FT-INTERACTING PROTEIN 1 is a novel protein that is involved in transporting florigen, a long-known mobile signal that induces flowering in plants in response to day length, from companion cells to sieve elements in the phloem of Arabidopsis.
Abstract
The capacity to respond to day length, photoperiodism, is crucial for flowering plants to adapt to seasonal change. The photoperiodic control of flowering in plants is mediated by a long-distance mobile floral stimulus called florigen that moves from leaves to the shoot apex. Although the proteins encoded by FLOWERING LOCUS T (FT) in Arabidopsis and its orthologs in other plants are identified as the long-sought florigen, whether their transport is a simple diffusion process or under regulation remains elusive. Here we show that an endoplasmic reticulum (ER) membrane protein, FT-INTERACTING PROTEIN 1 (FTIP1), is an essential regulator required for FT protein transport in Arabidopsis. Loss of function of FTIP1 exhibits late flowering under long days, which is partly due to the compromised FT movement to the shoot apex. FTIP1 and FT share similar mRNA expression patterns and subcellular localization, and they interact specifically in phloem companion cells. FTIP1 is required for FT export from companion cells to sieve elements, thus affecting FT transport through the phloem to the SAM. Our results provide a mechanistic understanding of florigen transport, demonstrating that FT moves in a regulated manner and that FTIP1 mediates FT transport to induce flowering.
Author Summary
The transition to flowering is the most dramatic phase change in flowering plants and is crucial for reproductive success. Such a transition from vegetative to reproductive growth is controlled by seasonal changes in day length. Studies originally performed in the 1930s were the first to suggest that day length is perceived by a plant's leaves; by contrast, flower formation takes place in the shoot apical meristem (the tip of the shoot that gives rise to plant organs, such as leaves and flowers). The term “florigen” was later proposed to describe a mobile floral stimulus that moves from leaves to the shoot apical meristem to induce flowering. It is only recently that FLOWERING LOCUS T (FT) in Arabidopsis, and its orthologs in various other plant species, was identified as being florigen, but how florigen is transported in plants remains completely unknown. Here, we report that a novel ER membrane protein, FT-INTERACTING PROTEIN 1 (FTIP1), interacts with FT in companion cells of the phloem (a specialized type of parenchyma cell in the phloem of the plant's vascular system) and mediates FT protein movement from companion cells to sieve elements (the conducting cells of the phloem), thus affecting FT transport to the shoot apical meristem in Arabidopsis. To our knowledge, this study reveals the first regulator that is required for florigen transport and offers new insights into possible florigen transport mechanisms in other flowering plants.
Introduction
The transition to flowering, which is crucial for the reproductive success, is the most dramatic phase change in flowering plants. Plants are able to adjust the timing of this transition in response to environmental conditions, such as photoperiod, temperature, and availability of nutrients. Classic experiments on the photoperiodic control of flowering in various plants have demonstrated that plant response to day length begins with the perception of photoperiod in leaves, followed by the transmission of a floral stimulus into the shoot apical meristem (SAM), where flowers are generated instead of leaves. Such mobile floral stimulus moving from leaves to the SAM was proposed as “florigen” in the 1930s [1]. Since then, tremendous efforts have been made to understand the molecular nature of this signal. Recent findings have suggested that the proteins encoded by FLOWERING LOCUS T (FT) in Arabidopsis and its orthologs in other plant species are part of the long-sought florigen [2]–[6].
FT encodes a member of the phosphatidylethanolamine-binding protein family and acts as a crucial regulator that relays flowering signals from the photoperiod pathway to floral meristem identity genes in Arabidopsis, which is a long-day (LD) facultative plant [7]–[10]. Under LDs, FT mRNA expression is activated by the CONSTANS (CO) transcriptional regulator in the vascular tissues of leaves and displays circadian rhythm [8],[11]–[14]. It has been suggested that long-distance movement of FT protein from leaves to the shoot apex through the phloem system plays a role in floral induction [2],[4],[5]. In the SAM, FT interacts with the bZIP transcription factor FLOWERING LOCUS D (FD), which in turn activates the downstream floral meristem identity genes such as APETALA1 (AP1) to initiate flower development [8],[9]. Despite the remarkable progress in elucidating FT function, it is so far completely unknown whether and how FT protein transport is regulated. As the abundance of native FT protein is too low to be detectable, it has been hypothesized that simple diffusion of FT protein from companion cells to sieve elements might not be sufficient for transporting FT to the SAM [15].
Here we show that an endoplasmic reticulum (ER) membrane protein, FT-INTERACTING PROTEIN 1 (FTIP1), is required for FT protein transport in Arabidopsis. Loss of function of FTIP1 exhibits late flowering under LDs, which is partly due to the compromised FT movement to the SAM. FTIP1 and FT have similar mRNA expression patterns and subcellular localization, and they interact in vivo in phloem companion cells. Furthermore, FTIP1 is required for FT export from companion cells to sieve elements, thus affecting FT transport through the phloem to the SAM. Our results provide a mechanistic understanding of florigen transport and demonstrate that FT protein moves in a regulated manner and that FTIP1 is involved in mediating the export of FT protein from phloem companion cells to induce flowering.
Results
FTIP1 Regulates Flowering Time under LDs
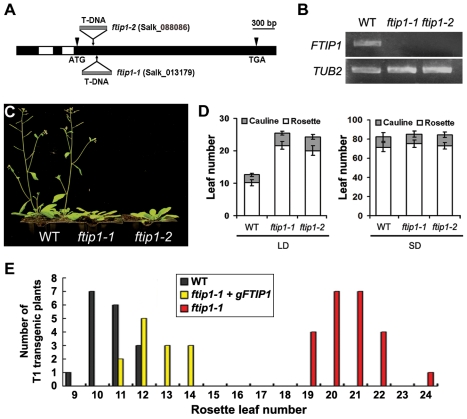
To understand how FT function is regulated, we performed yeast two-hybrid screening to identify proteins that interact with FT. Approximately 3 million yeast transformants were screened and 66 colonies were identified on the selective medium (Table S1), among which a partial sequence belonging to an unknown protein with three C2 domains and one phosphoribosyltransferase C-terminal domain (PRT_C) was isolated (Figure S1). The corresponding gene (At5g06850) was therefore named FT-INTERACTING PROTEIN 1 (FTIP1). We isolated two T-DNA insertional alleles, ftip1-1 (Salk_013179) and ftip1-2 (Salk_088086), from Arabidopsis Biological Resource Center (Figure 1A). The full-length FTIP1 transcript was undetectable in either homozygous mutant (Figure 1B). Both ftip1-1 and ftip1-2 flowered late under LDs, but not under short days (SDs) (Figure 1C,D; Table 1), suggesting that FTIP1 plays a role in mediating the effect of photoperiod on flowering. We transformed ftip1-1 with a genomic construct (gFTIP1) harboring a 5.1-kb FTIP1 genomic region including 2.1 kb of the upstream sequence, the 2.4-kb coding sequence, and 0.6 kb of the downstream sequence (Figure S2A). Most ftip1-1 gFTIP1 T1 transformants exhibited similar or slightly late flowering time as compared to wild-type plants (Figure 1E), demonstrating that FTIP1 is responsible for promoting flowering particularly under LDs.
Figure 1. FTIP1 promotes flowering under LDs.
(A) Schematic diagram showing the FTIP1 coding region and T-DNA insertion mutants. Exons and introns are indicated by black and white boxes, respectively. Two T-DNA insertion lines, ftip1-1 and ftip1-2, were obtained from Arabidopsis Biological Resource Center. (B) FTIP1 expression is undetectable in ftip1-1 or ftip1-2 by semi-quantitative PCR using the primers flanking T-DNA insertion sites (Table S2). (C) ftip1-1 and ftip1-2 show later flowering than wild-type plants at 35 d after germination under LDs. (D) Flowering time of ftip1-1 and ftip1-2 grown under LDs and SDs. Error bars indicate SD. (E) Distribution of flowering time in T1 transgenic plants carrying the FTIP1 genomic fragment (Figure S2A) in ftip1-1 background.
Table 1. Flowering time of transgenic and mutant plants.
| Genotypea | Number of Rosette Leavesb | Number of Cauline Leavesb | n |
| Experiment 1 | |||
| Wild type | 9.8±1.0 (8–11) | 2.5± 0.5 (2–3) | 25 |
| ftip1-1 | 21.8±1.0 (19–24) | 4.2±0.7 (3–5) | 20 |
| ftip1-2 | 20.2±1.3 (18–23) | 4.5±0.7 (3–6) | 20 |
| soc1-2 | 24.2±1.3 (22–26) | 4.6±0.8 (4–6) | 16 |
| soc1-2 ftip1-1 | 40.8±1.5 (39–43) | 8.0±0.9 (7–9) | 10 |
| co-1 | 18.2±1.3 (16–20) | 5.4±0.8 (4–6) | 20 |
| co-1 ftip1-1 | 34.7±3.7 (29–40) | 7.6±0.5 (7–8) | 15 |
| gi-1 | 48.7±4.2 (44–55) | 8.6±1.3 (7–11) | 14 |
| gi-1 ftip1-1 | 55.3±7.3 (43–64) | 10.0±2.2 (8–14) | 15 |
| ft-1 | 44.3±6.2 (36–54) | 8.8±1.3 (7–11) | 16 |
| ft-1 ftip1-1 | 59.2±2.9 (56–64) | 11.8±1.9 (9–14) | 14 |
| ft-10 | 51.8±3.0 (48–56) | 9.8±1.6 (8–12) | 15 |
| ft-10 ftip1-1 | 60.8±6.0 (52–67) | 11.6±2.0 (9–14) | 14 |
| Experiment 2 | |||
| Wild type | 10.2±1.1 (8–11) | 2.3±0.4 (2–3) | 20 |
| ftip1-1 | 21.6±1.0 (19–24) | 4.1±0.7 (3–5) | 20 |
| ftip1-1 gFTIP1 #3 (T3) | 10.4±1.1 (9–12) | 2.3±0.7 (2–3) | 20 |
| ftip1-1 gFTIP1 #11 (T3) | 10.3±1.1 (8–12) | 2.4±0.6 (2–3) | 20 |
| SUC2:FTIP1 ftip1-1 | 11.1±1.6 (9–13) | 2.4±0.4 (2–3) | 15 |
| FTIP1:4HA:FTIP1 ftip1-1 | 11.1±1.8 (9–13) | 2.4±0.4 (2–3) | 15 |
| FTIP1:FTIP1:GFP ftip1-1 | 12.9±1.3 (11–15) | 2.8±0.8 (2–4) | 15 |
| SUC2:FTIP1:GFP ftip1-1 | 13.1±1.6 (11–15) | 2.4±0.6 (2–4) | 15 |
| Experiment 3 | |||
| Wild type | 9.5±1.0 (8–11) | 2.6±0.5 (2–3) | 20 |
| ft-10 tsf-1 | 66.7±4.2 (59–70) | 10.0±1.4 (8–12) | 15 |
| ft-10 tsf-1 ftip1-1 | 67.3±3.5 (62–71) | 10.9±1.3 (9–13) | 13 |
| Experiment 4 | |||
| Wild type | 9.5±1.1 (8–11) | 2.5±0.6 (2–3) | 20 |
| ftip1-1 | 22.5±1.2 (19–24) | 3.9±0.9 (3–5) | 20 |
| KNAT1:FT | 6.6±0.8 (5–8) | 2.2±0.4 (2–3) | 25 |
| KNAT1:FT ftip1-1 | 6.9±1.0 (5–9) | 2.3±0.5 (2–3) | 25 |
| KNAT1:FT:GFP | 9.2±0.9 (7–11) | 2.5±0.2 (2–3) | 25 |
| KNAT1:FT:GFP ftip1-1 | 9.5±1.1 (8–11) | 2.7±0.4 (2–3) | 25 |
| SUC2:FT:GFP | 8.7±0.8 (7–10) | 2.8±0.6 (2–4) | 30 |
| SUC2:FT:GFP ft-10 | 21.5±1.4 (18–25) | 4.4±0.7 (3–6) | 24 |
| SUC2:FT:GFP ftip1-1 | 12.1±0.8 (11–13) | 3.3±0.5 (3–4) | 30 |
| SUC2:FT c | 3.1±0.4 (2–4) | 1.7±0.4 (1–2) | 30 |
| SUC2:FT ftip1-1 c | 3.7±0.6 (3–5) | 1.8±0.5 (1–3) | 30 |
| SUC2:FT ft-10 | 5.3±0.7 (4–6) | 1.9±0.6 (1–2) | 30 |
| SUC2:GFP:CO d | 4.0±0.4 (3–5) | 1.4±0.5 (1–2) | 30 |
| SUC2:GFP:CO ftip1-1 d | 4.8±0.4 (4–6) | 1.5±0.5 (1–2) | 30 |
| Experiment 5 | |||
| Wild type | 10.2±1.0 (8–11) | 2.7±0.5 (2–3) | 20 |
| SUC2:FT:9myc | 7.5±0.6 (6–9) | 2.5±0.5 (2–4) | 25 |
| SUC2:FT:9myc ft-10 | 20.4±1.3 (18–23) | 4.0±0.4 (3–6) | 20 |
| SUC2:FT:9myc ftip1-1 | 15.2±0.8 (14–17) | 4.3±0.6 (3–5) | 25 |
| Experiment 6 | |||
| Wild type | 10.8±1.2 (8–11) | 2.6±0.5 (2–3) | 20 |
| 35S:FTIP1 (line 2) | 16.2±0.7 (15–18) | 3.3±0.5 (3–4) | 16 |
| SUC2:FT:GFP 35S:FTIP1 | 12.6±2.2 (10–15) | 2.4±0.5 (2–3) | 13 |
All of the plants are in the same Columbia background and grown under LDs.
Flowering time is presented as average ± standard deviation (range).
The flowering time of SUC2:FT and SUC2:FT ftip1-1 is statistically different (p = 7.2×10−4).
The flowering time of SUC2:GFP:CO and SUC2:GFP:CO ftip1-1 is statistically different (p = 4.1×10−11). Statistical analyses were performed using a two-tailed unpaired Student's t test.
Gene Expression and Subcellular Localization of FTIP1
We tested FTIP1 expression in various tissues of wild-type plants using quantitative real-time PCR and found its highest expression in leaves and stems (Figure S3). To examine the detailed expression pattern of FTIP1, we generated a FTIP1:β-glucuronidase (GUS) reporter construct in which the same 2.1-kb FTIP1 upstream sequence included in gFTIP1 for the gene complementation test was fused to the GUS reporter gene (Figure S2A). We created 23 independent FTIP1:GUS lines, most of which showed similar GUS staining patterns. A representative line was selected to monitor the detailed expression pattern of FTIP1. FTIP1:GUS showed specific GUS staining in vascular tissues of various plant organs (Figure S2B–H). Notably, in developing seedlings during the floral transition occurring 7 d after germination, FTIP1:GUS and FT:GUS [14] shared similar GUS staining patterns in vascular tissues of cotyledons and rosette leaves, although the former had a relatively broad and intensive staining pattern (Figure 2A). A cross-section of a primary leaf vein revealed that FTIP1:GUS expression was specifically located in the phloem including companion cells (Figure 2B), which is similar to the FT:GUS expression pattern [14]. Neither FTIP1:GUS nor FT:GUS was expressed in the SAM (Figure 2C,D; Figure S2C) [14]. Furthermore, the late-flowering phenotype of ftip1-1 was rescued by the expression of FTIP1 coding sequence driven by the promoter of SUCROSE TRANSPORTER 2 (SUC2) (Figure 3A), which is active specifically in phloem companion cells [16]. These results suggest that FTIP1 functions in the phloem to promote flowering.
Figure 2. FTIP1 is expressed in vascular tissues.
(A) Comparison of GUS staining of FTIP1:GUS and FT:GUS grown under LDs for 7 to 11 d. (B) Cross-section of the primary vein of the first rosette leaf from an 11-d-old FTIP1:GUS seedling. Ph, phloem; Xy, xylem; SE, sieve element; CC, companion cell. Bar, 50 µm. (C) Longitudinal section through an 11-d-old FTIP1:GUS seedling. Bar, 100 µm. (D) A higher magnification of the area within the box indicated in (C). Asterisk indicates the shoot apical meristem.
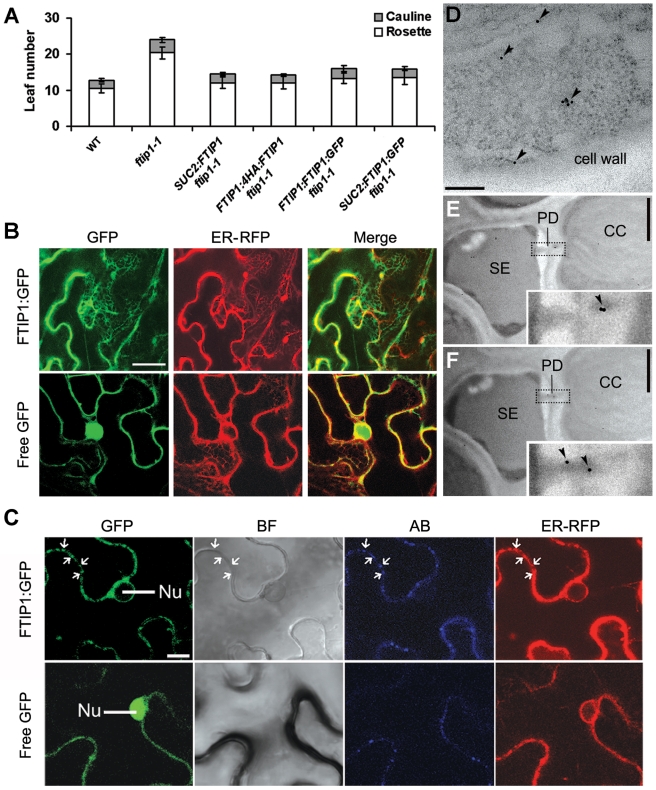
Figure 3. Subcellular localization of FTIP1.
(A) Flowering time of various transgenic plants grown under LDs. Error bars indicate SD. (B and C) Subcellular localization of FTIP1:GFP and free GFP in N. benthamiana leaf epidermal cells. (B) As compared to free GFP, FTIP1:GFP is mostly colocalized with an ER marker. (C) Both FTIP1:GFP and callose are enriched in the same regions at the cell wall (arrows). GFP, GFP fluorescence; ER-RFP, RFP fluorescence of an ER marker [17]; Merge, merge of GFP and RFP; BF, bright field image; AB, aniline blue staining; Nu, nucleus. Bars: (B), 20 µm; (C), 10 µm. (D–F) Analysis of 4HA:FTIP1 localization in CC-SE complexes in the first rosette leaves of 15-d-old FTIP1:4HA:FTIP1 ftip1-1 by immunogold electron microscopy. (D) 4HA:FTIP1 is localized in the phloem companion cell. Arrowheads indicate the locations of gold particles. (E,F) 4HA:FTIP1 is localized in the plasmodesma that connects a CC with a SE in two continuous sections. Arrowheads in insets show the location of gold particles in enlarged PD regions. SE, sieve element; CC, companion cell; PD, plasmodesma. Bars: (D), 250 nm; (E and F), 1 µm.
Given that FTIP1 functions in flowering time control, we investigated whether its expression is regulated by known flowering genetic pathways. FTIP1 expression was not regulated by photoperiod and did not exhibit an obvious circadian rhythm under LDs (Figure S4A,E). Similarly, vernalization treatment did not affect FTIP1 expression (Figure S4B), and GA treatment did not affect FTIP1 expression and the flowering phenotype of ftip1-1 (Figure S4C,D). In addition, FTIP1 expression was also not altered in several mutants tested in known flowering genetic pathways (Figure S5). These observations imply that flowering signals may not regulate FTIP1 function through affecting its mRNA levels.
Next, we examined the subcellular localization of FTIP1 through monitoring the signal of the green fluorescent protein (GFP) fused with FTIP1 under the control of FTIP1 or SUC2 promoter, respectively. Both constructs could rescue the late flowering phenotype of ftip1-1 (Figure 3A). However, we could not detect fluorescent signal from either SUC2:FTIP1:GFP ftip1-1 or FTIP1:FTIP1:GFP ftip1-1 transgenic lines, indicating that FTIP1 protein might be present at very low abundance in plant cells. Alternatively, we transiently expressed 35S:FTIP1:GFP with various fluorescent protein-tagged organelle markers in N. benthamiana leaf epidermal cells and found that FTIP1:GFP was mostly colocalized with an endoplasmic reticulum (ER) marker (Figure 3B,C; Figure S6) [17]. We did not observe FTIP1:GFP signals in the nucleus (Figure 3C). Notably, at the cell wall, FTIP1:GFP colocalized with callose deposition stained with aniline blue, which marks the position of plasmodesmata (Figure 3C).
To precisely localize FTIP1, we performed immunoelectron microscopy on an FTIP1:4HA:FTIP1 ftip1-1 transgenic line, in which FTIP1:4HA:FTIP1 was able to rescue the flowering defect of ftip1-1 (Figure 3A). The result revealed that 4HA:FTIP1 was specifically localized in phloem companion cells (Figure 3D) and plasmodesmata between companion cells and sieve elements (Figure 3E,F; Figure S7), where the ER membrane runs through.
FTIP1 Interacts with FT in Phloem Companion Cells
Several pieces of evidence, including the initial identification of FTIP1 as an FT interacting partner, similar tissue expression pattern of FTIP1 and FT, and similar late-flowering phenotype exhibited by ftip1 and ft mutants specifically under long days, point to a possible role of FTIP1 in mediating FT function in the control of flowering time. Thus, we further carried out a detailed analysis of the interaction between FTIP1 and FT. As revealed in our yeast two-hybrid screening, a truncated FTIP1 protein devoid of the PRT_C domain interacted with FT in both yeast two-hybrid and GST pull-down assays (Figure 4A–C), whereas no interaction was detected using the full-length FTIP1 (unpublished data). Since the PRT_C domain of FTIP1 was predicted to be a membrane-targeted domain according to a protein topology analysis (Figure S1), the full-length FTIP1 protein might not be in the membrane-bound state in yeast cells or under in vitro conditions and thus might undergo inappropriate folding, which prevents its interaction with FT. Alternatively, in yeast two-hybrid assay the full-length FTIP1 protein might be membrane-bound and unable to reconstitute a functional transcription factor in the yeast nucleus to drive the reporter gene expression.
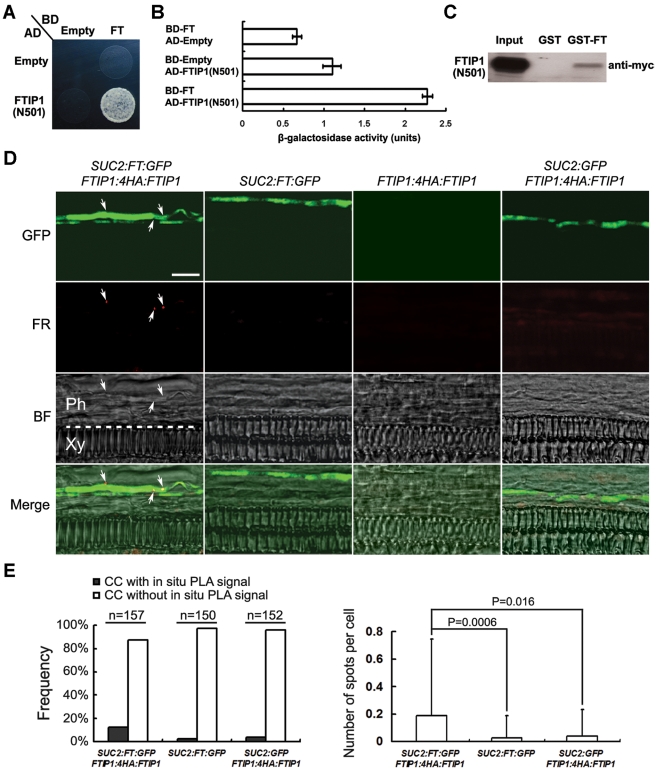
Figure 4. FTIP1 interacts with FT.
(A) Yeast two-hybrid assay of interaction between FT and the N-terminal region of FTIP1 (aa 1–501; N501), which contains three C2 domains (Figure S1A). Yeast cells were grown on SD-His/-Trp/-Leu medium supplemented with 30 mM 3-amino-1, 2, 4-triazole. (B) Quantification of the interaction between FT and FTIP1 (N501) in yeast by β-galactosidase assays. (C) In vitro pull-down assay of interaction between FT and FTIP1 (N501). “Input” indicates 5% of myc-labeled FTIP1 (N501) subjected to pull-down by GST and GST-FT. (D) In situ PLA detection of interaction between FT:GFP and 4HA:FTIP1 in phloem companion cells of an 11-d-old Arabidopsis leaf. Protein-protein interactions are visualized as small red spots indicated by arrows. The dotted line indicates the border between phloem and xylem. GFP, GFP fluorescence; FR, far red fluorescence; BF, bright field image; Merge, merge of GFP, FR, and BF; Ph, phloem; Xy, xylem. Bar, 10 µm. (E) Quantification of in situ PLA data. Statistical analysis was performed by counting the number of far red fluorescence signals (red spots) in the phloem companion cells that could be identified with the GFP signal. The left panel shows the frequency histogram of appearance of red spots found in phloem companion cells. The number of sections examined for each genotype is listed above the histogram. The right panel shows the average number of red spots per phloem companion cell. Statistical analysis was performed using a two-tailed unpaired Student's t test. The results are considered statistically significant at p<0.05.
We transiently expressed 35S:FTIP1:GFP with 35S:FT:RFP in N. benthamiana leaf epidermal cells and revealed that both FTIP1:GFP and FT:RFP were colocalized to ER connected to the nuclear envelope (Figure S8). However, in contrast to FTIP1:GFP, FT:RFP was also localized in the nucleus, which is consistent with a previous observation [9]. These results indicate that FTIP1 may not directly mediate FT function in transcriptional regulation of other target genes.
To test whether and how FT interacts with FTIP1 in vivo, we performed in situ Proximity Ligation Assay (PLA) [18], in which dual recognition of target proteins by pairs of affinity probes generates an amplifiable DNA reporter molecule that serves as a surrogate marker for interacting proteins, to examine the subcellular localization of FT and FTIP1 interaction at single-molecule resolution in the leaves of 11-d-old SUC2:FT:GFP; FTIP1:4HA:FTIP1 transgenic plants. PLA signals visualized as small red dots were specifically detected in the phloem companion cells of SUC2:FT:GFP; FTIP1:4HA:FTIP1, but barely in those transgenic plants containing only SUC2:FT:GFP, FTIP1:4HA:FTIP1, or SUC2:GFP; FTIP1:4HA:FTIP1 (Figure 4D,E). This result demonstrates that FT and FTIP1 physically interact in close proximity in phloem companion cells.
FTIP1 Controls the Export of FT Protein from Phloem Companion Cells to Sieve Elements
The findings on the interaction between FT and FTIP1, and FTIP1 localization to ER and plasmodesmata prompted us to hypothesize that FTIP1 may regulate FT export from phloem companion cells. To this end, we first examined whether FTIP1 affects FT transport to the SAM during the floral transition. We generated a SUC2:FT:GFP transgenic line as previously described [2]. As this transgenic allele could significantly rescue the late-flowering phenotype of the FT null mutant, ft-10 (Table 1), we further crossed this SUC2:FT:GFP allele with ftip1-1 and 35S:FTIP1. Confocal analysis of the distribution of FT:GFP fusion protein revealed that in 11-d-old seedlings, which were undergoing the floral transition, FT:GFP was clearly detected in the inner cone-like region of the SAM in wild-type background, but not in ftip1-1 (Figure 5A). In contrast, the distribution of free GFP protein was comparably undetectable in the inner region of the SAM in wild-type and ftip1-1 (Figure S9A), indicating a specific effect of FTIP1 on FT:GFP distribution in the SAM during the floral transition. In agreement with the above observations, SUC2:FT:GFP ftip1-1 flowered later than SUC2:FT:GFP (Table 1). Since the abundance of FT:GFP mRNA and protein in SUC2:FT:GFP was not downregulated in ftip1-1 (Figure 6A–C), the difference in FT:GFP distribution in the SAM between wild-type and ftip1-1 plants suggests a role of FTIP1 in regulating FT transport rather than FT mRNA or protein abundance.
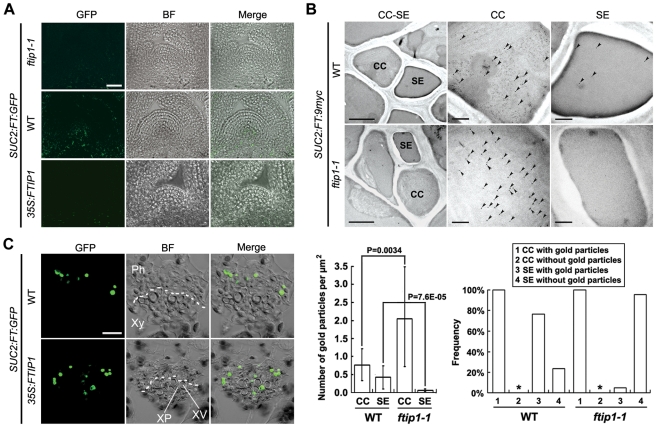
Figure 5. FTIP1 is required for FT protein transport.
(A) Confocal analysis of FT:GFP protein distribution in the apical region of 11-d-old SUC2:FT:GFP seedlings in different genetic backgrounds. Bar, 20 µm. (B) Analysis of FT:9myc distribution in CC-SE complexes in the first rosette leaves of 15-d-old SUC2:FT:9myc and SUC2:FT:9myc ftip1-1 seedlings by immunogold electron microscopy using anti-myc antibody. The upper left panels show the representative CC-SE complexes, while higher magnification views of CCs or SEs are shown in the upper middle or right panels, respectively. Arrowheads indicate the locations of gold particles. The lower left panel shows the quantification of FT:9myc immunogold signals in CCs and SEs of SUC2:FT:9myc (WT background) or SUC2:FT:9myc ftip1-1 (ftip1-1 background). The data are presented as the mean number of gold particles per µm2 plus or minus standard deviation. Statistical analysis was performed using a two-tailed unpaired Student's t test. The results are considered statistically significant at p<0.05. The lower right panel shows the frequency histogram of appearance of FT:9myc immunogold signals in CCs and SEs in all examined sections of SUC2:FT:9myc or SUC2:FT:9myc ftip1-1. Asterisks indicate that in all sections examined, the frequency we observed CCs without gold particles is zero. Bars: upper left panels, 2 µm; upper middle and right panels, 0.5 µm. (C) Confocal analysis of FT:GFP protein distribution in the primary vein of the first rosette leaves from 11-d-old SUC2:FT:GFP seedlings in different genetic backgrounds. The dotted lines indicate the approximate boarder between xylem and phloem. GFP, GFP fluorescence; BF, bright field image; Merge, merge of GFP and BF; CC, companion cell; SE, sieve element; Ph, phloem; Xy, xylem; XP, xylem parenchyma; XV, xylem vessel. Bar, 20 µm.
Figure 6. FTIP1 does not regulate FT mRNA or protein stability.
(A) Schematic diagrams showing native FT and transgenic SUC2:FT:GFP transcripts. The fragments labeled with a, b, and c indicate amplicons in real-time PCR analyses shown in (B). Fragments a, b, and c were amplified with primers FT-F and FT-R, FT(UTR)-F and FT(UTR)-R, and GFP-F1 and GFP-R1 (Table S1), respectively. (B) Examination of steady-state levels of FT or FT:GFP mRNA in wild-type and ftip1-1 backgrounds. Amplification of fragment a, which detects the amplicon in both native FT and transgenic FT:GFP transcripts, shows that native FT expression is downregulated in ftip1-1 versus wild-type, whereas total FT expression (including minor native FT and major transgenic FT:GFP expression) remains unchanged in SUC2:FT:GFP ftip1-1 versus SUC2:FT:GFP. Although the former indicates that FTIP1 affects the steady-state levels of native FT expression, the latter implies that FTIP1 does not directly affect FT mRNA stability. Amplification of fragment c, which only detects the amplicon in transgenic FT:GFP transcripts, further supports that FTIP1 does not directly affect FT mRNA stability as transgenic FT:GFP expression is not changed in ftip1-1. Amplification of fragment b, which only detects the amplicon in native FT transcripts, shows that native FT expression is also downregulated in SUC2:FT:GFP transgenic plants. 9-d-old seedlings grown under LDs were harvested for expression analysis by quantitative real-time PCR. Results were normalized against the expression of TUB2. Asterisks indicate that the expression of fragment c was undetectable in wild-type and ftip1-1 seedling. Error bars indicate SD. (C) Western blot analysis using anti-GFP antibody shows the comparable abundance of FT:GFP protein in wild-type and ftip1-1 plants. Ponceau S staining of the membrane is used as a loading control. (D) GUS staining of rosette leaves of 9-d-old FT:GUS and FT:GUS SUC2:FT seedling.
As FTIP1 was expressed in the phloem (Figure 2) and its protein was localized in phloem companion cells (Figure 3D–F; Figure 4D), we examined whether FTIP1 affects FT transport from companion cells to sieve elements in a newly created SUC2:FT:9myc line in wild-type and ftip1-1 backgrounds using immunoelectron microscopy (Figure 5B). This SUC2:FT:9myc transgenic allele substantially rescued the late-flowering phenotype of ft-10 (Table 1), indicating that FT:9myc retains the biological function of endogenous FT protein. Signals corresponding to FT:9myc could be specifically detected by anti-myc antibody in the phloem of the transgenic plants harboring SUC2:FT:9myc (Figure 5B; Figure S10). Quantitative analysis of labeling density of FT:9myc in companion cell-sieve element complexes showed that although all sections from SUC2:FT:9myc and SUC2:FT:9myc ftip1-1 displayed FT:9myc labeling in companion cells (Figure 5B, lower right panel), there was an approximate 3-fold enrichment of labeling density in ftip1-1 over wild-type background (Figure 5B, lower left panel). More importantly, we detected FT:9myc labeling in sieve elements in nearly 80% of the wild-type sections, whereas only 4% of ftip1-1 sections displayed FT:9myc labeling in sieve elements (Figure 5B, lower right panel). In addition, the labeling density of FT:9myc in sieve elements was much higher in wild-type than in ftip1-1 (Figure 5B, lower left panel). Thus, in the absence of FTIP1, FT:9myc accumulated in companion cells and its transport to sieve elements was compromised. In agreement with this result, SUC2:FT:9myc ftip1-1 displayed later flowering than SUC2:FT:9myc (Table 1). As ftip1-1 also delayed flowering in SUC2:FT and SUC2:GFP:CO where CO directly promotes the endogenous FT expression (Table 1) [12], it seems that FTIP1 similarly affects the promotive effect of untagged FT protein on flowering as other FT fusion proteins used in this study. These observations support that FTIP1 regulates FT export from phloem companion cells to sieve elements, thus affecting FT transport through the phloem to the SAM. Consistent with this conclusion, the early-flowering phenotype caused by expression of FT or FT:GFP under the control of the KNAT1 promoter, which is active in the SAM [12], was not affected by ftip1-1 (Table 1).
Unlike other flowering promoters, overexpression of FTIP1 surprisingly caused late flowering (Figure S11; Table 1). Confocal analysis showed that the expression of FT:GFP protein in the inner region of the SAMs of 11-d-old seedlings was substantially lower in 35S:FTIP1 than in wild-type plants (Figure 5A; Figure S9A). In the primary leaf vein, FT:GFP driven by the SUC2 promoter was exclusively detected in phloem companion cells in wild-type background, whereas in 35S:FTIP1, the distribution of FT:GFP signals was detected in both phloem companion cells and xylem parenchyma cells (Figure 5C). However, the free GFP driven by the SUC2 promoter remained in phloem companion cells of 35S:FTIP1 as compared to wild-type plants (Figure S9B). These observations demonstrate that that ectopic expression of FTIP1 specifically deregulates the transport of FT:GFP protein out of the phloem system, an effect previously shown for a viral movement protein [19]. This could compromise the eventual distribution of FT:GFP in the SAM of 35S:FTIP1 and thus delay flowering.
FTIP1 Is Involved in Feedback Regulation of FT mRNA Expression
During the floral transition, FT interacts with FD in the SAM to promote the expression of AP1 and other flowering genes such as SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) [8],[9],[20]. As expected, the expression of these genes was downregulated in ftip1-1 in which FT transport is defective (Figure S12A). Surprisingly, FT expression was also downregulated in ftip1-1, whereas the expression of CO, a direct upstream activator of FT, was not significantly changed (Figure S12). As FTIP1 protein is not localized in the nucleus, it is unlikely that FTIP1 directly controls FT transcription. To address whether FTIP1 could regulate the stability of FT transcripts, we compared the levels of FT transcripts generated from the native and SUC2:FT:GFP transgenic loci. Although steady-state levels of native FT expression were downregulated in ftip1-1, total FT expression including native FT and transgenic FT:GFP expression remained unchanged in SUC2:FT:GFP ftip1-1 (Figure 6A,B). In addition, the abundance of FT:GFP fusion protein remained unchanged in wild-type and ftip1-1 backgrounds (Figure 6C). These results suggest that FTIP1 may not be directly involved in regulating FT mRNA or protein stability. Meanwhile, we observed downregulation of native FT expression in SUC2:FT:GFP (Figure 6A,B) and reduced FT:GUS staining in SUC2:FT (Figure 6D). These results are in agreement with the observation in a previous study [2] implying that an excessive accumulation of FT protein in phloem companion cells caused by the SUC2 promoter might directly or indirectly result in a reduction in native FT mRNA expression through a negative feedback loop. This may explain the observed downregulation of native FT expression in ftip1-1, where defective export of FT protein causes accumulation of FT protein in phloem companion cells (Figure 5B).
Discussion
Our results have demonstrated that FTIP1 and FT share similar mRNA expression patterns and subcellular localization, and that they interact in vivo in phloem companion cells. During the floral transition, the FT:GFP accumulation at the SAM is compromised in ftip1 mutants, which eventually exhibit late flowering under LDs. Consistently, FTIP1 is required for FT:9myc export from phloem companion cells to sieve elements, thus affecting the flowering time of SUC2:FT:9myc. In addition, overexpression of FTIP1 causes the transport of FT:GFP out of the phloem system, which also results in late flowering. These observations suggest that FT protein moves from phloem companion cells to sieve elements in a regulated manner and that a subtle regulation of FTIP1 activity is indispensable for the export of FT protein from phloem companion cells to induce flowering.
We envisage that in addition to FTIP1 and FT, florigen transport should involve other relevant regulators. First, although the transport of FT:9myc protein from phloem companion cells to sieve elements in ftip1-1 is significantly compromised, it is not completely abolished. This implies either that there is a basal level of diffusion of FT protein or that FT transport also depends on other regulators that share a redundant function with FTIP1 in mediating FT export from phloem companion cells. Furthermore, previous examinations of the spatial distribution of FT:GFP fusion protein in both Arabidopsis and rice have shown that FT:GFP accumulates in the rib zone beneath the SAM in a conical shape [2],[3], indicating that the movement of FT protein from phloem to the rib zone is not a simple diffusion process. As FTIP1 is clearly not expressed in the whole SAM (Figure 2C,D), regulation of FT protein transport from the phloem stream to the rib zone might also involve other regulators. The requirement of other regulators for FT protein transport is supported by the genetic analysis showing that ft mutants display much later flowering than ftip1 (Table 1). Potential candidates for these regulators include FTIP1 homologs (Figure S13A) because some combinations of ftip1 with loss-of-function mutants of FTIP1 homologs show much later flowering than any single mutant (unpublished data).
Second, the late-flowering phenotype of ft mutants is further enhanced by ftip1-1 (Table 1), indicating that FTIP1 may be required for transporting other flowering molecules in addition to FT. A potential candidate could be TWIN SISTER OF FT (TSF), which encodes another phosphatidylethanolamine-binding protein with very high sequence similarity with FT [21],[22]. Mutation of TSF further enhances the late flowering of ft mutants, and the resulting double mutants fail to accelerate flowering in response to LD conditions [21],[22]. The expression domain of TSF also overlaps with that of FTIP1 [21]. Furthermore, loss of function of FTIP1 does not further delay flowering of ft-10 tsf-1 under LD conditions (Table 1). These data support that TSF functions redundantly with FT and could be another molecule whose transport is affected by FTIP1.
As both FTIP1 and FT proteins are localized to ER, regulation of FT movement by FTIP1 across the border between companion cells and sieve elements might be partly mediated by a continuous ER network within plasmodesmata [23],[24]. In plasmodesmata, the ER becomes appressed to form the central axial desmotubule surrounded by the plasma membrane continuum between adjacent cells [25]. Although it has been suggested that the desmotubule is not the main route for plasmodesmatal transport, some molecules are known to be transported through this channel [26]. In contrast, the space between the desmotubule and the plasma membrane, which is referred as the cytoplasmic sleeve, is the proposed place where the general trafficking of molecules and ions occurs [25]. Because FTIP1 possesses a membrane-targeted PRT_C domain (Figure S1) and is localized to plasmodesmata (Figure 3C,E,F), it is likely that the C-terminus of FTIP1 is anchored to the desmotubule. How FTIP1 is oriented in plasmodesmata is an important question as its N-terminus, which is included in the region that interacts with FT protein (Figure 4A–C), might face either the cytoplasmic sleeve or the interior of the desmotubule. Further addressing this question will help to identify the route where FT protein passes through plasmodesmata and other possible factors involved in FT transport. Based on the size of FT:GFP, the route through the cytoplasmic sleeve might be possible for FT transport as the current understanding is that molecules larger than 27 kDa do not move readily through desmotubule [23].
The presence of C2 domains and a transmembrane domain in FTIP1 and its close homologs in Arabidopsis makes them topologically resemble synaptotagmins (Figure S13A) that constitute a family of membrane-trafficking proteins widely found in plants and animals. In Arabidopsis, the synaptotagmin SYTA has been shown to regulate endosome recycling and movement protein-mediated trafficking of plant virus genomes through plasmodesmata [27]. Our finding on the function of FTIP1 in mediating FT export from phloem companion cells to sieve elements, together with the proposed SYTA function, implies that synaptotagmin-like proteins may serve as essential regulators that mediate the transport of macromolecules in plants. Another FTIP1-like gene, QUIRKY (QKY; At1g74720), has been suggested to contribute to plant organ organogenesis mediated by the receptor-like kinase STRUBBELIG [28], implying a role of QKY in intercellular signaling. As FTIP1-like proteins are highly conserved in the angiosperms (Figure S13B), further investigation of FTIP1 and its homologs might shed light on the conserved mechanisms underlying which flowering plants regulate cell-to-cell communication to coordinate the growth and development.
Materials and Methods
Plant Materials
Arabidopsis plants were grown at 22°C under long days (16 h light/8 h dark) or short days (8 h light/16 h dark). The mutants ftip1-1, ftip1-2, co-1, gi-1, ft-1 (Ler ft-1 introgressed into Col), ft-10, tsf-1, soc1-2, agl24-1, fve-3, and svp-41 are in Columbia (Col) background, while co-2, fca-1, fpa-1, and fve-1 are in Landsberg erecta (Ler) background.
Plasmid Construction and Plant Transformation
To construct 35S:FTIP1, the cDNA encoding FTIP1 was amplified with primers and cloned into pGreen-35S [29]. For the complementation test, a 5.1-kb FTIP1 genomic fragment (gFTIP1) was amplified and cloned into pHY105 [28]. Based on this construct, FTIP1:FTIP1:GFP and FTIP1:4HA:FTIP1 were generated using a modified QuikChange Site-Directed Mutagenesis approach [30]. The cDNAs encoding GFP and 4HA were amplified. The resulting PCR fragments were annealed to the methylated template plasmid DNA containing gFTIP1 and elongated with the PfuTurbo DNA polymerase (Stratagene). Upon DpnI digestion, the mutated plasmids containing either GFP or 4HA were recovered from E. coli transformation. To construct pGreen-SUC2 and pGreen-KNAT1, SUC2 and KNAT1 promoters were amplified from Col genomic DNA and cloned into pHY105 [28]. To construct SUC2:FTIP1, the cDNA encoding FTIP1 was amplified and cloned into pGreen-SUC2. Based on SUC2:FTIP1 and 35S:FTIP1, SUC2:FTIP1:GFP and 35S:FTIP1:GFP were generated using the same modified QuikChange Site-Directed Mutagenesis approach [30] for creating FTIP1:FTIP1:GFP. To construct SUC2:FT and KNAT1:FT, the cDNA encoding FT was amplified and cloned into pGreen-SUC2 and pGreen-KNAT1, respectively. Based on the constructs of SUC2:FT and KNAT1:FT, SUC2:FT:GFP, SUC2:FT:9myc and KNAT1:FT:GFP were generated using the same modified QuikChange Site-Directed Mutagenesis approach [30] for creating FTIP1:FTIP1:GFP. To construct 35S:FT:RFP, the cDNA encoding RFP was amplified and cloned into pGreen-35S to generate pGreen-35S-RFP. The cDNA encoding FT was subsequently amplified and cloned into pGreen-35S-RFP. To construct FTIP1:GUS, the 2.1-kb FTIP1 5′ upstream sequence was amplified and cloned into pHY107 [29]. All transgenic plants were created using the floral dip method [31] and screened by Basta on soil.
Yeast Two-Hybrid Assay
All vectors used in yeast two-hybrid assays were from Clontech. The coding sequence of FT was cloned into pGBKT7 to produce BD-FT, which was used as a bait to screen cDNA library (CD4-30 from ABRC) for identifying interacting proteins of FT. Selection was performed on medium lacking histidine, tryptophan, and leucine (SD-His/-Trp/-Leu) supplemented with 30 mM 3-amino-1, 2, 4-triazole. To verify the interaction between FT and FTIP1, various versions of FTIP1 coding sequences were cloned into pGADT7. The resulting vectors were co-transformed with BD-FT, and the transformed cells were selected on SD-His/-Trp/-Leu medium supplemented with 30 mM 3-amino-1, 2, 4-triazole. β-galactosidase assays were performed according to the Yeast Protocols Handbook (Clontech).
GST In Vitro Pull-Down Assay
The cDNA encoding FT was cloned into the pGEX-4T-1 vector (Pharmacia) and introduced into E. coli Rosetta (DE3) (Novagen). Transformed cells were cultured until the OD600 nm reached 0.6, and IPTG was added to a final concentration of 0.6 mM to start induction. After overnight induction at 16°C, cells were collected and homogenized with lysis buffer (10 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Triton-100, and 10 mM PMSF). The soluble GST fusion proteins were extracted and immobilized on glutathione sepharose beads (Amersham Biosciences) for subsequent pull-down assays. The FTIP1 N-terminal fragment containing the three C2 domains (N501) was cloned into pGBKT7 vector (Clontech). The resulting plasmid was added to the TNT T7 Quick Coupled Transcription/Translation Systems (Promega) to synthesize myc-FTIP1(N501) protein. The resulting fusion protein was then incubated with the immobilized GST and GST fusion proteins. Proteins retained on the beads were resolved by SDS-PAGE and detected with anti-myc antibody (Santa Cruz Biotechnology).
Transient Expression of Proteins in Nicotiana Benthamiana Leaf Epidermal Cells
The overnight Agrobacterium cultures with a desired expression vector (35S:FTIP1:GFP, various RFP- or CFP-tagged organelle markers, 35S:FT:RFP, or 35S:GFP) were harvested and resuspended with infiltration buffer (10 mM MES pH 5.6, 10 mM MgCl2, and 100 µM acetosyringone) with OD600 nm at 0.4. To compare protein localization, equal volumes of infiltration solutions containing different expression vectors were mixed together and incubated for 3 h at room temperature. Infiltration solutions were infiltrated into the abaxial surface of 3-wk-old tobacco (Nicotiana benthamiana) leaves with syringes. The leaves were examined 2 d after infiltration under a confocal microscope.
GUS Staining
Tissues were infiltrated with staining solution (50 mM sodium phosphate buffer, pH 7.0, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, and 0.5 mg/mL X-Gluc) in a vacuum chamber, and subsequently incubated with staining solution at 37°C overnight. For sectioning, samples were dehydrated through an ethanol series, an ethanol/histoclear series, and finally embedded in paraplast (Sigma). Samples were then orientated and sectioned at a thickness of 3 µm with a microtome.
Expression Analysis
Total RNA was isolated with RNeasy Plant Mini Kit (Qiagen) and reverse-transcribed with ThermoScript RT-PCR System (Invitrogen) according to the manufacturers' instructions. Real-time PCR was performed in triplicates on 7900HT Fast Real-Time PCR system (Applied Biosystems) with SYBR Green PCR Master Mix (Applied Biosystems). The difference between the cycle threshold (Ct) of the target gene and the Ct of TUB2 (ΔCt = Cttarget gene−Cttubulin) was used to obtain the normalized expression of target genes, which corresponded to 2−ΔCt. Expression analysis was performed with at least three biological replicates. Primers for real-time PCR are listed in Table S2.
In Vivo Protein Interaction Assay
Plant tissues were collected and fixed with ice-cold 4% paraformaldehyde (PFA; Sigma-Aldrich) at pH 7.0 in a vacuum chamber. A serial PFA/sucrose change was applied till the tissues were finally equilibrated in PFA with 20% sucrose. Tissues were then embedded in 1.5% agarose gel, placed onto the microtome tissue holder, and flash frozen with liquid nitrogen. Tissues were cut in cryo-microtome with 20 µm thickness, and the sections were placed on slides. After complete drying, the slides were rehydrated with 100 mM Tris pH 8, 50 mM EDTA, and permeabilized with proteinase K (1 µg/ml) in the same buffer for 10 min at room temperature. Slides were washed with 2 mg/ml glycine followed by washing with PBS solution. Chlorophyll molecules were subsequently removed by incubating the slides with 1∶1 acetone/methanol mixture twice for 5 min. After drying, slides were rehydrated with PBS and finally treated with 4% paraformaldehyde for 10 min followed by washing with PBS solution.
In situ Proximity Ligation Assay (PLA) was performed with Duolink kit (Olink Bioscience) with minor modifications. The above treated slides were firstly blocked with 2% Bovine Serum Albumin in 100 mM Tris pH 7.5, 150 mM NaCl, and 0.3% Triton X-100 for 45 min at 37°C, and probed with the mixture of anti-GFP and anti-HA antibodies diluted in the blocking solution (1∶60) for 45 min at 37°C. The slides were washed three times and probed with diluted PLUS and MINUS PLA probes for 1 h at 37°C and subsequently washed 5 times. The slides were further incubated with the ligation solution, washed, and subsequently incubated with the amplification-polymerase solution with all components provided in the kit. After signal amplification, the slides were washed and mounted with PBS solution for further observation.
Immunogold Transmission Electron Microscopy
Samples were fixed with paraformaldehyde-glutaraldehyde solution (2% and 2.5%, respectively) and imbedded with LR white medium (EMS). Ultra-thin sections (85 nm) were cut and mounted on nickel grids. The grids were blocked with 1% BSA in TTBS (20 mM Tris, 500 mM NaCl, and 0.05% Tween-20, pH 7.5) for 30 min and subsequently incubated with anti-HA or anti-myc antibody at 1∶5 (v/v) for 1 h at room temperature. The grids were washed with TTBS for three times and further incubated with 15 nm gold-conjugated goat anti-mouse antibody (EMS) that was diluted 1∶20 with blocking solution. After 40 min of incubation, the grids were washed with TTBS for three times and with distilled water twice. Tissue staining was performed with 2% uranyl acetate for 15 min at room temperature, and pictures were taken by transmission electron microscope (Jeol JEM-1230).
For quantitative analysis of immunogold labeling, micrographs of randomly photographed immunogold-labeled transverse sections of the first rosette leaves of 15-d-old seedlings with various genetic backgrounds were measured as previously reported [32]. The data were presented as the mean number of gold particles per µm2 plus or minus standard deviation. The projected cell area was measured by a LI-3100C area meter (Li-Cor). We analyzed 56 individual sections from eight different leaves of each genotype for calculating the density of gold particles over the projected cell area. Statistical analysis was performed using a two-tailed unpaired Student's t test. Two-tailed test results were considered statistically significant at p<0.05.
Supporting Information
Bioinformatic analysis of FTIP1 protein sequence. (A) Schematic drawing of the FTIP1 protein. Three C2 domains and the PRT_C domain are shown as red and blue boxes, respectively. The bar above the scheme indicates the fragment isolated from the yeast two-hybrid screening. (B) Topology prediction of the transmembrane region in FTIP1 using the TopPred program (http://mobyle.pasteur.fr/cgi-bin/portal.py?form=toppred).
(TIF)
FTIP1 is specifically expressed in vascular tissues. (A) Schematic diagrams of gFTIP1 and FTIP1:GUS constructs. A 5.1 kb FTIP1 genomic fragment (gFTIP1) including the 2.1 kb upstream sequence, 2.4 kb coding sequence (CDS), and 0.6 kb downstream sequence was able to rescue the late-flowering phenotype of ftip1-1 as shown in Figure 1E. To examine the detailed expression pattern of FTIP1, we generated the construct FTIP1:GUS, in which the same 2.1 kb FTIP1 upstream sequence included in gFTIP1 for the gene complementation test was fused to the GUS reporter gene. (B–H) GUS staining of various tissues of FTIP1:GUS. (B) A 3-d-old seedling. (C) The shoot apex of a 3-d-old seedling. Asterisk indicates the shoot apical meristem. (D) An inflorescence apex. (E) A cauline leaf with an auxiliary shoot. (F) A cross-section of an inflorescence stem. (G) An open flower. (H) A silique. Bars: (C), 20 µm; (F), 200 µm.
(TIF)
Quantitative real-time PCR analysis of FTIP1 expression in various tissues of wild-type plants. Plant tissues were collected from 40-d-old wild-type plants. Results were normalized against the expression of TUB2 based on three biological replicates. Error bars indicate SD.
(TIF)
FTIP1 mRNA expression is not regulated by photoperiod, GA, and vernalization pathways. (A) FTIP1 expression is not significantly changed in wild-type plants grown under long days (LDs) and short days (SDs). (B) FTIP1 expression is not affected by vernalization treatment. For vernalization treatment, seeds were grown on MS medium and vernalized at 4°C under low light condition for 8 wk. 9-d-old seedlings grown under LDs were harvested for expression analysis. (C) FTIP1 expression is not affected by gibberellin (GA) treatment. Wild-type plants grown under SDs were treated weekly with 100 µM GA. Seedlings treated for 1 wk (1 w) or 3 wk (3 w) were harvested for expression analysis. (D) ftip1-1 and wild-type plants exhibit similar flowering time in response to GA treatment. ftip1-1 and wild-type plants grown under SDs were treated weekly with 100 µM GA. (E) FTIP1 expression levels do not obviously oscillate within a 24-h cycle under LDs. 9-d-old wild-type plants grown under LDs were harvested at 2-h intervals over a 24-h period. Sampling time was expressed in hours as Zeitgeber time (ZT), which is the number of hours after the onset of illumination. The lowest expression level of each gene is set as 1. Gene expression in (A–C) and (E) was determined by quantitative real-time PCR and normalized against TUB2 levels. Error bars indicate SD.
(TIF)
FTIP1 mRNA expression is not obviously altered in various flowering time mutants. (A) FTIP1 expression in photoperiod-pathway mutants. (B) FTIP1 expression in autonomous-pathway mutants. (C) FTIP1 expression in several other flowering time mutants. 9-d-old wild-type and mutant seedlings grown under LDs were harvested for expression analysis by quantitative real-time PCR. Results were normalized against the expression of TUB2. Error bars indicate SD.
(TIF)
Subcellular colocalization of FTIP1:GFP and the ER marker in N. benthamiana leaf epidermal cells. Bar, 20 µm.
(TIF)
Control experiments for measuring 4HA:FTIP1 localization by immunogold electron microscopy. (A) Western blot analysis showing that the 4HA:FTIP1 protein is intact. As the crude extract did not generate any signal, the sample was enriched with anti-HA agarose conjugate and used for SDS-PAGE analysis. The membrane was probed with anti-HA antibody. Lane 1, wild-type seedlings; Lane 2, FTIP1:4HA:FTIP1 ftip1-1 seedlings. (B) Quantitative analysis of immunogold signals revealed by immunogold electron microscopy of FTIP1:4HA:FTIP1 ftip1-1 transgenic plants shows that anti-HA antibody could specifically recognize 4HA:FTIP1. The left panel shows the quantification of 4HA:FTIP1 immunogold signals or immunogold background signals in CC and PD of FTIP1:4HA:FTIP1 ftip1-1 probed with anti-HA antibody or mouse IgG control. The data are presented as the mean number of gold particles per µm2 with standard deviation. Statistical analysis was performed using a two-tailed unpaired Student's t test. The results are considered statistically significant at p<0.05. The middle and right panels show the frequency histograms of appearance of 4HA:FTIP1 immunogold signals and immunogold background signals in FTIP1:4HA:FTIP1 ftip1-1 probed with anti-HA antibody and mouse IgG, respectively. Asterisks indicate that in all sections examined using IgG control, the number and frequency of PD with gold particles are zero. (C) Immunogold electron microscopy of CC-SE complexes in wild-type plants using anti-HA antibody. Left panel, a representative CC-SE complex. Bar, 1 µm. Middle panel, density of immunogold background signals observed in CC and PD of wild-type plants probed with anti-HA antibody. Right panel, frequency histogram of appearance of immunogold background signals in CC and PD of wild-type plants probed with anti-HA antibody in all examined sections. Asterisks indicate that in all sections examined using anti-HA antibody, the number and frequency of PD with gold particles are zero. CC, companion cell; PD, plasmodesmata; SE, sieve element.
(TIF)
Colocalization of FTIP1:GFP and FT:RFP in N. benthamiana leaf epidermal cells. GFP, GFP fluorescence; RFP, RFP fluorescence; Merge, merge of GFP and RFP; Nu, nucleus. Bar, 10 µm.
(TIF)
Change in FTIP1 activity does not affect free GFP distribution. (A) Confocal analysis of free GFP protein distribution in the apical region of 11-d-old SUC2:GFP seedlings in different genetic backgrounds. Bar, 20 µm. (B) Confocal analysis of free GFP protein distribution in the primary vein of the first rosette leaves from 11-d-old SUC2: GFP seedlings in different genetic backgrounds. Bar, 20 µm. GFP, GFP fluorescence; BF, bright field image; Merge, merge of GFP and BF.
(TIF)
Control experiments for measuring FT:9myc localization by immunogold electron microscopy. (A) Analysis of FT:9myc distribution in CC-SE complexes of the phloem in the first rosette leaves of 15-d-old SUC2:FT:9myc and SUC2:FT:9myc ftip1-1 seedlings by immunogold electron microscopy using mouse IgG antibody. All tissues examined show similar background signals generated by IgG antibody. Left panel, representative CC-SE complexes from SUC2:FT:9myc and SUC2:FT:9myc ftip1-1 including higher magnification views of CCs and SEs. Bars: 2 µm in the left panels; 0.5 µm in the magnified views. Middle panel, density of immunogold background signals observed in CCs and SEs of SUC2:FT:9myc (WT background) and SUC2:FT:9myc ftip1-1 (ftip1-1 background). The data are presented as the mean number of immunogold background particles per µm2 with standard deviation. Right panel, frequency histogram of appearance of immunogold background signals in CCs and SEs in all examined sections. CC, companion cell; SE, sieve element. (B) Analysis of FT:9myc distribution in xylem parenchyma cells of the first rosette leaves of 15-d-old SUC2:FT:9myc and SUC2:FT:9myc ftip1-1 seedlings by immunogold electron microscopy using anti-myc antibody. The results show that anti-myc antibody does not generate non-specific signal in xylem parenchyma cells. Left panel, representative xylem parenchyma cells from SUC2:FT:9myc (WT background) and SUC2:FT:9myc ftip1-1 (ftip1-1 background). Bar, 2 µm. Middle panel, density of gold particles observed in xylem parenchyma cells. The data are presented as the mean number of gold particles per µm2 with standard deviation. Right panel, frequency histogram of appearance of immunogold signals in xylem parenchyma cells in all examined sections. XP, xylem parenchyma; XV, xylem vessel. (C) Analysis of immunogold background signals in CC-SE complexes of the phloem in the first rosette leaves of 15-d-old wild-type seedlings by immunogold electron microscopy using anti-myc antibody. All tissues examined show similar background signals. Left panel, a representative CC-SE complex including higher magnification views of CC and SE. Bars, 1 µm. Middle panel, density of immunogold background signals observed in CCs and SEs of wild-type plants probed with anti-myc antibody. Right panel, frequency histogram of appearance of immunogold background signals in CCs and SEs in all examined sections. CC, companion cell; SE, sieve element.
(TIF)
Overexpression of FTIP1 causes late flowering. (A) Distribution of flowering time in 35S:FTIP1 T1 transgenic plants. Among 28 independent lines generated, all of them show different degrees of late flowering. (B) Homozygous transgenic plants (T3 generation) of three representative 35S:FTIP1 lines consistently show late flowering. 35S:FTIP1 #1, 35S:FTIP1 #2, and 35S:FTIP1 #3 exhibit weak, moderate, and strong flowering phenotypes, respectively. Error bars indicate SD. (C) FTIP1 expression is elevated in 35S:FTIP1 lines. The degrees of late flowering in 35S:FTIP1 shown in (B) are not related to the elevated levels of FTIP1 in 35S:FTIP1 #1, 35S:FTIP1 #2, and 35S:FTIP1 #3. 9-d-old wild-type and transgenic seedlings grown under LDs were harvested for expression analysis by quantitative real-time PCR. Results were normalized against the expression of TUB2. Error bars indicate SD.
(TIF)
Expression of several key flowering genes in ftip1-1. (A) Expression of AP1, SOC1, and FT is downregulated in ftip1-1. 9-d-old wild-type and ftip1-1 seedlings grown under LDs were harvested for expression analysis. The gene expression in wild-type plants is set as 1. (B) CO expression is not significantly changed in ftip1-1 within a 24-h cycle under LDs. (C) FT expression is consistently downregulated in ftip1-1 within a 24-h cycle under LDs. In (B and C), 9-d-old wild-type and ftip1-1 seedlings grown under LDs were harvested at 2-h intervals over a 24-h period for expression analysis. Gene expression in (A–C) was determined by quantitative real-time PCR and normalized against TUB2 levels. Error bars indicate SD.
(TIF)
Phylogenetic analysis of FTIP1-like proteins. (A) Phylogenetic tree showing FTIP1 homologs and synaptotagmins in Arabidopsis. The phylogenetic tree was generated based on the protein alignment of FTIP1, its 16 Arabidopsis homologs, and three Arabidopsis synaptotagmins (SYTA, SYTB, and SYTC). The scale bar represents 0.1 amino acid substitution. (B) Phylogenetic analysis of FTIP1-like proteins in different plant species. The phylogenetic tree was constructed with the neighbor-joining algorithm using the program MEGA 5.05 based on the alignment of the amino acid sequences of FTIP1-like proteins. Each terminal node of the tree is labeled by the two-letter abbreviation of the corresponding species name and the unique identifier. Bootstrap values (>50%) in 500 replicates are indicated next to the nodes. Zm, Zea mays; Os, Oryza sativa; Mt, Medicago truncatula; Pt, Populus trichocarpa; Bd, Brachypodium distachyon.
(TIF)
List of potential FT-interacting proteins isolated from the yeast two-hybrid screening.
(PDF)
Primers used for expression analyses in this study.
(PDF)
Acknowledgments
We thank K. Goto for FT:GUS seeds, the NSF-funded Arabidopsis Biological Resource Center for SALK insertion lines produced by J. Ecker and colleagues, plant organelle markers produced by A. Nebenführ and colleagues, and Y. He, T. Ito, J. Dinneny, F. Berger, and J. Xu for critical reading of the manuscript.
Abbreviations
- AP1
APETALA1
- ER
endoplasmic reticulum
- FT
FLOWERING LOCUS T
- FTIP1
FT-INTERACTING PROTEIN 1
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- LDs
long days
- PLA
proximity ligation assay
- PRT_C
phosphoribosyltransferase C-terminal
- SAM
shoot apical meristem
- SDs
short days
- SOC1
SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1
- SUC2
SUCROSE TRANSPORTER 2
- TSF
TWIN SISTER OF FT
Footnotes
The authors have declared that no competing interests exist.
This work was supported by Academic Research Funds T208B3113 and MOE2011-T2-1-018 from the Ministry of Education, Singapore (http://www.moe.gov.sg), and the intramural resource support from National University of Singapore (http://www.nus.edu.sg) and Temasek Life Sciences Laboratory (http://www.tll.org.sg). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chailakhyan M. K. New facts in support of the hormonal theory of plant development. Compt Rend Acad Sci URSS. 1936;13:79–83. [Google Scholar]
- 2.Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 3.Tamaki S, Matsuo S, Wong H. L, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- 4.Mathieu J, Warthmann N, Kuttner F, Schmid M. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol. 2007;17:1055–1060. doi: 10.1016/j.cub.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Jaeger K. E, Wigge P. A. FT protein acts as a long-range signal in Arabidopsis. Curr Biol. 2007;17:1050–1054. doi: 10.1016/j.cub.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Lin M. K, Belanger H, Lee Y. J, Varkonyi-Gasic E, Taoka K, et al. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell. 2007;19:1488–1506. doi: 10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- 8.Wigge P. A, Kim M. C, Jaeger K. E, Busch W, Schmid M, et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- 9.Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- 10.Kardailsky I, Shukla V. K, Ahn J. H, Dagenais N, Christensen S. K, et al. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- 11.Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 12.An H, Roussot C, Suarez-Lopez P, Corbesier L, Vincent C, et al. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development. 2004;131:3615–3626. doi: 10.1242/dev.01231. [DOI] [PubMed] [Google Scholar]
- 13.Samach A, Onouchi H, Gold S. E, Ditta G. S, Schwarz-Sommer Z, et al. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288:1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- 14.Takada S, Goto K. Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell. 2003;15:2856–2865. doi: 10.1105/tpc.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giakountis A, Coupland G. Phloem transport of flowering signals. Curr Opin Plant Biol. 2008;11:687–694. doi: 10.1016/j.pbi.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Imlau A, Truernit E, Sauer N. Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell. 1999;11:309–322. doi: 10.1105/tpc.11.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson B. K, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 18.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius K. J, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 19.Itaya A, Ma F, Qi Y, Matsuda Y, Zhu Y, et al. Plasmodesma-mediated selective protein traffic between “symplasmically isolated” cells probed by a viral movement protein. Plant Cell. 2002;14:2071–2083. doi: 10.1105/tpc.003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo S. K, Chung K. S, Kim J, Lee J. H, Hong S. M, et al. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 2005;139:770–778. doi: 10.1104/pp.105.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005;46:1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- 22.Jang S, Torti S, Coupland G. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. 2009;60:614–625. doi: 10.1111/j.1365-313X.2009.03986.x. [DOI] [PubMed] [Google Scholar]
- 23.Martens H. J, Roberts A. G, Oparka K. J, Schulz A. Quantification of plasmodesmatal endoplasmic reticulum coupling between sieve elements and companion cells using fluorescence redistribution after photobleaching. Plant Physiol. 2006;142:471–480. doi: 10.1104/pp.106.085803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fitzgibbon J, Bell K, King E, Oparka K. Super-resolution imaging of plasmodesmata using three-dimensional structured illumination microscopy. Plant Physiol. 2010;153:1453–1463. doi: 10.1104/pp.110.157941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maule A. J. Plasmodesmata: structure, function and biogenesis. Curr Opin Plant Biol. 2008;11:680–686. doi: 10.1016/j.pbi.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Cantrill L. C, Overall R. L, Goodwin P. B. Cell-to-cell communication via plant endomembranes. Cell Biol Int. 1999;23:653–661. doi: 10.1006/cbir.1999.0431. [DOI] [PubMed] [Google Scholar]
- 27.Lewis J. D, Lazarowitz S. G. Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc Natl Acad Sci U S A. 2010;107:2491–2496. doi: 10.1073/pnas.0909080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fulton L, Batoux M, Vaddepalli P, Yadav R. K, Busch W, et al. DETORQUEO, QUIRKY, and ZERZAUST represent novel components involved in organ development mediated by the receptor-like kinase STRUBBELIG in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000355. doi: 10.1371/journal.pgen.1000355. doi: 10.1371/journal.pgen.1000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C, Zhou J, Bracha-Drori K, Yalovsky S, Ito T, et al. Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development. 2007;134:1901–1910. doi: 10.1242/dev.003103. [DOI] [PubMed] [Google Scholar]
- 30.Geiser M, Cebe R, Drewello D, Schmitz R. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques. 2001;31:88–92. doi: 10.2144/01311st05. [DOI] [PubMed] [Google Scholar]
- 31.Clough S. J, Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 32.Zechmann B, Zellnig G, Muller M. Immunocytochemical localization of glutathione precursors in plant cells. J Electron Microsc. 2006;55:173–181. doi: 10.1093/jmicro/dfl022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bioinformatic analysis of FTIP1 protein sequence. (A) Schematic drawing of the FTIP1 protein. Three C2 domains and the PRT_C domain are shown as red and blue boxes, respectively. The bar above the scheme indicates the fragment isolated from the yeast two-hybrid screening. (B) Topology prediction of the transmembrane region in FTIP1 using the TopPred program (http://mobyle.pasteur.fr/cgi-bin/portal.py?form=toppred).
(TIF)
FTIP1 is specifically expressed in vascular tissues. (A) Schematic diagrams of gFTIP1 and FTIP1:GUS constructs. A 5.1 kb FTIP1 genomic fragment (gFTIP1) including the 2.1 kb upstream sequence, 2.4 kb coding sequence (CDS), and 0.6 kb downstream sequence was able to rescue the late-flowering phenotype of ftip1-1 as shown in Figure 1E. To examine the detailed expression pattern of FTIP1, we generated the construct FTIP1:GUS, in which the same 2.1 kb FTIP1 upstream sequence included in gFTIP1 for the gene complementation test was fused to the GUS reporter gene. (B–H) GUS staining of various tissues of FTIP1:GUS. (B) A 3-d-old seedling. (C) The shoot apex of a 3-d-old seedling. Asterisk indicates the shoot apical meristem. (D) An inflorescence apex. (E) A cauline leaf with an auxiliary shoot. (F) A cross-section of an inflorescence stem. (G) An open flower. (H) A silique. Bars: (C), 20 µm; (F), 200 µm.
(TIF)
Quantitative real-time PCR analysis of FTIP1 expression in various tissues of wild-type plants. Plant tissues were collected from 40-d-old wild-type plants. Results were normalized against the expression of TUB2 based on three biological replicates. Error bars indicate SD.
(TIF)
FTIP1 mRNA expression is not regulated by photoperiod, GA, and vernalization pathways. (A) FTIP1 expression is not significantly changed in wild-type plants grown under long days (LDs) and short days (SDs). (B) FTIP1 expression is not affected by vernalization treatment. For vernalization treatment, seeds were grown on MS medium and vernalized at 4°C under low light condition for 8 wk. 9-d-old seedlings grown under LDs were harvested for expression analysis. (C) FTIP1 expression is not affected by gibberellin (GA) treatment. Wild-type plants grown under SDs were treated weekly with 100 µM GA. Seedlings treated for 1 wk (1 w) or 3 wk (3 w) were harvested for expression analysis. (D) ftip1-1 and wild-type plants exhibit similar flowering time in response to GA treatment. ftip1-1 and wild-type plants grown under SDs were treated weekly with 100 µM GA. (E) FTIP1 expression levels do not obviously oscillate within a 24-h cycle under LDs. 9-d-old wild-type plants grown under LDs were harvested at 2-h intervals over a 24-h period. Sampling time was expressed in hours as Zeitgeber time (ZT), which is the number of hours after the onset of illumination. The lowest expression level of each gene is set as 1. Gene expression in (A–C) and (E) was determined by quantitative real-time PCR and normalized against TUB2 levels. Error bars indicate SD.
(TIF)
FTIP1 mRNA expression is not obviously altered in various flowering time mutants. (A) FTIP1 expression in photoperiod-pathway mutants. (B) FTIP1 expression in autonomous-pathway mutants. (C) FTIP1 expression in several other flowering time mutants. 9-d-old wild-type and mutant seedlings grown under LDs were harvested for expression analysis by quantitative real-time PCR. Results were normalized against the expression of TUB2. Error bars indicate SD.
(TIF)
Subcellular colocalization of FTIP1:GFP and the ER marker in N. benthamiana leaf epidermal cells. Bar, 20 µm.
(TIF)
Control experiments for measuring 4HA:FTIP1 localization by immunogold electron microscopy. (A) Western blot analysis showing that the 4HA:FTIP1 protein is intact. As the crude extract did not generate any signal, the sample was enriched with anti-HA agarose conjugate and used for SDS-PAGE analysis. The membrane was probed with anti-HA antibody. Lane 1, wild-type seedlings; Lane 2, FTIP1:4HA:FTIP1 ftip1-1 seedlings. (B) Quantitative analysis of immunogold signals revealed by immunogold electron microscopy of FTIP1:4HA:FTIP1 ftip1-1 transgenic plants shows that anti-HA antibody could specifically recognize 4HA:FTIP1. The left panel shows the quantification of 4HA:FTIP1 immunogold signals or immunogold background signals in CC and PD of FTIP1:4HA:FTIP1 ftip1-1 probed with anti-HA antibody or mouse IgG control. The data are presented as the mean number of gold particles per µm2 with standard deviation. Statistical analysis was performed using a two-tailed unpaired Student's t test. The results are considered statistically significant at p<0.05. The middle and right panels show the frequency histograms of appearance of 4HA:FTIP1 immunogold signals and immunogold background signals in FTIP1:4HA:FTIP1 ftip1-1 probed with anti-HA antibody and mouse IgG, respectively. Asterisks indicate that in all sections examined using IgG control, the number and frequency of PD with gold particles are zero. (C) Immunogold electron microscopy of CC-SE complexes in wild-type plants using anti-HA antibody. Left panel, a representative CC-SE complex. Bar, 1 µm. Middle panel, density of immunogold background signals observed in CC and PD of wild-type plants probed with anti-HA antibody. Right panel, frequency histogram of appearance of immunogold background signals in CC and PD of wild-type plants probed with anti-HA antibody in all examined sections. Asterisks indicate that in all sections examined using anti-HA antibody, the number and frequency of PD with gold particles are zero. CC, companion cell; PD, plasmodesmata; SE, sieve element.
(TIF)
Colocalization of FTIP1:GFP and FT:RFP in N. benthamiana leaf epidermal cells. GFP, GFP fluorescence; RFP, RFP fluorescence; Merge, merge of GFP and RFP; Nu, nucleus. Bar, 10 µm.
(TIF)
Change in FTIP1 activity does not affect free GFP distribution. (A) Confocal analysis of free GFP protein distribution in the apical region of 11-d-old SUC2:GFP seedlings in different genetic backgrounds. Bar, 20 µm. (B) Confocal analysis of free GFP protein distribution in the primary vein of the first rosette leaves from 11-d-old SUC2: GFP seedlings in different genetic backgrounds. Bar, 20 µm. GFP, GFP fluorescence; BF, bright field image; Merge, merge of GFP and BF.
(TIF)
Control experiments for measuring FT:9myc localization by immunogold electron microscopy. (A) Analysis of FT:9myc distribution in CC-SE complexes of the phloem in the first rosette leaves of 15-d-old SUC2:FT:9myc and SUC2:FT:9myc ftip1-1 seedlings by immunogold electron microscopy using mouse IgG antibody. All tissues examined show similar background signals generated by IgG antibody. Left panel, representative CC-SE complexes from SUC2:FT:9myc and SUC2:FT:9myc ftip1-1 including higher magnification views of CCs and SEs. Bars: 2 µm in the left panels; 0.5 µm in the magnified views. Middle panel, density of immunogold background signals observed in CCs and SEs of SUC2:FT:9myc (WT background) and SUC2:FT:9myc ftip1-1 (ftip1-1 background). The data are presented as the mean number of immunogold background particles per µm2 with standard deviation. Right panel, frequency histogram of appearance of immunogold background signals in CCs and SEs in all examined sections. CC, companion cell; SE, sieve element. (B) Analysis of FT:9myc distribution in xylem parenchyma cells of the first rosette leaves of 15-d-old SUC2:FT:9myc and SUC2:FT:9myc ftip1-1 seedlings by immunogold electron microscopy using anti-myc antibody. The results show that anti-myc antibody does not generate non-specific signal in xylem parenchyma cells. Left panel, representative xylem parenchyma cells from SUC2:FT:9myc (WT background) and SUC2:FT:9myc ftip1-1 (ftip1-1 background). Bar, 2 µm. Middle panel, density of gold particles observed in xylem parenchyma cells. The data are presented as the mean number of gold particles per µm2 with standard deviation. Right panel, frequency histogram of appearance of immunogold signals in xylem parenchyma cells in all examined sections. XP, xylem parenchyma; XV, xylem vessel. (C) Analysis of immunogold background signals in CC-SE complexes of the phloem in the first rosette leaves of 15-d-old wild-type seedlings by immunogold electron microscopy using anti-myc antibody. All tissues examined show similar background signals. Left panel, a representative CC-SE complex including higher magnification views of CC and SE. Bars, 1 µm. Middle panel, density of immunogold background signals observed in CCs and SEs of wild-type plants probed with anti-myc antibody. Right panel, frequency histogram of appearance of immunogold background signals in CCs and SEs in all examined sections. CC, companion cell; SE, sieve element.
(TIF)
Overexpression of FTIP1 causes late flowering. (A) Distribution of flowering time in 35S:FTIP1 T1 transgenic plants. Among 28 independent lines generated, all of them show different degrees of late flowering. (B) Homozygous transgenic plants (T3 generation) of three representative 35S:FTIP1 lines consistently show late flowering. 35S:FTIP1 #1, 35S:FTIP1 #2, and 35S:FTIP1 #3 exhibit weak, moderate, and strong flowering phenotypes, respectively. Error bars indicate SD. (C) FTIP1 expression is elevated in 35S:FTIP1 lines. The degrees of late flowering in 35S:FTIP1 shown in (B) are not related to the elevated levels of FTIP1 in 35S:FTIP1 #1, 35S:FTIP1 #2, and 35S:FTIP1 #3. 9-d-old wild-type and transgenic seedlings grown under LDs were harvested for expression analysis by quantitative real-time PCR. Results were normalized against the expression of TUB2. Error bars indicate SD.
(TIF)
Expression of several key flowering genes in ftip1-1. (A) Expression of AP1, SOC1, and FT is downregulated in ftip1-1. 9-d-old wild-type and ftip1-1 seedlings grown under LDs were harvested for expression analysis. The gene expression in wild-type plants is set as 1. (B) CO expression is not significantly changed in ftip1-1 within a 24-h cycle under LDs. (C) FT expression is consistently downregulated in ftip1-1 within a 24-h cycle under LDs. In (B and C), 9-d-old wild-type and ftip1-1 seedlings grown under LDs were harvested at 2-h intervals over a 24-h period for expression analysis. Gene expression in (A–C) was determined by quantitative real-time PCR and normalized against TUB2 levels. Error bars indicate SD.
(TIF)
Phylogenetic analysis of FTIP1-like proteins. (A) Phylogenetic tree showing FTIP1 homologs and synaptotagmins in Arabidopsis. The phylogenetic tree was generated based on the protein alignment of FTIP1, its 16 Arabidopsis homologs, and three Arabidopsis synaptotagmins (SYTA, SYTB, and SYTC). The scale bar represents 0.1 amino acid substitution. (B) Phylogenetic analysis of FTIP1-like proteins in different plant species. The phylogenetic tree was constructed with the neighbor-joining algorithm using the program MEGA 5.05 based on the alignment of the amino acid sequences of FTIP1-like proteins. Each terminal node of the tree is labeled by the two-letter abbreviation of the corresponding species name and the unique identifier. Bootstrap values (>50%) in 500 replicates are indicated next to the nodes. Zm, Zea mays; Os, Oryza sativa; Mt, Medicago truncatula; Pt, Populus trichocarpa; Bd, Brachypodium distachyon.
(TIF)
List of potential FT-interacting proteins isolated from the yeast two-hybrid screening.
(PDF)
Primers used for expression analyses in this study.
(PDF)