound complexes were immunoblotted for HERC2 and RNF8. MM, molecular mass. (B) U2OS cells were transfected with plasmids encoding FLAG-tagged HERC2 fragments for 24 h, and SUMO binding was analyzed as in A. WB, Western blot. (C) Schematic depiction of conserved motifs in the SUMO-binding HERC2-F4 fragment. Residues mutated to disrupt the Zinc-coordinating ability of the ZZ finger are indicated. (D) Extracts of HEK293T cells transfected with the indicated Strep-HA-HERC2(2600–2900) constructs or empty vector for 24 h were incubated with SUMO1 or ubiquitin agarose. Bound complexes and inputs were analyzed by anti-HA immunoblotting. (E) ITC showing direct binding between the HERC2 ZZ domain and SUMO1/SUMO2. Purified SUMO1 or SUMO2 was titrated into the ITC sample cell at 10°C containing folded, recombinant HERC2 ZZ domain (see Fig. S3, B and C) until saturation was achieved. Heat effects (top) and cumulative heat effects (bottom) of the SUMO1/2-ZZ domain interaction are shown. Solid lines represent the single binding site model fit to the experimental data. ITC thermodynamic values are indicated.
Collectively, these data demonstrate that the ZZ Zinc finger motif in HERC2 is a novel bona fide SUMO-binding domain. The presence of this domain in some 20 other human proteins (Legge et al., 2004) suggests that it may also mediate SUMO-dependent protein interactions in other cellular processes. Several ubiquitin-binding Zinc finger motifs are known (Dikic et al., 2009). Our discovery of a SUMO-binding Zinc finger warrants the possible existence of additional SUMO-specific binding modules in cellular proteins.
The HERC2 ZZ domain promotes SUMO-dependent binding to RNF8
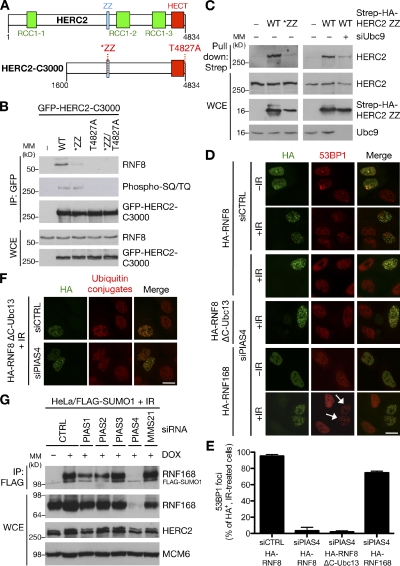
The presence of a SUMO-binding Zinc finger module in HERC2 prompted us to ask whether this motif might be functionally implicated in SUMOylation-dependent binding of HERC2 to RNF8. To this end, we used extended HERC2 fragments, which, unlike ectopic full-length HERC2, are well tolerated in cells (unpublished data) and in which mutations in both the ZZ motif and the T4827 phosphorylation site could be introduced (Fig. 4 A). As expected, T4827A mutation of this construct completely abolished binding to RNF8 (Fig. 4 B), confirming that even in the context of long HERC2 fragments, this phosphorylated residue provides the key RNF8-binding site. Notably, mutation of the SUMO-binding ZZ motif also severely compromised RNF8 binding but not other aspects of HERC2 function despite the fact that T4827 phosphorylation was not affected (Figs. 4 B and S3 [F–H]), suggesting that the SUMO-binding ability of HERC2 is necessary for efficient interaction with RNF8. The simultaneous requirement of HERC2 phosphorylation, SUMOylation, and SUMO-binding activity for its productive association with RNF8 could be explained by a conformational change in HERC2 induced by intramolecular interaction between the ZZ domain and the SUMO1-modified residues, which might facilitate more robust interaction with RNF8 in response to DSBs. The notion that HERC2 is selectively modified by SUMO1, to which the ZZ domain displays a clear binding preference (Fig. 3 E), is in good agreement with this idea. A similar mode of SUMO1-dependent regulation was described for the DNA mismatch repair enzyme thymine DNA glycosylase, in which an extensive conformational change mediated by intramolecular recognition of SUMO1-modified thymine DNA glycosylase by a SUMO-interacting motif modulates its enzymatic properties (Mohan et al., 2007; Smet-Nocca et al., 2011). In support of this hypothesis, WT but not mutated HERC2 ZZ domain could bind to endogenous HERC2 (Fig. 4 C), and this interaction was diminished in Ubc9-depleted cells, in which HERC2 SUMOylation is reduced (Figs. 4 C and S1 D), indicating that the ZZ domain bound specifically to SUMOylated HERC2. Although these observations are consistent with a SUMO-dependent intramolecular interaction within HERC2, we cannot rule out the existence of alternative mechanisms that could promote SUMO-dependent association between RNF8 and HERC2, but addressing this experimentally is severely hampered by the vast size of the 530-kD HERC2 protein. Regardless of its precise mechanistic function, the HERC2 ZZ domain is likely to promote important SUMO-mediated protein interactions within DSB repair foci.
Figure 4.
Roles of PIAS4-mediated SUMOylation of HERC2 and RNF168 in cellular DSB responses. (A) Schematic depiction of HERC2 C-terminal fragments (HERC2-C3000) used in B. (B) HEK293T cells transfected with the indicated versions of GFP-HERC2-C3000 for 24 h were exposed to IR and lysed 1 h later. GFP IPs were immunoblotted with the indicated antibodies. MM, molecular mass; WCE, whole-cell extract. (C) U2OS cells were left untreated or transfected with Ubc9-1 siRNA for 48 h, transfected with HERC2 ZZ (residues 2690–2770) constructs for an additional 24 h, and processed as in Fig. 2 A. (D) U2OS cells transfected with siRNAs for 48 h were transfected with the indicated HA-tagged constructs for an additional 24 h, exposed to 4 Gy IR, and fixed 1 h later. Cells were immunostained with HA and 53BP1 antibodies. Arrows indicate cells proficient for 53BP1 accumulation in IR-induced foci. siCTRL, control siRNA. Bar, 10 µm. (E) Quantification of data in D. At least 200 cells were counted for each treatment. Results depict the mean (±SD) of three independent experiments. (F) Cells were treated as in D and coimmunostained with HA and FK2 antibodies. Bar, 10 µm. (G) HeLa/FLAG-SUMO1 cells were transfected with the indicated siRNAs for 72 h and processed as in Fig. 1 C. DOX, doxycycline.
SUMOylation of both HERC2 and RNF168 promotes ubiquitin-mediated protein assembly at DSB sites
Finally, we asked whether the SUMOylation-dependent binding of HERC2 to RNF8 was sufficient to explain the requirement of PIAS4-mediated SUMOylation for execution of the ubiquitin-dependent DSB signaling response (Galanty et al., 2009; Morris et al., 2009). Because a key function of HERC2 SUMOylation appears to be to promote HERC2–RNF8 interaction and hence RNF8–Ubc13 complex formation (Fig. 2), we reasoned that a chimeric RNF8–Ubc13 protein, which mimics constitutive association between these proteins and overrides the requirement for HERC2 in the DSB response (Bekker-Jensen et al., 2010), might also complement loss of PIAS4 function. However, overexpression of RNF8–Ubc13 or RNF8 alone failed to rescue accumulation of ubiquitin conjugates and 53BP1 in IR-induced foci in PIAS4-depleted cells (Fig. 4, D–F). This suggests that PIAS4-mediated RNF8–HERC2 complex formation is not sufficient to trigger ubiquitin-dependent assembly of repair factors at DSB-flanking chromatin, indicating a requirement for at least one additional PIAS4-mediated SUMOylation event in this response. We speculated that SUMOylation of RNF168 could underlie this requirement. Indeed, unlike RNF8 or RNF8–Ubc13, overexpression of RNF168 efficiently restored 53BP1 foci formation in PIAS4-depleted cells (Fig. 4, D and E), suggesting that a key end point of PIAS4-dependent SUMOylation in the ubiquitin-dependent DSB response is to support generation of functional RNF168 at DSB sites, whereas 53BP1 SUMOylation by PIAS4 is dispensable for its accumulation in DSB repair foci. Because elevated levels of RNF168 effectively compensated for its lack of SUMOylation by PIAS4, we speculated that RNF168 SUMOylation could be involved in maintaining proper RNF168 expression levels in cells. Indeed, we observed a striking loss of RNF168 protein in cells depleted for PIAS4, but not a range of other SUMO E3 ligases, which do not strongly impair RNF168 SUMOylation (Figs. 4 G and S2 C). We previously reported similar effects in cells depleted for HERC2 (Bekker-Jensen et al., 2010), which is also needed for efficient RNF168 SUMOylation. PIAS4 depletion shortened the half-life of RNF168 protein and also significantly reduced RNF168 mRNA levels (Fig. S2, D and E). Thus, PIAS4 may directly support expression of RNF168 by SUMOylating it and indirectly via regulating RNF168 transcription.
In sum, our findings provide novel insight into the mechanistic aspects of how ubiquitin- and SUMO-dependent signaling mechanisms cooperate to promote retention of genome caretakers at DSB-flanking chromatin and reveal considerable complexity in the mechanisms governing DSB-inducible RNF8–HERC2 interaction, involving both phosphorylation and SUMOylation, and the SUMO-binding capability of HERC2.
Materials and methods
Plasmids and RNAi
To construct FLAG-SUMO1/2 plasmids, SUMO cDNAs including an N-terminal His tag were amplified by PCR and cloned into the BamHI–NotI sites of pcDNA5/FRT/TO-3xFLAG (Invitrogen). The GFP-SUMO1 plasmid was a gift from H. Yu (University of Texas Southwestern Medical Center, Dallas, TX). PIAS4 expression vector was generated by inserting PCR-amplified, full-length human PIAS4 cDNA into pcDNA4/TO (Invitrogen) containing an N-terminal Strep-HA tag. To generate siRNA-insensitive Strep-HA-PIAS4 (siR) constructs, the italicized silent mutations (GGAGTAAGAGCGGCCTGAA) were introduced into the PIAS4-1 siRNA target sequence in the PIAS4 coding region. Plasmids encoding HA-RNF8, HA-RNF168, HA-RNF8ΔC-Ubc13, and overlapping FLAG-tagged HERC2 fragments were previously described (Mailand et al., 2007; Doil et al., 2009; Bekker-Jensen et al., 2010). In brief, RNF8 and RNF168 were inserted into pcDNA3 (Invitrogen) containing an N-terminal HA tag, and an RNF8 fragment spanning amino acids 1–402 was inserted between the HA tag and Ubc13 in pEF-HA-Ubc13. Individual HERC2 fragments were cloned into pFLAG-CMV2 (Sigma-Aldrich). Plasmids encoding GFP-tagged HERC2-C3000 fragments (comprising amino acids 1600–4834 in human HERC2) were constructed in two successive cloning steps: first, a fragment corresponding to amino acids 3593–4834 of human HERC2 generated by overlapping PCR of the HERC2-F5 and -F6 constructs was inserted into the XhoI–PmeI sites of pEGFP-C1 (Takara Bio Inc.) to generate pEGFP-C1-HERC2(F5+F6). Subsequently, a fragment comprising amino acids 1600–3607 was amplified by overlapping PCR of the HERC2-F2, -F3, and -F4 fragments and subcloned into XhoI–NotI-digested pEGFP-C1-HERC2(F5+F6). For expression of the HERC2 ZZ domain, we inserted fragments comprising amino acids 2600–2900 or 2690–2770 (denoted ZZ) in HERC2 into pcDNA4/TO (Invitrogen) containing an N-terminal Strep-HA tag. For bacterial expression of HERC2 ZZ domain, the sequence encoding the core HERC2 ZZ domain (comprising amino acids 2702–2755) was amplified by PCR and inserted into pNIC28-Bsa4 by ligation-independent cloning. The ZZ (C2708S/C2711S) and TA (T4827A) point mutations in HERC2 constructs as well as the C342A mutation to generate catalytically inactive PIAS4 were introduced using the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies). All constructs were verified by sequencing. Plasmid transfections were performed using FuGene 6 (Roche) according to the manufacturer’s instructions. siRNA transfections were performed with Lipofectamine RNAiMAX (Invitrogen), as previously described (Mailand et al., 2007). siRNA target sequences used in this study were as follows: control (5′-GGGAUACCUAGACGUUCUA-3′), RNF8 (5′-UGCGGAGUAUGAAUAUGAA-3′), HERC2 (5′-GGAUGAUCAUGAAGAGUUA-3′), and Ubc9-1 (5′-UCGAACCACCAUUAUUUCACCCGAA-3′). siRNAs to SUMO E3 ligases were described in Galanty et al. (2009) and included the following: PIAS1 (5′-CGAAUGAACUUGGCAGAAA-3′), PIAS2 (5′-CUUGAAUAUUACAUCUUUA-3′), PIAS3 (5′-CCCUGAUGUCACCAUGAAA-3′), PIAS4-1 (5′-GGAGUAAGAGUGGACUGAA-3′), PIAS4-2 (5′-AGGCACUGGUCAAGGAGAA-3′), and MMS21 (5′-CUCUGGUAUGGACACACAGCU-3′).
Cell culture
Human U2OS osteosarcoma cells and HEK293T embryonic kidney cells were cultured in DME containing 10% FBS. To generate stable HeLa/FLAG-SUMO1/2 cell lines, HeLa/FRT/TRex cells (Invitrogen) were cotransfected with pcDNA5/FRT/TO-3xFLAG-SUMO1/2 and pOG44 and selected with hygromycin B according to the manufacturer’s protocol. To generate stable cell lines inducibly expressing Strep-HA-PIAS4 siR WT or catalytically inactive, U2OS cells were cotransfected with pcDNA6/TR (Invitrogen) and pcDNA4/TO-Strep-HA-PIAS4 siR constructs, and positive clones were selected by incubation in medium containing 400 µg/ml Zeocin and 5 µg/ml Blasticidin S (both from Invitrogen) for 14 d. The HEK293T/Strep-HA-Ubc13 cell line was generated by positive selection of HEK293T cells cotransfected with pcDNA6/TR and pcDNA4/TO-Strep-HA-Ubc13, as previously described (Bekker-Jensen et al., 2010). To induce DNA damage, cells were exposed to doses of 10 Gy IR and 25 J/m2 UV, unless stated otherwise.
Immunochemical methods
Immunoblotting, immunoprecipitation, Strep-Tactin pulldowns, and chromatin fractionation were performed as previously described (Mailand et al., 2006, 2007; Doil et al., 2009; Bekker-Jensen et al., 2010). In brief, cells were lysed in EBC buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 0.5% NP-40) supplemented with protease and phosphatase inhibitor cocktails (Sigma-Aldrich) and incubated on ice for 10 min, and lysates were cleared by centrifugation for 10 min at 20,000 rpm. Lysates were incubated with antibodies coupled to protein A/G Sepharose, FLAG agarose (Sigma-Aldrich), or Strep-Tactin Sepharose (IBA BioTAGnology) for 1.5 h on an end-over-end rotator at 4°C, washed five times with EBC buffer, and resuspended in 2× Laemmli sample buffer. To obtain chromatin-enriched fractions, cells were lysed in low-salt buffer (10 mM Hepes, pH 7.5, 10 mM KCl, and 0.05% NP-40). After centrifugation at 3,600 g, chromatin-associated proteins were released from the pellet by treatment with micrococcal nuclease. To detect SUMOylation of cellular proteins, cells were lysed in denaturing buffer (20 mM Tris, pH 7.5, 50 mM NaCl, 0.5% NP-40, 0.5% sodium deoxycholate, 0.5% SDS, and 1 mM EDTA) containing protease and phosphatase inhibitor cocktails (Sigma-Aldrich) and subjected to immunoprecipitation with FLAG agarose (Sigma-Aldrich) or GFP-TRAP (ChromoTek). Antibodies used in this study included the following: rabbit polyclonals to HERC2 (Bekker-Jensen et al., 2010), RNF8 (Mailand et al., 2007), HA and 53BP1 (Santa Cruz Biotechnology, Inc.), PIAS4, H2AX, γ-H2AX and phospho-(Ser/Thr) ATM/ATR substrate (Cell Signaling Technology), mouse monoclonals to conjugated ubiquitin (FK2; Millipore), FLAG (Sigma-Aldrich), Strep (IBA BioTAGnology), GFP (Roche), and goat polyclonals to Ubc9 (Abcam) and MCM6 (Santa Cruz Biotechnology, Inc.). Rabbit polyclonal antibody to RNF168 (Stewart et al., 2009) was a gift from D. Durocher (Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, Ontario, Canada).
Immunofluorescence staining and microscopy
Cells were fixed in 3% formaldehyde, permeabilized with PBS containing 0.2% Triton X-100 for 5 min, and incubated with primary antibodies diluted in DME for 1 h at room temperature. After staining with secondary antibodies (Alexa Fluor 488 and 568; Life Technologies) for 30 min, coverslips were mounted in VECTASHIELD mounting medium (Vector Laboratories) containing nuclear stain DAPI. Images were acquired with a confocal microscope (LSM 710; Carl Zeiss) mounted on an Axiovert 100M (Carl Zeiss) equipped with a Plan-Apochromat 40×/1.3 oil immersion objective using the following standard settings: DAPI and Alexa Fluor 488/GFP and 568 dyes were excited with 405-, 488-, and 546-nm laser lines, and emitted light was collected through 420–480-, 505–530, and 560–615-nm band-pass filters, respectively. Pinhole size was set to 1 Airy Unit or opened slightly for all wavelengths acquired if signal intensity was otherwise too low. Image acquisition and analysis were performed with Zen 2010 software. Raw images were exported as TIF files, and, if adjustments in image contrast and brightness were applied, identical settings were used on all images of a given experiment.
Purification and refolding of recombinant HERC2 ZZ domain
For bacterial expression of HERC2 ZZ domain, Escherichia coli BL21 (DE3) transformed with pNIC28-Bsa4-HERC2 ZZ were grown in Terrific Broth supplemented with 8 mg/ml glycerol and 50 µg/ml kanamycin, induced with 0.5 mM IPTG, and cultured at 18°C for 20 h. Cell pellets were disrupted in lysis buffer (300 mM NaCl, 50 mM NaP, pH 7.5, 10 mM imidazole, 10% glycerol, and 0.5 mM Tris(2-carboxyethyl)phosphine) supplemented with Complete Mini EDTA-free protease inhibitor (Roche), 1 mg/ml lysozyme, 0.1% n-dodecyl β-d-maltoside, and 50 U/ml Benzonase and lysed by passage through a homogenizer. After centrifugation, the protein was filtered through a 0.22-µm polyethersulfone bottle top filter and purified on the ÄKTAxpress system (GE Healthcare) at 4°C with the HiTrap Chelating columns (Gileadi et al., 2008). Subsequently, the ZZ domain was purified on 75-pg gel filtration columns (HiLoad 16/60 Superdex; GE Healthcare) equilibrated with buffer (150 mM NaCl and 50 mM NaP, pH 7.5). Eluted proteins were analyzed and verified by SDS-PAGE and mass spectrometry. The purified ZZ domain was reconstituted with Zn2+, as previously described (Omichinski et al., 1993), and verified by differential scanning calorimetry.
SUMO- and ubiquitin-binding assays
Cells lysed in EBC buffer supplemented with protease and phosphatase inhibitor cocktails were incubated with SUMO1/2 or ubiquitin immobilized on agarose beads (Enzo Life Sciences) for 1 h and washed extensively in EBC buffer, and bound complexes were resolved by SDS-PAGE and subjected to immunoblotting. For ITC assays, recombinant SUMO1/2 or ubiquitin (Boston Biochem) and the HERC2 ZZ domain were dialyzed extensively against 20 mM MES, pH 6.0. Protein concentrations were determined by UV spectroscopy and molar extinction coefficients at 280 nm of 4,470, 2,980, and 1,490 M−1 cm−1 for SUMO1, SUMO2, and the ZZ domain, respectively. The extinction coefficients were calculated as previously described (Gill and von Hippel, 1989). ITC experiments were performed at 10 or 25°C using an iTC200 instrument (MicroCal) by titrating 5-µl volumes of SUMO1/2 or ubiquitin into the ITC sample cell containing the ZZ domain until saturation was achieved. The heat of the reaction was obtained by integrating the peak after each injection of SUMO1/2 or ubiquitin and fitted to a model describing a single binding site (Wiseman et al., 1989), using software provided by the iTC200 manufacturer.
Online supplemental material
Fig. S1 demonstrates that HERC2 and RNF168 are covalently modified by SUMO1 under various experimental settings. Fig. S2 shows SUMOylation and phosphorylation of HERC2 fragments and impact of PIAS4 on RNF168 expression and stability. Fig. S3 shows biophysical characterization of the HERC2 ZZ domain, its SUMO- and ubiquitin-binding properties, and general impact of mutations in the ZZ domain on HERC2 functions. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201106152/DC1.
Acknowledgments
We thank Daniel Durocher and Hongtao Yu for providing reagents and Jiri Lukas and James G. Omichinski for helpful discussions.
This work was supported by grants from the Novo Nordisk Foundation, Danish Medical Research Council, the Danish Cancer Society, and the Lundbeck Foundation.
Footnotes
Abbreviations used in this paper:
- DSB
- double-strand break
- IP
- immunoprecipitate
- IR
- ionizing radiation
- ITC
- isothermal titration calorimetry
- WT
- wild type
References
- Bekker-Jensen S., Mailand N. 2010. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst.). 9:1219–1228 10.1016/j.dnarep.2010.09.010 [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S., Rendtlew Danielsen J., Fugger K., Gromova I., Nerstedt A., Lukas C., Bartek J., Lukas J., Mailand N. 2010. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell Biol. 12:80–86: 1–12 (published erratum appears in Nat. Cell Biol. 2010. 12:412) 10.1038/ncb2008 [DOI] [PubMed] [Google Scholar]
- Ciccia A., Elledge S.J. 2010. The DNA damage response: Making it safe to play with knives. Mol. Cell. 40:179–204 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devgan S.S., Sanal O., Doil C., Nakamura K., Nahas S.A., Pettijohn K., Bartek J., Lukas C., Lukas J., Gatti R.A. 2011. Homozygous deficiency of ubiquitin-ligase ring-finger protein RNF168 mimics the radiosensitivity syndrome of ataxia-telangiectasia. Cell Death Differ. 18:1500–1506 10.1038/cdd.2011.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I., Wakatsuki S., Walters K.J. 2009. Ubiquitin-binding domains - from structures to functions. Nat. Rev. Mol. Cell Biol. 10:659–671 10.1038/nrm2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D.H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 136:435–446 10.1016/j.cell.2008.12.041 [DOI] [PubMed] [Google Scholar]
- Galanty Y., Belotserkovskaya R., Coates J., Polo S., Miller K.M., Jackson S.P. 2009. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 462:935–939 10.1038/nature08657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F.R., Rosa J.L. 2005. The HERC proteins: Functional and evolutionary insights. Cell. Mol. Life Sci. 62:1826–1838 10.1007/s00018-005-5119-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gileadi O., Burgess-Brown N.A., Colebrook S.M., Berridge G., Savitsky P., Smee C.E., Loppnau P., Johansson C., Salah E., Pantic N.H. 2008. High throughput production of recombinant human proteins for crystallography. Methods Mol. Biol. 426:221–246 10.1007/978-1-60327-058-8_14 [DOI] [PubMed] [Google Scholar]
- Gill S.C., von Hippel P.H. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319–326 10.1016/0003-2697(89)90602-7 [DOI] [PubMed] [Google Scholar]
- Hecker C.M., Rabiller M., Haglund K., Bayer P., Dikic I. 2006. Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281:16117–16127 10.1074/jbc.M512757200 [DOI] [PubMed] [Google Scholar]
- Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 131:901–914 10.1016/j.cell.2007.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P., Bartek J. 2009. The DNA-damage response in human biology and disease. Nature. 461:1071–1078 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Chen J., Yu X. 2007. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 316:1202–1205 10.1126/science.1139621 [DOI] [PubMed] [Google Scholar]
- Kolas N.K., Chapman J.R., Nakada S., Ylanko J., Chahwan R., Sweeney F.D., Panier S., Mendez M., Wildenhain J., Thomson T.M., et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 318:1637–1640 10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M.F. 2008. Ataxia-telangiectasia: From a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell Biol. 9:759–769 10.1038/nrm2514 [DOI] [PubMed] [Google Scholar]
- Legge G.B., Martinez-Yamout M.A., Hambly D.M., Trinh T., Lee B.M., Dyson H.J., Wright P.E. 2004. ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module. J. Mol. Biol. 343:1081–1093 10.1016/j.jmb.2004.08.087 [DOI] [PubMed] [Google Scholar]
- Lindahl T., Barnes D.E. 2000. Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol. 65:127–133 10.1101/sqb.2000.65.127 [DOI] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Bartek J., Lukas J. 2006. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell. 23:307–318 10.1016/j.molcel.2006.06.016 [DOI] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 131:887–900 10.1016/j.cell.2007.09.040 [DOI] [PubMed] [Google Scholar]
- Matic I., Schimmel J., Hendriks I.A., van Santen M.A., van de Rijke F., van Dam H., Gnad F., Mann M., Vertegaal A.C. 2010. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell. 39:641–652 10.1016/j.molcel.2010.07.026 [DOI] [PubMed] [Google Scholar]
- Misteli T., Soutoglou E. 2009. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat. Rev. Mol. Cell Biol. 10:243–254 10.1038/nrm2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan R.D., Rao A., Gagliardi J., Tini M. 2007. SUMO-1-dependent allosteric regulation of thymine DNA glycosylase alters subnuclear localization and CBP/p300 recruitment. Mol. Cell. Biol. 27:229–243 10.1128/MCB.00323-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J.R., Solomon E. 2004. BRCA1 : BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum. Mol. Genet. 13:807–817 10.1093/hmg/ddh095 [DOI] [PubMed] [Google Scholar]
- Morris J.R., Boutell C., Keppler M., Densham R., Weekes D., Alamshah A., Butler L., Galanty Y., Pangon L., Kiuchi T., et al. 2009. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 462:886–890 10.1038/nature08593 [DOI] [PubMed] [Google Scholar]
- Omichinski J.G., Clore G.M., Schaad O., Felsenfeld G., Trainor C., Appella E., Stahl S.J., Gronenborn A.M. 1993. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 261:438–446 10.1126/science.8332909 [DOI] [PubMed] [Google Scholar]
- Panier S., Durocher D. 2009. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst.). 8:436–443 10.1016/j.dnarep.2009.01.013 [DOI] [PubMed] [Google Scholar]
- Polo S.E., Jackson S.P. 2011. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev. 25:409–433 10.1101/gad.2021311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smet-Nocca C., Wieruszeski J.M., Léger H., Eilebrecht S., Benecke A. 2011. SUMO-1 regulates the conformational dynamics of thymine-DNA Glycosylase regulatory domain and competes with its DNA binding activity. BMC Biochem. 12:4 10.1186/1471-2091-12-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B., Shao G., Lilli D.R., Culhane A.C., Moreau L.A., Xia B., Livingston D.M., Greenberg R.A. 2007. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 316:1198–1202 10.1126/science.1139516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G.S., Stankovic T., Byrd P.J., Wechsler T., Miller E.S., Huissoon A., Drayson M.T., West S.C., Elledge S.J., Taylor A.M. 2007. RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc. Natl. Acad. Sci. USA. 104:16910–16915 10.1073/pnas.0708408104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G.S., Panier S., Townsend K., Al-Hakim A.K., Kolas N.K., Miller E.S., Nakada S., Ylanko J., Olivarius S., Mendez M., et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 136:420–434 10.1016/j.cell.2008.12.042 [DOI] [PubMed] [Google Scholar]
- Wang B., Elledge S.J. 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. USA. 104:20759–20763 10.1073/pnas.0710061104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Matsuoka S., Ballif B.A., Zhang D., Smogorzewska A., Gygi S.P., Elledge S.J. 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 316:1194–1198 10.1126/science.1139476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman T., Williston S., Brandts J.F., Lin L.N. 1989. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179:131–137 10.1016/0003-2697(89)90213-3 [DOI] [PubMed] [Google Scholar]
- Wyman C., Kanaar R. 2006. DNA double-strand break repair: All’s well that ends well. Annu. Rev. Genet. 40:363–383 10.1146/annurev.genet.40.110405.090451 [DOI] [PubMed] [Google Scholar]