Abstract
The mechanisms underlying genetic susceptibility at loci discovered by genome-wide association study (GWAS) approaches in human cancer remain largely undefined. In this study we characterized the high-risk neuroblastoma association at the BRCA1-related locus, BARD1, showing that disease-associated variations correlate with increased expression of the oncogenically activated isoform, BARD1β. In neuroblastoma cells, silencing of BARD1β showed genotype-specific cytotoxic effects, including decreased substrate-adherent, anchorage-independent, and foci growth. In established murine fibroblasts, overexpression of BARD1β was sufficient for neoplastic transformation. BARD1β stabilized the Aurora family of kinases in neuroblastoma cells, suggesting both a mechanism for the observed effect and a potential therapeutic strategy. Together, our findings identify BARD1β as an oncogenic driver of high-risk neuroblastoma tumorigenesis, and more generally, they illustrate how robust GWAS signals offer genomic landmarks to identify molecular mechanisms involved in both tumor initiation and malignant progression. The interaction of BARD1β with the Aurora family of kinases lends strong support to the ongoing work to develop Aurora kinase inhibitors for clinically aggressive neuroblastoma.
Keywords: genome-wide association, neuroblastoma, BARD1, cancer susceptibility genes, functional genomics, oncogenes, genotype-phenotype correlations
Introduction
Genome-wide association studies (GWAS) have proven to be a powerful tool to identify susceptibility variants in complex disease, including cancer. However, the molecular mechanisms driving a majority of these signals remain undefined (1). In addition, GWAS efforts in cancer have largely focused on tumor initiation, and to date have not generally explored the role of implicated genes on tumor progression or clinical phenotype.
Neuroblastoma is a pediatric malignancy that arises from the developing sympathetic nervous system (2). It is the most frequently diagnosed neoplasm during infancy and accounts for up to 10% of childhood cancer mortality (3). Most neuroblastomas occur sporadically and approximately 50% of patients present with metastatic disease with survival rates below 50% despite dose-intensive chemoradiotherapy (2, 4). The genetic basis of neuroblastoma has recently come into focus, as we have discovered ALK as the major familial neuroblastoma gene and have utilized our SNP based GWAS to identify multiple genomic loci highly associated with sporadic neuroblastoma (5–10). These GWAS findings collectively provide the first evidence that common polymorphisms work in an additive fashion to influence neuroblastoma initiation. Moreover, a majority of these alleles are associated with specific neuroblastoma clinical subsets, strongly suggesting that the ultimate disease phenotype may be determined, at least in part, by initiating events.
One of the most significant and robustly replicated association signals enriched in the high-risk subset of neuroblastomas resides at the BRCA1-Associated RING Domain 1 gene (BARD1) locus at chromosome 2q35 (6, 10). BARD1 has classically been thought of as a tumor suppressor because it dimerizes with BRCA1 via their respective RING domains (11, 12). However, recent evidence suggests that increased expression of alternatively spliced BARD1 isoforms may be integral in driving cancer progression, independent of BRCA1 (11, 13–16). Here, we sought to explore the potential mechanisms of common genetic variation at the BARD1 locus in causing neuroblastoma and determine the extent to which BARD1 may also be involved in driving and maintaining neuroblastoma oncogenesis in established tumors.
Materials and Methods
Research samples and genotyping
Panel of fetal tissues and sympathetic ganglia RNA was obtained as previously described (7). Neuroblastoma tumor and constitutional samples were acquired from the Children’s Oncology Group. All genomic DNA was genotyped as previously described (7, 8, 10). LCL RNA was isolated from EBV transformed lymphocytes. Additional details can be found in supplementary methods.
Cell culture
Neuroblastoma cell lines (described in supplemental methods), RPE1-hTERT cells, and NIH 3T3 cells were regularly passaged in RPMI and routinely mycoplasma-tested and genotyped (AmpFISTR Identifiler kit, Applied Biosystems) to verify identity. HeLa cells were grown in DMEM. Additional details can be found in supplemental methods.
BARD1 CNV breakpoint cloning
Primers were designed using Primer 3. Genomic regions from constitutional DNA were PCR amplified and TA cloned into a pCR2.1-TOPO vector (Invitrogen) and sequenced. Primer sequences that span the BARD1 5′ CNV can be found in supplemental methods.
Real-time quantitative PCR validation of DNA copy number
Primers and probes were designed, synthesized; PCR reactions were set up and DNA copy number calculated as previously described (7).
Fetal ganglia and neuroblastoma cell PCR and BARD1 isoform sequencing
Total RNA from a panel of fetal sympathetic ganglia and neuroblastoma cell lines and tumors were prepared and PCR amplified as previously described (7, 9) using the following primers BARD1 F1: 5′-ATGGAACCGGATGGTC-3′ a n d B A R D 1 R 1 : 5′-CAGCTGTCAAGAGGAAGCAAC-3′, located in BARD1 exons 1 and 11, respectively. PCR products were TA cloned into a pCR2.1-TOPO vector (Invitrogen) sequenced.
Real-time quantitative RT-PCR for BARD1β expression
Primers and probes to assess BARD1β expression were designed using Primer Express 3.0 (Applied Biosystems, ABI) targeting the exon 1/4 boundary. Sequences can be found in the supplemental methods. Total LCL and neuroblastoma cell line RNA was prepared, PCR amplified and normalized to endogenous controls (TaqMan® assays, ABI) as previously described (7). Quantitative PCR for BARD1 exons 2/3 and BARD1 exons 10/11 was done similarly.
Affymetrix Human Exon 1.0 ST (HuEx) expression analysis
Total RNA from 251 primary neuroblastoma tumors was analyzed with the HuEx exon expression array (Affymetrix). Data from core probe set regions were normalized and summarized using RMA method (Affymetrix APT tools). Heat maps were generated with matrix2png software. Microarray data are available at the NCI TARGET data matrix (17).
Neuroblastoma tumor tissue microarray
The neuroblastoma TMA was constructed as previously described (18, 19). Immunohistochemical staining with antibodies against BARD1 exon 3 (PVC) and exon 4 (WFS) was done as previously described (13). Each tumor was evaluated for staining percentage and intensity (0/none to 3/intense) and a staining score was calculated (intensity × % cells; 0 to 300).
BARD1β siRNA knockdown in neuroblastoma cell lines
Neuroblastoma cells were plated in triplicate in a Real-time Excelligence system (F Hoffman La-Roche, Basal, Switzerland) and growth was monitored continuously as previously described (9). BARD1β targeting siRNA (Sequence: 3′-CUGCUCGCGUUGAUUUGAAUU-5′) was designed using the siDESIGN Center (Dharmacon) to target the exon 1/4 boundary of BARD1β and synthesized with ON-TARGET - Specificity Enhanced modifications. Cell transfection, growth inhibition calculation, and mRNA knockdown analysis was done as before (9, 10, 20) and described in detail in the supplementary methods.
Foci formation assay
NLF or Nb-Ebc1 neuroblastoma cells transfected with either BARD1β or NTC siRNA were plated in 100 mm2 dishes and grown for 3–5 weeks, fixed, stained with 0.4% crystal violet and photographed.
Soft agar tumorigenicity assay
NLF or Kelly neuroblastoma cells transfected with BARD1β or NTC siRNA were plated in soft agar in triplicate as previously described (21). Colonies were counted on the resulting JPEG images.
Apoptosis assay
NLF and Nb-Ebc1 neuroblastoma cells were transfected with BARD1β siRNA and caspase activities were measured with the Caspase-Glow 3/7 assay (Promega) and quantified relative to NTC siRNA transfected cells and further normalized to the cell number using the Cell Titer-Glo luminescent cell viability assay (Promega) performed in parallel. BARD1 transfected NIH-3T3 cells were plated in DMEM with 1% FBS and analyzed similarly.
Immunoblotting
Neuroblastoma cell lines and RPE1 cells (50 μg) were immunoblotted as previously described (9, 20). Antibodies targeting BARD1 Exon 4 (1:500, BL518, Bethyl Laboratories; A300-263A), BARD1 p25 (1:250) targeting the peptide sequence, MVAVPGTVAPRC, encoded in alternative ORF of exon 1 of BARD1β (14), Actin (1:2000, I-19; sc-1616), p53 (Cell Signaling, 2524), phospho-p53 (serine 15) (Cell Signaling, 9284), Aurora kinase A (Cell Signaling, 4718), and Aurora kinase B (BD Biosciences, 611082) antibodies were used.
BARD1 immunoprecipitation
Protein lysates (400 μg) from NLF cells were immunoprecipitated with BARD1 Exon 4 (BL518, Bethyl Laboratories, Inc.; A300-263A) and BARD1 p25 (targeting BARD1β) antibodies as previously described (14). Immunoprecipitation conditions are further described in the supplementary methods.
BARD1 forced over-expression in NIH-3T3 cells
NIH-3T3 cells were transfected with BARD1β pcDNA3 or FL-BARD1 pcDNA3 (a kind gift from Makiko Tsuzuki; construct synthesis further described in the supplementary methods) using the TransIT-3T3 transfection kit (Mirus Bio LLC), bulk selected, plated in triplicate in 1% FBS and growth was monitored as above (9).
Results
Genomic characterization of the BARD1 locus
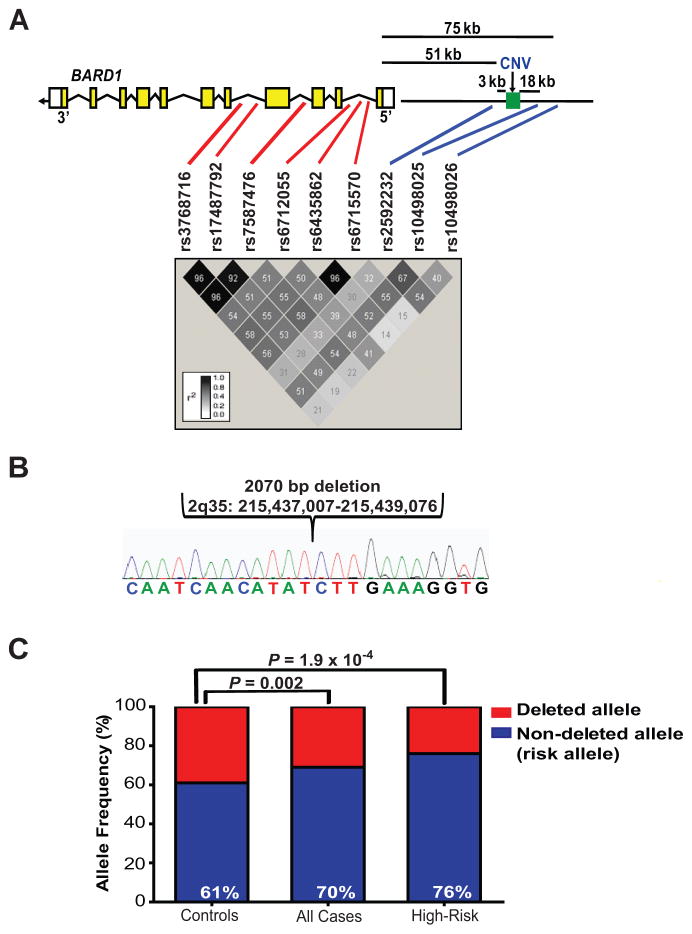
The neuroblastoma associated SNPs at chromosome 2q35 cover a 113 kb genomic region (Human Genome Build 36.3; Fig. 1A). One SNP upstream of BARD1, rs10498025, is in modest linkage disequilibrium (LD) to the intronic SNPs (r2 = 0.49–0.55). However, the other two SNPs (rs2592232 and rs10498026) are in weaker LD with these SNPs (r2 = 0.28–0.39 and r2 = 0.14–0.41, respectively; Fig. 1A), but have a similar degree of association with neuroblastoma, suggesting that more than one disease-contributing variant may exist at 2q35 (10).
Figure 1. Genomic characterization of the BARD1 locus on chromosome 2q35.
A, BARD1 exons are shown with the relative positions of the neuroblastoma associated SNPs and CNV. In red are SNPs discovered in the initial high-risk neuroblastoma discovery case-control series (6) and in blue are additional SNPs identified in an expanded case-control series (10). The LD plot was constructed with Haploview software using HapMap CEU data (Release 22). B, Chromatogram showing 2 kb deletion 5′ upstream of BARD1 (Human Genome Build 36.3). C, Histogram showing association of CNV with high-risk neuroblastoma (% = non-deleted allele, the risk allele). kb = kilobases.
We first sought evidence for copy number variants (CNVs) at this locus that may be in linkage to these SNPs and contribute to neuroblastoma susceptibility. First, we analyzed data generated from a high-resolution oligonucleotide array (22), which suggested the presence of a CNV upstream of BARD1. We sequenced this CNV breakpoint from the constitutional DNA of a neuroblastoma subject, identifying a 2 kb deletion (Fig. 1A and B), which is in strong LD with the neuroblastoma associated SNP rs10498026 (r2 = 0.64; eight other associated SNPs r2 = 0.12 – 0.23; (Supplementary Table S1). We next directly examined whether this CNV was associated with neuroblastoma using quantitative PCR in an independent case series (N = 255 Caucasian neuroblastoma cases, 275 Caucasian controls without cancer). This 2 kb allele (hereafter referred to as “non-deleted allele”) was found to be significantly overrepresented in neuroblastoma cases, specifically in the high-risk phenotype (Fig. 1C) and is the second most significantly associated common variant at this locus with an odds ratio (OR) of 2.00 (Supplementary Table S2). In a multivariate logistic regression analysis of high-risk cases versus controls in this series, the CNV remained significant (P = 0.009) after adjusting for the effect of the most significant SNP, rs7587476. These data suggest that this CNV represents an independent association signal with modest effect size and that more than one neuroblastoma causal variant exists at the BARD1 locus.
BARD1 SNP and CNV variations are associated with differential BARD1 isoform expression
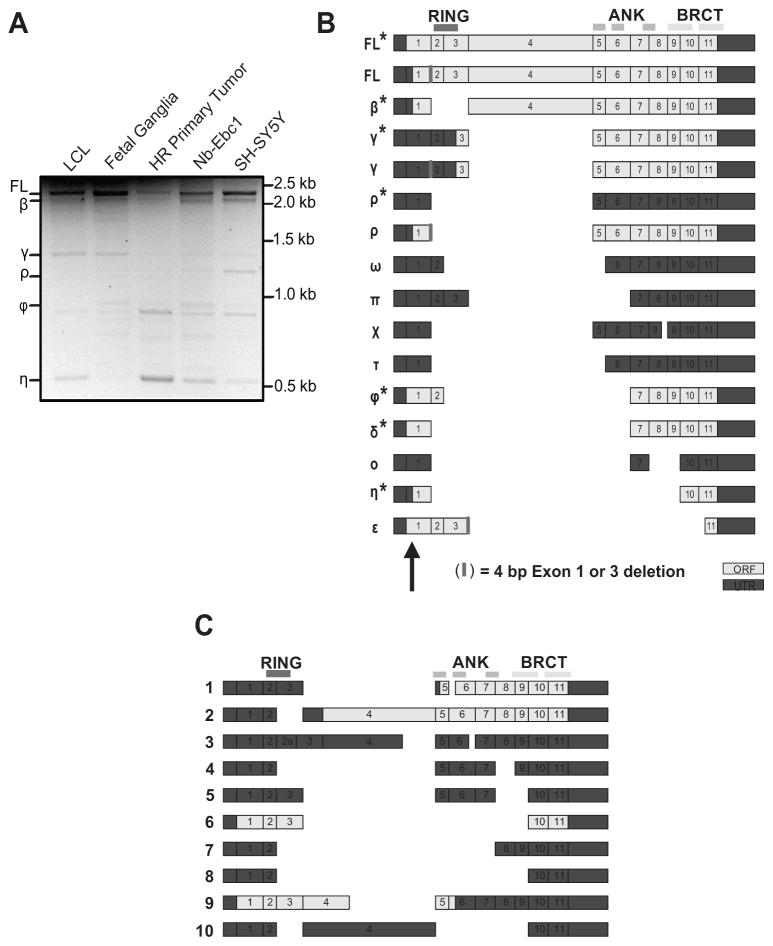
Given the genomic location of the neuroblastoma associated common variations (Fig. 1A) along with the growing evidence that SNPs may influence gene splicing (23), we next sought to fully characterize the BARD1 isoforms present in both normal and neuroblastoma cells. Using RT-PCR primers spanning the BARD1 coding region, we found multiple alternatively spliced isoforms in all cells (Fig. 2A). To identify the exon structure of these isoforms we first cloned and sequenced cDNA from fetal sympathetic ganglia tissue (20–24 weeks gestational age, N = 3), identifying FL BARD1 and 15 unique BARD1 isoforms (Fig. 2B). Nine of these isoforms are novel, nine maintain an open reading frame (ORF), and four (FL, β, ρ, and η) may use an alternative translational start site located upstream to the native AUG in exon 1 (Fig. 2B) (11, 13, 16). We also cloned BARD1 variants from four neuroblastoma cell lines (SKNAS, NB-Ebc1, SH-SY5Y, and NGP) and found that full length BARD1 and six of these BARD1 isoforms (β,γ,ρ,ϕ,δ,η) were also expressed in neuroblastoma cells (Fig. 2A and B), suggesting a possible mechanistic role in not only tumor initiation, but also malignant progression. Ten additional BARD1 isoforms were cloned only from neuroblastoma cells, six of which are novel (Fig. 2C).
Figure 2. Multiple BARD1 isoforms are expressed in fetal sympathetic ganglia and neuroblastoma cells.
A, Representative PCR using primers in BARD1 exons 1 and 11, showing multiple BARD1 splice variants. B, Exon structure of BARD1 variants identified in fetal sympathetic ganglia. BARD1 isoforms also expressed in neuroblastoma cells indicated with (*). C, Exon structure of additional BARD1 variants identified in neuroblastoma cells. BARD1 functional domains are shown at top. Exon 1 alternative start site indicated with arrow in B.
Considering that BARD1 was identified as a neuroblastoma susceptibility gene via a GWAS approach, we next investigated how neuroblastoma associated common variants correlated with BARD1 isoform expression. First, we examined the relationship between disease associated SNP variation and BARD1 exon expression by utilizing the SNP Express database that profiles BARD1 exon expression in peripheral blood mononuclear cells (PBMCs). A majority of the neuroblastoma associated SNP alleles correlated with differential expression of at least one BARD1 exon (P < 0.05). Most notably, six of the nine neuroblastoma associated SNPs were significantly correlated with decreased expression of BARD1 exons 2 and 3 (P = 8.9 × 10−4- 0.03), an expression pattern that is consistent with the putative oncogenic BARD1β isoform that splices out these two exons (14, 16). The 2q35 SNPs that lack association with neuroblastoma (rs3754546, rs7591615, and rs4234006) did not show a statistically significant similar relationship for exon 2 (P = 0.07, 0.10, and 0.54, respectively) or exon 3 (P = 0.42, 0.28, and 0.66, respectively).
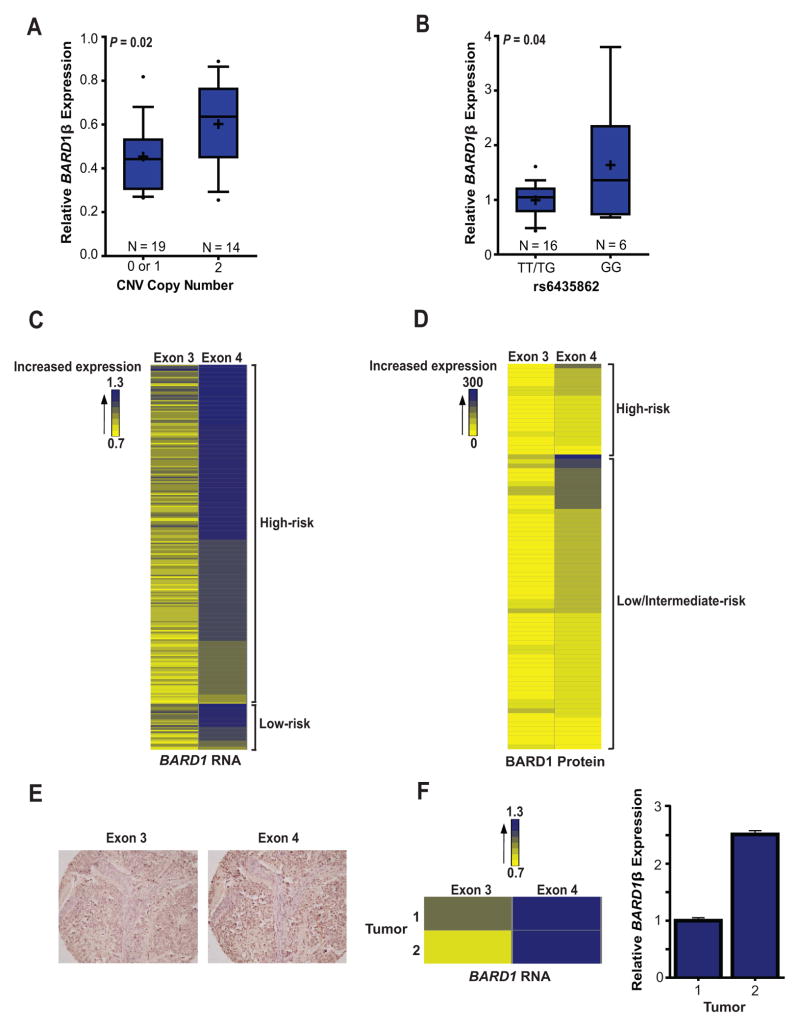
We next sought to directly determine the relationship of BARD1β expression to the neuroblastoma associated common SNP and CNV alleles. First, we analyzed the expression of the BARD1β isoform in lymphocyte cell lines (LCLs; N = 33) and found that LCLs with 2 copies of the non-deleted allele of the BARD1 5′ CNV (risk allele) had a significantly higher expression of BARD1β versus samples with 0 or 1 copy of this allele (Fig. 3A). This relationship was not seen with TaqMan® probes targeting the BARD1 exon 10/11 or exon 2/3 boundaries, suggesting that this common variation is directly associated with up-regulation of BARD1β. In addition, a trend towards increased expression of BARD1β in LCLs harboring a homozygous risk allele genotype (GG) at SNP rs6435862 was found, but did not reach statistical significance. The BARD1β isoform was also found to be ubiquitously expressed in a panel of fetal tissues, including eight fetal sympathetic ganglia from a wide-range of gestational ages (19–36 weeks) (Supplemental Fig. S1).
Figure 3. Neuroblastoma associated common variation is associated with increased BARD1β expression in both germline and neuroblastoma cells.
A and B, BARD1β expression was measured with a TaqMan® probe spanning the BARD1 exon 1/4 alternatively spliced boundary in LCLs (A) and neuroblastoma cell lines (B). The non-deleted CNV (A) and the “G” allele at SNP rs6435862 (B) are the risk alleles. C and D, Heat-maps displaying that neuroblastoma primary tumors express higher levels of exon 4 compared to exon 3, both at the RNA (C) and protein (D) level, consistent with the expression of BARD1β. E, Representative immunohistochemistry staining of a primary neuroblastoma tumor with BARD1 antibodies targeting epitopes in exon 3 or 4 (20x magnification). F, Tumor exon array data correlates well with direct measurement of BARD1β expression with an isoform specific TaqMan® probe. Box plots (A and B) represent the 25–75th percentile, error bars represent the 10–90th percentile, and crosses represent mean. Error bars in F represent SEM.
This correlation between neuroblastoma common risk alleles at the BARD1 locus with BARD1β expression was also found in primary neuroblastoma cells as we detected significantly increased BARD1β expression in neuroblastoma cells harboring a homozygous risk allele genotype (GG) at SNP rs6435862 (Fig. 3B). However, the complexity of the neuroblastoma genome with frequent chromosome 2q gain and hyperdiploidy prevented accurate 5′ BARD1 CNV copy number state precluding a similar CNV and BARD1β expression analysis in this sample set.
Finally, we analyzed BARD1 isoform expression in a set of neuroblastoma primary tumors using both an Affymetrix HuEx array (N = 251) and a non-overlapping neuroblastoma tumor tissue microarray (N = 119) (Fig. 3C–F, Supplemental Table S4). We first explored differential BARD1 exon expression at the RNA level, finding higher exon 4 expression compared to exon 3 expression, a pattern consistent with BARD1β expression (Fig. 3C). Probe sets within exon 2 of the BARD1 gene showed global poor hybridization kinetics, so these data were not considered. We also used antibodies raised against isoform-specific epitopes to examine BARD1β protein expression in neuroblastoma primary tumors (14). As shown in Fig. 3D and E and Supplementary Table S4, the majority of diagnostic human neuroblastomas also showed immunohistochemical staining patterns consistent with BARD1β isoform expression. This array data correlated well with direct TaqMan® measurement of BARD1β expression (Fig. 3F). Taken together, these results suggest that developing nervous tissues and a majority of neuroblastoma cells express the BARD1β isoform, and that its expression correlates with neuroblastoma associated common SNP and CNV variation at chromosome 2q35.
BARD1β has the characteristics of a neuroblastoma oncogene
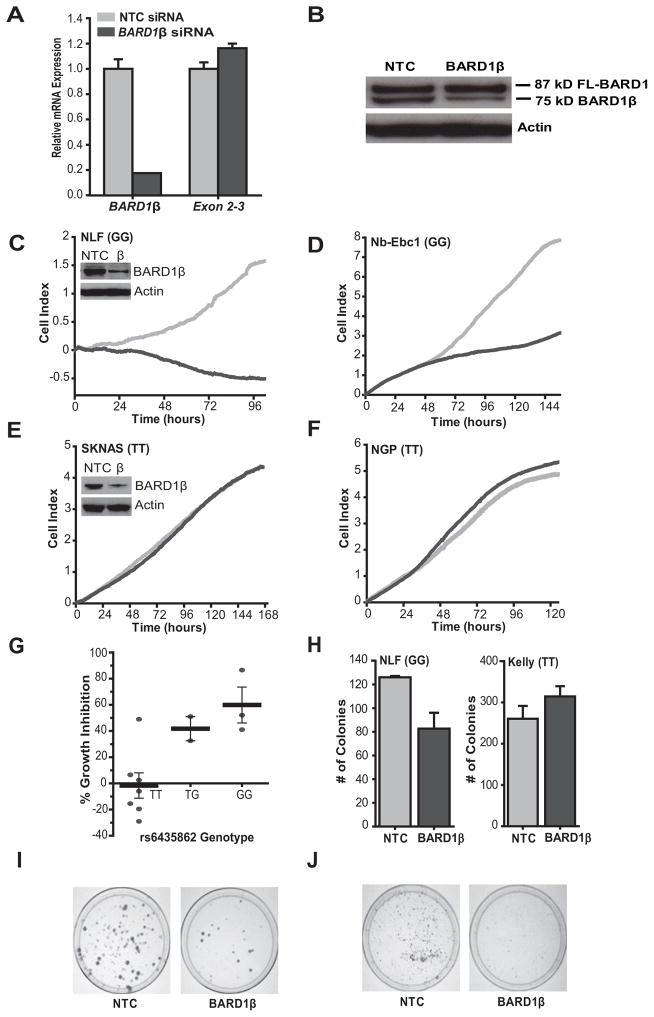
To determine whether high levels of BARD1β are functionally relevant in models of high-risk neuroblastoma, we examined the consequences of disrupting BARD1β expression with a specific short interfering RNA (siRNA) targeting the alternatively spliced exon 1/exon 4 boundary of the BARD1β mRNA (Fig. 4A and B). We transfected a panel of neuroblastoma cell lines (N = 12) and BARD1β knockdown showed differential effects on cellular proliferation that correlated with both the allelic state of SNP rs6435862, and the level of BARD1β expression (Fig. 4 and Supplementary Table S5). Neuroblastoma cells harboring at least one risk allele at SNP rs6435862 (GT or GG) and the highest BARD1β expression (N = 4; mean BARD1β expression = 2.02) showed the most significant inhibition of cell proliferation upon BARD1β knockdown (% growth inhibitionrange (% GIrange) = 41 – 86%, Prange = 0.0002 – 0.002) (Fig. 4C,D,G and Supplementary Table S5). BARD1β knockdown had no effect on cellular proliferation in a majority of neuroblastoma cell lines with a homozygous major allele genotype (TT) at SNP rs6435862 and lower BARD1β expression (N = 6, mean BARD1β expression = 1.05; % GIrange = (−) 29 - 6%, P > 0.5) (Fig. 4E–G and Supplementary Table S5).
Figure 4. BARD1β is an oncogenic driver in neuroblastoma cells harboring disease associated common risk variation with high BARD1β expression.
A and B, RNAi silencing of BARD1β is both efficient and selective shown by quantitative PCR (A) and Western blot with an antibody targeting BARD1 exon 4 epitopes (BL518; B) in NLF cells. C–F, Cellular growth for four representative neuroblastoma cell lines transfected with BARD1β (dark grey) and a negative control (NTC) siRNA (light grey). SNP rs6435862 genotype shown in parentheses (G = risk allele). C and D, NLF (C) and Nb-Ebc1 (D) show significant growth inhibition when BARD1β is silenced (P = 0.0002 and 0.002, respectively). E and F, SKNAS (E) and NGP (F) show no growth inhibition (P = 0.92 and 0.48, respectively). Positive control, PLK siRNA, not shown for clarity. Insets of (C) and (E) show comparable BARD1β knockdown by Western blot. G, Summary of percent growth inhibition across neuroblastoma cell line panel (N = 12) stratified by SNP rs6435862 genotype (P = 0.002 between TT and TG/GG genotypes). Each point represents mean of percent growth inhibition across two biological replicates. H, NLF (left, P = 0.03) but not Kelly (right, P = 0.24) cells have decreased anchorage independent growth in soft agar after BARD1β silencing. Histograms represent the mean number of colonies across three replicates ± SEM. I and J, BARD1β silencing in NLF (I) and Nb-Ebc1 (J) cells results in decreased clonogenic survival. Error bars (A and G) represent SEM.
In addition, we found that cell growth was also inhibited upon BARD1β knockdown in HeLa cells (Supplementary Fig. S2A, GIavg = 33%, P = 0.002), consistent with prior reports (14). However, BARD1β silencing did not affect cell growth in the non-transformed neuroblast control cell line, retinal pigment epithelial cells (RPE1-hTERT), despite a similar degree of BARD1β knockdown, suggesting that BARD1β only drives growth in malignant cells (Supplementary Fig. S2B and C; % GIavg = −18%).
To further investigate the oncogenicity of BARD1β in neuroblastoma, we studied how BARD1β silencing influenced growth in soft agar and clonogenic assays. Similarly, BARD1β silencing decreased growth in these assays in BARD1β dependent cells (NLF, Nb-Ebc1; GG risk genotype at SNP rs6435862), but not BARD1β independent cells (Kelly; TT protective genotype at SNP rs6435862) (Fig. 4H–J).
BARD1β knockdown in this cell line panel ranged from 23% (KCN) to nearly 100% (RPE1). Importantly, those cell lines showing growth inhibition had similar mRNA and protein knockdown to those with no phenotype (mean BARD1β mRNA knockdown = 62% and 56%, respectively, P = 0.62; and insets in Fig. 4C and E) and the level of BARD1β knockdown showed no correlation with the amount of growth inhibition (r2 = 0.017).
BARD1β blocks apoptosis in neuroblastoma cells
Given the role of BARD1 in mediating apoptosis, we next sought to determine whether high levels of BARD1β protect neuroblastoma cells from induction of programmed cell death. First, we found significant upregulation of the activity of the apoptotic pathway effectors caspase-3 and −7 after BARD1β silencing in NLF and Nb-Ebc1 cells (Fig. 5A and B). However, this apoptosis does not appear to be due to removal of a dominant negative effect of BARD1β on FL-BARD1’s role in phosphorylating and stabilizing p53 in response to DNA damage (11, 24–26), as BARD1β silencing did not change cellular levels of ser15 phosphorylated p53 (Fig. 5C). Similarly, the growth promoting effects of BARD1β appear to be largely independent of any antagonistic functions on the BARD1/BRCA1 driven homology-directed repair (HDR) of double stranded DNA breaks (27). Suppressing the level of PARP1 mRNA to < 20% of baseline in these BARD1β dependent NLF cells did not result in synthetic lethality as would be expected if a deficiency in the HDR pathway was present (27). Only a minor fraction of the growth inhibition observed in BARD1β silenced NLF cells was seen with PARP1 silencing (25% vs. 87% for BARD1β silencing; Fig. 5D, Fig. 4C and G). Taken together, these results suggest that the increase in neuroblastoma risk observed with increased BARD1β expression and evasion of apoptosis in neuroblastoma cells does not involve BARD1β driven defects in the HDR maintenance of genomic stability or interruption of the FL-BARD1/p53 phosphorylation interaction. However, given that total p53 levels decrease upon BARD1β silencing (Fig. 5C), this oncogenic protein may be exerting its effects by stabilization of p53 in a different manner.
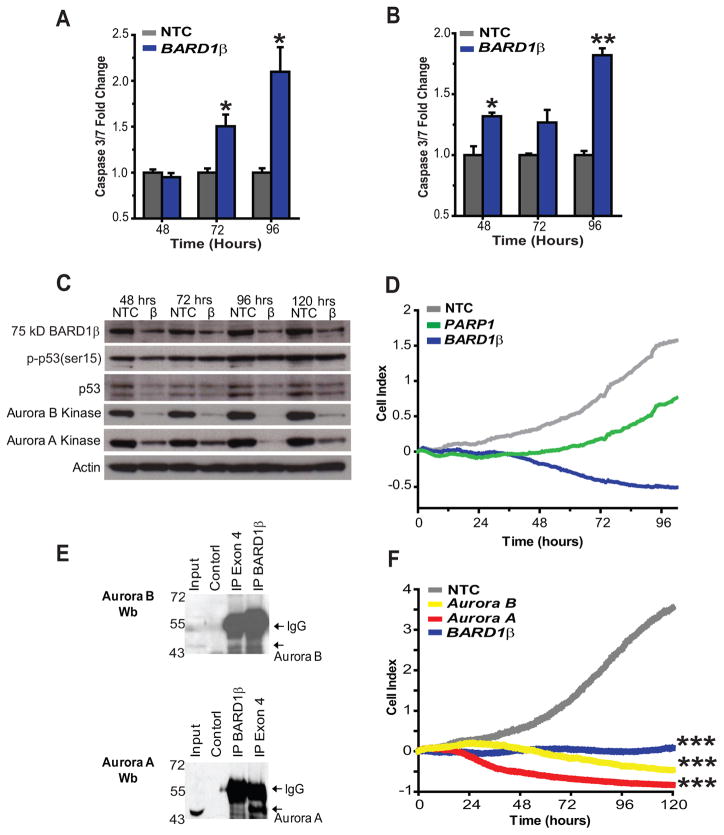
Figure 5. BARD1β blocks apoptosis in risk allele harboring neuroblastoma cells and complexes with Aurora kinase B.
A and B, Caspase-3 and −7 activity was significantly upregulated after BARD1β silencing in the two highest BARD1β expressing, risk allele harboring cell lines, Nb-Ebc1 (A) and NLF (B). C, BARD1β silencing in NLF cells leads to a decrease in the steady state levels of the Aurora kinases A and B, but not to changes in the levels of ser15-phosphorylated p53. D, A representative example of PARP1 silencing in NLF cells. E, Western blots with Aurora B (top) or A (bottom) kinase antibody in risk allele harboring NLF cells after immunoprecipitation using BARD1β targeting antibody (p25) or exon 4 targeting antibody (BL518). F, Parallel knockdown of both Aurora kinases A and B and BARD1β in NLF cells show similar growth inhibition. *p<0.05, **p<0.001, ***p<0.0001.
BARD1β binds to and stabilizes Aurora kinase B in neuroblastoma cells
The BARD1β isoform has previously been shown to bind and stabilize Aurora kinase B in HeLa cells (14). These findings coupled with the identification of Aurora kinase A as a MYCN stabilized, high-risk phenotype associated oncogene in neuroblastoma (2, 28), led us to investigate if there was interaction between BARD1β and the Aurora kinases in neuroblastoma. First, we performed co-immunoprecipitation using antibodies toward epitopes in exon 4 (BL518), common to both FL BARD1 and BARD1β, or to epitopes in the alternatively translated exon 1 of BARD1β (p25; (14)). In BARD1β dependent NLF cells, Aurora B co-precipitated with both BARD1 antibodies (Fig. 5E, top) and Aurora A co-precipitated with the exon 4 antibody (Fig. 5E, bottom). Moreover, BARD1β silencing in NLF cells showed that levels of both Aurora A and B kinase decreased in parallel with levels of BARD1β suggesting that this BARD1β/Aurora kinase interaction may stabilize the latter proteins (Fig. 5C). Finally, parallel knockdown of these Aurora kinases and BARD1β in NLF cells show similar growth inhibition (Fig. 5F). Taken together, these results suggest that the oncogenic function of BARD1β in neuroblastoma, at least in part, functions through the Aurora family of kinases.
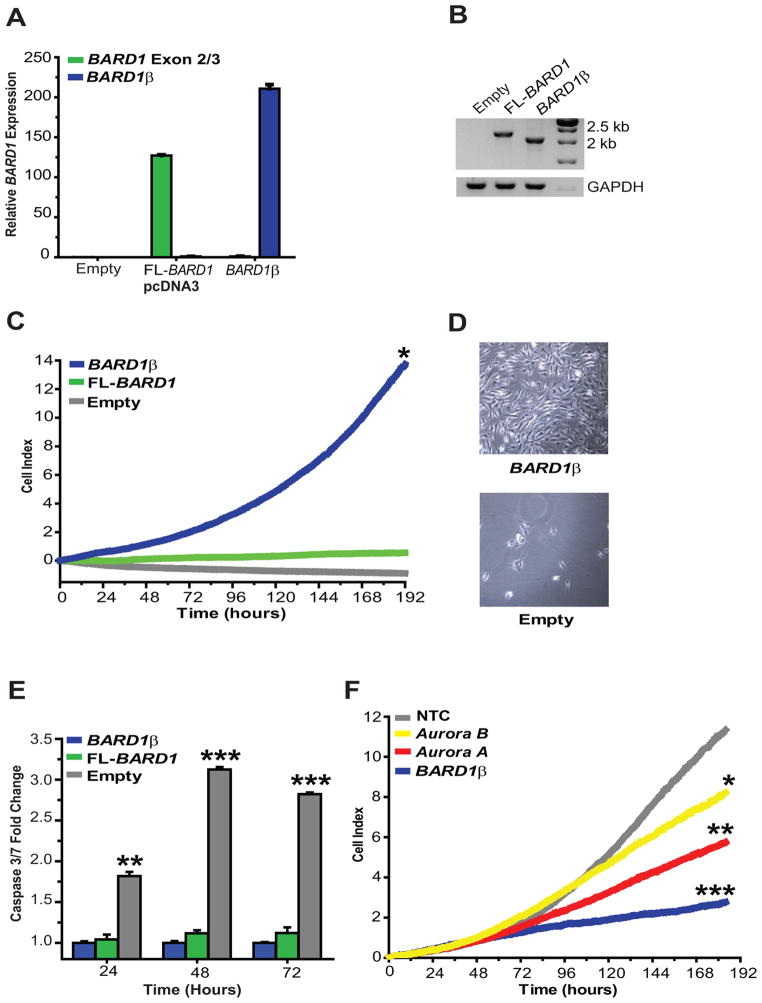
BARD1β transforms NIH-3T3 cells
We next investigated how over-expression of BARD1β and FL-BARD1 influenced the growth of non-transformed NIH-3T3 cells. NIH-3T3 cells were transfected with a BARD1β or FL-BARD1 cDNA resulting in stable clones that had a greater than 100-fold increase in BARD1 expression (Fig. 6A and B). Forced over-expression of BARD1β permitted cell proliferation under low serum (1% FBS) conditions protecting cells from apoptosis, while empty pcDNA3 transfected cells underwent apoptosis, and FL-BARD1 transfected cells were protected from apoptosis but unable to proliferate (Fig. 6C–E). This transformed phenotype was silenced when BARD1β was knocked down by RNAi and partially silenced when either Aurora A or B kinase were depleted (Fig. 6F). Importantly, no growth inhibition was observed when the Aurora kinases were silenced under full serum conditions (10% FBS; data not shown). These results confirm BARD1β as a neuroblastoma oncogene and lend support to our hypothesis that BARD1β acts via a mechanism involving the Aurora kinase family in neuroblastoma cells.
Figure 6. BARD1β transforms NIH-3T3 cells inducing serum independent cell proliferation and escape from apoptosis.
A and B, BARD1 mRNA overexpression in stably transfected NIH-3T3 cells measured by quantitative PCR (A) and RT-PCR (B). Taqman® probe targeting BARD1 exons 2/3 is a surrogate for FL-BARD1 expression. C and D, BARD1β stably transfected NIH-3T3 cells grow exponentially in low serum conditions with D representing growth after 1 week. E, Over-expression of BARD1β and FL-BARD1 induces NIH-3T3 cell escape from serum deprivation induced apoptosis. F, This transformed phenotype is lost when BARD1β is silenced with RNAi, and partially lost when either Aurora kinase A or B is silenced. *p<0.01, **p<0.001, ***p<0.0001.
Discussion
GWAS discoveries have the potential to help decipher the mechanistic basis of complex human disease and impact risk prediction. In cancer, there is additionally the question of whether or not susceptibility genes are also involved in tumor progression, and moreover, whether the GWAS approach has the potential to identify clinically relevant therapeutic targets. We have begun to address these goals in neuroblastoma by studying the GWAS identified, highly robust association signal at the BARD1 locus in both tumor and host genomes.
Our data suggest that the primary mechanism for increased neuroblastoma susceptibility at the BARD1 locus is via disease associated common variation’s impact on differential BARD1 exon expression, leading to the preferential formation of an oncogenic isoform, BARD1β. We show a clear correlation between genomic haplotypes associated with neuroblastoma and expression of BARD1β. These data are consistent with the growing evidence that the complex regulation of gene expression involves heritable common alleles and that variation in isoform expression influences cancer susceptibility (23, 29).
One of the major goals of this work was to determine if BARD1 alterations are further selected for during tumor progression and therefore we also studied BARD1β in neuroblastoma cells. The correlation of risk common variation with BARD1β expression also largely holds in primary neuroblastoma cells (Fig. 3B), which is quite striking considering the sheer complexity of the neuroblastoma genome (2). More importantly, however, we show that genetic manipulation of BARD1β has profound impact on neuroblastoma cell phenotype (Fig. 4–6). These data suggest that BARD1β may be an important oncogene and therapeutic target in high-risk neuroblastoma, one that would not likely have been discovered via standard genomic profiling efforts, and lend strong support to the ongoing work to develop Aurora kinase inhibitors for clinically aggressive neuroblastoma (28, 30, 31).
It is likely that additional BARD1 isoforms may also be involved in neuroblastoma tumorigenesis. For example, the δ-BARD1 isoform (Fig. 2B) is expressed in highly proliferative cytotrophoblasts and stabilizes the estrogen receptor α (ESR1), a transcription factor involved in neuronal differentiation (13, 15, 32). Further exploration of the biological implications of a subset of these isoforms in neuroblastoma is clearly needed (13, 16, 33).
Our multivariate regression analysis suggests multiple neuroblastoma causal variants at 2q35, including the BARD1 CNV, which is not surprising given the complexity of the LD block (Fig. 1A, Supplementary Table S1). However, how this CNV (or other causal variants) influences BARD1β expression has not yet been identified. We hypothesize that regulatory elements that act as either distal splicing enhancers of BARD1 or factors that increase mRNA transcription from the alternative start site in BARD1 exon 1 used to initiate BARD1β expression (Fig. 2) may be present in this CNV region. Regional resequencing and further integrated epigenetic, computational, and functional analysis will be needed to definitively identify the causal variants at 2q35 and the mechanisms by which they enhance BARD1β expression. One strong candidate regulatory protein for control of BARD1 splicing, that has binding motifs susceptible to SNP changes, is the neuronal specific, cancer associated, splicing modifier FOX2 (23, 34, 35). Neuroblastoma associated SNPs at 2q35 are in strong LD with multiple predicted FOX2 binding sites (35) and there are two FOX2 binding motifs located in and 235 base pairs 3′ downstream to BARD1 exon 3.
Future studies will also focus on defining the mechanism by which BARD1β stabilizes the Aurora kinases. Our data suggests that BARD1β may stabilize Aurora B kinase by scaffolding this protein as previously observed in HeLa cells (14), while the stabilization of Aurora kinase A likely acts via a different mechanism such as via phosphorylation or stabilization of a known or unknown Aurora A scaffolding protein (36, 37). Further in vivo and cell cycle specific in vitro functional analysis is needed to definitively define the tumorigenic potential of BARD1β and uncover the details of how this protein interacts with the Aurora kinases in neuroblastoma cells, work which may ultimately further support BARD1 expression as a biomarker for therapeutic vulnerability to Aurora kinase inhibition.
In summary, here we have identified BARD1β as an oncogenic protein in neuroblastoma, and translational potential exists due to interaction with and stabilization of the Aurora kinases. This work shows that post-GWAS functional genomics efforts have the potential to both identify cancer susceptibility mechanisms and therapeutically relevant oncogenic vulnerabilities that may be exploitable clinically.
Supplementary Material
Acknowledgments
Grant Support
The authors acknowledge the Children’s Oncology Group (COG) for providing neuroblastoma specimens and the Neuroblastoma TARGET consortium for tumor profiling. This work was supported in part by an HHMI Medical Student Research Fellowship (KB), an NIDDK Training Grant (KB), NIH Grants R01-CA124709 (JMM), U01-CA98543 to COG and the TARGET consortium, the Giulio D’Angio Endowed Chair (JMM), the Alex’s Lemonade Stand Foundation (JMM), Andrew’s Army Foundation (JMM), the SuperJake Foundation (JMM), the Abramson Family Cancer Research Institute (JMM), the Center for Applied Genomics (HH) at CHOP, and a Swiss NSF grant 31003A-110038 (IIF).
Footnotes
No author has a financial or other conflict of interest.
References
- 1.Freedman ML, Monteiro AN, Gayther SA, Coetzee GA, Risch A, Plass C, et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nature Genetics. 2011;43:513–8. doi: 10.1038/ng.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–11. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith MA, Seibel NL, Altekruse SF, Ries LA, Melbert DL, O’Leary M, et al. Outcomes for children and adolescents with cancer: challenges for the twenty-first century. J Clin Oncol. 2010;28:2625–34. doi: 10.1200/JCO.2009.27.0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matthay KK, Reynolds CP, Seeger RC, Shimada H, Adkins ES, Haas-Kogan D, et al. Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: a children’s oncology group study. J Clin Oncol. 2009;27:1007–13. doi: 10.1200/JCO.2007.13.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen le B, Diskin SJ, Capasso M, Wang K, Diamond MA, Glessner J, et al. Phenotype restricted genome-wide association study using a gene-centric approach identifies three low-risk neuroblastoma susceptibility Loci. PLoS Genet. 2011;7:e1002026. doi: 10.1371/journal.pgen.1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capasso M, Devoto M, Hou C, Asgharzadeh S, Glessner JT, Attiyeh EF, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41:718–23. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diskin SJ, Hou C, Glessner JT, Attiyeh EF, Laudenslager M, Bosse K, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–91. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maris JM, Mosse YP, Bradfield JP, Hou C, Monni S, Scott RH, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358:2585–93. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–5. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K, Diskin SJ, Zhang H, Attiyeh EF, Winter C, Hou C, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–20. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irminger-Finger I, Jefford CE. Is there more to BARD1 than BRCA1? Nat Rev Cancer. 2006;6:382–91. doi: 10.1038/nrc1878. [DOI] [PubMed] [Google Scholar]
- 12.Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–40. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Ryser S, Dizin E, Pils D, Krainer M, Jefford CE, et al. Oncogenic BARD1 isoforms expressed in gynecological cancers. Cancer Res. 2007;67:11876–85. doi: 10.1158/0008-5472.CAN-07-2370. [DOI] [PubMed] [Google Scholar]
- 14.Ryser S, Dizin E, Jefford CE, Delaval B, Gagos S, Christodoulidou A, et al. Distinct roles of BARD1 isoforms in mitosis: full-length BARD1 mediates Aurora B degradation, cancer-associated BARD1beta scaffolds Aurora B and BRCA2. Cancer Res. 2009;69:1125–34. doi: 10.1158/0008-5472.CAN-08-2134. [DOI] [PubMed] [Google Scholar]
- 15.Dizin E, Irminger-Finger I. Negative feedback loop of BRCA1-BARD1 ubiquitin ligase on estrogen receptor alpha stability and activity antagonized by cancer-associated isoform of BARD1. The international journal of biochemistry & cell biology. 2010;42:693–700. doi: 10.1016/j.biocel.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 16.Sporn JC, Hothorn T, Jung B. BARD1 expression predicts outcome in colon cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:5451–62. doi: 10.1158/1078-0432.CCR-11-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.NCI TARGET data matrix. [Web Page; Available from: http://target.nci.nih.gov/dataMatrix/TARGET_DataMatrix.html]
- 18.Winter C, Pawel B, Seiser E, Zhao H, Raabe E, Wang Q, et al. Neural cell adhesion molecule (NCAM) isoform expression is associated with neuroblastoma differentiation status. Pediatr Blood Cancer. 2008;51:10–6. doi: 10.1002/pbc.21475. [DOI] [PubMed] [Google Scholar]
- 19.Peddinti R, Zeine R, Luca D, Seshadri R, Chlenski A, Cole K, et al. Prominent microvascular proliferation in clinically aggressive neuroblastoma. Clin Cancer Res. 2007;13:3499–506. doi: 10.1158/1078-0432.CCR-07-0237. [DOI] [PubMed] [Google Scholar]
- 20.Cole KA, Huggins J, Laquaglia M, Hulderman CE, Russell MR, Bosse K, et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1012351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole KA, Attiyeh EF, Mosse YP, Laquaglia MJ, Diskin SJ, Brodeur GM, et al. A Functional Screen Identifies miR-34a as a Candidate Neuroblastoma Tumor Suppressor Gene. Mol Cancer Res. 2008;6:735–42. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–12. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, et al. Cancer- associated regulation of alternative splicing. Nat Struct Mol Biol. 2009;16:670–6. doi: 10.1038/nsmb.1608. [DOI] [PubMed] [Google Scholar]
- 24.Feki A, Jefford CE, Berardi P, Wu JY, Cartier L, Krause KH, et al. BARD1 induces apoptosis by catalysing phosphorylation of p53 by DNA-damage response kinase. Oncogene. 2005;24:3726–36. doi: 10.1038/sj.onc.1208491. [DOI] [PubMed] [Google Scholar]
- 25.Irminger-Finger I, Leung WC, Li J, Dubois-Dauphin M, Harb J, Feki A, et al. Identification of BARD1 as mediator between proapoptotic stress and p53-dependent apoptosis. Molecular cell. 2001;8:1255–66. doi: 10.1016/s1097-2765(01)00406-3. [DOI] [PubMed] [Google Scholar]
- 26.Fabbro M, Savage K, Hobson K, Deans AJ, Powell SN, McArthur GA, et al. BRCA1-BARD1 complexes are required for p53Ser-15 phosphorylation and a G1/S arrest following ionizing radiation-induced DNA damage. The Journal of biological chemistry. 2004;279:31251–8. doi: 10.1074/jbc.M405372200. [DOI] [PubMed] [Google Scholar]
- 27.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 28.Otto T, Horn S, Brockmann M, Eilers U, Schuttrumpf L, Popov N, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maris JM. Unholy matrimony: Aurora A and N-Myc as malignant partners in neuroblastoma. Cancer Cell. 2009;15:5–6. doi: 10.1016/j.ccr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Maris JM, Morton CL, Gorlick R, Kolb EA, Lock R, Carol H, et al. Initial testing of the aurora kinase A inhibitor MLN8237 by the Pediatric Preclinical Testing Program (PPTP) Pediatr Blood Cancer. 2010;55:26–34. doi: 10.1002/pbc.22430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loven J, Zinin N, Wahlstrom T, Muller I, Brodin P, Fredlund E, et al. MYCN-regulated microRNAs repress estrogen receptor-alpha (ESR1) expression and neuronal differentiation in human neuroblastoma. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1553–8. doi: 10.1073/pnas.0913517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuzuki M, Wu W, Nishikawa H, Hayami R, Oyake D, Yabuki Y, et al. A truncated splice variant of human BARD1 that lacks the RING finger and ankyrin repeats. Cancer letters. 2006;233:108–16. doi: 10.1016/j.canlet.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 34.Minovitsky S, Gee SL, Schokrpur S, Dubchak I, Conboy JG. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic acids research. 2005;33:714–24. doi: 10.1093/nar/gki210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–7. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitajima S, Kudo Y, Ogawa I, Tatsuka M, Kawai H, Pagano M, et al. Constitutive phosphorylation of aurora-a on ser51 induces its stabilization and consequent overexpression in cancer. PLoS ONE. 2007;2:e944. doi: 10.1371/journal.pone.0000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giubettini M, Asteriti IA, Scrofani J, De Luca M, Lindon C, Lavia P, et al. Control of Aurora-A stability through interaction with TPX2. J Cell Sci. 2011;124:113–22. doi: 10.1242/jcs.075457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.