Abstract
Cytotoxic T lymphocytes (CTL) induce apoptosis by engaging death receptors or by exocytosis of cytolytic granules containing granzyme (Gzm) proteases and perforin. The lamins, which maintain the structural integrity of the nuclear envelope, are cleaved by caspases during caspase-mediated apoptosis. Although death receptor engagement and GzmB activate caspases, CTL also induce apoptosis during caspase blockade. Both GzmA and GzmB directly and efficiently cleave laminB in vitro, in situ in isolated nuclei and in cells loaded with perforin and Gzms, even in the presence of caspase inhibitors. LaminB is cleaved by GzmA at concentrations of 3 nM, but GzmB is 50 times less active. GzmA cuts laminB at R392; GzmB cuts at the caspase VEVD231 site. Characteristic laminB fragments generated by Gzm proteolysis also are observed during CTL lysis, even in the presence of caspase inhibitors or in cells overexpressing bcl-2. Lamins A/C are direct substrates of GzmA, but not GzmB. GzmA and GzmB therefore directly target critical caspase substrates in caspase-resistant cells.
Cytotoxic T lymphocytes (CTL) and natural killer cells induce apoptosis primarily by exocytosis of specialized granules, which contain perforin (PFP) and Ser proteases, termed granzymes (Gzm). With PFP, either GzmA or GzmB, the most abundant Gzms in CTL, can independently induce cell death (reviewed in ref. 1). GzmB activates the caspase pathway of apoptosis by directly cleaving caspases 3 and 7 (2, 3). However, caspase-independent cell death is also initiated by PFP loading of Gzms or by CTL because cytolysis occurs during caspase blockade (4–7). GzmA initiates rapid caspase-independent death with single-strand DNA breaks but not oligonucleosomal fragmentation (8, 9). Caspase-independent apoptosis is important because many viruses and tumors can evade caspase-mediated cell death.
GzmA is a tryptase and GzmB cleaves after Asp or Met (10–13). Both proteases are highly specific and few intracellular substrates have been identified. GzmA cleaves IL-1β (14), the nucleosome assembly protein called SET, PHAP II, or TAF-Iβ (15), and histones (16). In addition to the effector caspases, GzmB directly cleaves the downstream caspase substrates, bid, nuclear matrix antigen (NuMA), the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs), and the caspase-associated DNase inhibitor (17–19).
The lamin intermediate filament proteins are the main structural components of the lamina underlying the inner nuclear membrane (20). Lamins A, B, and C are present in equal amounts in most mammalian cells. Lamins share a tripartite organization with a conserved central α-helical domain flanked by N- and C-terminal nonhelical domains of variable size and sequence. Lamins A and C are alternate splice products of the same gene. Lamin polymerization is required for nuclear reassembly after mitosis; nuclei assembled in vitro after lamin depletion are fragile. The lamina is a major chromatin-anchoring site of matrix-associated regions during interphase and is dynamically regulated by phosphorylation (21).
Lamin is proteolyzed during apoptosis induced by various agents (22–26). LaminB degradation in apoptotic thymocytes precedes DNA fragmentation (25). The sites of lamin A and B cleavage during caspase-mediated apoptosis were mapped to a conserved Asp residue in a consensus caspase cleavage sequence. LaminA is proteolysed by caspase 6 at the site VEID230 (26–28). The analogous caspase cleavage site in laminB VEVD231 also fits the consensus caspase 6 sequence (29). Overexpression of caspase-resistant lamin mutants or inhibition of lamin cleavage delays DNA fragmentation and nuclear condensation (26, 30). A nuclear lamina-associated chymotryptase, the Ca2+-regulated nuclear scaffold protease (CRNSP), possibly a proteosome component, also plays a role in lamin cleavage during apoptosis (30–33). Because disruption of the nuclear envelope may be essential, even for caspase-independent apoptosis, we investigated the fate of laminB after exposure to GzmA and GzmB in vitro, in isolated nuclei, and in intact cells. LaminB is a direct substrate of both Gzms, but lamins A/C are cleaved only by GzmA.
Materials and Methods
Cell Lines.
EL-4, K562, and Jurkat cells were grown in K medium (RPMI medium 1640 with 10% FCS/2 mM Glu/2 mM Hepes/100 units/ml penicillin/100 μg/ml streptomycin/50 μM β-mercaptoethanol); HeLa cells were grown in supplemented DMEM. Peptide-specific human CTL lines were generated by stimulating HLA A2.1 + peripheral blood mononuclear cells with 5 μg/ml human cytomegalovirus (CMV) antigenic peptide (NLVPMVATV; Tufts Core Facility, Boston) for 1 h at 37°C. Two days later 1 unit/ml IL-2 was added, and the cells were then fed every 2 days with IL-2-containing medium for 2 wk. Peptide-specific mouse CTL lines were generated similarly by peptide stimulation of RBC-lysed splenocytes from P14 mice transgenic for the T cell antigen receptor recognizing lymphocytic choriomeningitis virus gp33–41 (The Jackson Laboratory) (34) by using KAVYNFATM peptide (Tufts Core Facility). Human lymphokine-activated killer (LAK) cells were peripheral blood mononuclear cells cultured for 2 wk in K medium with 1,000 units/ml IL-2 (Chiron Oncology, Emeryville CA). HeLa cells were stably transfected with puromycin resistance or bcl-2 expression plasmid pJ436, a gift of J. Yuan, Harvard Medical School, Boston (35). Bcl-2 was overexpressed at least 100-fold more than in control plasmid transfected cells by immunoblot.
Antibodies and Reagents.
The 3,4 dichloroisocoumarin (DCI) and caspase inhibitors Z-DEVD-FMK, Z-VAD-FMK, and Ac-YAVD-CHO were from Calbiochem-Novabiochem (San Diego). Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK) and Nα-p-tosyl-l-phenylalanine chloromethyl ketone (TPCK) were from Sigma. A specific inhibitor of CRNSP, Z-APF-CMK, was from Bachem (King of Prussia, PA). Immunoblots were probed with mouse mAb 101-B7 to laminB (Oncogene Research Products, Cambridge MA), goat polyclonal Ab to C-terminal laminB peptide (Santa Cruz Biotechnology), mouse mAb to laminA/C [gift of F. McKeon, Harvard Medical School, Boston (20)], goat polyclonal glutathione S-transferase (GST) Ab (Amersham Pharmacia), rabbit bcl-2 Ab, rabbit ref-1 Ab (Santa Cruz Biotechnology), or mouse PHAPI mAb, generated in our laboratory. Ph-NHCONH-CiEtOIC, a GzmA-specific inhibitor (GAI), was a gift of J. Powers, Georgia Institute of Technology, Atlanta (15). Experiments performed under caspase inhibition (CI) were in the presence of 100 μM each Z-DEVD-FMK and Z-VAD-FMK, unless otherwise indicated. A mixture of protease inhibitors [50 μg/ml antipain/2 μg/ml aprotinin/40 μg/ml bestatin/60 μg/ml chymostatin/10 μg/ml E-64/1 μg/ml leupeptin/1 μg/ml pepstatin/1 mg/ml 4-(2-aminoethyl)benzenesulfonyl fluoride (Sigma)/200 μM GAI] was added at the end of assay incubations to terminate proteolysis. The same concentrations were used in inhibitor experiments. pET-30b(+) vector and BL21-competent cells were from Novogen. Plasmid DNA was prepared with the Qiagen (Chatsworth, CA) plasmid DNA purification kit. Protein concentrations were determined by bicinchoninic acid assay (Pierce).
Gzms and PFP.
Recombinant GzmA, inactive S-AGzmA, and GzmB were produced and PFP was purified from RNK-16 cells as described (8, 15, 36, 37). For loading, PFP was added at a sublytic concentration, determined for each cell as that needed to induce ≤10% lysis in 2 h (8). For each reaction 1 × 105 cells in 60 μl of Hanks' balanced salt solution with 1 mg/ml BSA, 1 mM CaCl2, and 1 mM MgCl2 were incubated with indicated amounts of Gzms and PFP at 37°C for indicated times. After adding protease inhibitors, cells were lysed in a 5× SDS-loading buffer.
Recombinant LaminB.
Sequences used for lamin self-assembly in the N and C termini were removed to produce soluble-truncated laminB corresponding to amino acids 143–551. Recombinant protein was engineered with C-terminal GST by using laminB cDNA (38) from K. Pollard (Scripps Clinic, La Jolla, CA). GST DNA, PCR amplified from pGEM-2K (Amersham Pharmacia), was inserted into the pET-30b(+) plasmid through NotI- and XhoI-cloning sites. Truncated LaminB sequence was PCR amplified by using primers containing BamHI and NotI restriction sites and cloned into the reconstructed plasmid through EcoRI- and NotI-cloning sites. LaminB-GST plasmid was used to transform BL21 cells, expression was induced with isopropyl β-d-thiogalactoside, and protein was purified by using glutathione Sepharose 4B (Amersham Pharmacia).
In Vitro Cleavage Assay.
LaminB-GST (1 μg) was incubated at 37°C for 1 h with indicated amounts of Gzms in 25 μl of 25 mM Tris⋅HCl (pH 8.0), 140 mM NaCl, and 5 mM glutathione. The reaction was stopped by adding DCI to 50 μM and 5 μl of 5× SDS-loading reducing buffer. Boiled samples were electrophoresed through SDS/PAGE gels and analyzed by immunoblot or staining with GelCode Blue (Pierce). Cleavage fragments blotted onto poly(vinylidene difluoride) membranes were analyzed for N-terminal sequence (Tufts Core Facility).
Cleavage Assay in Isolated Nuclei.
HeLa nuclei were isolated after cell lysis in 25 mM KCl, 20 mM Tris⋅HCl (pH 7.5), 5 mM MgCl2, and 0.2% Nonidet P-40 and washed twice with lysis buffer and once with lysis buffer without Nonidet P-40. Nuclei (1 × 106 in 100 μl of lysis buffer without Nonidet P-40) were incubated with indicated amounts of Gzms at 37°C for indicated times. The reaction was stopped by adding protease inhibitors and 5 × SDS-loading buffer.
CTL Assays.
Human targets were preincubated (1 × 105 in 50 μl of K medium) for 30 min at 37°C with 5 μg/ml Con A, before adding 5 × 105 human LAK or CMV-specific CTLs in 50 μl of K medium. Alternatively, mouse EL-4 cells (5 × 105 in 50 μl of K medium) were preincubated for 30 min at 37°C with 5 μg/ml gp33 peptide before adding 2.5 × 105 mouse CTLs in 50 μl of K medium. Cells were vortexed, pulse microfuged, and incubated for indicated times at 37°C. Boiled cell lysates were analyzed by immunoblot.
Immunoblot.
After electrophoresis through reducing SDS/PAGE gels, samples were transferred to nitrocellulose, probed with primary Ab at room temperature overnight, and washed three times with TBS containing 0.05% Tween-20. After secondary Ab incubation at room temperature for 3 h, blots were washed five times and developed with Luminol/Enhancer solution (Pierce). Cleavage was quantitated relative to the signal for ref-1-loading controls by using chemi doc and quanone software (Bio-Rad).
Immunofluorescence.
K562 cells, mixed with Gzms and/or PFP in the presence or absence of CI, were plated onto polyLys-coated slides and incubated at 37°C for 1 h. Cells, fixed on slides in methanol for 10 min at −20°C and air-dried, were washed twice with PBS and incubated at room temperature for 1 h with 5 μg/ml mAb 101-B 7, 50 μg/ml normal goat IgG and 100 μg/ml RNase I. After washing with PBS, slides were incubated with FITC-conjugated goat α-mouse IgG (DAKO, Carpinteria CA) at room temperature for 45 min and then soaked for 5 min in PBS containing 0.1 μg/ml propidium iodide. After further washes, slides were mounted by using the ProLong Antifade Kit (Molecular Probes) and examined with a Bio-Rad Radiance 2000 laser-scanning confocal microscope. Images were acquired by focusing on the central plane of each cell, defined by the greatest cell diameter and brightest staining.
Results
PFP Loading of Cells with GzmA or GzmB Induces LaminB Cleavage.
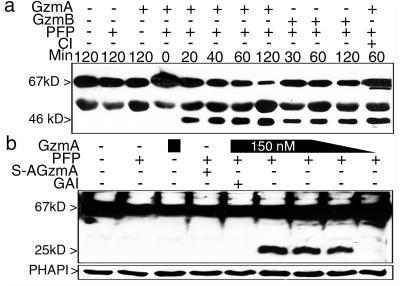
We first verified that laminB is cleaved when recombinant GzmB is PFP loaded into K562 cells (Fig. 1a). LaminB, a 586-aa protein, migrates with an apparent molecular mass of 67 kDa. Caspase cleavage after VEVD231 yields a 46-kDa C-terminal fragment visualized with mAb 101-B7. GzmA loading also cuts laminB to generate a similarly sized fragment. Because the GzmA fragment of laminB is the same size as the caspase cleavage product, we repeated the GzmA-loading experiment in the presence of CI. LaminB cleavage by GzmA was unimpaired by CI. GzmA-loaded cells also were analyzed with polyclonal Ab to the C terminus of laminB (Fig. 1b). The cleavage induced by GzmA occurs at an independent site to generate a 25-kDa C-terminal fragment compared to the 46-kDa C-terminal fragment in GzmB-treated cells (Fig. 2). Therefore the 46-kDa GzmA-cleaved fragment corresponds to the N terminus, whereas the 46-kDa caspase-cleaved fragment corresponds to the C terminus of laminB. Enzymatically inactive S-AGzmA does not induce laminB cleavage.
Figure 1.
PFP loading of GzmA or GzmB induces laminB cleavage. (a) GzmA or GzmB (1 μg) were loaded with PFP into K562 cells at 37°C for indicated times. Electrophoresed samples were probed with laminB mAb (101-B7), which recognizes full-length laminB and a 46-kDa cleaved product in cells treated with either GzmA or GzmB, as well as an unknown cross reactive protein migrating at 50 kDa. GzmA laminB cleavage was not blocked by preincubation with CI, maintained throughout the experiment. (b) GzmA loading of K562 cells generates a 25-kDa C-terminal laminB fragment at 90 min, visualized by using polyclonal Ab against the C terminus of laminB. Reprobing for the GzmA-interacting protein PHAPI (15) controlled for loading.
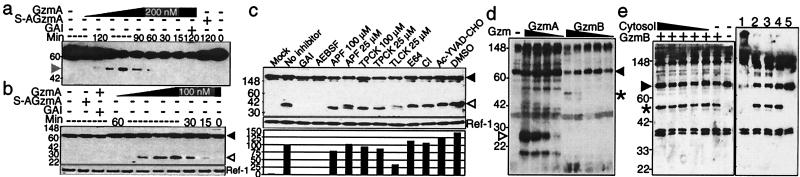
Figure 2.
GzmA and GzmB cleave laminB in isolated nuclei independently of caspase activation. (a and b) Four-fold dilutions of GzmA ranging from 0.4 to 200 nM were added to isolated HeLa nuclei for the indicated times. Full-length laminB is indicated by a black arrowhead, the C-terminal 25-kDa laminB fragment by a white arrowhead, and the N-terminal 46-kDa fragment by a gray arrowhead. LaminB degradation requires the active Ser protease because S-AGzmA does not induce cleavage, and the reaction is blocked by GAI. (c) Ser protease inhibitors, but not caspase or other protease inhibitors, reduce lamin cleavage in HeLa nuclei by GzmA. HeLa nuclei were incubated for 1 h with nothing (mock) or with 100 nM GzmA in the presence of no inhibitor, DMSO, or the indicated inhibitors. Blot was reprobed for the nuclear protein Ref-1 as a loading control to quantitate enzymatic cleavage. The percentage of cleavage with inhibition was normalized to that with uninhibited enzyme. (d) GzmB induces laminB degradation in isolated nuclei less efficiently than GzmA. The 46-kDa C-terminal fragment after GzmB cleavage (*) is different from the 25-kDa fragment generated by GzmA cleavage (white arrowhead). Serial 4-fold dilutions of GzmA (starting with 200 nM) or GzmB (starting with 310 nM) were added for 2 h. LaminB is fragmented with 3 nM GzmA but requires at least 155 nM GzmB. (e) LaminB cleavage in GzmB-treated nuclei is not enhanced by adding cytosol (1 × 10 7 cell equivalents at highest concentration followed by 4-fold serial dilutions) (Left). The Ser protease inhibitor DCI (lane 1), but not CI (lane 2) or DMSO (lane 3), blocks lamin cleavage by GzmB (Right). GzmB without inhibitors is shown in lane 4 and nuclei without GzmB in lane 5. Blot in a was probed with mAb 101-B7 and b--e with polyclonal laminB Ab.
GzmA Cleaves LaminB in Isolated Nuclei.
To determine whether laminB cleavage occurs in situ, we treated isolated HeLa nuclei with GzmA (Fig. 2 a and b). Identically sized 25- and 46-kDa cleavage products were visualized in isolated nuclei treated with as little as 3–12 nM GzmA beginning at 15 min. When the in situ assay was carried out in the presence of protease inhibitors, only Ser protease inhibitors reduced laminB cleavage (Fig. 2c). Inhibitors of the CRNSP, Z-APF-CMK, and Nα-p-tosyl-l-phenylalanine (TPCK) did not significantly block cleavage by GzmA. This, together with the fact that the reactions were carried out in Ca2+-free buffer, effectively rules out the involvement of CRNSP in GzmA-mediated laminB cleavage. Because treatment of isolated nuclei leads to cleavage, no cytoplasmic component is required for laminB proteolysis. These findings suggest that laminB is a direct GzmA substrate.
GzmB Cleaves LaminB in Nuclei Independently of Caspase Activation.
The caspase cleavage site of laminB (VEVD) is also an acceptable sequence for GzmB recognition in peptide library screening (29). To investigate whether GzmB also might cleave laminB directly, we treated HeLa nuclei with limiting dilutions of GzmA or GzmB. GzmB induces laminB cleavage in isolated nuclei less efficiently than GzmA (Fig. 2d). LaminB is cut by 3 nM GzmA, but the GzmB-induced fragment is not seen until 155 nM of enzyme is present. GzmB cleavage of laminB occurs even in the presence of CI, although the Ser protease inhibitor DCI blocks the cleavage (Fig. 2e). The addition of cytosol as a source of caspases does not enhance in situ cleavage, suggesting that direct cleavage occurs more efficiently and rapidly than via indirect activation of the caspase pathway. These findings suggest that GzmB cleaves laminB directly without caspase activation or involvement of other cytosolic components.
LaminB Is a Direct Substrate of GzmA and GzmB.
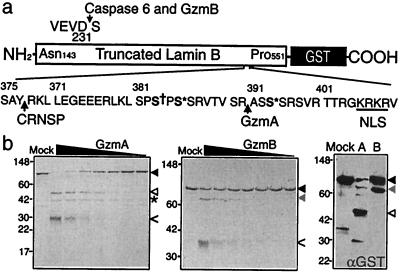
To test whether laminB is a direct substrate and identify the Gzm cleavage sites, we treated recombinant truncated laminB-GST fusion protein with recombinant Gzms (Fig. 3). (39) GzmA completely degraded the 88-kDa fusion protein to produce two dominant bands of 46 and 42 kDa. The 46-kDa band, which reacts with anti-GST Ab and is therefore the C-terminal fragment, was analyzed by N-terminal sequencing. R392 in the sequence VTVSR ASS is the GzmA target. The GzmA site is contained in a Ser-rich region, phosphorylated by protein kinase C and p34cdc2 (21, 40). It is also near the cleavage site at amino acid 377 recognized by CRNSP and is just proximal to the nuclear localization signal at amino acids 405–408. GzmB cuts the fusion protein to a 76-kDa C-terminal fragment, whose N-terminal sequence identifies the GzmB cleavage site after VEVD231, the caspase 6 site. Because mAb 101-B7 recognizes the N-terminal fragment (amino acids 1–392) after GzmA cleavage and the C-terminal fragment (amino acids 232–586) after GzmB cleavage, this mAb recognizes a determinant within amino acids 231–392 of laminB.
Figure 3.
LaminB-GST is cleaved in vitro by GzmA and GzmB. (a) LaminB (amino acids 143–551)-GST fusion protein was produced in Escherichia coli. Known sites of laminB cleavage by caspases after D231 and by the CRNSP after Y377 are indicated, as are the nuclear localization signal (NLS, underlined), and sites of phosphorylation by p34cdc2 (S† 383) and protein kinase C (S* 385, 395). Figure modified from ref. 39. (b) LaminB-GST protein (1 μg, black arrowhead) was incubated for 1 h at 37°C with 2-fold dilutions of GzmA (Left) or GzmB (Middle) beginning at a highest concentration of 1 μg and analyzed by SDS/PAGE and GelCode stain. Gzm bands are indicated by <. GzmA and GzmB generated cleavage fragments of laminB, indicated by white and gray arrowheads, respectively, were analyzed for N-terminal sequence. The * marks the 42-kDa N-terminal laminB fragment generated by GzmA; the N-terminal GzmB cleavage fragment was not visualized. The sequenced fragments contain the C terminus of the fusion protein because they stain with anti-GST by immunoblot (Right). The GzmA cleavage site is after R392 and the GzmB cleavage site is the known caspase site after D231. GzmA is more efficient than GzmB at cleaving laminB-GST.
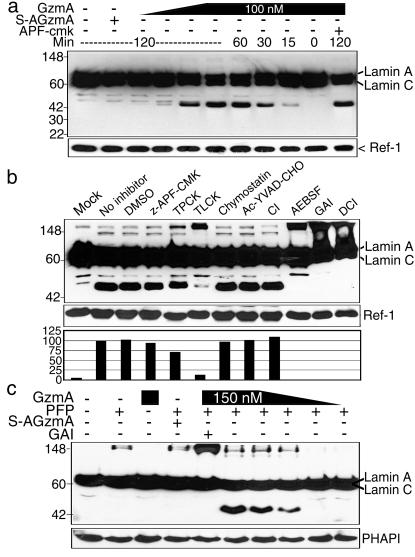
LaminsA/C Are Cleaved in Isolated Nuclei or Target Cells Loaded with GzmA.
Lamins A and C are alternate splice products of a gene with considerable homology to laminB. However the cleavage sites in laminB for caspase/GzmB and GzmA are modified in laminA/C from VEVD231 to VEID230 and from TVSR392 to SRGR401, respectively. We therefore looked to see whether GzmA loading of isolated HeLa nuclei results in laminA/C cleavage (Fig. 4 a and b). Both lamin A and C are degraded by GzmA in situ with similar kinetics and concentration requirement for cleavage as laminB. Moreover, laminA/C cleavage by GzmA is inhibited by Ser protease inhibitors, but not by CI or CRNSP inhibitors. LaminsA/C are not cleaved in situ by GzmB (not shown). Cleavage of laminsA/C was also evident in K562 cells PFP loaded with GzmA (Fig. 4c).
Figure 4.
LaminA/C are degraded in isolated HeLa nuclei by GzmA. GzmA-treated nuclei were analyzed by immunoblot probed with anti-laminA/C mAb and Ab to ref-1 as a control for loading. (a) LaminsA/C are degraded with similar kinetics as laminB (Fig. 2). Four-fold dilutions of GzmA from 1.5 to 100 nM were added for the indicated times. Cleavage is blocked by GAI and is not induced by inactive S-AGzmA. GzmB does not induce laminA/C cleavage (not shown). (b) Ser protease inhibitors, but not CI, reduce GzmA cleavage of laminA/C. HeLa nuclei were incubated for 1 h with nothing (mock) or with 100 nM GzmA in the presence of no inhibitor, DMSO, or the indicated inhibitors. The percentage of cleavage in the presence of inhibitors compared to uninhibited enzyme is graphed. (c) LaminA/C cleavage is detected 90 min after PFP loading 4-fold dilutions of active GzmA (0.2–150 nM) into K562 cells.
LaminB Is Fragmented During CTL Attack Even in the Presence of CI or bcl-2 Overexpression.
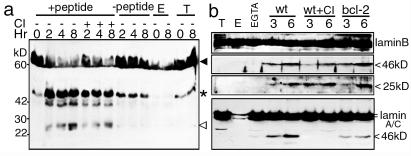
The ultimate test of the physiological relevance of a Gzm substrate is whether it is cleaved during CTL-mediated lysis. We therefore analyzed cell lysates, obtained after CTL-target cell incubation, by immunoblot probed with the polyclonal anti-laminB Ab. Murine lymphocytic choriomeningitis virus peptide-specific CTLs were mixed with peptide-decorated EL4 targets (Fig. 5a). Within 2 h, laminB C-terminal fragments of 46 kDa, characteristic of caspase/GzmB cleavage, and 25 kDa, characteristic of GzmA cleavage, were detected by immunoblot in peptide-treated targets but not in cultures to which no peptide was added. Minor degradation fragments of laminB of ≈42 kDa also were detected. The 25-kDa cleavage product of SET, another GzmA substrate (15), was also first detected around 2 h (not shown). When cells were mixed in the presence of EGTA to inhibit granule exocytosis, no laminB fragments were seen (not shown). Similar results were found in five independent experiments performed with effector:target ratios ranging from 0.5:1 to 2:1. When the CTL attack was carried out in the presence of CI, the expected GzmA and GzmB cleavage fragments were still present at comparable levels.
Figure 5.
LaminB is cleaved during CTL attack even in the presence of CI or bcl-2 overexpression. (a) Mouse peptide-specific CTL were incubated for the indicated times with an equal number of EL4 cells, preincubated with either medium or the cognate peptide. Lamin cleavage products expected for cleavage by GzmB (46 kDa, *) and GzmA (25 kDa, white arrowhead) are detected with Ab against the C terminus of laminB only in targets incubated with antigen. Although there is a background cross-reactive band in the target cells just below the expected 46-kDa caspase/GzmB fragment, the laminB cleavage signal is clearly enhanced in the presence of specific antigenic peptide. Additional minor laminB cleavage products of ≈42 kDa are also produced specifically during CTL attack. When target cells are preincubated with CI, maintained during the CTL attack, the cleavage products are still clearly seen. E and T indicate lysates of just effector CTL or target cells, respectively. (b) ConA-treated HeLa cells or HeLa cells overexpressing bcl-2 were incubated with human CMV-specific CTL and assayed for lamin cleavage by immunoblot and probed with polyclonal anti-laminB Ab or laminA/C. When granule exocytosis is blocked with EGTA, lamins are not cut. In the presence of Ca2+, both expected GzmA and caspase/GzmB fragments are detected. Bcl-2 overexpression does not inhibit lamin B cleavage and CI only partly reduces the appearance of the 46-kDa caspase/GzmB product. Bcl-2 and CI inhibit caspase-mediated laminA/C cleavage.
Results with mouse CTLs were tested with human CTL effectors against Con A-sensitized HeLa (Fig. 5b) or K562 targets (not shown). LaminB C-terminal fragments at 46 kDa (expected for GzmB/caspase) and 25 kDa (expected for GzmA) were detected within 3 h of initiating CTL attack. LaminB cleavage was not detected in the presence of EGTA or when the reaction was carried out in the presence of 10–50 μM DCI (not shown). Complete CI, verified by the absence of caspase-3 activation (not shown), only partly inhibits production of the 46-kDa caspase/GzmB fragment of laminB. Bcl-2 overexpression in HeLa cells does not inhibit the production of either fragment of laminB. Similar results were found in three experiments with LAK effector cells in place of CMV-specific CTLs (not shown). Therefore, in human cells, like mouse cells, direct caspase-independent GzmB cleavage of laminB occurs efficiently during CTL attack. However, production of the 46-kDa laminA/C caspase cleavage product was partially inhibited by bcl-2 expression and completely inhibited by CI. This provides additional evidence that laminA/C is not a direct substrate of GzmB. Despite the unambiguous detection of laminA/C cleavage after PFP loading of GzmA into K562 cells even in the presence of CI (not shown), no laminA/C fragments were detected after CTL or LAK attack of HeLa cells in the presence of CI.
Disruption of the Nuclear Lamina After Gzm Loading Visualized by Immunofluoresence Microscopy.
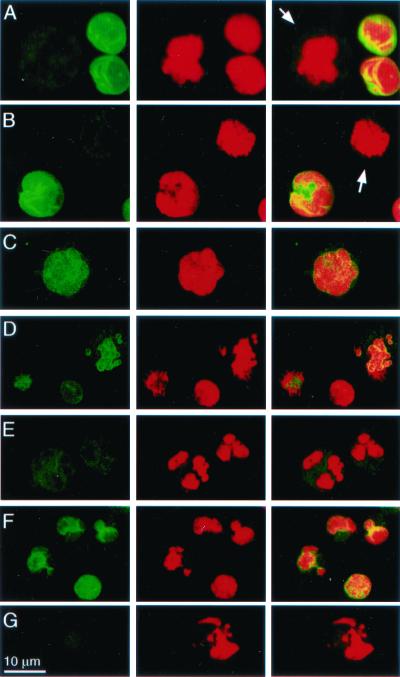
Lamin cleavage should disturb the cohesiveness of the nuclear lamina. We therefore looked at propidium iodide and laminB staining of K562 cells 1 h after PFP-loading Gzms (Fig. 6). No significant change in lamin staining occurs after treatment with PFP or Gzms (not shown) alone. However, after treatment with PFP and GzmA, laminB nuclear staining is greatly reduced, especially in cells with the most chromatin condensation. Confocal images show some heterogeneity of laminB staining, which probably reflects different snapshots along the path to apoptosis. The earliest change is coalescence of staining into a punctate pattern, followed by fainter staining as the chromatin condenses and the nuclei fragment. Some laminB also disperses to the cytoplasm. GzmB causes similar changes in laminB staining, although the extent of lamin degradation is less pronounced than after GzmA loading. Altered laminB staining after GzmB loading occurs even with CI.
Figure 6.
The nuclear lamina in K562 cells is disrupted after PFP-loading Gzms. K562 cells, mock-treated (A), or treated with PFP alone (B) or with PFP and GzmA (C–E) or GzmB (F and G) for 1 h, were stained with FITC-anti-laminB mAb 101-B7 (Left) or propidium iodide (Center); overlay (Right). Arrows in A and B indicate dissolution of the lamina and dispersion of laminB to the cytoplasm in mitotic cells. (C–E) Progressive changes in laminB degradation after GzmA treatment. Before chromatin changes are evident (C), lamin staining becomes punctate. As chromatin condensation and nuclear fragmentation progress, the nuclear lamina dissolves and lamin staining diminishes and disperses to the cytoplasm. K562 cells loaded with GzmB exhibit less pronounced changes in laminB staining. These changes persist even with CI (G).
Discussion
The lamins are cleaved by the Gzms directly in a physiologically significant manner during PFP loading or CTL-mediated lysis. GzmA is more efficient than GzmB at direct laminB proteolysis. However, disruption of the lamina after PFP loading of either GzmA or GzmB is clearly seen by immunofluorescence microscopy, even with CI. GzmA targets all of the lamins, but GzmB does not directly cleave lamins A/C, in agreement with a report (41) using in vitro transcribed and translated laminA/C. The GzmB cleavage site in laminB (VEVD) is altered to VEID in laminA/C. Peptide library screening predicts that this site in laminA/C is a good target for caspase 6, as has been shown experimentally. However, Ile in the P2 position is not recognized for cleavage by GzmB, nor are there alternate GzmB recognition sequences in laminA/C. The preferred recognition site for GzmA cleavage is unknown. The GzmA target sequence (TVSR ASSS) at R392 in laminB shares a basic residue at the P1 site, but has no other obvious similarity to the reported sites in IL-1β (APVR SLNC), the thrombin receptor (LDPR SFLL) (14), or the histones (H1: LGLK SLVS; H2b: PAPK KGSK) (16).
Granzyme proteolysis of lamins occurs in the presence of CI or in target cells that overexpress bcl-2. This is consistent with the fact that the GzmA apoptotic pathway is caspase-independent (8) and that cytolysis induced by GzmB occurs in the presence of caspase blockade (4–8). In human K562 (not shown) or HeLa targets, laminB is cleaved in a caspase-independent manner directly by GzmB because it is not blocked by CI or by bcl-2 overexpression. In human HeLa cells we did not detect laminA/C cleavage in the presence of CI, despite the fact that GzmA cleavage of laminA/C in situ and in PFP-loaded K562 cells is caspase-independent. However, the relative activity of the Gzm pathways, or of parts of the pathway, in different cells can vary. For example, we found little single-stranded DNA nicking and no detectable laminB cleavage in Jurkat cells loaded with GzmA, although they were efficiently lysed (not shown and ref. 8). Some of the variation in detection of lamin cleavage in different target cells might also be due to the technical challenge of detecting transient degradation products during CTL attack, in which most of the remaining cells in the analyzed lysates are effectors, which are resistant to cytolysis, and not targets.
Lamin cleavage during both caspase-dependent and -independent apoptosis suggests that nuclear lamina disruption is critical for apoptosis. Because chromatin is anchored to the lamins, lamin proteolysis may be required before chromatin can be adequately targeted by apoptotic DNases. In fact, lamin cleavage occurs before oligonucleosomal DNA fragmentation, and overexpression of caspase-resistant lamins delays nuclear condensation and DNA fragmentation during caspase-mediated apoptosis (24–26, 42). Moreover chromatin matrix attachment sites separate DNA into fragments of ≈50 kb, the size of the earliest DNA fragments cleaved in apoptosis (43), and the first DNA breaks during apoptosis are in the nuclear periphery near sites of chromatin attachment to the lamina (44).
The granzymes concentrate in the nucleus during CTL-mediated cell death. How they get into the nucleus is unclear because they lack recognized nuclear localization signals and translocation is ATP-independent (45, 46). One intriguing possibility is that disruption of the nuclear lamina by the granzymes facilitates nuclear entry because nuclear pore complexes are anchored to the nuclear membrane via the lamins.
Several reports have implicated the CRNSP in laminB degradation during apoptosis (30–33). This chymotryptase cleaves laminB at Y377 (31), near the GzmA cleavage site at R392, to produce a ≈21-kDa C-terminal fragment. The activation of CRNSP is blocked in apoptotic thymocytes by CI, bcl-2 overexpression, or chymotryptase inhibitors (32). Therefore, the chymotryptase is activated secondarily to caspase activation. Moreover, DNA fragmentation in apoptotic thymocytes is blocked by Nα-p-tosyl-l-phenylalanine (TPCK). This suggests that laminB cleavage by CRNSP facilitates DNA degradation in caspase-mediated apoptosis (33). During CTL lysis, GzmA cleavage of laminB may substitute for the endogenous CRNSP chymotryptase. Involvement of CRNSP in the direct lamin cleavage by Gzms has been ruled out by performing the in situ and in vitro experiments in Ca2+-free buffer and by the lack of effect of CRNSP inhibitors.
Caspase-independent apoptosis can be activated by CTLs or ceramide (4–6, 8, 47). A systematic study of the known substrates of the effector caspases during caspase-independent cell death may help distinguish substrates whose proteolysis is essential for apoptosis from incidental targets. The recent report of direct cleavage of bid, NuMA and DNA-PKcs by GzmB (17, 19) suggests that cleavage of these proteins, like laminB, may be required for completing the apoptotic program. Analysis of the fate of these proteins during GzmA loading may help determine whether this is indeed the case.
Acknowledgments
We thank D. Meshulam and Z. Xu for excellent technical support, K. Pollard for laminB cDNA, J. Yuan for bcl-2 plasmid, J. Powers for GAI, F. McKeon for laminA/C mAb, Chiron Oncology for IL-2, and P. Shankar and N. Manjunath for helpful suggestions. This work was supported by National Institutes of Health Grant AI45587.
Abbreviations
- CTL
cytotoxic T lymphocyte
- Gzm
granzyme
- PFP
perforin
- CI
caspase inhibitors
- CRNSP
Ca2+-regulated nuclear scaffold protease
- LAK
lymphokine-activated killer cell
- GAI
GzmA-specific inhibitor Ph-NHCONH-CiEtOIC
- GST
glutathione S-transferase
- DCI
3,4 dichloroisocoumarin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Henkart P A. Immunity. 1994;1:343–346. doi: 10.1016/1074-7613(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 2.Darmon A J, Nicholson D W, Bleackley R C. Nature (London) 1995;377:446–448. doi: 10.1038/377446a0. [DOI] [PubMed] [Google Scholar]
- 3.Yang X, Stennicke H R, Wang B, Green D R, Janicke R U, Srinivasan A, Seth P, Salvesen G S, Froelich C J. J Biol Chem. 1998;273:34278–34283. doi: 10.1074/jbc.273.51.34278. [DOI] [PubMed] [Google Scholar]
- 4.Sarin A, Williams M S, Alexander-Miller M A, Berzofsky J A, Zacharchuk C M, Henkart P A. Immunity. 1997;6:209–215. doi: 10.1016/s1074-7613(00)80427-6. [DOI] [PubMed] [Google Scholar]
- 5.Sarin A, Haddad E K, Henkart P A. J Immunol. 1998;161:2810–2816. [PubMed] [Google Scholar]
- 6.Sutton V R, Vaux D L, Trapani J A. J Immunol. 1997;158:5783–5790. [PubMed] [Google Scholar]
- 7.Anel A, Gamen S, Alava M A, Schmitt-Verhulst A M, Pineiro A, Naval J. J Immunol. 1997;158:1999–2006. [PubMed] [Google Scholar]
- 8.Beresford P J, Xia Z, Greenberg A H, Lieberman J. Immunity. 1999;10:585–594. doi: 10.1016/s1074-7613(00)80058-8. [DOI] [PubMed] [Google Scholar]
- 9.Shresta S, Graubert T A, Thomas D A, Raptis S Z, Ley T J. Immunity. 1999;10:595–605. doi: 10.1016/s1074-7613(00)80059-x. [DOI] [PubMed] [Google Scholar]
- 10.Pasternack M S, Eisen H N. Nature (London) 1985;314:743–745. doi: 10.1038/314743a0. [DOI] [PubMed] [Google Scholar]
- 11.Bleackley R C, Lobe C G, Duggan B, Ehrman N, Fregeau C, Meier M, Letellier M, Havele C, Shaw J, Paetkau V. Immunol Rev. 1988;103:5–20. doi: 10.1111/j.1600-065x.1988.tb00746.x. [DOI] [PubMed] [Google Scholar]
- 12.Jenne D E, Tschopp J. Immunol Rev. 1988;103:53–71. doi: 10.1111/j.1600-065x.1988.tb00749.x. [DOI] [PubMed] [Google Scholar]
- 13.Pham C T, Thomas D A, Mercer J D, Ley T J. J Biol Chem. 1998;273:1629–1633. doi: 10.1074/jbc.273.3.1629. [DOI] [PubMed] [Google Scholar]
- 14.Irmler M, Hertig S, MacDonald H R, Sadoul R, Becherer J D, Proudfoot A, Solari R, Tschopp J. J Exp Med. 1995;181:1917–1922. doi: 10.1084/jem.181.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beresford P J, Kam C M, Powers J C, Lieberman J. Proc Natl Acad Sci USA. 1997;94:9285–9290. doi: 10.1073/pnas.94.17.9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D, Pasternack M S, Beresford P J, Wagner L, Greenberg A H, Lieberman J. J Biol Chem. 2001;276:3683–3690. doi: 10.1074/jbc.M005390200. [DOI] [PubMed] [Google Scholar]
- 17.Andrade F, Roy S, Nicholson D, Thornberry N, Rosen A, Casciola-Rosen L. Immunity. 1998;8:451–460. doi: 10.1016/s1074-7613(00)80550-6. [DOI] [PubMed] [Google Scholar]
- 18.Thomas D A, Du C, Xu M, Wang X, Ley T J. Immunity. 2000;12:621–632. doi: 10.1016/s1074-7613(00)80213-7. [DOI] [PubMed] [Google Scholar]
- 19.Barry M, Heibein J A, Pinkoski M J, Lee S F, Moyer R W, Green D R, Bleackley R C. Mol Cell Biol. 2000;20:3781–3794. doi: 10.1128/mcb.20.11.3781-3794.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKeon F D, Kirschner M W, Caput D. Nature (London) 1986;319:463–468. doi: 10.1038/319463a0. [DOI] [PubMed] [Google Scholar]
- 21.Peter M, Heitlinger E, Haner M, Aebi U, Nigg E A. EMBO J. 1991;10:1535–1544. doi: 10.1002/j.1460-2075.1991.tb07673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufmann S H. Cancer Res. 1989;49:5870–5878. [PubMed] [Google Scholar]
- 23.Ucker D S, Meyers J, Obermiller P S. J Immunol. 1992;149:1583–1592. [PubMed] [Google Scholar]
- 24.Oberhammer F A, Hochegger K, Froschl G, Tiefenbacher R, Pavelka M. J Cell Biol. 1994;126:827–837. doi: 10.1083/jcb.126.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neamati N, Fernandez A, Wright S, Kiefer J, McConkey D J. J Immunol. 1995;154:3788–3795. [PubMed] [Google Scholar]
- 26.Rao L, Perez D, White E. J Cell Biol. 1996;135:1441–1455. doi: 10.1083/jcb.135.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi A, Alnemri E S, Lazebnik Y A, Fernandes-Alnemri T, Litwack G, Moir R D, Goldman R D, Poirier G G, Kaufmann S H, Earnshaw W C. Proc Natl Acad Sci USA. 1996;93:8395–8400. doi: 10.1073/pnas.93.16.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orth K, Chinnaiyan A M, Garg M, Froelich C J, Dixit V M. J Biol Chem. 1996;271:16443–16446. [PubMed] [Google Scholar]
- 29.Thornberry N A, Rano T A, Peterson E P, Rasper D M, Timkey T, Garcia-Calvo M, Houtzager V M, Nordstrom P A, Roy S, Vaillancourt J P, et al. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 30.Lazebnik Y A, Takahashi A, Moir R D, Goldman R D, Poirier G G, Kaufmann S H, Earnshaw W C. Proc Natl Acad Sci USA. 1995;92:9042–9046. doi: 10.1073/pnas.92.20.9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clawson G A, Norbeck L L, Hatem C L, Rhodes C, Amiri P, McKerrow J H, Patierno S R, Fiskum G. Cell Growth Differ. 1992;3:827–838. [PubMed] [Google Scholar]
- 32.McConkey D J. J Biol Chem. 1996;271:22398–22406. doi: 10.1074/jbc.271.37.22398. [DOI] [PubMed] [Google Scholar]
- 33.Zhivotovsky B, Gahm A, Orrenius S. Biochem Biophys Res Commun. 1997;233:96–101. doi: 10.1006/bbrc.1997.6411. [DOI] [PubMed] [Google Scholar]
- 34.Pircher H, Burki K, Lang R, Hengartner H, Zinkernagel R M. Nature (London) 1989;342:559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 35.Miura M, Zhu H, Rotello R, Hartwieg E A, Yuan J. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 36.Xia Z, Kam C M, Huang C, Powers J C, Mandle R J, Stevens R L, Lieberman J. Biochem Biophys Res Commun. 1998;243:384–389. doi: 10.1006/bbrc.1998.8102. [DOI] [PubMed] [Google Scholar]
- 37.Shi L, Kam C M, Powers J C, Aebersold R, Greenberg A H. J Exp Med. 1992;176:1521–1529. doi: 10.1084/jem.176.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollard K M, Chan E K L, Grant B J, Sullivan K F, Tan E M, Glass C A. Mol Cell Biol. 1990;10:2164–2175. doi: 10.1128/mcb.10.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimizu T, Cao C-X, Shao R-G, Pommier Y. J Biol Chem. 1998;273:8669–8674. doi: 10.1074/jbc.273.15.8669. [DOI] [PubMed] [Google Scholar]
- 40.Hocevar B A, Burns D J, Fields A P. J Biol Chem. 1993;268:7545–7552. [PubMed] [Google Scholar]
- 41.Pinkoski M J, Heibein J A, Barry M, Bleackley R C. Cell Death Differ. 2000;7:17–24. doi: 10.1038/sj.cdd.4400604. [DOI] [PubMed] [Google Scholar]
- 42.Weaver V M, Carson C E, Walker P R, Chaly N, Lach B, Raymond Y, Brown D L, Sikorska M. J Cell Sci. 1996;109:45–56. doi: 10.1242/jcs.109.1.45. [DOI] [PubMed] [Google Scholar]
- 43.Oberhammer F, Wilson J W, Dive C, Morris I D, Hickman J A, Wakeling A E, Walker P R, Sikorska M. EMBO J. 1993;12:3679–3684. doi: 10.1002/j.1460-2075.1993.tb06042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krystosek A. Histochem Cell Biol. 1999;111:265–276. doi: 10.1007/s004180050357. [DOI] [PubMed] [Google Scholar]
- 45.Trapani J A, Browne K A, Smyth M J, Jans D A. J Biol Chem. 1996;271:4127–4133. doi: 10.1074/jbc.271.8.4127. [DOI] [PubMed] [Google Scholar]
- 46.Blink E J, Trapani J A, Jans D A. Immunol Cell Biol. 1999;77:206–215. doi: 10.1046/j.1440-1711.1999.00817.x. [DOI] [PubMed] [Google Scholar]
- 47.Metkar S S, Anand M, Manna P P, Naresh K N, Nadkarni J J. Exp Cell Res. 2000;255:18–29. doi: 10.1006/excr.1999.4773. [DOI] [PubMed] [Google Scholar]