Ohsumi's work continues to provide much of the foundation for our current understanding of autophagy.
Ohsumi's work continues to provide much of the foundation for our current understanding of autophagy.
Autophagy is a process by which cellular components are captured into organelles called autophagosomes and then brought to the lysosome or vacuole to be broken down and recycled for other uses. It frequently comes into play during starvation, allowing cells to survive periods of privation.
Yoshinori Ohsumi
PHOTO COURTESY OF YOSHINORI OHSUMI
Yoshinori Ohsumi survived privation while growing up in post–World War II Japan and struggled to establish his own course of independent research. But he persevered and went on to make key early discoveries in the field of autophagy (1, 2). In his work, he has identified most of the proteins and pathways involved in the process (3), demonstrated how they are regulated by proteins that sense cells’ metabolic states (4), and started to outline the fine mechanistic details of autophagosome formation in yeast (5, 6). In spite of all his successes, he was surprised by our request for an interview but nevertheless happy to speak with us about his work from his office at Tokyo Institute of Technology.
“We realized we had almost the entire pathway in our hands.”
EARLY LEAPS
How did you decide to go into a research career?
I was probably influenced by my father, who was a professor of engineering at Kyushu University. I was familiar with academic life while I was growing up. But whereas my father worked in a very industrially oriented field, I was more interested in the natural sciences. In high school I was interested in chemistry, so I entered the University of Tokyo to learn chemistry. I quickly discovered chemistry wasn't so attractive to me, because the field was already quite established. But I was lucky, I think, because the early 1960s was the golden age of molecular biology. I decided I wanted to work on that instead.
There were not very many molecular biology labs in Japan at that time. I joined Dr. Kazutomo Imahori's lab as a graduate student to study protein synthesis in E. coli. Unfortunately, I did not get very good results in my work, and, when I had finished my graduate studies, I discovered it was very difficult to find a good position in Japan. So, on Dr. Imahori's advice, I took a postdoctoral position with Dr. Gerald Edelman at The Rockefeller University in New York.
What did you work on there?
That was the hardest time in my life. [Laughs] As a graduate student I had worked on E. coli, but in Dr. Edelman's lab I switched to working on mammalian cell and developmental biology. I was supposed to establish a system for in vitro fertilization in mice, but I did not know very much about early embryology and I had only a very small number of eggs to work with. I grew very frustrated. Then, one and a half years later, Mike Jazwinski joined Edelman's lab, and I decided to work with him instead on studying DNA duplication in yeast. That was another huge leap for me, but it was also my first introduction to yeast cells, which I have worked with ever since.
Finally, I was offered a position as a junior professor in Yasuhiro Anraku's lab at the University of Tokyo and was able to return to Japan.
FIRST ADVANCE
Is that when you first started working on the yeast vacuole?
At that time, many people were studying the transport of ions and small molecules at the plasma membrane, but few people had started looking at transport across other organelle membranes. The vacuole was thought to be just a garbage can in the cell, and not very many people were interested in its physiology, so I thought it would be good to study transport in the vacuole because I would not have much competition. Another reason I chose to study vacuole physiology is that, while I was in Dr. Edelman's lab, we had tried to isolate nuclei from yeast cells, and along the way we discovered that it was easy to get pure preparations of vacuoles. Using these preparations, I was able to find many active transport systems in the vacuolar membrane, including the vacuolar-type ATPase that pumps protons into the vacuole. So, I had a little bit of success, and eventually I finally got my own laboratory. But by that time I was 43 years old. I would not say I had a very successful career up to that point; I had many difficulties, but I mostly caused them myself. [Laughs]
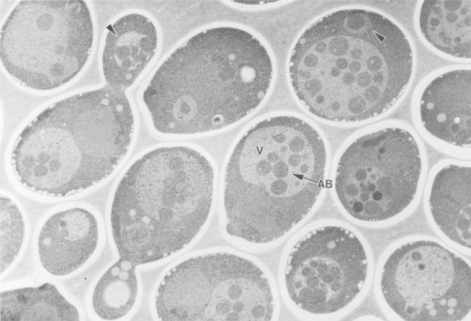
Ohsumi and colleagues found that vesicles accumulate in the vacuole of starving yeast cells (1).
© 1992, THE ROCKEFELLER UNIVERSITY PRESS
You were working on your own when you first observed the autophagic process…
I wanted to work on a different subject from what I had been doing in Dr. Anraku's lab, so I decided to study the lytic function of the vacuole. At that time, nothing was known about what is degraded in the vacuole and how.
I had a very simple idea: the vacuole can be detected under the light microscope, and it was already considered to be a garbage compartment where protein degradation takes place. So, I thought it would be easy to observe morphological change in the vacuoles of cells that were undergoing lots of degradation. Cell differentiation processes require lots of protein degradation, so I looked at vacuolar proteinase-deficient mutants, which cannot sporulate as normal cells do under nitrogen-starvation conditions, to see if I could observe any changes to vacuolar structure.
One thing I want to say is that I like to observe cells under a microscope. The microscope can tell us some quite important information about each cell, especially about the vacuole, which is quite easy to see. So, I was looking at the mutants under the microscope, and I saw that after 30 minutes of starvation many vesicles appeared and accumulated in the vacuole. I also knew a friendly electron microscopist and was lucky to observe autophagosome formation and vacuole fusion. That was the starting point of my work on autophagy in yeast.
NEXT STEPS
You also characterized many of the genes involved in autophagy…
In 1991, one of my first two graduate students—a wonderful student—performed a very laborious screen, observing individual mutants under the microscope. Using this approach, she found the first autophagy-defective mutant, which at the time we called apg1-1 but is now called atg1. I expected that we could use this type of screen to find many more autophagy genes, because autophagy is such a complex phenomenon, and, indeed, we found 14 atg mutants in this way.
I had quite a small lab, only three people, when we started the genetic analysis of ATG genes. I was afraid it would take a very long time to complete our analysis, but the yeast genome sequence was published around that time, so we were able to quickly clone many of the genes. But except for ATG1, which encodes a protein kinase, all the other ATG genes were novel genes; their amino acid sequence did not tell us very much about what they did.
Ohsumi worked alone in his first lab; now he has plenty of company.
PHOTO COURTESY OF YOSHINORI OHSUMI
One breakthrough was made by Noboru Mizushima, a medical doctor who joined my lab as a postdoc. He showed that Atg12 is a ubiquitin-like protein that is conjugated to Atg5. Atg7 is an E1 enzyme, and Atg10 is an E2 enzyme, so we realized we had almost the entire pathway in our hands. We published that result in Nature without any serious objections, and that was the first major success in my lab. [Laughs] There is also another, similar pathway that results in the addition of Atg8, another ubiquitin-like protein, to the phospholipid phosphatidylethanolamine.
“I am not very competitive, so I always look for a new subject to study, even if it is not so popular.”
We now have a better understanding of what the individual Atg proteins do, but we still don't understand how they drive autophagosome formation. We want to understand how new autophagosomal membrane appears, how it elongates, and how it seals to become an autophagosome. We are also working to analyze the structural biology of Atg proteins and to understand how they interact with each other to form complexes. These complexes are very transient, so it is difficult to study them. Those are the basic questions we're working on now.
Do you have advice for scientists who are struggling, as you did early in your career?
Unfortunately, these days, at least in Japan, young scientists want to get a stable job, so they are afraid to take risks. Most people decide to work on the most popular field because they think that is the easiest way to get a paper published. But I am just the opposite of that. I am not very competitive, so I always look for a new subject to study, even if it is not so popular. If you start from some sort of basic, new observation, you will have plenty to work on.
References
- 1.Takeshige K., et al. 1992. J. Cell Biol. 119:301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba M., et al. 1994. J. Cell Biol. 124:903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsukada M., Ohsumi Y.. 1993. FEBS Lett. 333:169–174. [DOI] [PubMed] [Google Scholar]
- 4.Noda T., Ohsumi Y.. 1998. J. Biol. Chem. 273:3963–3966. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N., et al. 1998. Nature. 395:395–398. [DOI] [PubMed] [Google Scholar]
- 6.Nakatogawa H., et al. 2012. Autophagy. 8:177–186. [DOI] [PubMed] [Google Scholar]