SUMMARY
Pathogenic microbes often modulate phytohormone physiology in the host to their advantage. We previously showed that the Pseudomonas syringae effector protein AvrB perturbs hormone signaling exemplified by an up-regulation of jasmonic acid (JA) response gene expression and enhances plant susceptibility. Here we show that AvrB does so by interacting with the Arabidopsis mitogen activated protein kinase MAP KINASE 4 (MPK4) and HSP90 chaperone components. AvrB induces MPK4 activation and this is directly promoted by the HSP90 chaperone. A previously identified AvrB-interacting protein, RIN4, also is required for AvrB to perturb hormone signaling and induce plant susceptibility, likely by acting down-stream of MPK4. These findings uncover a novel pathway through which a bacterial effector protein stimulates host processes to the benefit of the P. syringae bacterium.
INTRODUCTION
Plant hormones play a vital role not only in growth, development, and responses to abiotic stresses, but also in plant immunity. Salicylic acid (SA), jasmonic acids (JA), and ethylene (ET) play a particularly important role in plant disease resistance to diverse pathogenic microbes (Spoel and Dong, 2008). The activation of JA and ethylene signaling in Arabidopsis is required for defenses against herbivores and necrotrophic microbial pathogens, such as Botrytis cinerea and Alternaria brassicicola, but often renders plants more susceptible to biotrophic and hemibiotrophic pathogens, such as Hyaloperonospora parasitica and Pseudomonas syringae (Robert-Seilaniantz et al., 2007). Conversely, SA primarily regulates disease resistance to biotrophic pathogens, but contributes to susceptibility to necrotrophic pathogens. The two pathways often antagonize each other, although synergism occurs under certain circumstances (Mur et al., 2006). Recent data indicate that the SA and JA signaling pathways are subject to regulation by other phytohormones, forming a complex regulatory network (Spoel and Dong, 2008).
Not surprisingly, many plant pathogens perturb hormonal homeostasis or signaling to aid in the infection of plants (Robert-Seilaniantz et al., 2007). Some do so by synthesizing phytohormones or compounds that functionally mimic phytohormones, while others secrete effector proteins into the host cell to modulate hormone biosynthesis or responses. P. syringae is known to use multiple strategies to alter plant hormone physiology. For example, some P. syringae strains produce a phytotoxin called coronatine, which structurally and functionally mimics JA-isoleucine, an active form of JA, to assist bacterial entry into plant tissues (Melotto et al., 2006). In addition, this bacterium injects a large repertoire of effector proteins into the plant cells to enhance host susceptibility (Cunnac et al., 2009). While many of these effectors are known to directly inhibit plant immune signaling pathways (Zhou and Chai, 2008), several effector proteins have been reported to alter hormone responses in plants. For example, transgenic expression of AvrRpt2 enhances auxin response gene expression (Chen et al., 2007). Likewise, transgenic expression of AvrPtoB increases ABA content in plants (Torres-Zabala et al., 2007). In addition, several P. syringae effectors are capable of inducing JA-response gene expression (He et al., 2004). However, our knowledge of how these proteins alter plant hormone signaling is quite limited.
The P. syringae effector AvrB is known to trigger disease resistance in soybean plants carrying Rpg1b (Ashfield et al., 2004) and Arabidopsis plants carrying RPM1 (Grant et al., 2005). In Arabidopsis, AvrB directly associates with the RPM1-interacting protein RIN4 and induces RIN4 phosphorylation in plants (Mackey et al., 2002). Consistent with a role in inducing protein phosphorylation, the crystal structure of AvrB has features resembling protein kinases (Lee et al., 2004; Desveaux et al., 2007), although the biochemical function of AvrB remains to be elucidated.
In susceptible plants lacking the cognate resistance genes, AvrB assists P. syringae colonization in the host. In soybean plants lacking the corresponding resistance gene Rpg1b, AvrB enhances P. syringae virulence through an unknown mechanism (Ashfield et al., 1995). On Arabidopsis plants, avrB induces the transcription of a JA response gene in Arabidopsis (He et al., 2004). Expression of AvrB as a transgene in Arabidopsis plants lacking RPM1 enhances plant susceptibility to a nonpathogenic strain of P. syringae bacteria (Shang et al., 2006). Both AvrB-induced JA response gene expression and susceptibility in Arabidopsis requires COI1, a JA receptor (Yan et al., 2009), indicating that AvrB enhances plant susceptibility by perturbing hormone signaling. Our previous work also showed that the AvrB transgene-induced susceptibility in Arabidopsis requires RAR1 (Shang et al., 2006), a co-chaperone for HSP90. AvrB can interact with RAR1 in vivo, although it was not determined if this interaction occurs directly or indirectly. These results suggest that HSP90 and its client proteins are involved in the AvrB-induced JA signaling.
In this study, we show that MPK4 is regulated by HSP90. AvrB directly interacts with RAR1, and this enables AvrB to associate with HSP90 and MPK4 in vivo, consequently promoting MPK4 kinase activation. MPK4 directly interacts with RIN4 and phosphorylates RIN4 in vitro. Molecular and genetic evidence indicated that HSP90, MPK4 and RIN4 form a new pathway through which AvrB perturbs hormone signaling and induce plant susceptibility.
RESULTS
HSP90, but Not TAO1 and RPM1, Are Required for AvrB-Induced Susceptibility
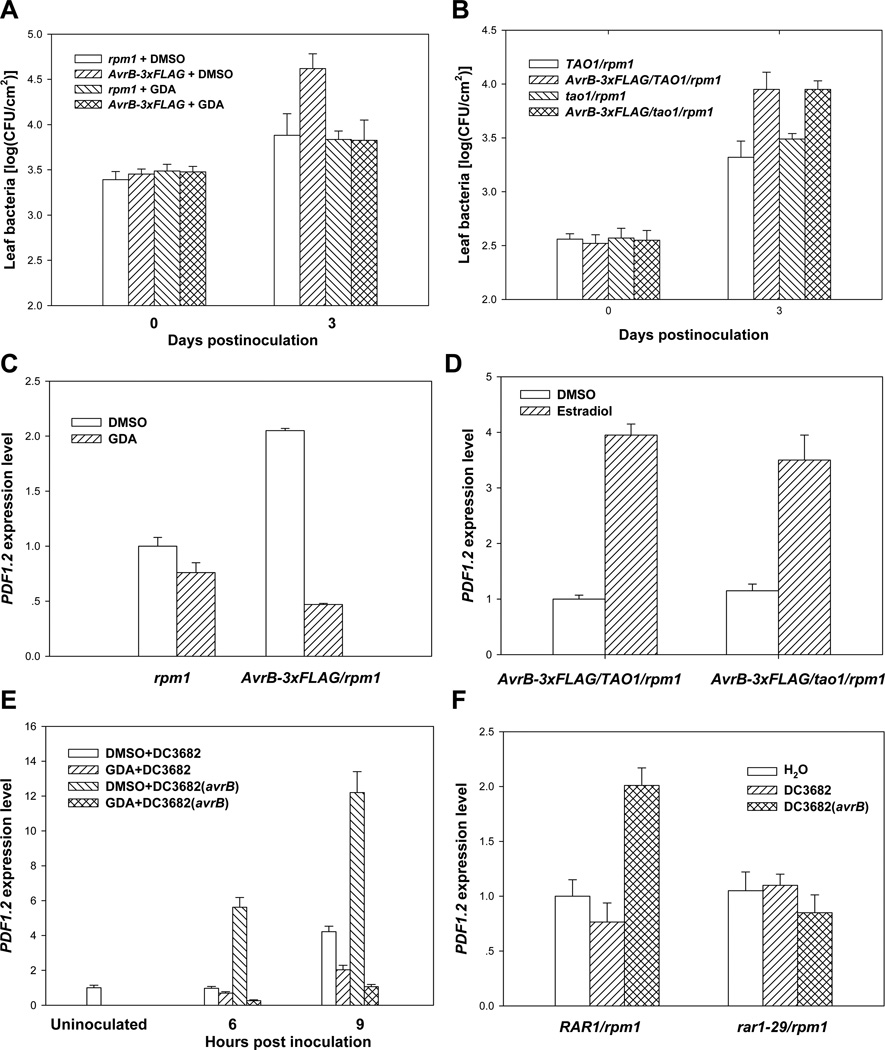
Because RAR1 and another protein, SGT1, are co-chaperones for HSP90 (Takahashi et al., 2003; Hubert et al., 2003), we reasoned that the HSP90 complex may be required for AvrB to enhance plant susceptibility. We therefore tested if the AvrB-induced plant susceptibility required HSP90. As we showed previously (Shang et al., 2006), expression of the AvrB transgene in the rpm1 mutant background enhanced the growth of the nonpathogenic P. syringae hrcC mutant strain in plants (Figure 1A). Co-infiltration of geldanamycin (GDA), a specific inhibitor of HSP90, did not affect the AvrB accumulation (Figure S1A), but abolished the AvrB transgene-induced bacterial growth in AvrB-3xFLAG/rpm1 plants (Figure 1A). GDA does not have deleterious effects on the P. syringae bacterium at the concentration used for the experiment (Figure S1B), indicating that HSP90 is required for the AvrB-induced susceptibility to the bacterium. It was shown recently that, in addition to RPM1, another nucleotide-binding, leucine-rich-repeat (NB-LRR) resistance protein called TAO1 recognizes AvrB and triggers weak resistance responses in Arabidopsis (Eitas et al., 2008). Because the stability of NB-LRR proteins often is regulated by HSP90 and RAR1 (Holt III et al., 2005; Azevedo et al., 2006), the observed AvrB activity could be caused indirectly by the function of TAO1. We therefore generated AvrB-3xFLAG/tao1/rpm1 plants by crossing the tao1-11 mutant (Eitas et al., 2008) to the AvrB-3xFLAG/rpm1 transgenic line. The AvrB transgene enhanced susceptibility to P. syringae in both AvrB-3xFLAG/tao1/rpm1 and AvrB-3xFLAG/TAO1/rpm1 plants (Figure 1B), indicating that neither TAO1 nor RPM1 is required for the AvrB-induced plant susceptibility.
Figure 1. HSP90 Chaperone Components, but Not TAO1, Are Required for AvrB-Induced Susceptibility and PDF1.2 Expression.
(A) GDA inhibits AvrB-3xFLAG transgene-induced susceptibility. (B) TAO1 is not required for AvrB-induced susceptibility. Plants of the indicated genotypes were pre-treated with estradiol, infiltrated with hrcC mutant bacteria in the presence of GDA or buffer (DMSO), and bacterial populations within leaves determined at the indicated times (A and B). (C) GDA abolishes PDF1.2 induction by the AvrB-3xFLAG transgene. (D) TAO1 is not required for PDF1.2 induction by the AvrB-3xFLAG transgene. (E) GDA inhibits PDF1.2 induction by bacterially delivered AvrB. (F) RAR1 is required for PDF1.2 induction by bacterially delivered AvrB. Arabidopsis plants of the indicated genotype were infiltrated with estradiol for 24 hours (C and D) or the indicated bacterial strains (E and F) for 6 and 9 hours (E) or 6 hours (F), and RNA isolated for quantitative RT PCR. For experiments in C and E, GDA or buffer (DMSO) was co-infiltrated into the leaves. The PDF1.2 expression was determined by real-time RT-PCR. Data in are representative of three independent experiments with similar results.
JA Responses, but Not JA Biosynthesis, Is Induced by AvrB Transgene
We earlier showed that AvrB induces the expression of RAP2.6, a JA response gene (He et al., 2004). To further determine the perturbation of JA signaling by AvrB, we examined expression of two additional JA response genes PDF1.2 and THI2.1 (Robert-Seilaniantz et al., 2007) in plants expressing the AvrB transgene. Estradiol-induced expression of the AvrB transgene led to 2–6 fold induction of PDF1.2 (Figures 1C and 1D) and 15–30 fold induction of THI2.1 transcripts (Figures S1C and S1D) in AvrB/rpm1 plants compared to rpm1 plants. Further more, expression of AvrB resulted in ~5 fold increase of anthocyanin (Figure S1E), a phenotype associated with elevated JA signaling (Shan et al., 2009). To determine if the elevated JA responses in AvrB transgenic plants was caused by increased JA accumulation or signaling, we measured JA content in plants expressing AvrB. Figure S1F shows that the expression of AvrB does not alter JA content in the plants. Together these results demonstrate that the AvrB-induces JA responses, but not JA biosynthesis.
The HSP90 Chaperone, but Not TAO1, Is Required for AvrB-Induced JA Responses
To determine the involvement of the HSP90 chaperone in the perturbation of hormone signaling, we focused on JA response genes and examined the expression of PDF1.2 and THI2.1 in AvrB-3xFLAG/rpm1 transgenic plants upon GDA-treatment. The GDA-treatment completely abolished the AvrB-induced PDF1.2 and THI2.1 expression (Figures 1C and S1C). Likewise, the AvrB-induced THI2.1 and anthocyanin accumulation are also abolished in AvrB/rar1-29/rpm1 plants (Figures S1D and S1E), indicating an essential role for RAR1. In contrast, PDF1.2 was induced normally by the AvrB transgene in the tao1/rpm1 background, indicating that the effect of AvrB on JA responses is independent of TAO1 and RPM1 (Figure 1D).
Because the AvrB protein delivered from the P. syringae bacterium is expected to exist at a very low level in the host cell, the observed PDF1.2 induction by the AvrB transgene might have resulted from the over accumulation of AvrB. To unequivocally determine if the HSP90 complex is required for AvrB to induce PDF1.2 expression during bacterial infection, we inoculated rpm1 plants with DC3682 (avrB) along with GDA. While DC3682 (avrB) strongly induced PDF1.2 expression 6 and 9 hours post inoculation in buffer-treated plants, it failed to induce PDF1.2 expression in the presence of GDA (Figure 1E). Similarly, we examined PDF1.2 expression in rar1-29/rpm1 upon the inoculation of DC3682 (avrB). The avrB-induced PDF1.2 expression was completely blocked in the rar1-29/rpm1 mutant background (Figure 1F), further confirming a role of RAR1 in AvrB-induced JA signaling (Shang et al., 2006). Together these results demonstrate that the effect of AvrB on JA responses requires the HSP90 chaperone components, but not TAO1.
AvrB Interacts with HSP90 through RAR1
We previously showed that AvrB can interact with RAR1 in vivo in a co-immunoprecipitation (co-IP) assay (Shang et al., 2006). Because RAR1 is a zinc-containing protein, the relatively high concentration of EDTA in our buffer system may lead to a release of free cysteine residues potentially impacting the specificity in protein-protein interactions. We therefore performed the co-IP experiment in the absence of EDTA. Figure S2 shows that the AvrB-RAR1 interaction occurs in the absence of EDTA, further supporting the specificity of interaction.
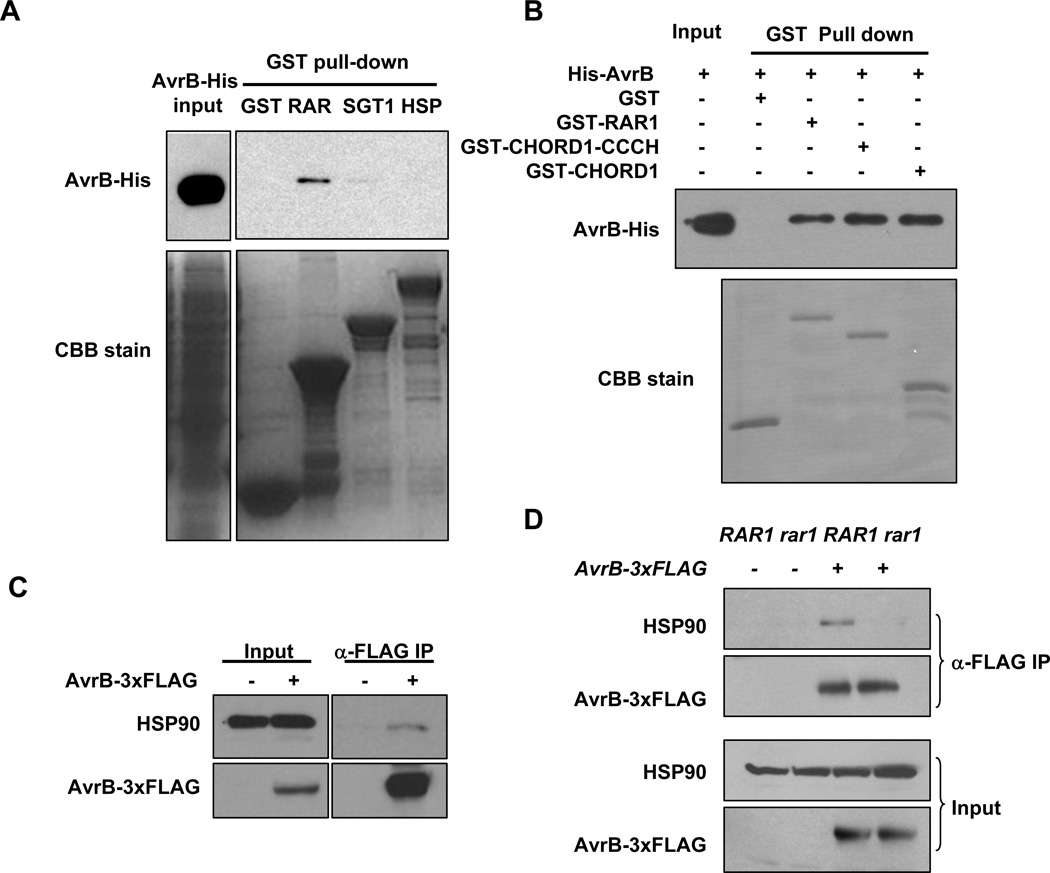
We next used an in vitro pull-down assay to determine if AvrB directly interacts with individual components of the HSP90 complex. AvrB directly interacted with RAR1, but not HSP90 or GST (Figure 2A). A weak interaction was also detected between AvrB and SGT1b under longer exposure. The RAR1 protein is consisted of an N terminal CHORD1 domain and a C terminal CHORD2 domain connected by a CCCH domain (Shirasu, 2009). The two CHORD domains are known to interact with HSP90 and SGT1, respectively. GST pull-down assays showed that the N terminal CHORD1 domain is sufficient to interact with AvrB (Figure 2B).
Figure 2. AvrB Interacts with the HSP90 Chaperone.
(A) AvrB directly interacts with RAR1. An equal amount of AvrB-His recombinant protein was incubated with bacterial lysates containing GST or GST-tagged RAR1, SGT1b, or HSP90. The presence of AvrB-His in the GST pull-down was detected by immunoblot using an anti-His antibody. Coomassie Brilliant Blue (CBB) staining shows the amounts of GST-fusion proteins. (B) CHORD1 domain is sufficient for interaction with AvrB. GST-tagged full-length and truncated RAR1 were incubated with equal amount of His-AvrB in a GST pull-down assay, the presence of His-AvrB in the protein complex was detected by anti-His immunoblot. (C) AvrB interacts with HSP90 in plants. Arabidopsis rpm1 plants with (+) or without (−) the AvrB-3xFLAG transgene were induced with estradiol, and protein extract was immunoprecipitated with an agarose-conjugated anti-FLAG antibody. Amounts of HSP90 proteins in the immune complex were determined by immunoblot using anti-HSP90 antibodies. (D) RAR1 is required for AvrB-HSP90 interaction in vivo. Co-immunoprecipitation assay for AvrB-HSP90 interaction was conducted as in (C) using Arabidopsis rpm1 (RAR1) or rpm1/rar1-29 (rar1) plants containing (+) or lacking (−) the AvrB-3xFLAG transgene.
Co-IP experiment showed that HSP90 was capable of interacting with AvrB in plants (Figure 2C). Importantly, the AvrB-HSP90 interaction was largely abolished in the rar1-29 background (Figure 2D), indicating that the interaction is specific. Together with previous findings on AvrB-RAR1 interaction (Shang et al., 2006), these results indicate that AvrB is associated with the HSP90 complex, primarily through a direct interaction with RAR1.
AvrB Interacts with and Induces Phosphorylation of MPK4
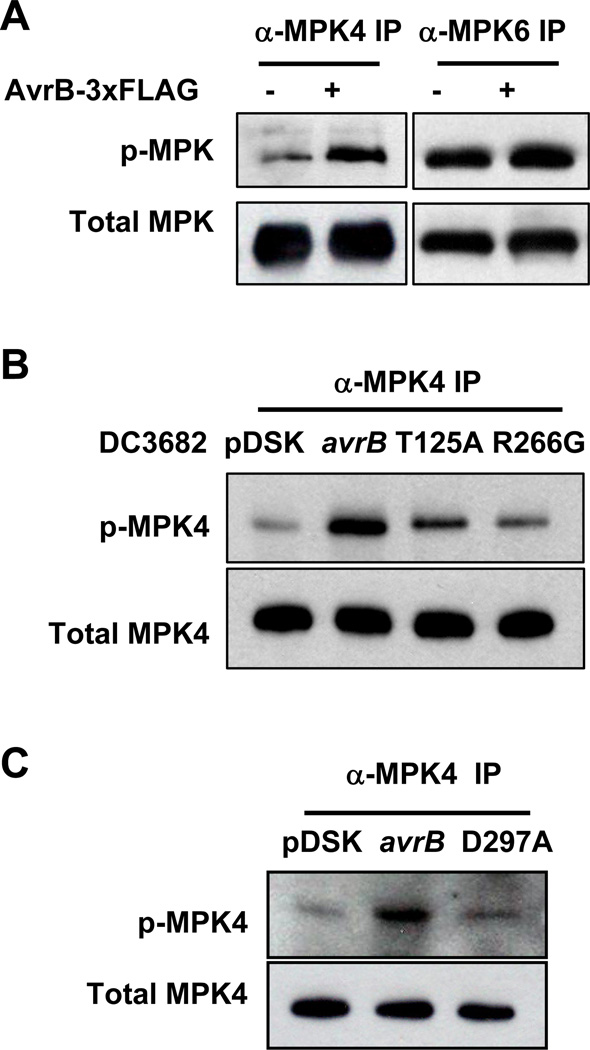
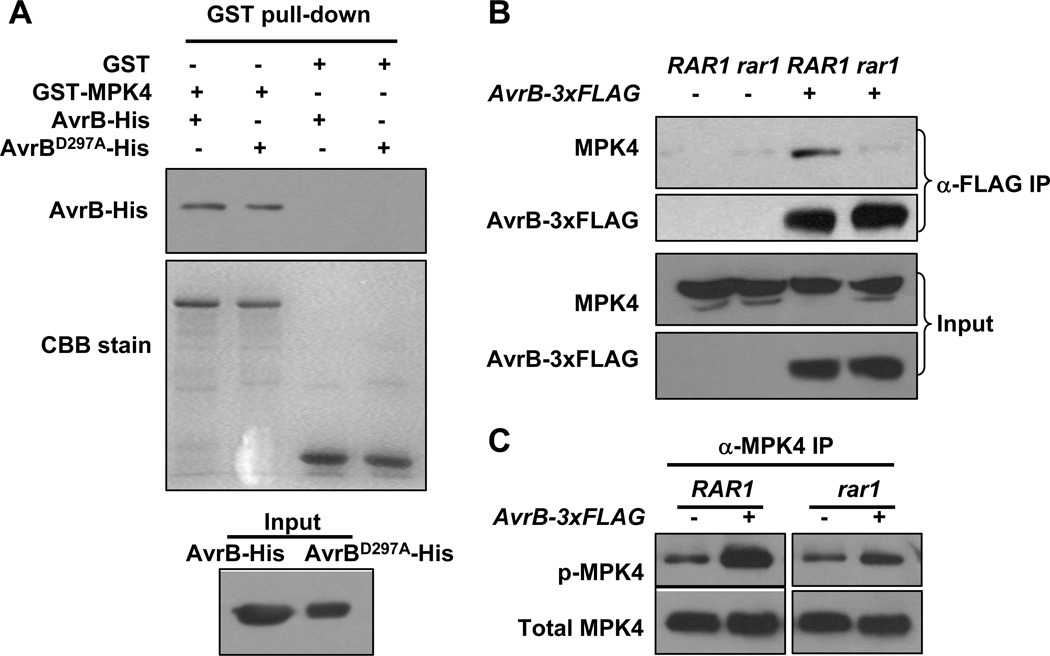
Transgenic expression of AvrB suppresses Pathogen/Microbe-Associated Molecular Pattern (PAMP/MAMP)-induced responses (Shang et al., 2006), which are known to involve the activation of the MAP kinases MPK4, MPK3 and MPK6 (Bittel and Robatzek, 2007). MPK6 and MPK3 are thought to positively regulate disease resistance to P. syringae, whereas MPK4 negatively impact resistance to P. syringae (Petersen et al., 2000), likely through its regulation of multiple hormone pathways. We therefore determined if expression of an AvrB-3xFLAG transgene affected MPK phosphorylation. Specific antibodies against MPK4 and MPK6 were used to immunoprecipitate MPK4 and MPK6 proteins from control plants (rpm1) or plants expressing the AvrB transgene (AvrB-3xFLAG/rpm1), and then the level of dual phosphorylation on MPK4 and MPK6 was determined by immunoblot using anti-phospho-ERK1 antibodies. The AvrB transgene specifically enhanced the phosphorylation of MPK4, but not MPK6 (Figures 3A and S3A). The AvrB-stimulated phosphorylation of MPK4 correlated with greater MPK4 kinase activity as indicated by stronger phosphorylation of myelin basic protein (MBP; Figure S3B). We further tested if AvrB delivered from the bacterium similarly induced MPK4 phosphorylation in plants. While Arabidopisis rpm1 plants inoculated with the strain carrying en empty vector showed a MPK4 phosphorylation slightly above the background level (Figure S3C), plants inoculated with DC3682 (avrB) exhibited strong MPK4 phosphorylation (Figures 3B and S3C). The DC3682 (pDSK)-induced MPK4 phosphorylation was significantly less than observed in a prior report (Qiu et al., 2008), likely because of different time points were used as PAMP-triggered MPK activation is known to be transient and time-sensitive. Ong and Innes (2006) showed that AvrB enhances the growth of P. syringae on susceptible cultivars of soybean and that this virulence activity is blocked by specific amino acid substitutions at threonine 125 (Thr125Ala), arginine 266 (Arg266Gly), and aspartate 297 (Asp297Ala). We tested if these mutants were able to induce MPK4 phosphorylation. The avrB-induced MPK4 phosphorylation was diminished by the avrBT125A, avrBR266G and avrBD297A substitutions (Figs. 3B and 3C), indicating that the induction of MPK4 phosphorylation is correlated with the virulence function of AvrB.
Figure 3. AvrB Directly Interacts with and Induces Phosphorylation of MPK4.
(A) AvrB-3xFLAG induces MPK4 phosphorylation. Specific anti-MPK antibodies were used to immunoprecipitate MPK4 and MPK6 proteins from estradiol-treated rpm1 plants with (+) or without (−) the estradiol-inducible AvrB-3xFLAG transgene, and the level of dual phosphorylation on MPK4 and MPK6 was determined by immunoblot using anti-phospho-ERK1 antibodies. Equal loading of immunoprecipitated proteins was confirmed by immunoblot using anti-MPK4 and anti-MPK6 antibodies. (B) and (C) Bacterially-delivered AvrB induces MPK4 phosphorylation. Arabidopsis rpm1 plants were inoculated with 108 CFU/ml DC3682 carrying an empty vector (pDSK), WT avrB, avrBT125A, avrBR266G or avrBD297A mutant plasmids for 6 hrs, and the phosphorylation state of the immunoprecipitated MPK4 was determined by immunoblot using anti-phospho-ERK1 antibodies.
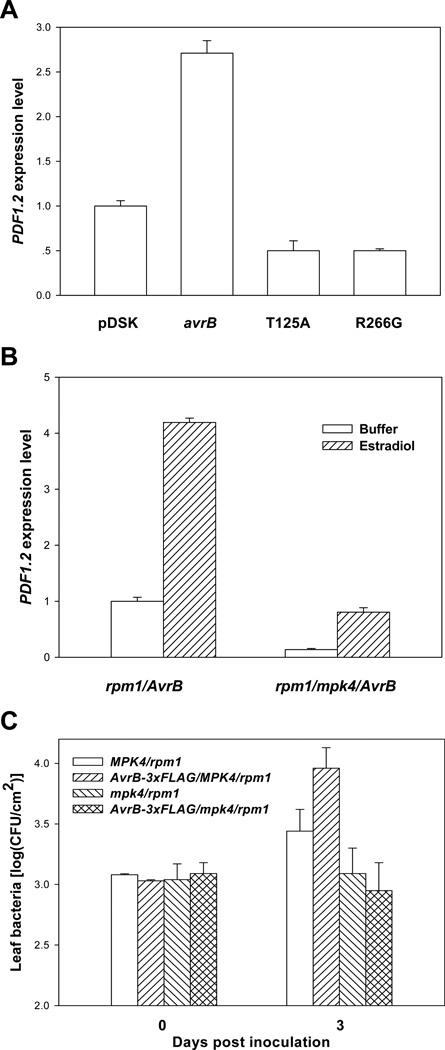
We next determined if avrB mutant forms that are unable to induce MPK4 phosphorylation are capable of inducing PDF1.2 expression. While the bacterial strain carrying WT avrB induced PDF1.2 expression by ~3 fold compared to the strain carrying an empty vector, the strains carrying avrBT125A and avrBR266G were completely unable to induce PDF1.2 expression (Figure 4A), indicating that the virulence function of AvrB in soybean is correlated with its ability to induce both MPK4 phosphorylation and JA signaling in Arabidopsis.
Figure 4. MPK4 Is Required for AvrB-Induced PDF1.2 Expression and Susceptibility.
(A) The avrBT125A and avrBR266G mutations abolish JA-signaling activity. Arabidopsis rpm1 plants were inoculated with DC3682 carrying an empty vector (pDSK), WT avrB, avrBT125A, or avrBR266G. RNA was isolated 6 hr post inoculation, and the expression of PDF1.2 was determined by real-time RT-PCR. (B) MPK4 is required for PDF1.2 induction by the AvrB-3xFLAG transgene. Plants of the indicated genotypes were treated with estradiol for 24 hrs, and RNA was isolated for real-time RT-PCR. (C) The AvrB-3xFLAG transgene does not induce susceptibility in the mpk4 mutant. Plants of the indicated genotypes were pre-treated with estradiol, inoculated with the P. syringae hrcC mutant strain, and the bacterial population in the leaf was determined at the indicated times. The data shown are representative of two independent experiments with similar results.
To further determine a role of MPK4 in the AvrB-induced JA responses and susceptibility in plants, we crossed the AvrB-3xFLAG/rpm1 transgenic line (Shang et al., 2006) with the mpk4 mutant (Petersen et al., 2000) to generate AvrB-3xFLAG/MPK4/rpm1 and AvrB-3xFLAG/mpk4/rpm1 plants. Figure 4B shows that the PDF1.2 expression was reduced to background level in AvrB-3xFLAG/mpk4/rpm1 plants, suggesting that MPK4 is required for the AvrB-induced JA responses. Furthermore, the AvrB transgene enhanced the growth of a nonpathogenic P. syringae mutant hrcC in AvrB-3xFLAG/MPK4/rpm1 but not AvrB-3xFLAG/mpk4/rpm1 plants (Figure 4C). Together these results are consistent with a role of MPK4 in the AvrB-induced disease susceptibility and hormone signaling.
AvrB Interacts with MPK4
The involvement of MPK4 in AvrB-induced JA signaling and disease susceptibility prompted us to test protein-protein interaction between AvrB and MPK4. In vitro pull-down experiments showed that AvrB is capable of interacting with MPK4 (Figure 5A). The structure of AvrB suggests that AvrB may act as an enzyme related to protein kinases, with amino acid residue D297 as a potential active site. This residue was previously shown to be required for AvrB virulence function, but not its ability to interact with RIN4 (Ong and Innes, 2006). The AvrBD297A substitution also did not affect AvrB-MPK4 interaction, suggesting that this residue may be specifically required for the activation of MPK4 phosphorylation, but not protein-protein interaction.
Figure 5. AvrB Interacts with MPK4.
(A) AvrB interacts with MPK4 in vitro. Equal amounts of AvrB-His or AvrBD297A-His were incubated with GST or GST-MPK4, and the presence of AvrB after GST pull-down was detected by immunoblot using anti-His antibody. (B) AvrB-3xFLAG interacts with MPK4 in plants in a RAR1-dependent manner. RAR1/rpm1 (RAR1) or rar1-29/rpm1 (rar1) plants with or without the AvrB-3xFLAG transgene were induced with estradiol, and protein extracts were immunoprecipitated with agarose-conjugated anti-FLAG antibody. The presence of MPK4 in the immune complex was detected by immunoblot using anti-MPK4 antibodies. (C) RAR1 is required for AvrB-3xFLAG-induced MPK4 phosphorylation. Plants of the indicated genotypes were induced with estradiol, and the phosphorylation state of immunoprecipitated MPK4 was determined by immunoblot using anti-phospho-ERK1 antibodies.
Co-IP assays showed that transgenic AvrB can interact with MPK4 in vivo (Figures 5B and S4). Because RAR1 is required for AvrB function, we tested if RAR1 plays a role in AvrB-MPK4 interaction in vivo. The association of AvrB with MPK4 was largely impaired in AvrB-3xFLAG/rar1-29/rpm1 plants (Figure 5B), indicating that the interaction is specific and that RAR1 is required for stable AvrB-MPK4 interaction in plants. Furthermore, the AvrB-induced phosphorylation, but not the basal phosphorylation of MPK4, was compromised in AvrB-3xFLAG/rar1-29/rpm1 plants (Figure 5C). These results suggest that AvrB, upon the association with RAR1, interacts with and induces the phosphorylation of MPK4 in vivo.
HSP90 Positively Regulates MPK4 Activity
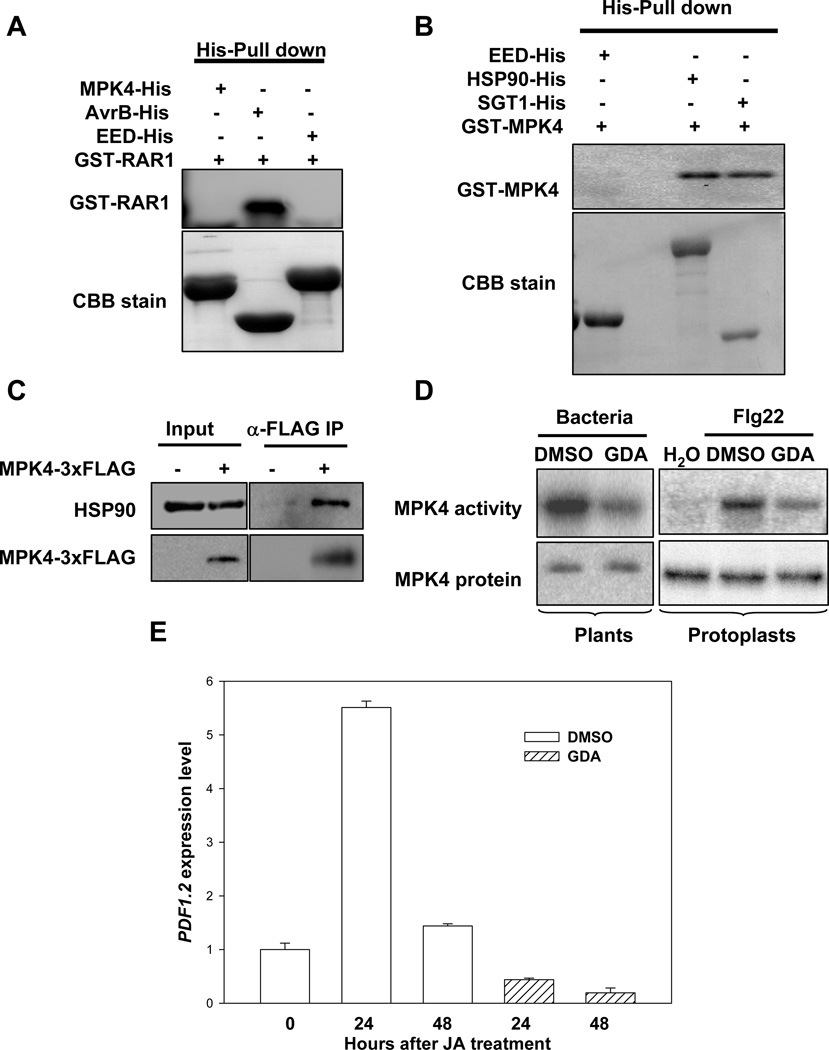
HSP90 is a molecular chaperone responsible for the maturation of a large number of signaling proteins, particularly protein kinases (Pearl and Prodromou, 2006). The requirement of RAR1 in AvrB-induced phosphorylation of MPK4 prompted us to test if HSP90 interacts with and regulates MPK4. A pull-down assay failed to detect a direct interaction between MPK4 and RAR1 in vitro (Figure 6A). However, MPK4 is capable of interacting with HSP90 and SGT1b protein in vitro (Figure 6B). To determine if MPK4 is capable of interacting with the HSP90 in vivo, we generated NP-MPK4-3xFLAG transgenic plants in which MPK4-3xFLAG was expressed under the control of MPK4 native promoter. A co-IP assay showed that MPK4-3xFLAG protein indeed interacted with the endogenous HSP90 protein (Figure 6C).
Figure 6. HSP90 Promotes MPK4 Activation.
(A) MPK4 does not interact with RAR1 in vitro. AvrB-His and GST-RAR1 were expressed in E. coli and purified proteins used in a pull-down assay. The presence of GST-RAR1 was detected by immunoblot using anti-RAR1 antibodies. The AvrB-His protein was used as a positive control. A His-tagged human WD-repeat protein EED (Han et al., 2007) was included as a negative control. (B) MPK4 interacts with SGT1b and HSP90 in vitro. GST-MPK4, SGT1b-His, and HSP90-His proteins were expressed in E. coli. The presence of GST-MPK4 was detected by immunoblot using anti-MPK4 antibodies. (C) MPK4 interacts with HSP90 in plants. Protein extracts from transgenic plants carrying NP-MPK4-3xFLAG were immunoprecipitated with an agarose-conjugated anti-FLAG antibody, and the presence of HSP90 was determined by immunoblot using anti-HSP90 antibodies. The presence of MPK4-3xFLAG was determined by immunoblot using anti-MPK4 antibodies. (D) GDA inhibits PAMP-induced MPK4 activation. WT Arabidopsis plants (Col-0) were infiltrated with hrcC bacteria in the presence of GDA or buffer (DMSO). Kinase activity of the immunoprecipitated MPK4 protein was determined using an in vitro kinase assay employing myelin basic protein (MBP) as a substrate. To assess MPK4 activation in protoplasts, MPK4-3xFLAG was expressed in protoplasts prepared from WT plants, treated with GDA or buffer (DMSO), and induced with flg22. The MPK activity of the immunoprecipitated MPK4-3xFLAG was determined using the in vitro kinase assay with MBP as a substrate. (E) GDA inhibits MeJA-induced PDF1.2 expression. WT plants were pretreated with GDA or buffer (DMSO) prior to the application of MeJA. PDF1.2 expression was determined using real-time RT-PCR.
We next determined if HSP90 plays a role in MPK4 kinase activity following normal induction by PAMPs. WT Arabidopsis plants were co-infiltrated with GDA and the hrcC mutant strain, which is considered to carry a collection of PAMPs, and the endogenous MPK4 protein was isolated by immunoprecipitation. An in vitro kinase assay demonstrated that the MPK4 activity was greatly reduced in GDA-treated plants (Figure 6D). We further tested the role of HSP90 in MPK4 activation by flg22, a well studied PAMP, by transiently expressing MPK4-3xFLAG in protoplasts. While the control protoplasts treated with flg22 showed strong MPK4 activity, protoplasts exposed to GDA showed a marked reduction of MPK4 activity. Furthermore, JA-induced PDF1.2 expression was completely abolished by GDA treatment (Figure 6E). Together these results indicated that MPK4 is directly regulated by HSP90, with the latter playing a critical role in regulating MPK4 activity and JA signaling.
RIN4 Positively Regulates PDF1.2 Expression Downstream of MPK4
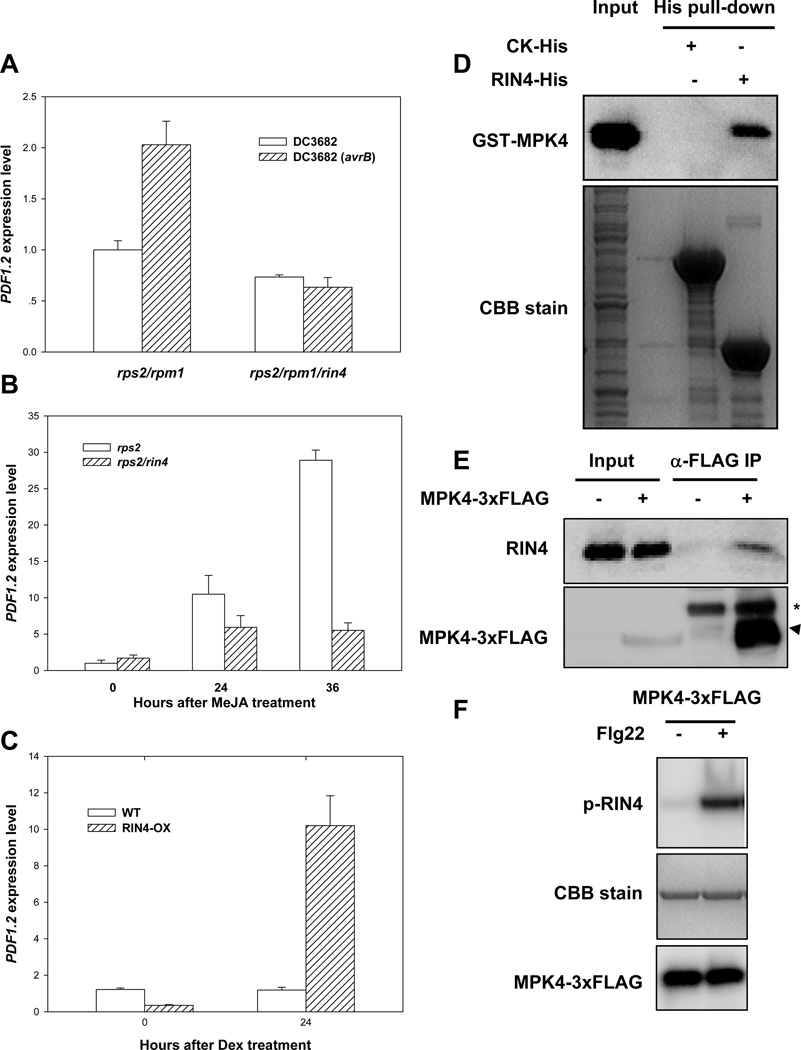
Because AvrB is known to interact with RIN4 (Mackey et al., 2002), and RIN4 negatively regulates disease resistance to P. syringae (Kim et al., 2005), we asked if RIN4 is involved in AvrB-induced PDF1.2 expression and plant susceptibility. Figure 7A shows that avrB was able to induce PDF1.2 expression in rps2/rpm1 mutant plants, but not rps2/rpm1/rin4 mutant plants, indicating that RIN4 is required for AvrB to induce JA signaling. We next transformed the AvrB transgene in rps2/rin4 mutant plants. The transgenic plants did not show enhanced growth of the P. syringae hrcC bacteria (Figure S5A). Together these results are consistent with a role of RIN4 in the AvrB-induced PDF1.2 expression and plant susceptibility. To test if RIN4 normally plays a role in JA responses, we treated rps2/rin4 mutant plants with MeJA and examined PDF1.2 expression. Figure 7B shows that the JA-induced PDF1.2 expression was significantly compromised in rps2/rin4 mutant compared to the rps2 control plants. To further determine a role of RIN4 in JA responses, we examined a transgenic line that overexpresses RIN4 (Kim et al., 2005). Figure 7C shows that plants overexpressing RIN4 constitutively expressed PDF1.2. Together these results indicate that RIN4 positively modulates JA responses.
Figure 7. RIN4 Mediates PDF1.2 Induction Down-Stream of MPK4.
(A) RIN4 is required for PDF1.2 induction by bacterially delivered AvrB. Plants of the indicated genotypes were infiltrated with the indicated bacterial strains and RNA was isolated 6 hr later for gene expression analysis. (B) RIN4 is required for JA-induced PDF1.2 expression. Plants of the indicated genotypes were treated with MeJA at the indicated times before RNA isolaton. (C) Overexpression of RIN4 constitutively activates PDF1.2 expression. WT or RIN4 transgenic (RIN4-ox) plants were treated with dexmethosome as described (Kim et al., 2005) for the indicated times before RNA was isolated for PDF1.2 expression analysis. (D) RIN4 interacts with MPK4 in vitro. Recombinant GST-MPK4 protein was incubated with bacterial lysates containing RIN4-His or CK-His (negative control as in Fig. 7A). (E) RIN4 interacts with MPK4 in plants. Protein extract from transgenic plants carrying NP-MPK4-3xFLAG was immunoprecipitated with an agarose-conjugated anti-FLAG antibody, and the presence of RIN4 in the immune complex was determined by immunoblot using anti-RIN4 antibodies. The presence of MPK4-3xFLAG was determined by immunoblot using anti-FLAG antibodies. Arrow head indicates MPK4-3xFLAG, whereas asterisk indicates IgG heavy chain from the anti-FLAG antibody used in immunoprecipitation. (F) MPK4 phosphorylates RIN4 in vitro. MPK4-3xFLAG was stimulated with (+) or without (−) flg22 in protoplasts, immunoprecipitated with anti-FLAG antibody, and the isolated MPK4-3xFLAG protein was incubated with recombinant RIN4 protein in an in vitro kinase assay. RIN4 phosphorylation (p-RIN4) was detected by autoradiography. CBB stain indicates amount of RIN4 protein in the gel.
Because both RIN4 and MPK4 interacted with AvrB and were required for the AvrB-induced JA responses, we tested if RIN4 and MPK4 interact. A GST pull-down assay showed that RIN4 and MPK4 indeed interacted in vitro (Figure 7D), and a co-IP assay showed that they interacted in vivo (Figure 7E). We next asked if MPK4 is capable of phosphorylating RIN4 in vitro. The MPK4-3xFLAG was expressed in protoplasts, induced by flg22, and MPK4-3xFLAG was isolated by anti-FLAG immunoprecipitation. The isolated MPK4 strongly phosphorylated recombinant RIN4 protein (Figures 7F and S5B), suggesting that RIN4 is a substrate for MPK4. In the rps2/rin4 mutant plants, the DC3682 (avrB) bacteria induced MPK4 phosphorylation normally (Figure S5C), indicating that RIN4 is not required for the AvrB-induced MPK4 phosphorylation. Taken together, these results suggest that RIN4 acts downstream of MPK4.
DISCUSSION
The study presented here shows that AvrB enhances plant susceptibility by promoting the phosphorylation of MPK4, which subsequently perturbs hormone signaling. Genetic and biochemical studies showed that this process is assisted by the molecular chaperone HSP90 and its co-chaperone RAR1. The previously identified AvrB-interacting protein RIN4 appears to act down-stream of MPK4 to regulate JA responses. Thus, the results uncover an unexplored regulatory mechanism for MPK4 that is actively promoted by a bacterial effector protein to enhance plant susceptibility.
Our observation that the plasmamembrane localized AvrB protein (Nimchuk et al., 2000) interacts with MPK4 is consistent with previous reports on MPK4. MPK4 is activated by MAP kinase kinases MKK1 and MKK2 during normal signaling in plants (Qiu et al., 2008; Gao et al., 2008). MKK1-MPK4 and MKK2-MPK4 complexes are located in both plasmamembrane and nucleus (Gao et al., 2008). It is important to note that the AvrB-induced MPK4 phosphorylation and JA responses occurs when AvrB is delivered by the P. syringae bacterium. AvrRpm1, another P. syringae effector known to interact with RIN4 (Mackey et al., 2002), does not appear to induce MPK4 phosphorylation (Qiu et al., 2008), suggesting that the induction of MPK4 phosphorylation is specific to AvrB. Several lines of evidence indicate that the AvrB-induced MPK4 phosphorylation is biologically significant. The loss of MPK4 renders plants unable to express PDF1.2 in response to JA (Petersen et al., 2000), it also prevents transgenic AvrB from inducing PDF1.2 expression and susceptibility to bacteria. Furthermore, AvrB mutants that are compromised in virulence function were unable to induce MPK4 phosphorylation. These mutants are also unable to induce JA responses.
Previous studies have indicated that MPK4 indirectly impact multiple hormone signaling pathways (Brodersen et al., 2006; Petersen et al., 2000). mpk4 plants show elevated SA signaling but a lack of JA-induced expression of PDF1.2 and are defective in the expression of a subset of ET pathway genes. The mechanism by which MPK4 regulates these pathways is not well understood. MPK4 has also been shown to sequester the transcription factor WRKY33 in the nucleus (Qiu et al., 2008). Treatment of plants with P. syringae or flg22 leads to a release of WRKY33 which then activate the transcription of a small number of genes including PAD3, which encodes a cytochrome P450 monoxygenase required for the biosynthesis of camalexin (Zhou et al., 1999). However, camalexin is not required for P. syringae resistance (Glazebrook and Ausubel, 1994), and WRKY33 does not appear to account for the hormone-response genes regulated by MPK4 (Qiu et al., 2008). mpk4 mutants over-accumulate EDS1 and PAD4 transcripts (https://www.genevestigator.com/gv; Zimmermann et al., 2004), which likely explains the overaccumulation of EDS1 protein in mpk4 (Brodersen et al., 2006). The JA-induced PDF1.2 expression can be restored by introducing eds1 mutation into mpk4 plants, suggesting that MPK4 modulates hormone signaling through EDS1 and PAD4. However, the interpretation is complicated by complex cross-talks among SA, JA and ethylene pathways. Indeed, EDS1 and PAD4 only accounted for some, but not all, MPK4-regulated ET response genes (Brodersen et al., 2006), suggesting that additional mechanisms remain to be found. It remains to be determined if the AvrB-induced PDF1.2 expression and disease susceptibility involve EDS1 and PAD4.
The AvrB-induced phosphorylation of MPK4 is consistent with the possibility that AvrB is related to protein kinases (Lee et al., 2004; Desveaux et al., 2007). R266 of AvrB makes direct contact with nucleotide, whereas D297 is a potential active site of AvrB. Both residues are required for MPK4 phsosphorylation. An intriguing possibility is that AvrB functionally mimics MKKs to phosphorylate MPK4. However, attempts to detect MPK4 phosphorylation by AvrB in vitro were unsuccessful. While a direct phosphorylation of MPK4 by AvrB cannot be ruled out, it is equally possible that AvrB may enhance the activity of upstream kinases, such as MKK1 and MKK2, to induce MPK4 phosphorylation.
HSP90 is a molecular chaperone responsible for the maturation and stability of a large number of signaling proteins in animals and fungi. In plants, the HSP90 complex is known to regulate resistance protein stability, but a role in regulating other signaling proteins has not been demonstrated. Our analysis demonstrated that Arabidopsis MPK4 interacts with HSP90, and MPK4 kinase activity is promoted by HSP90.
Our results showed that AvrB can directly interact with RAR1, MPK4, and RIN4, as indicated by our in vitro pull-down and co-IP assays. It is not clear if AvrB interacts with the three proteins simultaneously or sequentially. It remains to be determined if different domains in AvrB are involved in the interaction with different proteins. Nonetheless, the AvrB appears to interact with a CHORD domain in RAR1, and this interaction is required for AvrB-HSP90 and AvrB-MPK4 interactions in vivo. Furthermore, the AvrB-induced MPK4 phospohrylation also requires RAR1. Interestingly, RAR1 is known to enhance SGT1-HSP90 interaction (Boter et al., 2007). It is possible that RAR1 plays an important role in assisting protein-protein interactions in the HSP90 chaperone complex to promote the maturation of HSP90-associated proteins. These results are consistent with our findings that RAR1 is required for AvrB to induce plant susceptibility (Shang et al., 2006) and JA responses (this study). RAR1 does not appear to affect basal MPK4 phosphorylation. Instead, it specifically recruits AvrB to the HSP90 complex to regulate MPK4 phosphorylation.
RIN4 is known to be targeted by AvrB to trigger RPM1 resistance. Our results indicate that the AvrB-RIN4 interaction also contributes to AvrB-induced JA responses and plant susceptibility. RIN4 is required for JA-induced expression of PDF1.2, and RIN4 overexpression resulted in increased PDF1.2 expression in the absence of JA, indicating that RIN4 positively modulates JA responses. AvrB has been reported to induce RIN4 phosphorylation (Mackey et al., 2002). We were unable to detect RIN4 phosphorylation in vivo either in the presence or absence of AvrB for reasons unknown, which prevented us from testing whether MPK4 is required for AvrB-induced phosphorylation of RIN4. Nonetheless, our protein-protein interaction results indicated that RIN4, MPK4, and AvrB may exist in the same protein complex. It is formally possible that the AvrB-induced RIN4 phosphorylation is mediated by MPK4. This is supported by the in vitro phosphorylation of RIN4 by MPK4 in this study. Consistent with the idea that RIN4 acts downstream of MPK4, the rin4 mutant does not affect AvrB induces MPK4 phosphorylation.
Taken together, our results indicate that RAR1, HSP90, MPK4, and RIN4 constitute a new pathway modulating JA signaling and, possibly, other hormone signaling. This pathway is directly targeted by AvrB to increase plant susceptibility to P. syringae bacteria. These findings are consistent with the proposal that RIN4 is a virulence target for AvrB to enhance plant susceptibility (Dangl and Jones, 2001) and that the resistance protein RPM1 has evolved to “guard” RIN4 to trigger disease resistance in response to AvrB.
Several recent reports showed that P. syringae effectors, including HopM1, HopU1, AvrPto, AvrPtoB, and HopAI1 (Nomura et al., 2006, Fu et al., 2007, He et al., 2006; Xiang et al., 2008; Gohre et al. 2008; Gimenez-Ibanez et al., 2009, Zhang et al., 2007), directly block immune responses by inhibiting or degrading host proteins required for plant immunity. The results presented here illustrate how a P. syringae effector protein promotes a host pathway to induce inappropriate defenses that, in turn, make the plant more susceptible to P. syringae.
EXPERIMENTAL PROCEDURES
Arabidopsis Mutants and Transgenic Lines
The following Arabidopsis materials were used in this study: rpm1 (formerly described as rps3-1; Bisgrove et al., 1994), rar1-29 (Shang et al., 2006), tao1-11 (Eitas et al., 2008), AvrB-3xFLAG/rpm1 (Shang et al., 2006), mpk4 (Petersen et al., 2000), rps2/rpm1/rin4 (kim et al,. 2005), rps2/rin4 (kim et al,. 2005), and Dex::RIN4 (kim et al,. 2005). All materials are in Col-0 background except for mpk4, which is in Ler background..
Construction of Transgenic Plants Expressing MPK4-3xFLAG under the Control of MPK4 Native Promoter
The MPK4 coding sequence was PCR-amplified from cDNA using primers 5’-AGACTCGAGATGTCGGCGGAGAGTTGTTTCG-3’ and 5’-ATAACTAGTCACTGAGTCTTGAGGATTGAAC-3’, and inserted between the XhoI and Csp45I sites of pUC19-35S-FLAG-RBS (Li et al. 2005) to generate pUC19-35S-MPK4-FLAG-RBS. A 0.9kb upstream sequence of MPK4 was PCR-amplified using primers 5’-GAGGAATTCTCAATCGGTGCTAAGCTATAAC-3’ and 5’-TGTGGTACCCGGAGCAAAATTCCTCACAAC-3’. The 35S promoter of pUC19-35S-MPK4-FLAG-RBS was replaced with the MPK4 upstream fragment to generate pUC19-NP-MPK4-FLAG-RBS. Then the NP-MPK4-FLAG-RBS was transferred to pCAMBIA1300 with the EcoRI and SalI sites. The construct was transformed into Arabidopsis plants (Col-0) by Agrobacterium-mediated transformation. Transgenic plants were selected on Murashige and Skoog plates containing 25 mg/L hygromycin.
Construction of AvrB-3xFLAG/rar1-29/rpm1, AvrB-3xFLAG/mpk4/rpm1 and AvrB-3xFLAG/tao1/rpm1
An estradiol-inducible AvrB transgenic line AvrB-3xFLAG/rpm1 (Shang et al., 2006) was crossed to mpk4 and tao1-11 mutant lines to generate AvrB-3xFLAG /mpk4/rpm1 and AvrB-3xFLAG/tao1/rpm1 lines. The corresponding control lines AvrB-3xFLAG /MPK4/rpm1 and AvrB-3xFLAG/TAO1/rpm1 were siblings of the AvrB-3xFLAG /mpk4/rpm1 and AvrB-3xFLAG/tao1/rpm1 mutant lines. The presence of AvrB-3xFLAG and sgt1b, mpk4 and tao1 mutations was confirmed by PCR-based genotyping. The mpk4 mutation was caused by a transposon insertion (Petersen et al., 2000). The presence of WT MPK4 sequence was verified by using primers 5’- TTGAAGTTCTCTCTCTGCGG-3’ and 5’-GTATGTTCCTCCTCTTCGTC-3’. The presence of the transposon was verified by using primers 5’-TATGACTGGGCACAACAGAC-3’ and 5’-TATGTCCTGATAGCGGTCCG-3’. The tao1-11 mutant carries a T-DNA insertion. The presence or absence of the WT TAO1 sequence was verified by using primers 5’- CCCCTAAAGTTGGTTTTGAGC-3’ and 5’-AAATCAGGAAGCTCCTTCAGG-3’. The presence of the AvrB-3xFLAG transgene was verified by using primers 5’-CAGTAAGTCGAATACGCCTGAA-3’ and 5’-AAATCGGAAGATATTGCTTGTC-3’. F2 plants that were heterozygous for mpk4 were inoculated with P. syringae DC3000 (avrB) at 108 CFU/ml, and plants failed to develop HR were identified as homozygous rpm1 plants. Plants carrying AvrB-3xFLAG that were homozygous for mpk4 were then identified in the F3 generation. To generate AvrB-3xFLAG/rar1-29/rpm1 plants, the AvrB-3xFLAG was introduced into rar1-29/rpm1 plants (Shang et al., 2006) by transformation, and a line expressing a similar amount of AvrB-3xFLAG compared to the AvrB-3xFLAG/rpm1 transgenic line was used for experiments.
Bacterial Strains
Bacterial strains used in this study include DC3682 and a hrcC mutant derived from DC3000 (He et al., 2004, Yuan et al., 1996). WT avrB, avrBT125A, avrBR266G and avrBD297A mutant plasmids (Ong and Innes, 2006) were introduced into DC3682 in this study.
Plant Treatment
Five-week-old plants were sprayed once with 20 µM estradiol solution containing 0.01% silwet L-77 to induce the transgenic AvrB-3xFLAG expression. For bacterial delivery of AvrB, plants were infiltrated with DC3682 bacteria carrying avrB at 2×106 CFU/mL (for gene expression assay) or 2 ×108 CFU/mL (for MPKs activation assay). For JA-treatment, plants were infiltrated with 50 µM MeJA. To inhibit HSP90 activity, leaves were infiltrated with 2.5 µM GDA in 0.1% DMSO. 0.1% DMSO was used as control.
Bacterial Growth Assay
Five-week-old plants pre-induced with estradiol for 48 hr were infiltrated with hrcC mutant bacteria (5×105 CFU/ml), and bacterial populations were determined at the indicated times. Each data point consisted of at least six replicates. To inhibit HSP90 activity, GDA was mixed with the bacterial suspension prior to infiltration. To determine whether GDA alone affects bacteria growth, P. syringae hrcC bacteria was grown in luquid KB medium with GDA, and optical density at OD600 was measured hourly during the exponential phase.
Quantitative RT-PCR
The transcript levels of PDF1.2 and THI2.1 were determined by real-time RT-PCR using SYBR Premix Ex Taq™ kit (TaKaRa) following the manufacturer’s instructions. Actin was used as a reference gene. Primers 5’-GGTGTCATGGTTGGTATGGGTC-3’ and 5’-CCTCTGTGAGTAGAACTGGGTGC-3’ were used for PDF1.2, 5’-TTGGGTAAACGCCATTCTCG-3’ and 5’-ACATTGTTCCGACGCTCCAT-3’ were used for THI2.1, and primers 5’-TGGTGGAAGCACAGAAGTTG-3’ and 5’-GATCCATGTTTGGCTCCTTC-3’ were used for actin.
Constructs for GST- and His- Fusion Proteins
To construct GST-fusion plasmids, coding sequences were PCR-amplified from cDNA and inserted into pGEX-6P-1 (GE Healthcare Life Science) between BamHI and Sal I (for GST-HSP90.1, GST-MPK4, and GST-RAR1), EcoRI and SmaI (for GST-SGT1b) or EcoRI and XhoI ( for GST-CHORDI-CC and GST-CHORDI ). To construct His-fusion plasmids, coding sequences were inserted into pET30a (company name) between BamHI and SalI (for MPK4-His and HSP90-His), KpnI and EcoRI (for SGT1b-His) or KpnI and SalI (for AvrB-His), or BamHI and SalI (for RIN4-His). PCR primers used for these constructs were: 5’-ATTGAATTCGCCAAGGAATTAGCAGAGAAAG-3’ and 5’-ATGCCCGGGATACTCCCACTTCTTGAGCTCC-3’ for GST-SGT1b, 5’-CTCGGATCCATGGAAGTAGGATCTGCAACG-3’ and 5’-ATTGTCGACGACCGCCGGATCAGGGCTGCTG-3’ for GST-RAR1, 5’-AGCGAATTCATGGAAGTAGGATCTGCAAC-3’ and 5’-ATCCTCGAGTCATTGATTAATGTCTATCAC-3’ for GST-CHORDI-CC, 5’-AGCGAATTCATGGAAGTAGGATCTGCAAC-3’ and 5’-ATCCTCGAGTCAAACTGGTTTCTCAGTTGTG-3’ for GST-CHORDI, 5’-TAGGATCCGTTGCGATGGCGGATGTTCAG-3’ and 5’-TTAGTCGACTTCCTCCATCTTGCTCTC-3’ for GST-HSP90.1 and HSP90.1-His, 5’- ATAGGATCCATGTCGGCGGAGAGTTGTTTCG-3’ and 5’-ATAGTCGACCACTGAGTCTTGAGGATTGAAC-3’ for GST-MPK4 and MPK4-His, 5’-AGAGGTACCATGGCCAAGGAATTAGCAGAG-3’ and 5’-ATGGAATTCATACTCCCACTTCTTGAGCTCC-3’ for SGT1b-His, 5’-ATCGGTACCATGGGCTGCGTCTCGTC-3’ and 5’-ATAGTCGACAAAGCAATCAGAATCTAGC-3’ for AvrB-His, 5’-GACGGATCCATGGCACGTTCGAATGTACC-3’ and 5’-GAGGTCGACTTTTCCTCCAAAGCCAAAGC-3’ for RIN4-His.
GST Pull-Down and His Pull-Down Assays
For GST pull-down assays, bacterial cells expressing GST-fusion protein were lysed in a buffer containing: 25 mM Tris, PH7.5, 150 mM NaCl, 3 mM DTT. Approximately 20 mg bacterial lysate was incubated with 30 µL glutathione agarose beads in a centrifuge tube for 30 min, and washed once with GST wash buffer: 25 mM Tris, PH7.5, 150 mM NaCl, 3 mM DTT and 0.2% triton x-100. 20 mg bacterial lysate containing His-tagged protein was then added to the agarose beads and incubated for 1 hr and washed three times with the wash buffer. The bound protein was eluted with 100 µl 15 mM reduced GSH (25mM Tris, PH9.0). Approximately 1% of input and one fifth of eluted protein were subjected to immunoblot analysis.
For His pull-down assays, approximately 20 mg bacterial lysate containing His-tagged protein was incubated with 30 µL Ni-NTA agarose (QIAGEN) in a centrifuge tube for 30 min, washed once with His wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole pH 8.0). The agarose beads were then incubated with 20 mg bacterial lysate containing GST-tagged protein for 1 hr, and washed three times with the His wash buffer. The bound protein was eluted with 100 µl elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole pH 9.0). Approximately 1% of input and one fifth of eluted protein100 µl were subjected to immunoblot analysis.
Protoplast Preparation and Transfection
Protoplast preparation and transfection were essentially as described (Li et al., 2005), except that the transfected protoplasts were incubated in W5 medium (154 mM NaCl, 125 mM CaCl2, 5mM KCl, and 2 mM MES pH 5.7) instead of 0.4 M mannitol.
Immunoprecipitation
To isolate MPK4 and MPK6 proteins for MPK phosphorylation assays, five-week old plants were treated with estradiol for 36 hr to induce AvrB-3xFLAG. Protein was extracted with IP buffer (50 mM HEPES, pH7.5, 50 mM NaCl, 10 mM EDTA, 0.1% Triton, 1mM DTT, 2 mM NaF, and protease inhibitor cocktail; Roche). Approximately 2 mg total protein lysate was incubated for 1 hr with 3 µg anti-MPK4 or anti-MPK6 antibodies at 4°C followed by incubation for 1 hr with 20 µl fast flow protein A agarose beads (Upstate). After washing the beads four times with the IP buffer, the proteins were eluted by boiling in 60 µl Laemmli loading buffer. For MPK4 activity assays, plants were infiltrated with 2×108 CFU/ml hrcC mutant bacteria along with GDA and incubated for 6 hr prior to protein extraction and immunoprecipitation. Protein A agarose beads containing MPK4 were used for MPK activity assays.
FLAG-tagged proteins were isolated by immunoprecipitation as described (Shang et al., 2006). To detect AvrB-3xFLAG-HSP90 interaction in plants, AvrB-3xFLAG/rpm1 transgenic plants were sprayed with estradiol 48 hr prior to protein extraction. To detect MPK4-3xFLAG-HSP90 interaction in plants, protein was extracted from NP-MPK4-3xFLAG transgenic plants for immunoprecipitation. To isolate MPK4-3xFLAG protein from protoplasts, protoplasts were transfected with the 35S::MPK4-3xFLAG plasmid and then induced with 100 nM flg22 15 min prior to immunoprecipitation. Where indicated, GDA or 0.1% DMSO was added 3 hr prior to flg22 treatment.
To isolate HA-tag proteins, approximately 200 µg total protein extract was incubated at 4°C for 1 hr with 2 µg mouse anti-HA monoclonal antibody (TianGen) followed by incubation with fast flow protein A agarose beads (Upstate) for 2 hours. The beads were washed six times with IP buffer, and the bound protein was eluted by boiling in 60 µl Laemmli loading buffer.
MPK Activity Assay
The activity of MPK4 and MPK4-3xFLAG protein was determined by using myelin basic protein as a substrate. 4 µl protein A agarose beads containing IgG-MPK4 complex or 2 µL MPK4-3xFLAG eluted from anti-FLAG immunoprecipitation were incubated with 1 µg MBP in 20 µl reaction buffer containing: 50 mM HEPES, PH7.4, 10mM MgCl2, 1mM DTT, 50 uM ATP and 1 µCi γ-32P ATP. Reactions were allowed to proceed for 30 min at 30°C and terminated by adding Laemmli buffer and boiling for 5 min. The phosphorylation on MBP was determined by electrophoresis and autoradiography.
Immunoblot Analysis
Primary antibodies used for immunoblots included rabbit anti-MPK4 and anti-MPK6 antibodies (Sigma), rabbit anti-HSP90 (Chen et al., 2009), rabbit anti-RAR1 antibodies (Shang et al., 2006), mouse anti-HA monoclonal antibody (TianGen), mouse anti-FLAG monoclonal antibody (Sigma), and rabbit anti-phospho-ERK1 antibodies (Cell signaling). Secondary antibodies included HRP-conjugated mouse anti-rabbit light chain monoclonal antibody (for total and phosphorylated MPK4 and MPK6; CHEMICON), HRP-conjugated goat anti-rabbit antibodies (for HSP90 and RAR1; Sigma), HRP-conjugated goat anti-mouse antibodies (for HA-tagged proteins; Sigma). In co-immunoprecipitation experiments, approximately 1% of input and one third of eluted protein were subjected to immunoblot analysis.
Protein samples were electrophoresed through a 10% SDS/PAGE gel. Protein was electrotransferred to an Immobilon P membrane (Millipore). Immunodetection was performed with a 1:5,000 dilution of anti-RAR1, anti-MPK4, anti-MPK6, anti-HSP90, anti-HA, and anti-FLAG antibodies or a 1:2,500 dilution of anti-phospho-ERK1 or anti-RAR1 antibodies. The blot was then hybridized with HRP-conjugated secondary antibody and visualized with ECL or ECL Plus Western blotting detection reagents (GE Healthcare, Amersham™), following manufacturer’s instructions.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Eric Hulob, John Mundy, David Mackey and Jeffery Dangl for providing Arabidopsis mutants and Jijie Chai, Daoxin Xie for critical reading of the manuscript. R.W. Innes was supported by the National Institutes of Health, National Institute of General Medical Sciences (GM045461). J.-M. Zhou was supported by a grant from the Chinese Ministry of Science and Technology (2003-AA210080).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ashfield T, Keen NT, Buzzell RI, Innes RW. Soybean resistance genes specific for different Pseudomonas syringue avirwlence genes are allelic, or closely linked, at the RPG1 locus. Genetics. 1995;141:1594–1604. doi: 10.1093/genetics/141.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW. Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell. 2004;16:309–318. doi: 10.1105/tpc.016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noel L, Sadanandom A, Casais C, Parker J, Shirasu K. Role of SGT1 in resistance protein accumulation in plant immunity. Embo J. 2006;25:2007–2016. doi: 10.1038/sj.emboj.7601084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 2002;295:2073–2076. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- Bittel P, Robatzek S. Microbe-associated molecular patterns (MAMPs) probe plant immunity. Curr. Opin. Plant Biol. 2007;10:335–341. doi: 10.1016/j.pbi.2007.04.021. [DOI] [PubMed] [Google Scholar]
- Boter M, Amigues B, Peart J, Breuer C, Kadota Y, Casais C, Moore G, Kleanthous C, Ochsenbein F, Shirasu K, et al. Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell. 2007;19:3791–3804. doi: 10.1105/tpc.107.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Bjorn Nielsen H, Zhu S, Newman MA, Shokat KM, Rietz S, Parker J, Mundy J. Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 2006;47:532–546. doi: 10.1111/j.1365-313X.2006.02806.x. [DOI] [PubMed] [Google Scholar]
- Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, Cai R, Zuo J, Tang X, Li X, et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell. 2009;21:2527–2540. doi: 10.1105/tpc.108.065193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc. Natl. Acad. Sci. USA. 2007;104:20131–20136. doi: 10.1073/pnas.0704901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnac S, Lindeberg M, Collmer A. Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr. Opin. Microbiol. 2009;12:53–60. doi: 10.1016/j.mib.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bogre L, Grant M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007;26:1434–1443. doi: 10.1038/sj.emboj.7601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D, Singer AU, Wu AJ, McNulty BC, Musselwhite L, Nimchuk Z, Sondek J, Dangl JL. Type III effector activation via nucleotide binding, phosphorylation, and host target interaction. PLoS Pathog. 2007;3:e48. doi: 10.1371/journal.ppat.0030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitas TK, Nimchuk ZL, Dangl JL. Arabidopsis TAO1 is a TIR-NB-LRR protein that contributes to disease resistance induced by the Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. USA. 2008;105:6475–6480. doi: 10.1073/pnas.0802157105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, et al. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–288. doi: 10.1038/nature05737. [DOI] [PubMed] [Google Scholar]
- Gao M, Liu J, Bi D, Zhang Z, Cheng F, Chen S, Zhang Y. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008;18:1190–1198. doi: 10.1038/cr.2008.300. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 2009;19:423–429. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Ausubel F. Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA. 1994;91:8955–8959. doi: 10.1073/pnas.91.19.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohre V, Spallek T, Haweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 2008;18:1824–1832. doi: 10.1016/j.cub.2008.10.063. [DOI] [PubMed] [Google Scholar]
- Gopalan S, Bauer DW, Alfano JR, Loniello AO, He SY, Collmer A. Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- Han Z, Xing X, Hu M, Zhang Y, Liu P, Chai J. Structural basis of EZH2 recognition by EED. Structure. 2007;15:1306–1315. doi: 10.1016/j.str.2007.08.007. [DOI] [PubMed] [Google Scholar]
- He P, Chintamanani S, Chen Z, Zhu L, Kunkel BN, Alfano JR, Tang X, Zhou JM. Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J. 2004;37:589–602. doi: 10.1111/j.1365-313x.2003.01986.x. [DOI] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nurnberger T, Sheen J. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell. 2006;125:563–575. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- Holt BF, III, Belkhadir Y, Dangl JL. Antagonistic Control of Disease Resistance Protein Stability in the Plant Immune System. Science. 2005;309:929–932. doi: 10.1126/science.1109977. [DOI] [PubMed] [Google Scholar]
- Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 2003;22:5679–5689. doi: 10.1093/emboj/cdg547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MG, da Cunha L, McFall AJ, Belkhadir Y, DebRoy S, Dangl JL, Mackey D. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Lee CC, Wood MD, Ng K, Andersen CB, Liu Y, Luginbuhl P, Spraggon G, Katagiri F. Crystal structure of the type III effector AvrB from Pseudomonas syringae. Structure. 2004;12:487–494. doi: 10.1016/j.str.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Li X, Lin H, Zhang W, Zou Y, Zhang J, Tang X, Zhou JM. Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl. Acad. Sci. USA. 2005;102:12990–12995. doi: 10.1073/pnas.0502425102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Kenton P, Atzorn R, Miersch O, Wasternack C. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 2006;140:251–261. doi: 10.1104/pp.105.072348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk Z, Marois E, Kjemtrup S, Leister RT, Katagiri F, Dangl JL. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell. 2000;101:353–363. doi: 10.1016/s0092-8674(00)80846-6. [DOI] [PubMed] [Google Scholar]
- Nomura K, Debroy S, Lee YH, Pumplin N, Jones J, He SY. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- Ong LE, Innes RW. AvrB mutants lose both virulence and avirulence activities on soybean and Arabidopsis. Mol. Microbiol. 2006;60:951–962. doi: 10.1111/j.1365-2958.2006.05162.x. [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, et al. Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103:1111–1120. doi: 10.1016/s0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- Qiu JL, Zhou L, Yun BW, Nielsen HB, Fiil BK, Petersen K, et al. Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 2008;148:212–222. doi: 10.1104/pp.108.120006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Navarro L, Bari R, Jones JD. Pathological hormone imbalances. Curr. Opin. Plant Biol. 2007;10:372–379. doi: 10.1016/j.pbi.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Shan X, Zhang Y, Peng W, Wang Z, Xie D. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J Exp Bot. 2009 doi: 10.1093/jxb/erp223. xxx. [DOI] [PubMed] [Google Scholar]
- Shang Y, Li X, Cui H, He P, Thilmony R, Chintamanani S, Zwiesler-Vollick J, Gopalan S, Tang X, Zhou JM. RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. USA. 2006;103:19200–19205. doi: 10.1073/pnas.0607279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol. 2009;60:139–164. doi: 10.1146/annurev.arplant.59.032607.092906. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X. Making sense of hormone crosstalk during plant immune responses. Cell Host & Microbe. 2008;3:248–351. doi: 10.1016/j.chom.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Casais C, Ichimura K, Shirasu K. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2003;100:11777–11782. doi: 10.1073/pnas.2033934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, et al. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 2008;18:74–80. doi: 10.1016/j.cub.2007.12.020. [DOI] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, Cheng Z, Peng W, Luo H, Nan F, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis Microarray Database and Analysis Toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host & Microbe. 2007;1:175–185. doi: 10.1016/j.chom.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Zhou JM, Chai J. Plant pathogenic bacterial type III effectors subdue host responses. Curr. Opin. Microbiol. 2008;11:179–185. doi: 10.1016/j.mib.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Glazebrook J. Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome p450 monooxygenase. Plant Cell. 1999;11:2419–2428. doi: 10.1105/tpc.11.12.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.