Abstract
Nitrogenase is a two-component enzyme that catalyzes the nucleotide-dependent reduction of N2 to 2 NH3. This process involves three redox-active metal-containing cofactors including a [4Fe-4S] cluster, an eight-iron P cluster and a seven-iron plus molybdenum FeMo-cofactor, the site of substrate reduction. A deficit-spending model for electron transfer has recently been proposed that incorporates protein conformational gating that favors uni-directional electron transfer among the metalloclusters for activation of the substrate binding site. Also reviewed is a proposal that each of the metal clusters cycles through only two redox states of the metal-sulfur core as the system accumulates the multiple electrons required for substrate binding and reduction. In particular, it was suggested that as FeMo-cofactor acquires the four electrons necessary for optimal binding of N2, each successive pair of electrons is stored as an Fe-H−-Fe bridging hydride, with the FeMo-cofactor metal-ion core retaining its resting redox state. We here broaden the discussion of stable intermediates that might form when FeMo-cofactor receives an odd number of electrons.
Introduction
Nitrogenase is the complex, two-component metalloenzyme that catalyzes the nucleotide-dependent reduction of N2 and protons [1–3]. The physiologically relevant reaction (8H+ + 8e− + N2 → H2 + 2NH3), as well as the multi-electron reduction of other artificial substrates, such as acetylene, requires both catalytic partner proteins and the hydrolysis of two MgATP for each electron delivered to the substrate. In the absence of N2 or artificial substrates, the enzyme continues to catalyze ATP hydrolysis and proton reduction yielding H2. Key questions related to nitrogenase catalysis are framed in Figures 1 and 2 and include: What is the nature of inter- and intra-molecular electron transfer events associated with the process?; How is nucleotide binding and hydrolysis linked to electron transfer?; Where and how are the multiple electrons necessary for substrate activation and reduction accumulated within the system?; and Where do substrates bind? In this brief review, we summarize recent advances in understanding some of the features of the nitrogenase catalytic mechanism.
Figure 1.
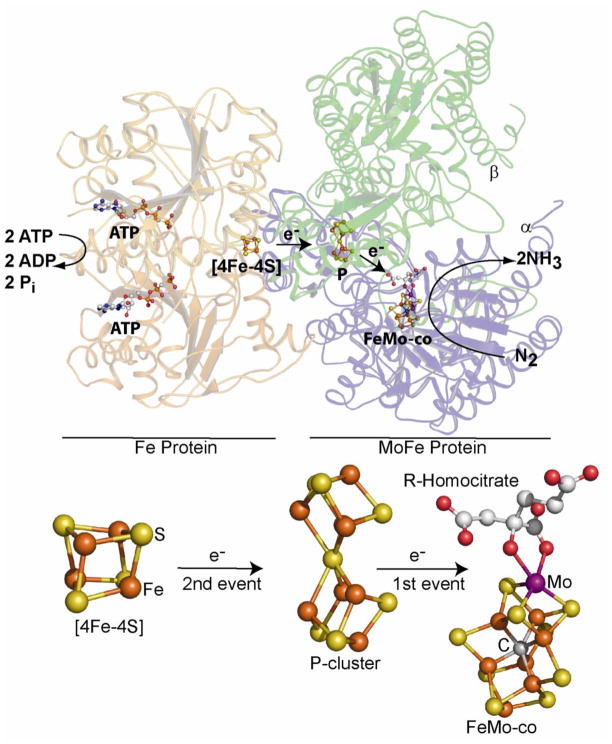
Nitrogenase proteins and cofactors. Shown are the Fe protein (left) and an αϐ-unit of the MoFe protein (right) along with two ATP molecules, the [4Fe-4S] cluster of the Fe protein, and the P cluster and the FeMo-cofactor of the MoFe protein. Atom colors are Fe in rust, S in yellow, C in gray, O in red, and Mo in purple. This figure was made using the computer program PyMol from the PDB file 2AFK.
Figure 2.
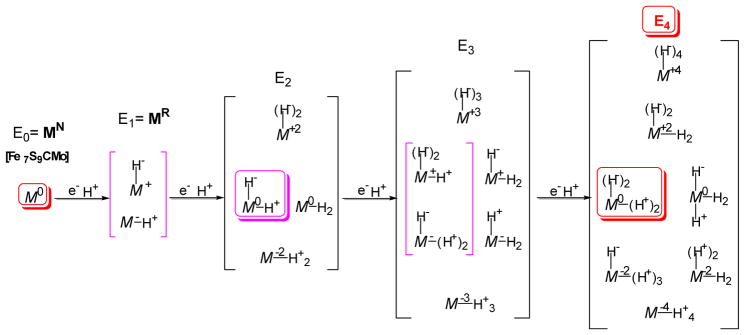
The possible redox states available to the metal-ion core of the FeMo-cofactor at each stable intermediate of the Lowe-Thorneley scheme during activation by sequential addition to the MoFe protein of up to four [e−/H+]. The symbol Mk represents the FeMo-cofactor metal core with the superscript indicating the oxidation state of the metal core relative to that of resting-state FeMo-cofactor. Red symbols represent states that have been characterized by ENDOR spectroscopy; magenta boxes represent the proposed core alternatives in stable Lowe-Thorneley intermediates.
Nitrogenase electron transfer reactions
During catalysis, the Fe protein [4Fe-4S] cluster cycles between 1+ and 2+ redox states in a dynamic process that involves the association and dissociation of the Fe protein and the MoFe protein and the obligate hydrolysis of two MgATP for each net electron transfer [3,4]. As will become relevant later in our discussion, at least three, and probably four, electrons must be accumulated within the MoFe protein before N2 can bind to the active site FeMo-cofactor [5].
Previous studies established that delivery of an electron to nitrogenase during catalysis involves two types of electron transfer events (Figure 1): one such event is intermolecular electron transfer between the [4Fe-4S]2+/1+ cluster of the Fe protein and the [8Fe-7S] “P-cluster” located within the MoFe protein; the other is intramolecular electron transfer between the P cluster and FeMo-cofactor that provides the substrate reduction site [5,6]. These studies did not, however, establish the order of events. Two plausible models, designated here as “sequential” and “deficit-spending”, can be used to describe the net electron delivery process that occurs during nitrogenase catalysis [7]. A sequential model invokes initial electron transfer from the Fe protein’s [4Fe-4S] cluster in the 1+ oxidation state to the P cluster, followed by an electron transfer from the reduced P cluster to the FeMo-cofactor. Although intuitively satisfying, the sequential model is undermined by the finding that all of the Fe atoms in the resting state of the P cluster, designated PN, are in the ferrous oxidation state [8,9]. Thus, operation of the sequential model would demand that the P cluster have the capacity to transiently exist in a super-reduced oxidation state that appears chemically untenable and for which there is no precedent in known biological FeS clusters [10–13]. In a deficit-spending model, interaction of the Fe protein and the MoFe protein elicits an initial electron transfer event involving intramolecular delivery of an electron from the P cluster to the FeMo-cofactor [7]. This situation would result in the P cluster having a “deficit” of one electron, designated P1+, relative to the all-ferrous PN resting state. The deficit could then be erased in a second step involving reduction of P1+ to yield PN through intermolecular delivery of an electron from the Fe protein’s [4Fe-4S]1+ cluster.
There is evidence that the P1+ state is chemically accessible and could be mechanistically relevant because it can be readily generated by treating the PN form of the resting state MoFe protein with oxidizing reagents [8]. The oxidation of the PN cluster to the P1+ state has been observed to be accompanied by a ligation change of the cluster, with the P1+ state having a Ser-O− group ligated to a Fe atom [14].
The overall rate of electron delivery within the normal nitrogenase system (rate constant k ~ 100 s−1) can be measured with stopped-flow spectrometry by monitoring the change in optical absorption associated with oxidation of the Fe protein’s [4Fe-4S] cluster [15–17]. Until recently, however, differentiation between the sequential and deficit-spending models could not be achieved because it has not been possible to resolve the inter- and intra-molecular electron transfer events. This technical challenge was overcome when it was fortuitously discovered that a MoFe protein having an amino acid substitution for a residue that provides a reversible P cluster ligand (ϐ-188Ser→Cys) is catalytically competent and is significantly populated in the P1+ state [18]. Stopped-flow analysis using this substituted MoFe protein revealed that electron transfer is resolved into two phases [7]. These phases include a fast step (k > 1700 s−1), which can be directly correlated with reduction of P1+ to yield PN (the second electron transfer step shown in Figure 1), and a slow step (the first electron transfer step shown in Figure 1) whose rate constant (k = 168 s−1) is the same as the overall rate constant observed in the normal system. The slow electron transfer phase is reasonably assigned to electron transfer between the PN cluster and the FeMo-cofactor because this step is eliminated, without affecting the capacity for P1+ reduction when a form of the substituted MoFe protein that does not contain the FeMo-cofactor is used in stopped-flow experiments. These observations are consistent with the deficit-spending model in which the P cluster cycles only through two redox states, PN and P1+, during catalysis.
Other stopped-flow experiments have revealed that the fast electron transfer event associated with the reduction of P1+ is unaffected by osmotic pressure [19]. In contrast, the overall electron transfer process in wild-type MoFe protein and the slow electron transfer step in the variant, now suggested to be associated with intramolecular electron transfer between the P cluster and the FeMo-cofactor, are responsive to changes in the solution osmotic pressure [7,19]. These data indicate that large protein conformational changes (> 800 Å2) precede and gate this rate-limiting electron transfer reaction. To ensure unidirectional electron flow, such conformational changes are likely to be ephemeral, which could explain why no insight along these lines has yet been gained by examination of the crystal structures of Fe protein-MoFe protein complexes [20]. Nonetheless, recent studies do hint at a possible role for amino acids within the MoFe protein as participants in gating conformational changes associated with electron transfer events. For example, substitution of histidine for the tyrosine residue located between the P cluster and FeMo-cofactor at the ϐ-98 position within the MoFe protein results in a MoFe protein that can reduce hydrazine, a nitrogenase substrate and possible mimic of an N2 reduction intermediate, in the absence of the Fe protein [21]. Such reduction only occurs if a powerful small molecule electron donor, such as ligated Eu(II), is used as a substitute for the Fe protein. In this case, it could be that the single amino acid change induces a conformation of the MoFe protein, usually achieved only transiently during docking with the Fe protein that permits substrate reduction. The relative roles of ATP binding and hydrolysis in these processes remains unclear. The recent work is consistent with the need for ATP binding to allow the electron transfer events, but it appears that ATP hydrolysis is not necessary for electron transfer, but rather may be involved in the steps that lead to the dissociation of the Fe protein from the MoFe protein [7]. Clearly, more work is needed to define the roles of ATP and how the energy of hydrolysis is captured in the mechanism. The features of the ϐ-98 substituted MoFe protein provide a starting point for future studies that will be aimed at better defining how the Fe protein, ATP binding, and hydrolysis specifically work to control nitrogenase catalysis.
FeMo-cofactor and substrate reduction
The resting state of the FeMo-cofactor is paramagnetic and exhibits a characteristic EPR signature making it amenable to characterization by a host of advanced spectroscopic techniques [2,22]. In principle, it should also be possible to characterize paramagnetic intermediates formed as the system advances through the catalytic cycle. However, such analyses have historically been frustrated by three technical problems. First, no substrates are known to bind to the resting state of the MoFe protein [3]. Instead, the active site must be activated for substrate reduction by the accumulation of multiple electrons [5]. Second, because nitrogenase has the capacity to reduce protons, the availability of which cannot be controlled, the system is continuously returned to the resting state. Third, because multiple electrons are required for the reduction of any substrate, and electron delivery cannot be synchronized, there are multiple redox states that exist under turnover conditions. These problems have been overcome, at least in part, by using a combination of approaches that involve freeze-quenching the system under turnover conditions [23], the discovery of conditions, or altered forms of the MoFe protein having amino acid substitutions that favor accumulation of intermediate states, and the development of a step-annealing procedure for determining the number of electrons accumulated by a trapped intermediate [24]. The application of these approaches has resulted in the identification of intermediate states involved in the reduction of protons [23], alkynes [25], hydrazine and diazene [26], and N2 [27], as well as interaction of the inhibitor, CO [28], with the FeMo-cofactor under turnover conditions. These studies have recently been reviewed [2].
As already mentioned, multiple electrons must accumulate within the MoFe protein before it has the capacity to bind any substrate. For example, careful kinetic studies have revealed that no fewer than three and probably four electrons must be accumulated within the system before N2 is able to bind and become activated for reduction, whereas only two electrons are required for the activation of alkyne reduction [5]. The ability of different substrates to bind the active site at different redox states explains the unusual non-reciprocity in the mutual inhibition patterns for the reduction of different substrates by nitrogenase [29]. Because electrons are delivered one at a time from the Fe protein to the MoFe protein, Thorneley and Lowe developed a convenient shorthand for describing the successive states formed under turnover conditions [5]. In the Thorneley-Lowe scheme, the resting state of the system is designated as E0, and the most stable intermediate states resulting from the sequential entry of electrons via nucleotide-dependent Fe protein delivery are denoted E1, E2, E3, and so forth.
In the first section of this review, we described evidence for a deficit-spending model where each single electron transfer results in the net delivery of an electron to the FeMo-cofactor, while leaving the P cluster in the all-ferrous PN state. Such a transient, gated process provides an elegant way to ensure unidirectional flow of electrons for substrate binding and reduction. However, given that the system must achieve the E3 or E4 state before N2 binding can occur, important questions about the nitrogenase mechanism include where and how those electrons accumulate. Figure 2 presents alternative states of the FeMo-cofactor, represented in the figure as “M”, that might exist as the system accumulates the protons and electrons necessary to achieve the E4 state. As can be seen, the metal core of the FeMo-cofactor in the En states formed during activation to E4 might be reduced or oxidized by up to four electron equivalents, depending on whether the accumulated electrons reside on the metal ions, on bound H2, or on bound hydrides. Like the E0 resting state, the E2 and E4 states are also paramagnetic and, therefore, tractable to analysis by advanced electron spin techniques such as 1,2H and metal-ion (57Fe, 95Mo) ENDOR spectroscopy. This feature turned out to be of paramount significance in the analysis of the system.
During the course of the analysis of MoFe proteins having amino acid substitutions that make the active site more accessible [30,31], such that larger alkyne substrates can be reduced, a substituted form of the MoFe protein (α-70Val→Ile) was isolated that denies access of all substrates, except protons, to the active site [23]. When samples of this substituted MoFe protein were freeze-quenched under turnover conditions, they were found to exist in a paramagnetic state indicating that they had accumulated an even number of electrons relative to the E0 state and, thus, most likely represented the E2 or E4 state. Subsequent temperature annealing experiments established that two H2 atoms could be successively released from the trapped state, clearly indicating that the trapped form represents the E4 state having accumulated 4 electrons and 4 protons [24]. Proton [23] and 95Mo [32] ENDOR spectroscopic analysis of this trapped state revealed the presence of two metal-bound hydrides, most likely Fe-H-Fe fragments [32]. In aggregate, these findings permitted an electron inventory analysis [33] that surprisingly revealed the formal redox level of the metal-sulfur core of the FeMo-cofactor in the E4 state to be the same as that of the resting E0 state [34]. Namely, the four accumulated electrons reside not on the metal ions but, instead, must be located on the two Fe-bound hydrides. These studies, together with similar analyses of the system having a trapped alkyne reduction intermediate [33] or having the CO inhibitor bound to the metal-sulfur core [33] lead to the remarkable conclusion that throughout the nitrogenase catalytic cycle the FeMo-cofactor might cycle through only a single redox couple connecting two formal redox levels of the metal-ion core [34]: one corresponding to the resting state, designated M0 in Figure 2, and the other to the one-electron reduced state of the metal-sulfur core, designated M−.
The addition of one electron/proton to the MoFe protein results in the E1 state experimentally observed in Mossbauer experiments [9]. It was presumed that this state contains the reduced metal-ion core of FeMo-co, denoted M− in Fig 2, with the proton bound to sulfur. Upon delivery of the second electron/proton to form E2, we proposed that the metal-sulfur core of the FeMo-cofactor shuttles both electrons to one proton to form an Fe-H-Fe hydride, leaving the second proton bound to sulfur and the core at the resting-state, M0, redox level. A subsequent, analogous two-stage process would then yield the E4 state, which has two Fe-H-Fe hydrides, two sulfur-bound protons, and the core at the resting-state, M0, redox level. The E4 state is optimized for N2 binding, during which one of the Fe-H-Fe fragments plus a bound proton combine for the obligate evolution of one molecule of H2 as required by the Thorneley-Lowe kinetic model. Activation thus occurs without formation of a highly reduced metal-ion core.
An alternative description of the E1 and E3 states can also be considered. If hydride formation is energetically dominant, then upon addition of an electron/proton to the metal-ion core of the resting state (redox level, E0), the resulting reduced core M− might occur only as a transient that converts to a stable E1 intermediate that contains a bridging hydride bound to an oxidized metal-ion core, M+. Addition of the second electron/proton would then yield E2 with a hydride bound to the M0 core plus a sulfur-bound proton, as described above. Analogously, two subsequent stages of electron/proton addition would generate E4 with its two bridging hydrides, two protons bound to sulfur, and M0 core, poised for N2 binding.
Regardless of which redox couple, M0/M+ or M0/M−, is operative in the stable intermediates throughout the catalytic cycle, or whether FeMo-co utilizes a different couple at different stages (e.g., activation to form E4 vs hydrogenation of N2), the proposal that the eight metal ions of the FeMo-cofactor core access no more than two, and likely just one, redox couple, when forming stable intermediates during the eight-electron catalytic cycle might seem counterintuitive. However, other proteins routinely utilize only a single redox couple of those available to their [Fe-S] clusters, including the Fe protein [4Fe-4S] cluster and the eight-iron P cluster [35]. If the FeMo-cofactor does not utilize multiple redox couples during catalysis, then why is it constructed from so many metal ions? Our results suggest that hydride binding and substrate reduction require at least two adjacent Fe ions, and it is further likely that catalysis is modulated by the linkage of these Fe ions to a hemilabile anionic atom C that is centrally located within the metal-sulfur core of the FeMo-cofactor [36,37]. Formation of such a face and the incorporation of C could not occur with a smaller cluster, as no less than a trigonal prism of six Fe ions is needed to generate these structural features. In the FeMo-cofactor, the trigonal prismatic core of six Fe ions plus C is capped, and likely ‘tuned’, by two “anchor” ions--one Fe plus a Mo, or a V or Fe in the alternative nitrogenases.
Another noteworthy advance in the field is the discovery that both the Mo-dependent and V-dependent nitrogenases have the ability to reduce CO, albeit at extremely low levels, to yield CH3 and short-chain alkenes [38,39]. While an extremely poor substrate, CO is a very powerful inhibitor of the reduction of all nitrogenase substrates, except for proton reduction. Spectroscopic analyses have revealed there are two distinct CO binding sites within nitrogenase with each having a characteristic EPR signature [40]. It seems reasonable to expect that further analysis of the binding of CO by using the advanced spectroscopic approaches already described, together with the manipulation of the enzyme’s capacity to catalyze short-chain alkene formation, could provide considerable insight concerning exactly where and how substrates and inhibitors interact with the nitrogenase active site. Also of relevance is the intriguing possibility for using the nitrogenase catalytic mechanism to guide the design of inorganic catalysts for hydrocarbon formation using CO feedstocks.
Summary
This commentary summarizes recent studies and proposals that explain how the nitrogenase active site becomes primed for substrate binding. These advances, together with an emergent model for the reaction pathway that describes the reduction intermediates bound to the FeMo-cofactor during the catalytic process [26], now provide the basis for fresh experimental approaches aimed at fully describing the nitrogenase catalytic mechanism. A noteworthy aspect of the recent work summarized here is that the proposed models are consistent with, and grounded by, decades of kinetic, structural, and spectroscopic studies by many investigators.
Highlights.
The order of electron transfers between nitrogenase metal clusters is described.
The core of each nitrogenase metal cluster cycles through a single redox couple.
Hydrides bridging Fe ions of FeMo-cofactor ‘store’ reducing equivalents.
Acknowledgments
The authors thank the National Institutes of Health for support (GM59087 to LCS and DRD and HL63203 and HL13531 to BMH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Lance C. Seefeldt, Email: lance.seefeldt@usu.edu.
Brian M. Hoffman, Email: bmh@chemistry@northwestern.com.
Dennis R. Dean, Email: deandr@vt.edu.
References
- 1.Seefeldt LC, Hoffman BM, Dean DR. Mechanism of Mo-dependent nitrogenase. Annu Rev Biochem. 2009;78:701–722. doi: 10.1146/annurev.biochem.78.070907.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffman BM, Dean DR, Seefeldt LC. Climbing nitrogenase: toward a mechanism of enzymatic nitrogen fixation. Acc Chem Res. 2009;42:609–619. doi: 10.1021/ar8002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgess BK, Lowe DJ. Mechanism of molybdenum nitrogenase. Chem Rev. 1996;96:2983–3012. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 4.Hageman RV, Burris RH. Nitrogenase and nitrogenase reductase associate and dissociate with each catalytic cycle. Proc Natl Acad Sci US A. 1978;75:2699–2702. doi: 10.1073/pnas.75.6.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thorneley R, Lowe D. Molybdenum Enzymes. Wiley-Interscience Publications; 1985. Kinetics and mechanism of the nitrogenase enzyme; pp. 221–284. [Google Scholar]
- 6.Smith BE, Lowe DJ, Chen GX, O’Donnell MJ, Hawkes TR. Evidence on intramolecular electron transfer in the MoFe protein of nitrogenase from Klebsiella pneumoniae from rapid-freeze electron-paramagnetic-resonance studies of its oxidation by ferricyanide. Biochem J. 1983;209:207–213. doi: 10.1042/bj2090207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7••.Danyal K, Dean DR, Hoffman BM, Seefeldt LC. Electron transfer within nitrogenase: evidence for a deficit-spending mechanism. Biochemistry. 2011;50:9255–9263. doi: 10.1021/bi201003a. Using a MoFe protein with an amino acid substitution near the P cluster and stopped-flow spectroscopy, the authors separate the two nitrogenase electron transfer events, allowing assignment of the order of electron transfer events. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindahl PA, Papaefthymiou V, Orme-Johnson WH, Münck E. Mössbauer studies of solid thionin-oxidized MoFe protein of nitrogenase. J Biol Chem. 1988;263:19412–19418. [PubMed] [Google Scholar]
- 9.Yoo SJ, Angove HC, Papaefthymiou V, Burgess BK, Münck E. Mössbauer study of the MoFe protein of nitrogenase from Azotobacter vinelandii using selective 57Fe enrichment of the M-centers. J Am Chem Soc. 2000;122:4926–4936. [Google Scholar]
- 10.Mouesca J-M, Noodleman L, Case DA. Analysis of the 57Fe hyperfine coupling constants and spin states in nitrogenase P-clusters. Inorg Chem. 1994;33:4819–4830. [Google Scholar]
- 11.Surerus KK, Hendrich MP, Christie PD, Rottgardt D, Orme-Johnson WH, Münck E. Mössbauer and integer-spin EPR of the oxidized P-clusters of nitrogenase: POX is a non-Kramers system with a nearly degenerate ground doublet. J Am Chem Soc. 1992;114:8579–8590. [Google Scholar]
- 12.Angove HC, Yoo SJ, Münck E, Burgess BK. An all-ferrous state of the Fe protein of nitrogenase. Interaction with nucleotides and electron transfer to the MoFe protein. J Biol Chem. 1998;273:26330–26337. doi: 10.1074/jbc.273.41.26330. [DOI] [PubMed] [Google Scholar]
- 13.Chakrabarti M, Deng L, Holm RH, Münck E, Bominaar EL. The modular nature of all-ferrous edge-bridged double cubanes. Inorg Chem. 2010;49:1647–1650. doi: 10.1021/ic902050k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peters JW, Stowell MH, Soltis SM, Finnegan MG, Johnson MK, Rees DC. Redox-dependent structural changes in the nitrogenase P-cluster. Biochemistry. 1997;36:1181–1187. doi: 10.1021/bi9626665. [DOI] [PubMed] [Google Scholar]
- 15.Thorneley RN, Lowe DJ. The mechanism of Klebsiella pneumoniae nitrogenase action. Pre-steady-state kinetics of an enzyme-bound intermediate in N2 reduction and of NH3 formation. Biochem J. 1984;224:887–894. doi: 10.1042/bj2240887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe DJ, Thorneley RN. The mechanism of Klebsiella pneumoniae nitrogenase action. Pre-steady-state kinetics of H2 formation. Biochem J. 1984;224:877–886. doi: 10.1042/bj2240877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowe DJ, Thorneley RN. The mechanism of Klebsiella pneumoniae nitrogenase action. The determination of rate constants required for the simulation of the kinetics of N2 reduction and H2 evolution. Biochem J. 1984;224:895–901. doi: 10.1042/bj2240895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan JM, Christiansen J, Dean DR, Seefeldt LC. Spectroscopic evidence for changes in the redox state of the nitrogenase P-cluster during turnover. Biochemistry. 1999;38:5779–5785. doi: 10.1021/bi982866b. [DOI] [PubMed] [Google Scholar]
- 19•.Danyal K, Mayweather D, Dean DR, Seefeldt LC, Hoffman BM. Conformational gating of electron transfer from the nitrogenase Fe protein to MoFe protein. J Am Chem Soc. 2010;132:6894–6895. doi: 10.1021/ja101737f. The authors analyze rates of electron transfer under different osmotic pressures to infer that a major protein conformational gate controls electron transfer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tezcan FA, Kaiser JT, Mustafi D, Walton MY, Howard JB, Rees DC. Nitrogenase complexes: multiple docking sites for a nucleotide switch protein. Science. 2005;309:1377–1380. doi: 10.1126/science.1115653. [DOI] [PubMed] [Google Scholar]
- 21•.Danyal K, Inglet BS, Vincent KA, Barney BM, Hoffman BM, Armstrong FA, Dean DR, Seefeldt LC. Uncoupling nitrogenase: catalytic reduction of hydrazine to ammonia by a MoFe protein in the absence of Fe protein-ATP. J Am Chem Soc. 2010;132:13197–13199. doi: 10.1021/ja1067178. A single amino acid substitution in the MoFe protein is shown to partially mimic the effects of Fe protein-ATP binding, allowing the MoFe protein to reduce substrates without the Fe protein, instead using small molecule reductants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orme-Johnson WH, Hamilton WD, Jones TL, Tso MY, Burris RH, Shah VK, Brill WJ. Electron paramagnetic resonance of nitrogenase and nitrogenase components from Clostridium pasteurianum W5 and Azotobacter vinelandii OP. Proc Natl Acad Sci USA. 1972;69:3142–3145. doi: 10.1073/pnas.69.11.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Igarashi RY, Laryukhin M, Dos Santos PC, Lee H-I, Dean DR, Seefeldt LC, Hoffman BM. Trapping H- bound to the nitrogenase FeMo-cofactor active site during H2 evolution: characterization by ENDOR spectroscopy. J Am Chem Soc. 2005;127:6231–6241. doi: 10.1021/ja043596p. [DOI] [PubMed] [Google Scholar]
- 24.Lukoyanov D, Barney BM, Dean DR, Seefeldt LC, Hoffman BM. Connecting nitrogenase intermediates with the kinetic scheme for N2 reduction by a relaxation protocol and identification of the N2 binding state. Proc Natl Acad Sci USA. 2007;104:1451–1455. doi: 10.1073/pnas.0610975104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benton PMC, Laryukhin M, Mayer SM, Hoffman BM, Dean DR, Seefeldt LC. Localization of a substrate binding site on the FeMo-cofactor in nitrogenase: trapping propargyl alcohol with an alpha-70-substituted MoFe protein. Biochemistry. 2003;42:9102–9109. doi: 10.1021/bi034595x. [DOI] [PubMed] [Google Scholar]
- 26•.Lukoyanov D, Dikanov SA, Yang Z-Y, Barney BM, Samoilova RI, Narasimhulu KV, Dean DR, Seefeldt LC, Hoffman BM. ENDOR/HYSCORE studies of the common intermediate trapped during nitrogenase reduction of N2H2, CH3N2H, and N2H4 support an alternating reaction pathway for N2 reduction. J Am Chem Soc. 2011;133:11655–11664. doi: 10.1021/ja2036018. In these studies, spectroscopic evidence is presented that favor an alternating mechanism for N2 hydrogenation in contrast to a distal mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barney BM, Lukoyanov D, Igarashi RY, Laryukhin M, Yang T-C, Dean DR, Hoffman BM, Seefeldt LC. Trapping an intermediate of dinitrogen (N2) reduction on nitrogenase. Biochemistry. 2009;48:9094–9102. doi: 10.1021/bi901092z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H-I, Hales BJ, Hoffman BM. Metal-ion valencies of the FeMo cofactor in CO-inhibited and resting state nitrogenase by 57Fe Q-band ENDOR. J Am Chem Soc. 1997;119:11395–11400. [Google Scholar]
- 29.Hwang JC, Chen CH, Burris RH. Inhibition of nitrogenase-catalyzed reductions. Biochim Biophys Acta. 1973;292:256–270. doi: 10.1016/0005-2728(73)90270-3. [DOI] [PubMed] [Google Scholar]
- 30.Dos Santos PC, Mayer SM, Barney BM, Seefeldt LC, Dean DR. Alkyne substrate interaction within the nitrogenase MoFe protein. J Inorg Biochem. 2007;101:1642–1648. doi: 10.1016/j.jinorgbio.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dos Santos PC, Igarashi RY, Lee H-I, Hoffman BM, Seefeldt LC, Dean DR. Substrate interactions with the nitrogenase active site. Acc Chem Res. 2005;38:208–214. doi: 10.1021/ar040050z. [DOI] [PubMed] [Google Scholar]
- 32•.Lukoyanov D, Yang Z-Y, Dean DR, Seefeldt LC, Hoffman BM. Is Mo involved in hydride binding by the four-electron reduced (E4) intermediate of the nitrogenase MoFe protein? J Am Chem Soc. 2010;132:2526–2527. doi: 10.1021/ja910613m. Using isotopically labeled MoFe protein, ENDOR spectroscopy, and nitrogenase trapped by rapid freezing, it is concluded that the two bound hydrides each bridge two Fe ions of FeMo-cofactor, and Mo is not involved. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Lee H-I, Sørlie M, Christiansen J, Yang T-C, Shao J, Dean DR, Hales BJ, Hoffman BM. Electron inventory, kinetic assignment (E(n)), structure, and bonding of nitrogenase turnover intermediates with C2H2 and CO. J Am Chem Soc. 2005;127:15880–15890. doi: 10.1021/ja054078x. Using isotopically labeled MoFe protein, novel spectroscopic methods, and nitrogenase trapped during turnover, it is concluded that the metal-sulfur core of FeMo-cofactor in the 4-electron reduced state is in the same oxidation state as the resting FeMo-cofactor, highlighting the importance of metal bound hydrides in the nitrogenase mechanism. It is proposed that the FeMo-cofactor core accesses only a single redox couple. [DOI] [PubMed] [Google Scholar]
- 34.Doan PE, Telser J, Barney BM, Igarashi RY, Dean DR, Seefeldt LC, Hoffman BM. (57)Fe ENDOR spectroscopy and “electron inventory” analysis of the nitrogenase E(4) intermediate suggest the metal-ion core of FeMo-cofactor cycles through only one redox couple. J Am Chem Soc. 2011;133:17329–17340. doi: 10.1021/ja205304t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beinert H, Holm RH, Münck E. Iron-sulfur clusters: nature’s modular, multipurpose structures. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 36.Lancaster KM, Roemelt M, Ettenhuber P, Hu Y, Ribbe MW, Neese F, Bergmann U, DeBeer S. X-ray emission spectroscopy evidences a central carbon in the nitrogenase iron-molybdenum cofactor. Science. 2011;334:974–977. doi: 10.1126/science.1206445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spatzal T, Aksoyoglu M, Zhang L, Andrade SLA, Schleicher E, Weber S, Rees DC, Einsle O. Evidence for interstitial carbon in nitrogenase FeMo cofactor. Science. 2011;334:940. doi: 10.1126/science.1214025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z-Y, Dean DR, Seefeldt LC. Molybdenum nitrogenase catalyzes the reduction and coupling of CO to form hydrocarbons. J Biol Chem. 2011;286:19417–19421. doi: 10.1074/jbc.M111.229344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee CC, Hu Y, Ribbe MW. Vanadium nitrogenase reduces CO. Science. 2010;329:642. doi: 10.1126/science.1191455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis LC, Henzl MT, Burris RH, Orme-Johnson WH. Iron-sulfur clusters in the molybdenum-iron protein component of nitrogenase. Electron paramagnetic resonance of the carbon monoxide inhibited state. Biochemistry. 1979;18:4860–4869. doi: 10.1021/bi00589a014. [DOI] [PubMed] [Google Scholar]