Abstract
The era of “Personalized Medicine,” guided by individual molecular variation in DNA, RNA, expressed proteins and other forms of high volume molecular data brings new requirements and challenges to the design and implementation of Electronic Health Records (EHRs). In this article we describe the characteristics of biomolecular data that differentiate it from other classes of data commonly found in EHRs, enumerate a set of technical desiderata for its management in healthcare settings, and offer a candidate technical approach to its compact and efficient representation in operational systems.
Keywords: Electronic Health Records, Genomics, Knowledge representation, Data compression
1. Introduction
High throughput technologies for analyzing DNA, DNA methylation, RNA, proteins and other biologically important molecules are an essential infrastructure for the nascent era of clinical care that is tailored to one's unique ‘molecular self.’ The availability of low-cost complete genome sequences portends a flood of molecular sequence data being generated in clinical care contexts, and the need to efficiently store, display, and use that data for healthcare purposes including patient-specific clinical decision support [1,2].
The majority of common diseases have their roots in biomolecular structures and interactions; although these molecules and interactions that make up human physiology are highly regular, specialized, redundant and fault-tolerant, their complexity and variety in the body and within and between individuals is staggering. Approximately 1.5% of the 3 billion base pairs in the human genome code for proteins [3], and each of those 45 million base pairs can acquire polymorphisms, many of them non-fatal. Further complicating the picture, the approximately 50 trillion cells in the body may undergo a total of ten quadrillion cell divisions during a human lifespan, each carrying with it further risk of genetic damage. Each of the 200 known cell types has its own gene expression profile in healthy and diseased states that may also demonstrate secular changes.
While the basic genome of individuals is likely the first, most complex source of data to challenge current EHR structures, other “omics” data such as gene expression profiles are already being used in clinical decision-making [4, 5]. The structure of unitary observations (e.g., single base pairs of DNA) is simple. However, the volume and complexity of the data and its annotation is large enough to have important implications for its storage and use within EHR systems. Based on consideration of the nature of the data, and the state of genomic science and clinical care, we sought to describe a set of desirable functional characteristics for any EHR that will incorporate individual molecular variation into the provision of healthcare services.
2. Materials and Methods
The content of this manuscript was assembled for presentation and refined by interdisciplinary group discussion at an invited workshop on “Integration of Genetic Test Results into Electronic Medical Records” convened by the National Heart Lung and Blood Institute, and held in Bethesda, MD on August 2-3, 2011.
3. Results
Table 1 presents a set of seven desiderata for the integration of genomic and other high volume biomolecular data into EHRs. We offer these functional characteristics both as a conceptual guideline for the design or extension of EHRs, and for their potential utility as evaluation criteria for the ‘meaningful use’ of EHRs to manage these types of data. The explanation and reasoning behind these desiderata are provided here.
Table 1.
Desiderata for the Integration of Genomic and other high volume biomolecular data into EHRs
| 1. Maintain separation of primary molecular observations from the clinical interpretations of those data |
| 2. Support lossless data compression from primary molecular observations to clinically manageable subsets. |
| 3. Maintain linkage of molecular observations to the laboratory methods used to generate them |
| 4. Support compact representation of clinically actionable subsets for optimal performance |
| 5. Simultaneously support human-viewable formats and machine-readable formats in order to facilitate implementation of decision support rules. |
| 6. Anticipate fundamental changes in the understanding of human molecular variation |
| 7. Support both individual clinical care and discovery science |
1. Maintain separation of primary molecular observations from the clinical interpretations of those data
A common current practice for the reporting of genetic variation by clinical laboratories is to acquire a large number of molecular observations via high throughput technologies, such as solid state chips that measure hundreds to thousands of molecular variants. Laboratories then deliver into the health record a report in document format on paper, through an electronic interface between the laboratory and EHR or as open standard for document exchange such as PDF, that cites only a small number of the observations made combined with professional interpretation of the significance of those observations. This parallels the current reporting practice in diagnostic medicine, pathology and radiology. This practice is potentially limiting in the emerging era of personalized medicine in three respects. First, it embodies a lossy sampling approach where only a subset of data is reported via a filter of professional opinion (albeit guided by then-current scientific evidence), and the remainder of the primary observation data is either discarded or held inaccessible. Second, it renders the primary data in a document format that is optimized only for human interpretation and ill-suited to the use of computer-based decision support rules. Third, it represents a point-in-time interpretation in an immature field of clinical science that is rapidly changing. The vast majority of molecular variants are currently of unknown significance, but it can be reasonably expected that determinations of significance or lack thereof will be assigned to increasing numbers of variants as genomic science evolves. Thus, the separation of primary observations from their interpretations, and the ability to update and improve those interpretations at a later date, will more significantly impact biomolecular data than other common clinical data types. Novel approaches to dynamic reporting of clinical laboratory genotyping and associated genomic knowledge bases are currently being developed [6,7]
2. Support lossless data compression from primary molecular observations to clinically manageable subsets
The large volume of each individual's DNA, protein and related data — hundreds of gigabytes to terabytes in its raw form --- exceeds the capacity of commonly available network bandwidth and disk storage in healthcare settings. In the absence of a major advance in data storage and transmission capabilities, this large volume of data will need to be compressed. Other high volume digital datasets do exist in healthcare, notably digital radiography and computed tomography, and specialized digital infrastructures (such as Picture Archiving and Communications Systems – PACS [8]) have been developed to store and display these data. A variety of data compression algorithms and image representation formats have been developed to accommodate the efficient transfer and viewing of clinical digital images. Most of these formats offer ‘lossy’ compression (reduction in file size associated with removal of data such that the ability to faithfully reconstruct all of the content of the original large volume source image is sacrificed). Since the key features of clinical images are often not exquisitely dependent upon single pixel level detail, this is a robust and useful approach to data compression for many types of health-related images.
In contrast, changing even a single letter of the ‘genetic alphabet’ (i.e. a point mutation) may dramatically affect human physiology. In some cases the significance of such changes is well known, as demonstrated by sickle cell disease and other inherited disorders [9].
Therefore, any sufficient data compression approach needs to be able to produce a fully accurate copy of the original sequence.
3. Maintain linkage of molecular observations to the laboratory methods used to generate them
Measurement technologies for DNA sequence and expressed proteins are rapidly evolving, and all are constrained by non-zero error rates and “blind spots” representing biological phenomena that are not detectable by the method. For the foreseeable future, the laboratory instruments, chemistry and methods used to obtain high throughput molecular measurements such as single nucleotide polymorphism (SNP) arrays, exome sequences and full genome sequences will continue to evolve, with successive generations of instruments having different strengths and weaknesses. Genomic sequence data representation standards such as the Genome Variation Format (GVF) [10] and the Human Genome Variation Society's nomenclature for the description of sequence variants [11] are being proposed to provide common coordination across sequencing platforms. For this reason it will be essential that EHRs maintain provenance that links molecular observations with the laboratory methods used to generate those observations. This binding of methods with results is a structural component of widely used laboratory data standards such as LOINC [12], and is a feature of proposed and evolving data standards such as HL7 [13].
4. Support compact representation of clinically actionable subsets for optimal performance
An important functionality of EHRs is the ability to rapidly find, assemble, and display the relevant clinical data for individual patients and groups of patients. Since the amount of molecular sequence data that currently has demonstrated clinical significance is a tiny fraction of the full genome and proteome, and it is neither computationally feasible nor desirable to query or analyze one's entire genome in real time to support healthcare-related decisions such as drug prescribing or diagnostic test ordering. EHR systems need to access and display relevant information, and/or recognize and act upon clinically relevant molecular patterns with sub-second response times [14]. These requirements for speed and efficiency make the creation of compact, derived forms of data representing the underlying molecular variation an attractive technical option in EHR systems. These derived observations can be efficiently represented as short “keywords” or codes representing a physiologic state. For example, the observation that an individual has a minor allele variant such as CYP2C19*2, that is associated with altered metabolism of commonly prescribed drugs, can be represented by a compact code of just a few unique alphanumeric characters or a global unique identifier from structured vocabulary/ontology such as the Clinical Bioinformatics Ontology (CBO) [15].
5. Simultaneously support human-viewable formats and machine-readable formats in order to facilitate implementation of decision support rules
In its simplest form, a single observation such as the value of a single nucleotide polymorphism, is recognizable upon inspection by a healthcare professional, and as noted above, genotyping results are commonly displayed as laboratory report documents.
However, molecular variation data introduces into clinical practice volumes of data whose complexity routinely exceeds the bounds of unaided human cognition [16]. The rapidly expanding literature on the association between molecular variation patterns and clinical phenomena [17] makes it difficult for even genetic medicine specialists to stay current, and far exceeds the interpretive capacity of most non-specialist providers. Consequently, perhaps more than for any class of clinical data that has preceded it into the EHR, molecular variation data will benefit from the implementation of clinical decision support rules that are designed to recognize key patterns (such as DNA variation that predicts altered drug response) and guide practitioners via patient-specific alerts and reminders at clinically relevant times. The inherently cryptic nature of genetic polymorphisms lobbies for systems approaches that guide not only specialists, but also providers who “do not know what they do not know” with respect to clinically important molecular variation. [18]
6. Anticipate fundamental changes in the understanding of human molecular variation
Designing EHR capacity based on the expectation that an individual has a single, unique genome will be insufficient to accommodate the actual data requirements for EHRs . This premise, commonly associated with the genome contained in germline (i.e. heritable) DNA, needs to at a minimum be modified to accommodate diseases such as cancer, in which somatic mutations occur [1, 19]. Thus, EHR systems need to anticipate circumstances such as a “unique genome for each metastasis”. The state of the germline DNA is generally inferred through sampling of leukocyte DNA, generally from blood, and less often from saliva. Emerging evidence that the DNA represented in the leukocytes may undergo structural changes as a result of normal aging [20] also suggests that as genomic science unfolds, EHRs may need to store multiple genome-scale datasets over an individual's lifetime. Other cell-, tissue- , organ-, and disease-specific genetic variations over time may yet be discovered. The oft-cited use case of storing 3 billion bases of DNA (which in reality is a minimum of 6 billion, since humans are diploid organisms) even when supplemented with data on copy number and splicing variation is only a starting point for a much larger universe of person-specific molecular variation data.
7. Support both individual clinical care and discovery science
Historically, the intersection of clinical care and biomedical research has been relatively minimal, as evidenced by the small fraction of eligible patients who enroll in clinical trials. [21, 22] Genomic science has unprecedented requirements for large numbers of individuals, each of whom has available large numbers of molecular observations, in order to confront the ‘curse of dimensionality’ (i.e. the expected false discovery rate of patterns that arise by chance when thousands to millions of simultaneous observations are made). Thus, to advance clinical science as rapidly and robustly as possible, the ability to support genomic discovery science as a secondary use of data acquired for person-specific care is at least a compelling opportunity, if not a social obligation to future generations. Such uses will be modulated by issues of consent and privacy, however a research focus on human molecular variation, and the emerging capability to measure that variation at all locations where it occurs in the genome and proteome, makes each individual's genome potentially a uniquely valuable research resource. Well-structured genomic information within the EHR will expedite secondary use of that data to support new discovery.
Discussion: A technical approach to lossless data compression for biomolecular sequences in EHRs
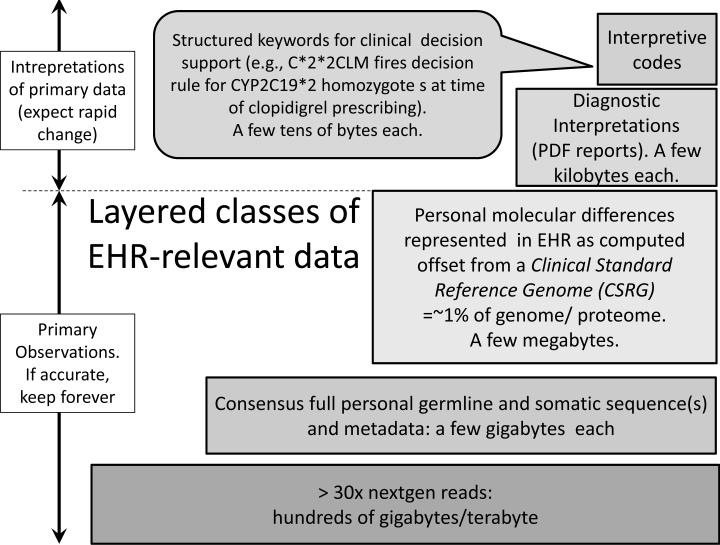
Figure 1 presents a size hierarchy of genomic data types that are of relevance to EHRs and may need to be stored and analyzed for optimal, individualized care. The current technologies such as ‘next generation’ (nextgen) DNA sequencing, with automated repeated observations of a single nucleotide base in order to assemble a consensus DNA sequence, generates primary data that occupies hundreds of gigabytes or more of disk space. Like the technology itself, the analytical software that interprets these data to generate a consensus sequence of a few gigabytes is evolving rapidly, so there is a need to preserve the source files for potential future re-analysis.
Fig. 1.
Layered classes and data sizes of EHR-relevant genomic data.
The layered approach to increasingly compact representations of lossless data compression shown in Figure 1 can benefit from an essential feature of human biology, which is that we are much more alike than we are different at a molecular level. First approximations of 1 to 4 million differences [23] contained in a roughly 3 billion nucleotide genome suggest that EHRs can achieve two orders of magnitude (100 fold) data size reduction by representing personal nucleotide and/or protein sequences as the difference between the individual and what we propose calling a “Clinical Standard Reference Genome” (CSRG). Such a sequence would not need to represent any biological reality and would best serve its purpose if it generated the smallest set of differences across a large number of complete human genomes. This would be achieved by inclusion of the most common allele at each locus in the CSRG, without regard to actual clinical, ethnic or racial data. While various groups generating sequence data use already this mode for data compression, a single standard does not currently exist. As with the binding of molecular observations to the methods used to generate them, revisions of the CSRG to reflect evolving knowledge or technology are easily managed by applying and recording a unique version identifier for each iteration. Generation and widespread use of a CSRG would be a boon to the ability to store and interpret biomolecular sequence data in EHRs without data loss relative to the source observations.
Molecular sequence data will be an important component of EHRs
The volume and complexity of the data are key features
Desiderata for representing this data in EHRs are presented
A technical approach to its efficient representation is offered
Acknowledgements
Supported in part by NIH grant 5RC2GM092618-02 (D. Masys, P.I.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ding L, Wendl MC, Koboldt DC, Mardis ER. Analysis of next-generation genomic data in cancer: accomplishments and challenges. Hum Mol Genet. 2010 Oct 15;19(R2):R188–96. doi: 10.1093/hmg/ddq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Human genome: Genomes by the thousand. Nature. 2010 Oct;467:1026–1027. doi: 10.1038/4671026a. [DOI] [PubMed] [Google Scholar]
- 3.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001 Feb 16;291(5507):1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 4.Slodkowska EA, Ross JS. MammaPrint 70-gene signature: another milestone in personalized medical care for breast cancer patients. Expert Rev Mol Diagn. 2009 Jul;9(5):417–22. doi: 10.1586/erm.09.32. [DOI] [PubMed] [Google Scholar]
- 5.Roepman P, Horlings HM, Krijgsman O, Kok M, Bueno-de-Mesquita JM, Bender R, Linn SC, Glas AM, van de Vijver MJ. Microarray-based determination of estrogen receptor, progesterone receptor, and HER2 receptor status in breast cancer. Clin Cancer Res. 2009 Nov 15;15(22):7003–11. doi: 10.1158/1078-0432.CCR-09-0449. [DOI] [PubMed] [Google Scholar]
- 6.Aronson SJ, Clark EH, Babb LJ, Baxter S, Barwell LM, Funke BH, Hernandez AL, Joshi VA, Lyon E, Parthum AR, Russell FJ, Varugheese M, Venman TC, Rehm HL. The GeneInsight Suite: a platform to support laboratory and provider use of DNA-based genetic testing. Hum Mutat. 2011 May;32(5):532–6. doi: 10.1002/humu.21470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jing X, Kay S, Marley T, Hardiker NR, Cimino JJ. Incorporating personalized gene sequence variants, molecular genetics knowledge, and health knowledge into an EHR prototype based on the Continuity of Care Record standard. Journal of Biomedical Informatics. 2011 Oct 11; doi: 10.1016/j.jbi.2011.09.001. doi:10.1016/j.jbi.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choplin R. Picture archiving and communication systems: an overview. Radiographics. 1992 Jan;12:127–129. doi: 10.1148/radiographics.12.1.1734458. [DOI] [PubMed] [Google Scholar]
- 9.Steinberg MH, Rodgers GP. Pathophysiology of sickle cell disease: Role of cellular and genetic modifiers. Seminars in Hematology. 2001 Oct;38:299–306. doi: 10.1016/s0037-1963(01)90023-x. [DOI] [PubMed] [Google Scholar]
- 10.Reese MG, Moore B, Batchelor C, Salas F, Cunningham F, Marth GT, Stein L, Flicek P, Yandell M, Eilbeck K. A standard variation file format for human genome sequences. Genome Biology. 2010;11:R88. doi: 10.1186/gb-2010-11-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. [5 December 2011];Nomenclature for the description of sequence variants. http://www.hgvs.org/mutnomen/.
- 12. [6 September 2011];Logical Observation Identifiers Names and Codes. http://www.loinc.org.
- 13.HL7 Clinical Genomics working group [6 September 2011]; http://www.hl7.org/Special/committees/clingenomics.
- 14.Bates DW, Kuperman GJ, Wang S, Gandhi T, Kittler A, Volk L, Spurr C, Khorasani R, Tanasijevic M, Middleton B. Ten commandments for effective clinical decision support: making the practice of evidence-based medicine a reality. J Am Med Inform Assoc. 2003 Nov-Dec;10(6):523–30. doi: 10.1197/jamia.M1370. Epub 2003 Aug 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deshmukh V, Hoffman M, Arnoldi C, Bray B, Mitchell J. Efficiency of CYP2C9 Genetic Test Representation for Automated Pharmacogenetic Decision Support. Methods Inf. Med. 2009;48(3):282–290. doi: 10.3414/ME0570. [DOI] [PubMed] [Google Scholar]
- 16.Masys DR. Effects of Current And Future Information Technologies on the Health Care Workforce. Health Affairs. 2002 Sep-Oct;21(5):33–41. doi: 10.1377/hlthaff.21.5.33. [DOI] [PubMed] [Google Scholar]
- 17.Hindorff LA, Junkins HA, Hall PN, Mehta JP, Manolio TA. [6 September 2011];A Catalog of Published Genome-Wide Association Studies. Available at: www.genome.gov/gwastudies.
- 18.Hoffman MA. The Genome Enabled EMR. J. Biomed. Info. 2007 Feb;40(1):44–6. doi: 10.1016/j.jbi.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Indran IR, Tufo G, Pervaiz S, Brenner C. Recent advances in apoptosis, mitochondria and drug resistance in cancer cells. Biochim Biophys Acta. 2011 Jun;1807(6):735–45. doi: 10.1016/j.bbabio.2011.03.010. Epub 2011 Mar 29. [DOI] [PubMed] [Google Scholar]
- 20.Geigl JB, Langer S, Barwisch S, Pfleghaar K, Lederer G, Speicher MR. Analysis of Gene Expression Patterns and Chromosomal Changes Associated with Aging. Cancer Res. 2004 Dec 1;64(23):8550–7. doi: 10.1158/0008-5472.CAN-04-2151. [DOI] [PubMed] [Google Scholar]
- 21.Peto V, Coulter A, Bond A. Factors affecting general practitioners’ recruitment of patients into a prospective study. Fam Pract. 1993 Jun;10(2):207–11. doi: 10.1093/fampra/10.2.207. [DOI] [PubMed] [Google Scholar]
- 22.Lara PN, Jr, Paterniti DA, Chiechi C, Turrell C, Morain C, Horan N, Montell L, Gonzalez J, Davis S, Umutyan A, Martel CL, Gandara DR, Wun T, Beckett LA, Chen MS., Jr Evaluation of factors affecting awareness of and willingness to participate in cancer clinical trials. J Clin Oncol. 2005 Dec 20;23(36):9282–9. doi: 10.1200/JCO.2005.02.6245. [DOI] [PubMed] [Google Scholar]
- 23.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, Lin Y, MacDonald JR, Pang AW, Shago M, Stockwell TB, Tsiamouri A, Bafna V, Bansal V, Kravitz SA, Busam DA, Beeson KY, McIntosh TC, Remington KA, Abril JF, Gill J, Borman J, Rogers YH, Frazier ME, Scherer SW, Strausberg RL, Venter JC. The Diploid Genome Sequence of an Individual Human. PLoS Biol. 2007 Sep 04;5(10):e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]