Abstract
Ataxia is a neurological disorder characterized by loss of control of body movements. Spinocerebellar ataxia (SCA), previously known as autosomal dominant cerebellar ataxia, is a biologically robust group of close to 30 progressive neurodegenerative diseases. Six SCAs, including the more prevalent SCA1, SCA2, SCA3, and SCA6 along with SCA7 and SCA17 are caused by expansion of a CAG repeat that encodes a polyglutamine tract in the affected protein. How the mutated proteins in these polyglutamine SCAs cause disease is highly debated. Recent work suggests that the mutated protein contributes to pathogenesis within the context of its “normal” cellular function. Thus, understanding the cellular function of these proteins could aid in the development of therapeutics.
Individuals suffering from ataxia are at first clumsy and unable to walk steadily, and have slurred speech. Patients can eventually lose the ability to swallow and breathe in a coordinated fashion, which can be fatal. Ataxia results from variable degeneration of neurons in the cerebellum, brain stem, spinocerebellar tracts, and their afferent/efferent connections. Such neurodegenerations can be due to a variety of clinical conditions, including multiple sclerosis, a brain tumor, alcoholism, or an inherited genetic defect. There are over 50 different forms of inherited ataxias that strike during childhood or adulthood (Taroni and DiDonato, 2004; Manto, 2005). Among the inherited forms of ataxia are several recessive forms of ataxia that include Friedreich ataxia—the most common inherited form of ataxia that affects mitochondrial function, AVED ataxia due to vitamin E deficiency, and ataxia telangiectasia affecting regulation of the cell cycle. Spinocerebellar ataxias, or SCAs, are inherited in an autosomal-dominant pattern. There are ∼30 different types of SCA identified to date but causative mutations have been identified for only half of them. Among these are the ataxias caused by mutations in protein kinase C (SCA14) and fibroblast growth factor 14. Most of the SCAs are caused by an abnormal expansion of a CAG repeat sequence that encodes for an expanded tract of polyglutamine (polyQ) residues within the mutated protein. It is the polyQ SCAs that are the subject of this review.
The cellular and molecular mechanisms responsible for pathogenesis of the polyQ neurodegenerative diseases are a matter of spirited discussions. For the most part these discussions center on two major points. The first focuses on to what extent the polyglutamine tract alone drives pathogenesis. It is clear that expanded polyQ peptides are toxic, with early studies indicating that disease might stem from the proteolytic production of such peptides (Marsh et al., 2000). Yet, only for Huntington’s disease is there evidence that proteolytic cleavage and the generation of a toxic polyQ peptide might be required for pathogenesis (Davies et al., 1997; DiFiglia et al., 1997; Graham et al., 2006). The second point is whether the tendency of mutant polyQ proteins to aggregate is responsible for the disease-associated neurodegeneration. PolyQ expansion renders the protein more prone to aggregation and formation of inclusion bodies that are a pathological hallmark of these disorders including the polyQ SCAs (Orr and Zoghbi, 2007). Yet, the extent to which these inclusions cause disease continues to be a complex issue. There are several studies demonstrating that in mouse models disease severity can be disassociated from presence of inclusions (Klement et al., 1998; Saudou et al., 1998; Cummings et al., 1999), Moreover, there are data indicating that inclusions may be protective, perhaps by sequestration of the mutant protein (Cummings et al., 1999; Arrasate et al., 2004; Bowman et al., 2007).
More recent studies in both patients and model systems support the concept that the native/normal functions of the polyQ proteins are required for development of disease. Most notably, in the polyQ disease spinobulbar muscular atrophy (SBMA), where disease is caused by a polyQ expansion in the androgen receptor (La Spada et al., 1991), both androgen binding and nuclear translocation of mutant androgen receptor are required for development of the SBMA-associated motor neuron disease (Katsuno et al., 2002; Takeyama et al., 2002; Nedelsky et al., 2010). Moreover, disruption of androgen receptor interaction with its normal transcriptional coregulators suppresses disease in a Drosophila model of SBMA (Nedelsky et al., 2010). In the case of the polyQ SCA SCA1, interactions with its native partners are required for disease in a mouse model of SCA1 (Lim et al., 2008). Furthermore, a SCA-like neurological phenotype can be induced in mice that overexpress wild-type ataxin-1 with a single amino acid substitution outside of the polyQ tract that enhances its interaction with a native partner (Duvick et al., 2010).
Glutamine-rich regions are found in the activation domains characteristic of a large family of highly conserved transcription activators exemplified by specificity factor 1 (Sp1). Within the multiprotein TFIID transcription complex, the glutamine rich regions of these activators function as protein–protein interaction domains (Saluja et al., 1998) Thus, not surprisingly, an alteration in neuronal transcription is a prominent model for polyQ pathogenesis. Very recently, a study in Caenorhabditis elegans revealed another function associated with a glutamine-rich protein, control of nonapoptotic cell death. Blum et al. (2012) found that the C. elegans polyQ protein pqn-41 is critical for the onset of linker-cell death during normal reproductive system development. Death of linker cells shares several morphological features seen in neurons undergoing polyQ-induced degeneration, e.g., formation of intracellular aggregates of the polyQ protein. It remains to be determined the extent to which neurodegeneration in diseases such as the polyQ SCAs is related to the nonapoptotic linker cell death regulated by pgn-41.
An emerging concept is that understanding the polyQ SCA diseases will require an appreciation of the role these proteins have in basic cellular processes (Orr, 2001). Thus, this review focuses on those polyQ SCAs (SCA1, SCA2, SCA3, SCA6, SCA7, and SCA17) where the cellular processes involving the affected protein are being uncovered. These functions span cellular processes from those at the cell membrane and in the cytoplasm to functions in the nucleus. Understanding the cell biology of these disorders is providing crucial insight into the pathogenesis of these SCAs. It is also of note that the cerebellum is one of the better understood neuronal networks in the brain. Thus, the SCAs offer a paradigm for moving from human genetics to cell biology to pathophysiology, a point exemplified by recent pathophysiological studies on mouse models showing that ataxia is associated with alterations in Purkinje cell innervation in SCA1 (Barnes et al., 2011; Hourez et al., 2011), and with a decrease in Purkinje cell intrinsic firing rate in SCA3 (Shakkottai et al., 2011).
SCA1: Signal transduction from the cytoplasm to the nucleus
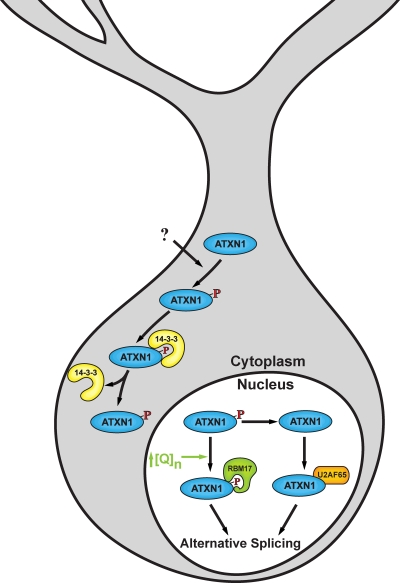
SCA1 is typically a late-onset fatal progressive neurodegenerative disease caused by the expansion of a polyQ tract within the ataxin-1 (ATXN1) protein (Orr et al., 1993). In the brain, cerebellar Purkinje neurons are a prominent pathological target of mutant ATXN1. ATXN1 is located in the cytoplasm and nucleus (Servadio et al., 1995), with the wild-type ATXN1 able to shuttle back and forth between these two subcellular compartments (Fig. 1). The dynamics of ataxin-1 within the nucleus is altered by expansion of the polyQ tract (Krol et al., 2008). Although mutant ATXN1 with an expanded polyQ tract is able to enter the nucleus, its ability to be transported back into the cytoplasm is dramatically reduced (Irwin et al., 2005). Also, the length of the polyQ tract negatively affects SUMOylation of ATXN1, which occurs when ATXN1 is able to be transported to the nucleus (Riley et al., 2005).
Figure 1.
An ATXN1 cellular pathway. ATXN1 is phosphorylated at S776 in the cytoplasm upon which it forms a complex with 14-3-3. Association with 14-3-3 blocks dephosphorylation of pS776 and transport of ATXN1 to the nucleus. Thus, nuclear transport of ATXN1 requires dissociation from 14-3-3, the regulation of which has yet to be determined. In the nucleus, ATXN1 is involved in alternative splicing by virtue of its interaction with RBM17 when phosphorylated on S776, or with U2AF65 after dephosphorylation of pS776. ↑[Q]n indicates that an increased interaction of expanded polyQ ATXN1 with RBM17 is hypothesized to be critical for driving disease in cerebellar Purkinje cells.
Other cellular molecules with which ATXN1 normally interacts include RNA (Yue et al., 2001), and several regulators of transcription, SMRT (Tsai et al., 2004), Capicua (Lam et al., 2006), Gfi-1 (Tsuda et al., 2005), and the Rora–Tip60 complex (Serra et al., 2006). In the case of Capicua and the Rora–Tip60 complex, mutant ATXN1 seems to enhance their degradation. For Capicua, loss of ATXN1 decreases its steady-state level (Lam et al., 2006). ATXN1 also interacts with two RNA-splicing factors, RBM17 (Lim et al., 2008) and U2AF65 (de Chiara et al., 2009).
Two evolutionarily conserved regions within ATXN1 drive these interactions; the 120-amino acid AXH domain (de Chiara et al., 2003; Chen et al., 2004), which interacts with Capicua (Lam et al., 2006), Rora–Tip60 (Gehrking et al., 2011), Gfi-1 (Tsuda et al., 2005) and ATXN1L (Bowman et al., 2007), and a 12-amino acid segment, residues 768–780 toward the C terminus of ATXN1. This C-terminal segment contains three overlapping functional motifs and an endogenous site of phosphorylation at Ser776 (S776; Emamian et al., 2003; Huttlin et al., 2010). The functional motifs in the C-terminal region include a nuclear localization sequence (NLS), amino acids 771–774 (Klement et al., 1998), a 14-3-3 binding motif, amino acids 774–778 (Chen et al., 2003), and a UHM ligand motif (ULM), amino acids 771–776 (de Chiara et al., 2009)
It is increasingly apparent that the amino acid stretch, encompassing residues 771–778 in the C-terminal portion of ATXN1, plays a key role in its function and the dysfunction associated with SCA1. First, inactivation of the NLS in ATXN1 and preventing mutant ATXN1 from entering the nucleus of neurons blocks the ability of ATXN1 with an expanded polyQ tract to cause neurodegeneration in vivo (Klement et al., 1998). Second, Ser776, immediately adjacent to the NLS, is an endogenous site of phosphorylation (Emamian et al., 2003; Huttlin et al., 2010). Phosphorylation of S776 stabilizes ATXN1 because the phospho-resistant Ala776 (A776) ATXN1 is far less stable than either S776 or phospho-mimicking Asp776 (D776) ATXN1 (Jorgensen et al., 2009; Lai et al., 2011). Studies indicate that S776 and perhaps its phosphorylation has a crucial role in regulating interactions of wild-type and mutant ATXN1 with at least three cellular proteins, the phospho-serine/phospho-threonine binding protein 14-3-3 (Chen et al., 2003), a regulator of many signal transduction pathways (Morrison, 2009), and splicing factors RBM17 (Lim et al., 2008) and U2AF65 (de Chiara et al., 2009).
The interaction of ATXN1 with RBM17 increases with increasing length of the polyQ tract whereas the interaction of RBM17 with ATXN1-A776 is decreased dramatically regardless of polyQ tract length (Lim et al., 2008). Interestingly, replacing S776 with an aspartic acid reside results in a form of ATXN1-30Q, ATXN1[30Q]-D776, that has an enhanced interaction with RBM17 similar to that seen with ATXN1[82Q]-S776. Moreover, expression of ATXN1[30Q]-D776 in Purkinje neurons of transgenic mice results in a disease similar to that seen with ATXN1[82Q]-S776 (Duvick et al., 2010). Importantly, ATXN1[82Q]-A776, which interacts weakly with RBM17 in tissue culture cells, is not pathogenic when expressed in Purkinje cells (Emamian et al., 2003). These results indicate a direct link between pathogenicity of mutant ATXN1 and its interaction with RBM17 and suggest that the phosphorylation state of serine 776 is critical for the strength of this interaction.
Together, the data support a model for an ATXN1 cellular pathway that involves its phosphorylation at S776 in the cytoplasm, with perhaps dopamine D2 receptor signaling having a role in regulating ATXN1-S776 phosphorylation by PKA (Jorgensen et al., 2009; Hearst et al., 2010). Binding of 14-3-3 to ATXN1 protects pS776 from being dephosphorylated and subsequently degraded. Because the 14-3-3 binding site is immediately adjacent to the NLS in ATXN1, 14-3-3 binding masks the NLS and blocks entry of ATXN1 into the nucleus (Lai et al., 2011). Thus, 14-3-3 would be released from ATXN1, presumably by some regulated process, for ATXN1 to be transported into the nucleus. Once in the nucleus critical interactions are with splicing factors RBM17 and U2AF65 because the relative activity of these pathways is regulated by the length of the polyQ tract in ATXN1 and phosphorylation status of S776 (Fig. 1). Because the extent to which ATXN1 interacts with RBM17 varies with length of the polyQ tract within wild-type alleles (Lim et al., 2008), one can speculate that ATXN1 normally functions to regulate the balance between the RBM17 and U2AF65 pathways. As the polyQ expands into the pathogenic range, the RBM17 pathway becomes abnormally favored (Fig. 1).
A second pathway in which ATXN1 participates is an association with the transcription repressor Capicua (Cic). The biological effect on Cic of ATXN1 polyQ expansion is complex. First, overexpression of Cic was protective (Lam et al., 2006) in a fly model of SCA1, whereas partial loss of Cic prolonged life span in mice (Fryer et al., 2011). It seems that for some Cic gene targets expanded ATXN1 causes a loss of function, whereas at other targets polyQ-expanded ATXN1 enhances Cic binding inducing a state of hyper-repression. In the mouse, it is postulated that the hyper-repressive effect of expanded polyQ ATXN1 is toxic (Fryer et al., 2011). Intriguingly, mild exercise perhaps involving the Cic pathway in the brainstem improves survival in a mouse model of SCA1 in absence of improving the cerebellar motor dysfunction (Fryer et al., 2011). Although more vigorous motor activity that engages the cerebellum might restore motor function, one also needs to consider that distinct ATXN1 complexes, i.e., cellular pathways, contribute to disease in different regions of the CNS. Such a hypothesis is supported by the ability of a partial loss of 14-3-3 to improve cerebellar phenotypes and not brainstem phenotypes (Jafar-Nejad et al., 2011).
SCA2: RNA metabolism and regulation of translation
Like other SCAs, SCA2 patients present with limb and gait ataxia (Gwinn-Hardy et al., 2000; Shan et al., 2001). The polyQ protein affected in SCA2, ATXN2, as well as closely related homologues are implicated in range of biological functions, including cell specification, actin filament formation, receptor-mediated signaling, and secretion (Satterfield et al., 2002; Huynh et al., 2003; Wiedemeyer et al., 2003).
Evidence indicates that ATXN2 is involved in this diverse group of functions by virtue of it having a role in cytoplasmic RNA-related functions, in particular the regulation of translation (Fig. 2). First of all, the primary sequence of ATXN2 contains Lsm (like-Sm) motifs known to function in RNA splicing and mRNA decay in the cytoplasm (Neuwald and Koonin, 1998). In addition, the ATXN2 sequence contains a polyA-binding protein (PABP) interacting motif PAM2 (Kozlov et al., 2001). Consequently, ATXN2 interacts with PABP (Ciosk et al., 2004; Ralser et al., 2005). ATXN2 also interacts with a putative RNA protein called ataxin-2–binding protein (Shibata et al., 2000). Studies indicate that important subcellular sites for the regulation of translation by ATXN2 are stress granules and P-bodies, the main cellular compartments for regulating mRNA degradation, and translation (Eulalio et al., 2007; Buchan and Parker, 2009). In addition to the PABP, another stress granule and P-body component with which ATXN2 interacts is the DEAD/H-box RNA helicase DDX6 (Nonhoff et al., 2007). This study also found that ATXN2 has a role in assembly of stress granules and P-bodies. Recently, McCann et al. (2011) found that Drosophila Atx2 is required for repression of several miRNA target mRNAs, suggesting that ATXN2 also functions in miRNA pathways.
Figure 2.
Proposed functions of ATXN2 in the regulation of mRNA translation. The SCA2-encoded protein, ATXN2, is thought to have a role in mRNA metabolism as a component of translating polysomes, cytoplasmic stress granules, P-bodies, and the miRNA pathway. One intriguing possibility would be for ATXN2 to function in the trafficking of RNA/RNPs between actively translating polysomes and inactive stress granules and P-bodies.
Additional data linking ATXN2 to regulation of translation comes from studies on the homologues of ATXN2 in model organisms. For example, in yeast there is a genetic interaction between Pab1, a gene that promotes translation, and the yeast ATXN2 homologue Pbp1. Deletion of Pbp1 suppresses lethality due to loss of Pab1 (Mangus et al., 1998). Furthermore, overexpression of Pbp1 in yeast results in the same phenotype as seen inhibiting translation with cycloheximide (Dunn and Jensen, 2003). In C. elegans partial loss of function of the ATXN2 homologue ATX-2 alters the abundance of certain proteins without altering the level of their mRNAs, indicating that ATX-2 functions in the regulation of translation in C. elegans. More direct evidence that ATXN2 functions in translation comes from the Drosophila homologue of ATXN2, ATX2. By sucrose gradient centrifugation, ATX2 assembles with polyribosomes in a manner dependent on the Lsm and PAM2 motifs (Satterfield and Pallanck, 2006). This study also found that ATXN2 in transfected human HEK293 cells co-sediments with polyribosomes.
Thus, there are considerable data indicating that ATXN2 interacts with various components of the cellular machinery involved in the regulation of mRNA translation (Fig. 2). ATXN2’s association with components of translational active polysomes as well as translational silent RNA granules suggests a role in the regulated trafficking of mRNA between these structures that allows a cell to tailor translation to meet the needs of changes in its environment. Intriguingly, in a Drosophila model of synaptic plasticity, long-term olfactory habituation (LTH), Atx2 functions along with the miRNA pathway components Me31B and Ago1 to generate the synaptic plasticity associated with LTH that is linked to translational repression (McCann et al., 2011). Unfortunately, there is little data indicating whether and at what points in the cellular translational pathway SCA2-associated polyQ expansion alters ATXN2 function.
There is a suggestion that the polyQ tract length in ATXN2 affects risk for another neurodegenerative disorder, amyotrophic lateral sclerosis (ALS), providing additional evidence linking ATXN2 to RNA metabolism. Mutations in the gene encoding 43-kD TAR DNA-binding protein (TDP-43) are associated with some sporadic and familial forms of ALS (Lagier-Tourenne and Cleveland, 2009; Pesiridis et al., 2009). Normally predominantly in the nucleus, in pathological material TDP-43 localizes to cytoplasmic inclusions, leading to the hypothesis that an altered subcellular localization of TDP-43 is pathogenic. TDP-43 is a heterogeneous nuclear riboprotein, hnRNP, which associates with single-stranded nucleic acids and is implicated in several aspects of RNA metabolism, including transcription, alternative splicing, and RNA stability. Three recent studies report that TDP-43 binds to multiple RNA targets in the brain with a preference for UG-rich intronic regions (Polymenidou et al., 2011; Sephton et al., 2011; Tollervey et al., 2011). In an unbiased screen for modifiers of TDP-43 toxicity in yeast, the yeast ATXN2 homologue Pbp1 enhanced TDP-43 toxicity (Elden et al., 2010). Atx2 was also found to modify TDP-43 toxicity in Drosophila. In yeast and mammalian cells, TDP-43 and ATXN2 interact in a fashion dependent on RNA (Elden et al., 2010). Further linking ATXN2 with TDP-43 is their observation that ATXN2 is mislocalized in ALS motor neurons. Taking the functional relationship between ATXN2 and TDP-43 one step further, Elden et al. (2010) show that intermediate expansions (27–33 glutamines) are significantly higher in ALS patents than in neurologically normal controls. The association between longer wild-type SCA2 repeat alleles with an increased risk for ALS was recently supported by a study by Van Damme et al. (2011). This association is interesting in two respects. First, it indicates that ATXN2 is a relatively common susceptibility gene for ALS. In addition, this relationship between risk for ALS and intermediate alleles of ATXN2 suggests that variation in the polyQ tract, within the wild-type range, affects its function—perhaps through some aspect of RNA metabolism impacted by both ATXN2 and TDP-43.
SCA3: A regulator of the ubiquitin–proteasome pathway
SCA3 is considered to be the most common SCA worldwide including the United States. Also known as Machado-Joseph disease, SCA3 is caused by the expansion of a polyQ stretch in ataxin-3 (ATXN3; Kawaguchi et al., 1994). ATXN3 is associated with two cellular processes: transcription repression (Li et al., 2002; Evert et al., 2006) and protein homeostasis (Reina et al., 2010). It remains unclear whether these two processes are linked and the extent to which one or both drive SCA3 pathogenesis.
Supporting the nucleus as the site of pathogenesis in SCA3 are studies using cell (Tait et al., 1998; Chai et al., 1999; Perez et al., 1999; Fujigasaki et al., 2000) and mouse models (Bichelmeier et al., 2007), which show that ATXN3 is more toxic in the nucleus than in the cytoplasm. In addition, ATXN3 interacts with several transcription regulators including TAFII130 (Shimohata et al., 2000; Takahashi et al., 2001), CBP (McCampbell et al., 2000; Chai et al., 2002), RAD23 (Wang et al., 2000), and NCoR, HDAC3, and HDAC6 (Burnett and Pittman, 2005; Evert et al., 2006). More recently, heat shock and oxidative stress were found to increase the nuclear localization of wild-type and mutant ATXN3 (Reina et al., 2010). This observation raises an interesting question regarding the nuclear function of ATXN3; perhaps it functions to regulate expression of genes that encode components of the cellular stress response.
Besides modulating the cellular stress response by regulating gene expression, ATXN3 likely regulates the ubiquitin–proteasome system directly. Considerable data show that ATXN3 can function as a deubiqutinating enzyme (DUB; Burnett et al., 2003; Scheel et al., 2003; Winborn et al., 2008). ATXN3 contains at its N terminus a Josephin domain (JD) that has ubiquitin protease activity (Burnett et al., 2003; Scheel et al., 2003) and up to three ubiquitin-interacting motifs (UIMs) that bind ubiquitin (Burnett et al., 2003; Chai et al., 2004). The number of UIMs varies due to alternative splicing, with the three UIM isoforms predominating in the brain (Harris et al., 2010). Both the JD and UIMs of ATXN3 are critical for its ability in Drosophila to protect neurons from the toxicity of other mutant polyQ proteins (Warrick et al., 2005). ATXN3 selectively edits Lys63 linkages in mixed ubiquitin chains (Winborn et al., 2008). In contrast to the role of Lys48-linked chains in targeting proteins to the proteasome for degradation, Lys63-linked chains are thought to convey regulatory information to the proteins. Lastly, the three-dimensional structure of the JD from ATXN3 demonstrates a tight connection between polyubiquitin binding and the DUB activity of ATXN3 (Mao et al., 2005; Nicastro et al., 2005).
In a recent study, Scaglione et al. (2011) showed that ATXN3 along with the initiator E2 Ube2w regulates the ability of the E3 ligase C terminus of Hsc70-interacting protein (CHIP) to ubiquitinate its substrates. Monoubiquitination of CHIP by Ube2w promotes the interaction of CHIP with ATXN3, which in turn through its DUB activity restricts the length of ubiquitin chains attached to CHIP substrates. In addition, once substrate ubiquitination is completed ATXN3 deubquitinates CHIP, thereby stopping the reaction. In terms of the role this pathway might play in SCA3/MJD pathogenesis, Scaglione et al. (2011) found that polyQ expansion in ATXN3 increases its affinity for CHIP that could underlie the decreased activity of this neuroprotective E3 seen in a mouse model of SCA3.
Thus, a substantive body of data supports a role for ATXN3 in regulating the ubiquitin–proteasome system, most likely in editing polyubiquitin chains. It is worth noting that the ubiquitin–proteasome system, besides dealing with misfolded proteins, has a role in transcription by regulating the assembly and disassembly of transcription complexes that accompany each cycle of promoter activation/inactivation (Muratani and Tansey, 2003). As described above, nuclear localization of wild-type and mutant ATXN3 is increased by cellular stress. Thus, it is tempting to speculate that the nuclear and cytoplasmic functions of ATXN3 are subcellular variations on the same theme. Perhaps the ability of ATXN3 to remove monoubiquitin side chains from yet to be identified nuclear proteins—as it does with CHIP (Scaglione et al., 2011)—is key to proper ubiquitin-dependent regulation of gene expression in the nucleus. In this model, it is proposed that like its ability to regulate the Ube2w/CHIP ubiquitination cycle, ATXN3 associates with E2 ligases to regulate gene expression through its DUB activity to promote a ubiquitination/deubiquitination cycle on yet to be identified transcription factors. Analogous to the Ube2W/CHIP/ATXN3 ubiquitination cycle presented by Scaglione et al. (2011), ATXN3 might also regulate the ubiquitination of transcription factors critical for setting up a proper assembly/disassembly cycle of transcription complexes (Fig. 3). In this model, there are transcription factors that upon ubiquitination by an E2 ligase would promote the recruitment of ATXN3 to the transcriptional complex on DNA, thus stabilizing the complex and stimulating transcription. ATXN3 regulates the length of the ubiquitin side chain on the transcription factor, keeping it within a critical length. When transcription is to cease, a signal would induce ATXN3 to deubiquitinate the transcription factor as it does with CHIP (Scaglione et al., 2011), thereby destabilizing the complex.
Figure 3.
A model for ATXN3 in the editing of Ub chains on nuclear targets. The model combines the recent work of Scaglione et al. (2011) with earlier studies showing that ATXN3, a deubiquitinating enzyme, has a role in regulating gene expression. Like the proposed role of ATXN3 in the Ube2W/CHIP ubiquitination cycle of misfolded proteins, it is postulated that ATXN3 also regulates a ubiquitination cycle involved in assembly/disassembly of transcription complexes in the nucleus. ↑[Q]n indicates an increased interaction of expanded polyQ ATXN3 with a yet to be identified nuclear protein involved in regulation of transcription.
Recent studies on the DUB activity of the ATXN3 orthologue in C. elegans, atx-3, speak to the biological relevance of its role in regulating the ubiquitin–proteasome system. ATXN3 interacts with the mammalian p97 protein, a chaperone-like AAA ATPase, to regulate the degradation of misfolded ER proteins (Wang et al., 2006). In C. elegans the p97 homologue is CDC-48. Worms deficient for both cdc-82 and atx-3 have increased protein stability and increased longevity (Kuhlbrodt et al., 2011). Thus, it seems that ATX-3 is a critical regulator of lifespan in worms. One can anticipate that as the chain-editing roles of ATXN3 are better defined in mammalian systems, its role in other physiologically relevant pathways will be forthcoming.
SCA6: A membrane channel with a connection to the nucleus?
At first, pathogenesis of SCA6 seemed to be a relatively straightforward channelopathy. This disease is due to an expansion of a polyQ tract in the C terminus of the α1A transmembrane subunit of the P/Q-type voltage-gated calcium channel CACNA1A (Riess et al., 1997; Zhuchenko et al., 1997). This channel is known to be abundant in Purkinje cells of the cerebellum (Kulik et al., 2004) and is the mutational target in several other inherited neurological disorders (Pietrobon, 2002). Initial studies indicated that SCA6 was a disorder due to P/Q channel dysfunction (Matsuyama et al., 1999; Restituito et al., 2000; Toru et al., 2000).
However, two additional findings complicate the picture. First, electrophysiological analysis of Sca684Q knock-in mice failed to find a change in the intrinsic properties of Purkinje cells (Watase et al., 2008). Kordasiewicz et al. (2006), noting earlier evidence that the polyQ-containing cytoplasmic C terminus is cleaved to form a stable peptide (Sakurai et al., 1996; Kubodera et al., 2003), examined the α1A C terminus in more detail. Intriguingly, Kordasiewicz et al. (2006) found that the α1A C-terminal peptide localizes to Purkinje cell nuclei in wild-type mice and humans. Moreover, the C-terminal peptide with an expanded polyQ tract is toxic to tissue culture cells with toxicity being dependent on its nuclear localization. These results support a pathogenic model for SCA6 where cleavage of the polyQ-containing C terminus of the α1A transmembrane subunit and its subsequent translocation to the nucleus contributes to disease. More generally, these data suggest that like certain other membrane proteins involved in signaling, e.g., βAPP, p75, Notch, and the α1C subunit of L-type calcium channels (De Jongh et al., 1994; Bland et al., 2003; Jung et al., 2003), in response to a stimulus at the membrane, the C terminus of the α1A calcium channel subunit is cleaved and translocates to the nucleus to regulate transcription.
Nevertheless, in the absence of direct evidence that fragment production is critical for disease, one should be cautious in discarding SCA6 as a calcium channelopathy given the vast amount of data implicating altered calcium signaling in the SCAs. Evidence for alterations in calcium signaling and/or handling is reported for SCA1 (Lin et al., 2000; Vig et al., 2001; Serra et al., 2004), SCA2 (Liu et al., 2009), and SCA3 (Chen et al., 2008). In addition, recent data show that SCA15, a non-polyQ SCA, is due to a mutation in the InsP3 receptor 1 gene encoding a receptor that functions in the release of Ca2+ from intracellular stores (van de Leemput et al., 2007).
SCA7 and SCA17: Transcription regulators
Besides progressive cerebellar degeneration, patients with SCA7 uniquely display retinal degeneration. ATXN7 functions as a component of the SAGA (Spt–Ada–Gcn5 acetyltransferase) complex (Helmlinger et al., 2006). SAGA regulates transcription via its two histone-modifying activities, the Gcn5 histone acetyltransferase and the Usp22 deubiquitinase. Although ATXN7 has no enzymatic activity it is reported to anchor the Usp22 component to the SAGA complex (Köhler et al., 2008; Zhao et al., 2008). It is the Gcn5 histone acetyltransferase (HAT) activity of SAGA that seems to be affected by the presence of ATXN7 with an expanded polyQ tract. Whether mutant ATXN7 decreases HAT activity (McMahon et al., 2005; Palhan et al., 2005) or increases HAT activity (Helmlinger et al., 2006) remains unclear. The reported increase in HAT activity with mutant ATXN7 is based on an in vivo analysis in the retina, whereas the studies showing a loss of activity either used human ATXN7 in yeast or transfected 293T cells. Perhaps differences between model systems and/or tissue underlie the discrepancies regarding the effect that mutant ATXN7 has on Gcn5 HAT activity. It is worth noting that a mutant ATXN5-induced gain in Gcn5 HAT activity is in line with the autosomal-dominant inheritance pattern of SCA7.
SCA17 exemplifies a paradox facing essentially all inherited neurodegenerative diseases: how can mutations in a widely expressed gene cause selective damage to the nervous system? In SCA17, polyQ expansion that occurs in the TATA-binding protein (TBP), an essential component in the initiation of transcription of all three RNA polymerases, causes a neurodegenerative disease that in addition to ataxia can include dementia and epilepsy (Koide et al., 1999; Rolfs et al., 2003). Importantly, individuals homozygous for mutant SCA17 alleles survive to adulthood, indicating that normal TBP function during development is not impaired by polyQ expansion (Zühlke et al., 2003; Toyoshima et al., 2004). In a series of studies using a transgenic mouse model of SCA17, Xioa-Jiang Li and colleagues found evidence indicating that polyQ expansion in TBP alters its ability to bind DNA and transcription regulators, perhaps leading to down-regulation of a select set of genes, e.g., HSPB1 and TrkA (Friedman et al., 2007, 2008; Shah et al., 2009). Thus, for SCA7 and SCA17, the implication is that polyQ expansion specifically impacts the ability of these regulators of transcription to control expression of a subset of genes—ones critical for neuronal function—while not impacting their ability to regulate most other genes.
The ataxia interactome
To what extent might the cellular processes for the polyQ-mediated SCAs reflect molecular pathways shared among the inherited cerebellar ataxias in general? A framework for addressing this question comes from an ataxia protein–protein interaction network, “ataxia” interactome, developed by Lim et al. (2006). The interactome was generated by yeast two-hybrid screens using 23 ataxia-causing proteins, cDNAs of proteins for 11 recessive and 12 dominant ataxias including the polyQ SCAs, and 31 ataxia-associated proteins, cDNAs for proteins that either were known to interact with an ataxia-causing protein or paralogues of such proteins. The study showed that many of the ataxia-causing proteins share interacting partners, supporting the idea that the disease phenotypes shared among the ataxias are due to common molecular pathways. Biological processes significantly enriched in the ataxia network included transcription regulation, import of proteins to nucleus, RNA splicing, and ubiquitination. Correspondingly, cellular components containing proteins enriched in the ataxia interactome include the nucleoplasm, nuclear membrane, and spliceosome complex. Among the enhanced molecular functions are transcription factor activity and kinase inhibitors. The high degree of overlap in the cellular/molecular themes in ataxia interactome with those discussed in the previous paragraph indicate that as the pathogenesis of these disorders is further understood, understanding of the SCAs more broadly will be forthcoming.
Potential approaches to therapy
Clearly for the polyQ SCAs there remains much to learn before a targeted approach to treatment, one based on correcting an altered function that drives pathology, is in sight. Yet, there are a couple of encouraging points worth noting. First, there is evidence that at least for SCA1 and two other polyQ disorders, Huntington’s disease and spinalbulbar muscular atrophy, pathology can be reversed even after disease is underway (Yamamoto et al., 2000; Chevalier-Larsen et al., 2004; Zu et al., 2004). In the case of SCA1, the data indicate that the ability to recover normal function decreases with age and progression of disease. This provides support for the widely held notion that the sooner one intervenes therapeutically, the better the chance of having a positive impact. The second key point is that for these SCAs, as well as other polyQ disorders, pathogenesis requires the presence of the mutant protein. Thus, decreasing the amount of mutant protein becomes a viable approach to therapy even in the absence of an understanding of the downstream effects that lead to disease.
One approach shown to be effective in reducing expression of mutant ATXN1 in vivo is the delivery to the cerebellum of AAV vectors expressing an shRNA directed against ATXN1 (Xia et al., 2004). Mutant ATXN1 expression was reduced such that motor function was significantly improved and Purkinje cell morphology was normalized in transduced cells. Although this SCA1 study used an shRNA that would target both wild-type and mutant ATXN1, allele-specific strategies where the mutant allele would be preferentially targeted are under development for several of the polyQ disorders including SCA3 (Alves et al., 2008). In this regard, selective targeting of the polyQ-expressing slice isoform of Cav2.1 should be applicable to SCA6 (Tsou et al., 2011).
That these diseases require expression of the mutant protein for disease to manifest itself has an additional implication in terms of a potential approach to therapy. The term proteostasis has been applied to the protein homeostatic network of cellular pathways that regulate the overall quality control of proteins that normally make up the proteome of each cell (Balch et al., 2008). There are numerous examples that the ability of this system to adapt to normal metabolic demands becomes compromised with aging and with the presence of misfolded mutant proteins such as those associated with many of the inherited neurodegenerative diseases (Voisine et al., 2010). For example, using a C. elegans model of Machado-Joseph disease (SCA3), the age-associated mutant ATXN3-induced phenotypes were suppressed by down-regulation of the Igf1-like pathway and activation of heat shock factor 1 (Teixeira-Castro et al., 2011). Such results provide support to the use of regulators of protein homeostasis as therapeutic agents in diseases like the polyglutamine ataxias.
Conclusions
The polyQ SCAs, as well as other inherited neurodegenerative diseases, are rich ground for investigation, one where cell biologists can make seminal contributions. The genes for these disorders are identified, yet how the proteins function normally and how this function is altered in disease remains to be established. Understanding the cell biology that underlies each affected protein and how it is altered in each disorder will be crucial for the development of therapeutic strategies. Furthermore, just as the study of genetic mutants in model organisms has made novel and fundamental contributions to basic cell biological processes, so can the study of inherited human neurodegenerative diseases such as the SCAs.
Acknowledgments
The author greatly appreciates the artistic talent of Jillian Frisch.
The research presented from the author was supported by National Institutes of Health/National Institute of Neurological Disorders and Stroke grants NS22920-23 and NS45667-09.
Footnotes
Abbreviations used in this paper:
- ALS
- amyotrophic lateral sclerosis
- ATXN
- ataxin
- CHIP
- C terminus of Hsc70-interacting protein
- DUB
- deubiquitinating enzyme
- NLS
- nuclear localization sequence
- polyQ
- polyglutamine
- SCA
- spinocerebellar ataxia
References
- Alves S., Nascimento-Ferreira I., Auregan G., Hassig R., Dufour N., Brouillet E., Pedroso de Lima M.C., Hantraye P., Pereira de Almeida L., Déglon N. 2008. Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado-Joseph disease. PLoS ONE. 3:e3341 10.1371/journal.pone.0003341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., Finkbeiner S. 2004. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 431:805–810 10.1038/nature02998 [DOI] [PubMed] [Google Scholar]
- Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. 2008. Adapting proteostasis for disease intervention. Science. 319:916–919 10.1126/science.1141448 [DOI] [PubMed] [Google Scholar]
- Barnes J.A., Ebner B.A., Duvick L.A., Gao W., Chen G., Orr H.T., Ebner T.J. 2011. Abnormalities in the climbing fiber-Purkinje cell circuitry contribute to neuronal dysfunction in ATXN1[82Q] mice. J. Neurosci. 31:12778–12789 10.1523/JNEUROSCI.2579-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichelmeier U., Schmidt T., Hübener J., Boy J., Rüttiger L., Häbig K., Poths S., Bonin M., Knipper M., Schmidt W.J., et al. 2007. Nuclear localization of ataxin-3 is required for the manifestation of symptoms in SCA3: in vivo evidence. J. Neurosci. 27:7418–7428 10.1523/JNEUROSCI.4540-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland C.E., Kimberly P., Rand M.D. 2003. Notch-induced proteolysis and nuclear localization of the Delta ligand. J. Biol. Chem. 278:13607–13610 10.1074/jbc.C300016200 [DOI] [PubMed] [Google Scholar]
- Blum E.S., Abraham M.C., Yoshimura S., Lu Y., Shaham S. 2012. Control of nonapoptotic developmental cell death in Caenorhabditis elegans by a polyglutamine-repeat protein. Science. 335:970–973 10.1126/science.1215156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman A.B., Lam Y.C., Jafar-Nejad P., Chen H.K., Richman R., Samaco R.C., Fryer J.D., Kahle J.J., Orr H.T., Zoghbi H.Y. 2007. Duplication of Atxn1l suppresses SCA1 neuropathology by decreasing incorporation of polyglutamine-expanded ataxin-1 into native complexes. Nat. Genet. 39:373–379 10.1038/ng1977 [DOI] [PubMed] [Google Scholar]
- Buchan J.R., Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 36:932–941 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett B.G., Pittman R.N. 2005. The polyglutamine neurodegenerative protein ataxin 3 regulates aggresome formation. Proc. Natl. Acad. Sci. USA. 102:4330–4335 10.1073/pnas.0407252102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett B.G., Li F., Pittman R.N. 2003. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum. Mol. Genet. 12:3195–3205 10.1093/hmg/ddg344 [DOI] [PubMed] [Google Scholar]
- Chai Y., Koppenhafer S.L., Shoesmith S.J., Perez M.K., Paulson H.L. 1999. Evidence for proteasome involvement in polyglutamine disease: localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum. Mol. Genet. 8:673–682 10.1093/hmg/8.4.673 [DOI] [PubMed] [Google Scholar]
- Chai Y., Shao J., Miller V.M., Williams A., Paulson H.L. 2002. Live-cell imaging reveals divergent intracellular dynamics of polyglutamine disease proteins and supports a sequestration model of pathogenesis. Proc. Natl. Acad. Sci. USA. 99:9310–9315 10.1073/pnas.152101299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y., Berke S.S., Cohen R.E., Paulson H.L. 2004. Poly-ubiquitin binding by the polyglutamine disease protein ataxin-3 links its normal function to protein surveillance pathways. J. Biol. Chem. 279:3605–3611 10.1074/jbc.M310939200 [DOI] [PubMed] [Google Scholar]
- Chen H.-K., Fernandez-Funez P., Acevedo S.F., Lam Y.C., Kaytor M.D., Fernandez M.H., Aitken A., Skoulakis E.M.C., Orr H.T., Botas J., Zoghbi H.Y. 2003. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 113:457–468 10.1016/S0092-8674(03)00349-0 [DOI] [PubMed] [Google Scholar]
- Chen Y.W., Allen M.D., Veprintsev D.B., Löwe J., Bycroft M. 2004. The structure of the AXH domain of spinocerebellar ataxin-1. J. Biol. Chem. 279:3758–3765 10.1074/jbc.M309817200 [DOI] [PubMed] [Google Scholar]
- Chen X., Tang T.-S., Tu H., Nelson O., Pook M., Hammer R., Nukina N., Bezprozvanny I. 2008. Deranged calcium signaling and neurodegeneration in spinocerebellar ataxia type 3. J. Neurosci. 28:12713–12724 10.1523/JNEUROSCI.3909-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier-Larsen E.S., O’Brien C.J., Wang H., Jenkins S.C., Holder L., Lieberman A.P., Merry D.E. 2004. Castration restores function and neurofilament alterations of aged symptomatic males in a transgenic mouse model of spinal and bulbar muscular atrophy. J. Neurosci. 24:4778–4786 10.1523/JNEUROSCI.0808-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R., DePalma M., Priess J.R. 2004. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development. 131:4831–4841 10.1242/dev.01352 [DOI] [PubMed] [Google Scholar]
- Cummings C.J., Reinstein E., Sun Y., Antalffy B., Jiang Y.-H., Ciechanover A., Orr H.T., Beaudet A.L., Zoghbi H.Y. 1999. Mutation of the E6-AP ubiquitin ligase reduces nuclear inclusion frequency while accelerating polyglutamine-induced pathology in SCA1 mice. Neuron. 24:879–892 10.1016/S0896-6273(00)81035-1 [DOI] [PubMed] [Google Scholar]
- Davies S.W., Turmaine M., Cozens B.A., DiFiglia M., Sharp A.H., Ross C.A., Scherzinger E., Wanker E.E., Mangiarini L., Bates G.P. 1997. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 90:537–548 10.1016/S0092-8674(00)80513-9 [DOI] [PubMed] [Google Scholar]
- de Chiara C., Giannini C., Adinolfi S., de Boer J., Guida S., Ramos A., Jodice C., Kioussis D., Pastore A. 2003. The AXH module: an independently folded domain common to ataxin-1 and HBP1. FEBS Lett. 551:107–112 10.1016/S0014-5793(03)00818-4 [DOI] [PubMed] [Google Scholar]
- de Chiara C., Menon R.P., Strom M., Gibson T.J., Pastore A. 2009. Phosphorylation of S776 and 14-3-3 binding modulate ataxin-1 interaction with splicing factors. PLoS ONE. 4:e8372 10.1371/journal.pone.0008372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jongh K.S., Colvin A.A., Wang K.K., Catterall W.A. 1994. Differential proteolysis of the full-length form of the L-type calcium channel alpha 1 subunit by calpain. J. Neurochem. 63:1558–1564 10.1046/j.1471-4159.1994.63041558.x [DOI] [PubMed] [Google Scholar]
- DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., Aronin N. 1997. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 277:1990–1993 10.1126/science.277.5334.1990 [DOI] [PubMed] [Google Scholar]
- Dunn C.D., Jensen R.E. 2003. Suppression of a defect in mitochondrial protein import identifies cytosolic proteins required for viability of yeast cells lacking mitochondrial DNA. Genetics. 165:35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick L., Barnes J., Ebner B., Agrawal S., Andresen M., Lim J., Giesler G.J., Zoghbi H.Y., Orr H.T. 2010. SCA1-like disease in mice expressing wild-type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 67:929–935 10.1016/j.neuron.2010.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden A.C., Kim H.-J., Hart M.P., Chen-Plotkin A.S., Johnson B.S., Fang X., Armakola M., Geser F., Greene R., Lu M.M., et al. 2010. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 466:1069–1075 10.1038/nature09320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emamian E.S., Kaytor M.D., Duvick L.A., Zu T., Tousey S.K., Zoghbi H.Y., Clark H.B., Orr H.T. 2003. Serine 776 of ataxin-1 is critical for polyglutamine-induced disease in SCA1 transgenic mice. Neuron. 38:375–387 10.1016/S0896-6273(03)00258-7 [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8:9–22 10.1038/nrm2080 [DOI] [PubMed] [Google Scholar]
- Evert B.O., Araujo J., Vieira-Saecker A.M., de Vos R.A., Harendza S., Klockgether T., Wüllner U. 2006. Ataxin-3 represses transcription via chromatin binding, interaction with histone deacetylase 3, and histone deacetylation. J. Neurosci. 26:11474–11486 10.1523/JNEUROSCI.2053-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman M.J., Shah A.G., Fang Z.H., Ward E.G., Warren S.T., Li S., Li X.J. 2007. Polyglutamine domain modulates the TBP-TFIIB interaction: implications for its normal function and neurodegeneration. Nat. Neurosci. 10:1519–1528 10.1038/nn2011 [DOI] [PubMed] [Google Scholar]
- Friedman M.J., Wang C.E., Li X.J., Li S. 2008. Polyglutamine expansion reduces the association of TATA-binding protein with DNA and induces DNA binding-independent neurotoxicity. J. Biol. Chem. 283:8283–8290 10.1074/jbc.M709674200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer J.D., Yu P., Kang H., Mandel-Brehm C., Carter A.N., Crespo-Barreto J., Gao Y., Flora A., Shaw C., Orr H.T., Zoghbi H.Y. 2011. Exercise and genetic rescue of SCA1 via the transcriptional repressor Capicua. Science. 334:690–693 10.1126/science.1212673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujigasaki H., Uchihara T., Koyano S., Iwabuchi K., Yagishita S., Makifuchi T., Nakamura A., Ishida K., Toru S., Hirai S., et al. 2000. Ataxin-3 is translocated into the nucleus for the formation of intranuclear inclusions in normal and Machado-Joseph disease brains. Exp. Neurol. 165:248–256 10.1006/exnr.2000.7479 [DOI] [PubMed] [Google Scholar]
- Gehrking K.M., Andresen J.M., Duvick L., Lough J., Zoghbi H.Y., Orr H.T. 2011. Partial loss of Tip60 slows mid-stage neurodegeneration in a spinocerebellar ataxia type 1 (SCA1) mouse model. Hum. Mol. Genet. 20:2204–2212 10.1093/hmg/ddr108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.K., Deng Y., Slow E.J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., et al. 2006. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 125:1179–1191 10.1016/j.cell.2006.04.026 [DOI] [PubMed] [Google Scholar]
- Gwinn-Hardy K., Chen J.Y., Liu H.C., Liu T.Y., Boss M., Seltzer W., Adam A., Singleton A., Koroshetz W., Waters C., et al. 2000. Spinocerebellar ataxia type 2 with parkinsonism in ethnic Chinese. Neurology. 55:800–805 [DOI] [PubMed] [Google Scholar]
- Harris G.M., Dodelzon K., Gong L., Gonzalez-Alegre P., Paulson H.L. 2010. Splice isoforms of the polyglutamine disease protein ataxin-3 exhibit similar enzymatic yet different aggregation properties. PLoS ONE. 5:e13695 10.1371/journal.pone.0013695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearst S.M., Lopez M.E., Shao Q., Liu Y., Vig P.J.S. 2010. Dopamine D2 receptor signaling modulates mutant ataxin-1 S776 phosphorylation and aggregation. J. Neurochem. 114:706–716 10.1111/j.1471-4159.2010.06791.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D., Hardy S., Eberlin A., Devys D., Tora L. 2006. Both normal and polyglutamine- expanded ataxin-7 are components of TFTC-type GCN5 histone acetyltransferase- containing complexes. Biochem. Soc. Symp. 73:155–163 [DOI] [PubMed] [Google Scholar]
- Hourez R., Servais L., Orduz D., Gall D., Millard I., de Kerchove d’Exaerde A., Cheron G., Orr H.T., Pandolfo M., Schiffmann S.N. 2011. Aminopyridines correct early dysfunction and delay neurodegeneration in a mouse model of spinocerebellar ataxia type 1. J. Neurosci. 31:11795–11807 10.1523/JNEUROSCI.0905-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin E.L., Jedrychowski M.P., Elias J.E., Goswami T., Rad R., Beausoleil S.A., Villén J., Haas W., Sowa M.E., Gygi S.P. 2010. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 143:1174–1189 10.1016/j.cell.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh D.P., Yang H.T., Vakharia H., Nguyen D., Pulst S.M. 2003. Expansion of the polyQ repeat in ataxin-2 alters its Golgi localization, disrupts the Golgi complex and causes cell death. Hum. Mol. Genet. 12:1485–1496 10.1093/hmg/ddg175 [DOI] [PubMed] [Google Scholar]
- Irwin S., Vandelft M., Pinchev D., Howell J.L., Graczyk J., Orr H.T., Truant R. 2005. RNA association and nucleocytoplasmic shuttling by ataxin-1. J. Cell Sci. 118:233–242 10.1242/jcs.01611 [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad P., Ward C.S., Richman R., Orr H.T., Zoghbi H.Y. 2011. Regional rescue of spinocerebellar ataxia type 1 phenotypes by 14-3-3epsilon haploinsufficiency in mice underscores complex pathogenicity in neurodegeneration. Proc. Natl. Acad. Sci. USA. 108:2142–2147 10.1073/pnas.1018748108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen N.D., Andresen J.M., Lagalwar S., Armstrong B., Stevens S., Byam C.E., Duvick L.A., Lai S., Jafar-Nejad P., Zoghbi H.Y., et al. 2009. Phosphorylation of ATXN1 at Ser776 in the cerebellum. J. Neurochem. 110:675–686 10.1111/j.1471-4159.2009.06164.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K.M., Tan S., Landman N., Petrova K., Murray S., Lewis R., Kim P.K., Kim D.S., Ryu S.H., Chao M.V., Kim T.W. 2003. Regulated intramembrane proteolysis of the p75 neurotrophin receptor modulates its association with the TrkA receptor. J. Biol. Chem. 278:42161–42169 10.1074/jbc.M306028200 [DOI] [PubMed] [Google Scholar]
- Katsuno M., Adachi H., Kume A., Li M., Nakagomi Y., Niwa H., Sang C., Kobayashi Y., Doyu M., Sobue G. 2002. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 35:843–854 10.1016/S0896-6273(02)00834-6 [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y., Okamoto T., Taniwaki M., Aizawa M., Inoue M., Katayama S., Kawakami H., Nakamura S., Nishimura M., Akiguchi I., et al. 1994. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 8:221–228 10.1038/ng1194-221 [DOI] [PubMed] [Google Scholar]
- Klement I.A., Skinner P.J., Kaytor M.D., Yi H., Hersch S.M., Clark H.B., Zoghbi H.Y., Orr H.T. 1998. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 95:41–53 10.1016/S0092-8674(00)81781-X [DOI] [PubMed] [Google Scholar]
- Köhler A., Schneider M., Cabal G.G., Nehrbass U., Hurt E. 2008. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat. Cell Biol. 10:707–715 10.1038/ncb1733 [DOI] [PubMed] [Google Scholar]
- Koide R., Kobayashi S., Shimohata T., Ikeuchi T., Maruyama M., Saito M., Yamada M., Takahashi H., Tsuji S. 1999. A neurological disease caused by an expanded CAG trinucleotide repeat in the TATA-binding protein gene: a new polyglutamine disease? Hum. Mol. Genet. 8:2047–2053 10.1093/hmg/8.11.2047 [DOI] [PubMed] [Google Scholar]
- Kordasiewicz H.B., Thompson R.M., Clark H.B., Gomez C.M. 2006. C-termini of P/Q-type Ca2+ channel alpha1A subunits translocate to nuclei and promote polyglutamine-mediated toxicity. Hum. Mol. Genet. 15:1587–1599 10.1093/hmg/ddl080 [DOI] [PubMed] [Google Scholar]
- Kozlov G., Trempe J.F., Khaleghpour K., Kahvejian A., Ekiel I., Gehring K. 2001. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl. Acad. Sci. USA. 98:4409–4413 10.1073/pnas.071024998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol H.A., Krawczyk P.M., Bosch K.S., Aten J.A., Hol E.M., Reits E.A. 2008. Polyglutamine expansion accelerates the dynamics of ataxin-1 and does not result in aggregate formation. PLoS One. 3:e1503 10.1371/journal.pone.0001503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubodera T., Yokota T., Ohwada K., Ishikawa K., Miura H., Matsuoka T., Mizusawa H. 2003. Proteolytic cleavage and cellular toxicity of the human alpha1A calcium channel in spinocerebellar ataxia type 6. Neurosci. Lett. 341:74–78 10.1016/S0304-3940(03)00156-3 [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K., Janiesch P.C., Kevei É., Segref A., Barikbin R., Hoppe T. 2011. The Machado-Joseph disease deubiquitylase ATX-3 couples longevity and proteostasis. Nat. Cell Biol. 13:273–281 10.1038/ncb2200 [DOI] [PubMed] [Google Scholar]
- Kulik A., Nakadate K., Hagiwara A., Fukazawa Y., Luján R., Saito H., Suzuki N., Futatsugi A., Mikoshiba K., Frotscher M., Shigemoto R. 2004. Immunocytochemical localization of the alpha 1A subunit of the P/Q-type calcium channel in the rat cerebellum. Eur. J. Neurosci. 19:2169–2178 10.1111/j.0953-816X.2004.03319.x [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C., Cleveland D.W. 2009. Rethinking ALS: the FUS about TDP-43. Cell. 136:1001–1004 10.1016/j.cell.2009.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S., O’Callaghan B., Zoghbi H.Y., Orr H.T. 2011. 14-3-3 Binding to ataxin-1(ATXN1) regulates its dephosphorylation at Ser-776 and transport to the nucleus. J. Biol. Chem. 286:34606–34616 10.1074/jbc.M111.238527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam Y.C., Bowman A.B., Jafar-Nejad P., Lim J., Richman R., Fryer J.D., Hyun E.D., Duvick L.A., Orr H.T., Botas J., Zoghbi H.Y. 2006. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 127:1335–1347 10.1016/j.cell.2006.11.038 [DOI] [PubMed] [Google Scholar]
- La Spada A.R., Wilson E.M., Lubahn D.B., Harding A.E., Fischbeck K.H. 1991. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 352:77–79 10.1038/352077a0 [DOI] [PubMed] [Google Scholar]
- Li F., Macfarlan T., Pittman R.N., Chakravarti D. 2002. Ataxin-3 is a histone-binding protein with two independent transcriptional corepressor activities. J. Biol. Chem. 277:45004–45012 10.1074/jbc.M205259200 [DOI] [PubMed] [Google Scholar]
- Lim J., Hao T., Shaw C., Patel A.J., Szabó G., Rual J.-F., Fisk C.J., Li N., Smolyar A., Hill D.E., et al. 2006. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 125:801–814 10.1016/j.cell.2006.03.032 [DOI] [PubMed] [Google Scholar]
- Lim J., Crespo-Barreto J., Jafar-Nejad P., Bowman A.B., Richman R., Hill D.E., Orr H.T., Zoghbi H.Y. 2008. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 452:713–718 10.1038/nature06731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Antalffy B., Kang D., Orr H.T., Zoghbi H.Y. 2000. Polyglutamine expansion down-regulates specific neuronal genes before pathologic changes in SCA1. Nat. Neurosci. 3:157–163 10.1038/81814 [DOI] [PubMed] [Google Scholar]
- Liu J., Tang T.S., Tu H., Nelson O., Herndon E., Huynh D.P., Pulst S.M., Bezprozvanny I. 2009. Deranged calcium signaling and neurodegeneration in spinocerebellar ataxia type 2. J. Neurosci. 29:9148–9162 10.1523/JNEUROSCI.0660-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus D.A., Amrani N., Jacobson A. 1998. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol. 18:7383–7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manto M.-U. 2005. The wide spectrum of spinocerebellar ataxias (SCAs). Cerebellum. 4:2–6 10.1080/14734220510007914 [DOI] [PubMed] [Google Scholar]
- Mao Y., Senic-Matuglia F., Di Fiore P.P., Polo S., Hodsdon M.E., De Camilli P. 2005. Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain. Proc. Natl. Acad. Sci. USA. 102:12700–12705 10.1073/pnas.0506344102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J.L., Walker H., Theisen H., Zhu Y.-Z., Fielder T., Purcell J., Thompson L.M. 2000. Expanded polyglutamine peptides alone are intrinsically cytotoxic and cause neurodegeneration in Drosophila. Hum. Mol. Genet. 9:13–25 10.1093/hmg/9.1.13 [DOI] [PubMed] [Google Scholar]
- Matsuyama Z., Wakamori M., Mori Y., Kawakami H., Nakamura S., Imoto K. 1999. Direct alteration of the P/Q-type Ca2+ channel property by polyglutamine expansion in spinocerebellar ataxia 6. J. Neurosci. 19:RC14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCampbell A., Taylor J.P., Taye A.A., Robitschek J., Li M., Walcott J., Merry D., Chai Y., Paulson H., Sobue G., Fischbeck K.H. 2000. CREB-binding protein sequestration by expanded polyglutamine. Hum. Mol. Genet. 9:2197–2202 10.1093/hmg/9.14.2197 [DOI] [PubMed] [Google Scholar]
- McCann C., Holohan E.E., Das S., Dervan A., Larkin A., Lee J.A., Rodrigues V., Parker R., Ramaswami M. 2011. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc. Natl. Acad. Sci. USA. 108:E655–E662 10.1073/pnas.1107198108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon S.J., Pray-Grant M.G., Schieltz D., Yates J.R., III, Grant P.A. 2005. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc. Natl. Acad. Sci. USA. 102:8478–8482 10.1073/pnas.0503493102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D.K. 2009. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 19:16–23 10.1016/j.tcb.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M., Tansey W.P. 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4:192–201 10.1038/nrm1049 [DOI] [PubMed] [Google Scholar]
- Nedelsky N.B., Pennuto M., Smith R.B., Palazzolo I., Moore J., Nie Z., Neale G., Taylor J.P. 2010. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron. 67:936–952 10.1016/j.neuron.2010.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A.F., Koonin E.V. 1998. Ataxin-2, global regulators of bacterial gene expression, and spliceosomal snRNP proteins share a conserved domain. J. Mol. Med. 76:3–5 10.1007/s109-1998-8098-0 [DOI] [PubMed] [Google Scholar]
- Nicastro G., Menon R.P., Masino L., Knowles P.P., McDonald N.Q., Pastore A. 2005. The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition. Proc. Natl. Acad. Sci. USA. 102:10493–10498 10.1073/pnas.0501732102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonhoff U., Ralser M., Welzel F., Piccini I., Balzereit D., Yaspo M.-L., Lehrach H., Krobitsch S. 2007. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell. 18:1385–1396 10.1091/mbc.E06-12-1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H.T. 2001. Beyond the Qs in the polyglutamine diseases. Genes Dev. 15:925–932 10.1101/gad.888401 [DOI] [PubMed] [Google Scholar]
- Orr H.T., Zoghbi H.Y. 2007. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 30:575–621 10.1146/annurev.neuro.29.051605.113042 [DOI] [PubMed] [Google Scholar]
- Orr H.T., Chung M.Y., Banfi S., Kwiatkowski T.J., Jr, Servadio A., Beaudet A.L., McCall A.E., Duvick L.A., Ranum L.P., Zoghbi H.Y. 1993. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat. Genet. 4:221–226 10.1038/ng0793-221 [DOI] [PubMed] [Google Scholar]
- Palhan V.B., Chen S., Peng G.-H., Tjernberg A., Gamper A.M., Fan Y., Chait B.T., La Spada A.R., Roeder R.G. 2005. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc. Natl. Acad. Sci. USA. 102:8472–8477 10.1073/pnas.0503505102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M.K., Paulson H.L., Pittman R.N. 1999. Ataxin-3 with an altered conformation that exposes the polyglutamine domain is associated with the nuclear matrix. Hum. Mol. Genet. 8:2377–2385 10.1093/hmg/8.13.2377 [DOI] [PubMed] [Google Scholar]
- Pesiridis G.S., Lee V.M., Trojanowski J.Q. 2009. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum. Mol. Genet. 18(R2):R156–R162 10.1093/hmg/ddp303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrobon D. 2002. Calcium channels and channelopathies of the central nervous system. Mol. Neurobiol. 25:31–50 10.1385/MN:25:1:031 [DOI] [PubMed] [Google Scholar]
- Polymenidou M., Lagier-Tourenne C., Hutt K.R., Huelga S.C., Moran J., Liang T.Y., Ling S., Sun E., Wancewicz E., Mazur C., et al. 2011. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 14:459–468 10.1038/nn.2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralser M., Albrecht M., Nonhoff U., Lengauer T., Lehrach H., Krobitsch S. 2005. An integrative approach to gain insights into the cellular function of human ataxin-2. J. Mol. Biol. 346:203–214 10.1016/j.jmb.2004.11.024 [DOI] [PubMed] [Google Scholar]
- Reina C.P., Zhong X., Pittman R.N. 2010. Proteotoxic stress increases nuclear localization of ataxin-3. Hum. Mol. Genet. 19:235–249 10.1093/hmg/ddp482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restituito S., Thompson R.M., Eliet J., Raike R.S., Riedl M., Charnet P., Gomez C.M. 2000. The polyglutamine expansion in spinocerebellar ataxia type 6 causes a beta subunit-specific enhanced activation of P/Q-type calcium channels in Xenopus oocytes. J. Neurosci. 20:6394–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riess O.L., Schöls L., Bottger H., Nolte D., Vieira-Saecker A.M., Schimming C., Kreuz F., Macek M., Jr, Krebsová A., Klockgether T., et al. 1997. SCA6 is caused by moderate CAG expansion in the alpha1A-voltage-dependent calcium channel gene. Hum. Mol. Genet. 6:1289–1293 10.1093/hmg/6.8.1289 [DOI] [PubMed] [Google Scholar]
- Riley B.E., Zoghbi H.Y., Orr H.T. 2005. SUMOylation of the polyglutamine repeat protein, ataxin-1, is dependent on a functional nuclear localization signal. J. Biol. Chem. 280:21942–21948 10.1074/jbc.M501677200 [DOI] [PubMed] [Google Scholar]
- Rolfs A., Koeppen A.H., Bauer I., Bauer P., Buhlmann S., Topka H., Schöls L., Riess O. 2003. Clinical features and neuropathology of autosomal dominant spinocerebellar ataxia (SCA17). Ann. Neurol. 54:367–375 10.1002/ana.10676 [DOI] [PubMed] [Google Scholar]
- Sakurai T., Westenbroek R.E., Rettig J., Hell J., Catterall W.A. 1996. Biochemical properties and subcellular distribution of the BI and rbA isoforms of alpha 1A subunits of brain calcium channels. J. Cell Biol. 134:511–528 10.1083/jcb.134.2.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saluja D., Vassallo M.F., Tanese N. 1998. Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol. Cell. Biol. 18:5734–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterfield T.F., Pallanck L.J. 2006. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum. Mol. Genet. 15:2523–2532 10.1093/hmg/ddl173 [DOI] [PubMed] [Google Scholar]
- Satterfield T.F., Jackson S.M., Pallanck L.J. 2002. A Drosophila homolog of the polyglutamine disease gene SCA2 is a dosage-sensitive regulator of actin filament formation. Genetics. 162:1687–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudou F., Finkbeiner S., Devys D., Greenberg M.E. 1998. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 95:55–66 10.1016/S0092-8674(00)81782-1 [DOI] [PubMed] [Google Scholar]
- Scaglione K.M., Zavodszky E., Todi S.V., Patury S., Xu P., Rodriguez-Lebrón E., Fischer S., Konen J., Djarmati A., Peng J., et al. 2011. Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase CHIP. Mol. Cell. 43:599–612 10.1016/j.molcel.2011.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel H., Tomiuk S., Hofmann K. 2003. Elucidation of ataxin-3 and ataxin-7 function by integrative bioinformatics. Hum. Mol. Genet. 12:2845–2852 10.1093/hmg/ddg297 [DOI] [PubMed] [Google Scholar]
- Sephton C.F., Cenik C., Kucukural A., Dammer E.B., Cenik B., Han Y., Dewey C.M., Roth F.P., Herz J., Peng J., et al. 2011. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J. Biol. Chem. 286:1204–1215 10.1074/jbc.M110.190884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra H.G., Byam C.E., Lande J.D., Tousey S.K., Zoghbi H.Y., Orr H.T. 2004. Gene profiling links SCA1 pathophysiology to glutamate signaling in Purkinje cells of transgenic mice. Hum. Mol. Genet. 13:2535–2543 10.1093/hmg/ddh268 [DOI] [PubMed] [Google Scholar]
- Serra H.G., Duvick L., Zu T., Carlson K., Stevens S., Jorgensen N., Lysholm A., Burright E., Zoghbi H.Y., Clark H.B., et al. 2006. RORalpha-mediated Purkinje cell development determines disease severity in adult SCA1 mice. Cell. 127:697–708 10.1016/j.cell.2006.09.036 [DOI] [PubMed] [Google Scholar]
- Servadio A., Koshy B., Armstrong D.B., Antalffy B., Orr H.T., Zoghbi H.Y. 1995. Expression analysis of the ataxin-1 protein in tissues from normal and spinocerebellar ataxia type 1 individuals. Nat. Genet. 10:94–98 10.1038/ng0595-94 [DOI] [PubMed] [Google Scholar]
- Shah A.G., Friedman M.J., Huang S., Roberts M., Li X.J., Li S. 2009. Transcriptional dysregulation of TrkA associates with neurodegeneration in spinocerebellar ataxia type 17. Hum. Mol. Genet. 18:4141–4152 10.1093/hmg/ddp363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakkottai V.G., do Carmo Costa M., Dell’Orco J.M., Sankaranarayanan A., Wulff H., Paulson H.L. 2011. Early changes in cerebellar physiology accompany motor dysfunction in the polyglutamine disease spinocerebellar ataxia type 3. J. Neurosci. 31:13002–13014 10.1523/JNEUROSCI.2789-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan D.E., Soong B.W., Sun C.M., Lee S.J., Liao K.K., Liu R.S. 2001. Spinocerebellar ataxia type 2 presenting as familial levodopa-responsive parkinsonism. Ann. Neurol. 50:812–815 10.1002/ana.10055 [DOI] [PubMed] [Google Scholar]
- Shibata H., Huynh D.P., Pulst S.M. 2000. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum. Mol. Genet. 9:1303–1313 10.1093/hmg/9.9.1303 [DOI] [PubMed] [Google Scholar]
- Shimohata T., Nakajima T., Yamada M., Uchida C., Onodera O., Naruse S., Kimura T., Koide R., Nozaki K., Sano Y., et al. 2000. Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat. Genet. 26:29–36 10.1038/79139 [DOI] [PubMed] [Google Scholar]
- Tait D., Riccio M., Sittler A., Scherzinger E., Santi S., Ognibene A., Maraldi N.M., Lehrach H., Wanker E.E. 1998. Ataxin-3 is transported into the nucleus and associates with the nuclear matrix. Hum. Mol. Genet. 7:991–997 10.1093/hmg/7.6.991 [DOI] [PubMed] [Google Scholar]
- Takahashi J., Tanaka J., Arai K., Funata N., Hattori T., Fukuda T., Fujigasaki H., Uchihara T. 2001. Recruitment of nonexpanded polyglutamine proteins to intranuclear aggregates in neuronal intranuclear hyaline inclusion disease. J. Neuropathol. Exp. Neurol. 60:369–376 [DOI] [PubMed] [Google Scholar]
- Takeyama K., Ito S., Yamamoto A., Tanimoto H., Furutani T., Kanuka H., Miura M., Tabata T., Kato S. 2002. Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron. 35:855–864 10.1016/S0896-6273(02)00875-9 [DOI] [PubMed] [Google Scholar]
- Taroni F., DiDonato S. 2004. Pathways to motor incoordination: the inherited ataxias. Nat. Rev. Neurosci. 5:641–655 10.1038/nrn1474 [DOI] [PubMed] [Google Scholar]
- Teixeira-Castro A., Ailion M., Jalles A., Brignull H.R., Vilaça J.L., Dias N., Rodrigues P., Oliveira J.F., Neves-Carvalho A., Morimoto R.I., Maciel P. 2011. Neuron-specific proteotoxicity of mutant ataxin-3 in C. elegans: rescue by the DAF-16 and HSF-1 pathways. Hum. Mol. Genet. 20:2996–3009 10.1093/hmg/ddr203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., Konig J., Hortobagyi T., Nishimura A.L., Zupunski V., et al. 2011. Characterizing the RNA targets and position dependent splicing regulation by TDP-43. Nat. Neurosci. 14:452–458 10.1038/nn.2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toru S., Murakoshi T., Ishikawa K., Saegusa H., Fujigasaki H., Uchihara T., Nagayama S., Osanai M., Mizusawa H., Tanabe T. 2000. Spinocerebellar ataxia type 6 mutation alters P-type calcium channel function. J. Biol. Chem. 275:10893–10898 10.1074/jbc.275.15.10893 [DOI] [PubMed] [Google Scholar]
- Toyoshima Y., Yamada M., Onodera O., Shimohata M., Inenaga C., Fujita N., Morita M., Tsuji S., Takahashi H. 2004. SCA17 homozygote showing Huntington’s disease-like phenotype. Ann. Neurol. 55:281–286 10.1002/ana.10824 [DOI] [PubMed] [Google Scholar]
- Tsai C.-C., Kao H.-Y., Mitzutani A., Banayo E., Rajan H., McKeown M., Evans R.M. 2004. Ataxin 1, a SCA1 neurodegenerative disorder protein, is functionally linked to the silencing mediator of retinoid and thyroid hormone receptors. Proc. Natl. Acad. Sci. USA. 101:4047–4052 10.1073/pnas.0400615101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou W.L., Soong B.-W., Paulson H.L., Rodríguez-Lebrón E. 2011. Splice isoform-specific suppression of the CaV2.1 variant underlying spinocerebellar ataxia type 6. Neurobiol. Dis. 43:533–542 10.1016/j.nbd.2011.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H., Jafar-Nejad H., Patel A.J., Sun Y., Chen H.-K., Rose M.F., Venken K.J.T., Botas J., Orr H.T., Bellen H.J., Zoghbi H.Y. 2005. The AXH domain in mammalian/Drosophila Ataxin-1 mediates neurodegeneration in spinocerebellar ataxia 1 through its interaction with Gfi-1/Senseless proteins. Cell. 122:633–644 10.1016/j.cell.2005.06.012 [DOI] [PubMed] [Google Scholar]
- Van Damme P., Veldink J.H., van Blitterswijk M., Corveleyn A., van Vught P.W.J., Thijs V., Dubois B., Matthijs G., van den Berg L.H., Robberecht W. 2011. Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology. 76:2066–2072 10.1212/WNL.0b013e31821f445b [DOI] [PubMed] [Google Scholar]
- van de Leemput J., Chandran J., Knight M.A., Holtzclaw L.A., Scholz S., Cookson M.R., Houlden H., Gwinn-Hardy K., Fung H.C., Lin X., et al. 2007. Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genet. 3:e108 10.1371/journal.pgen.0030108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vig P.J., Subramony S.H., McDaniel D.O. 2001. Calcium homeostasis and spinocerebellar ataxia-1 (SCA-1). Brain Res. Bull. 56:221–225 10.1016/S0361-9230(01)00595-0 [DOI] [PubMed] [Google Scholar]
- Voisine C., Pedersen J.S., Morimoto R.I. 2010. Chaperone networks: tipping the balance in protein folding diseases. Neurobiol. Dis. 40:12–20 10.1016/j.nbd.2010.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Sawai N., Kotliarova S., Kanazawa I., Nukina N. 2000. Ataxin-3, the MJD1 gene product, interacts with the two human homologs of yeast DNA repair protein RAD23, HHR23A and HHR23B. Hum. Mol. Genet. 9:1795–1803 10.1093/hmg/9.12.1795 [DOI] [PubMed] [Google Scholar]
- Wang Q., Li L., Ye Y. 2006. Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J. Cell Biol. 174:963–971 10.1083/jcb.200605100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick J.M., Morabito L.M., Bilen J., Gordesky-Gold B., Faust L.Z., Paulson H.L., Bonini N.M. 2005. Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol. Cell. 18:37–48 10.1016/j.molcel.2005.02.030 [DOI] [PubMed] [Google Scholar]
- Watase K., Barrett C.F., Miyazaki T., Ishiguro T., Ishikawa K., Hu Y., Unno T., Sun Y., Kasai S., Watanabe M., et al. 2008. Spinocerebellar ataxia type 6 knockin mice develop a progressive neuronal dysfunction with age-dependent accumulation of mutant CaV2.1 channels. Proc. Natl. Acad. Sci. USA. 105:11987–11992 10.1073/pnas.0804350105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemeyer R., Westermann F., Wittke I., Nowock J., Schwab M. 2003. Ataxin-2 promotes apoptosis of human neuroblastoma cells. Oncogene. 22:401–411 10.1038/sj.onc.1206150 [DOI] [PubMed] [Google Scholar]
- Winborn B.J., Travis S.M., Todi S.V., Scaglione K.M., Xu P., Williams A.J., Cohen R.E., Peng J., Paulson H.L. 2008. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J. Biol. Chem. 283:26436–26443 10.1074/jbc.M803692200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Mao Q., Eliason S.L., Harper S.Q., Martins I.H., Orr H.T., Paulson H.L., Yang L., Kotin R.M., Davidson B.L. 2004. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med. 10:816–820 10.1038/nm1076 [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Lucas J.J., Hen R. 2000. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell. 101:57–66 10.1016/S0092-8674(00)80623-6 [DOI] [PubMed] [Google Scholar]
- Yue S., Serra H., Zoghbi H.Y., Orr H.T. 2001. The SCA1 protein, ataxin-1, has RNA-binding activity that is inversely affected by the length of its polyglutamine tract. Hum. Mol. Genet. 10:25–30 10.1093/hmg/10.1.25 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Lang G., Ito S., Bonnet J., Metzger E., Sawatsubashi S., Suzuki E., Le Guezennec X., Stunnenberg H.G., Krasnov A., et al. 2008. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell. 29:92–100 10.1016/j.molcel.2007.12.011 [DOI] [PubMed] [Google Scholar]
- Zhuchenko O., Bailey J., Bonnen P., Ashizawa T., Stockton D.W., Amos C., Dobyns W.B., Subramony S.H., Zoghbi H.Y., Lee C.C. 1997. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat. Genet. 15:62–69 10.1038/ng0197-62 [DOI] [PubMed] [Google Scholar]
- Zu T., Duvick L.A., Kaytor M.D., Berlinger M.S., Zoghbi H.Y., Clark H.B., Orr H.T. 2004. Recovery from polyglutamine-induced neurodegeneration in conditional SCA1 transgenic mice. J. Neurosci. 24:8853–8861 10.1523/JNEUROSCI.2978-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zühlke C.H., Spranger M., Spranger S., Voigt R., Lanz M., Gehlken U., Hinrichs F., Schwinger E. 2003. SCA17 caused by homozygous repeat expansion in TBP due to partial isodisomy 6. Eur. J. Hum. Genet. 11:629–632 10.1038/sj.ejhg.5201018 [DOI] [PubMed] [Google Scholar]