Abstract
4-hydroxynonenal (4-HNE) alters numerous proteomic and genomic processes. Understanding chemical mechanisms of 4-HNE interactions with biomolecules and their respective stabilities may lead to new discoveries in biomarkers for numerous diseases of oxidative stress. Collision-induced dissociation (CID) and electron transfer dissociation (ETD) MS/MS were utilized to examine the stability of a 4-HNE-Cys Michael adduct. CID conditions resulted in the neutral loss of 4-HNE, also known as a retro-Michael addition reaction (RMA). Consequently, performing ETD fragmentation on this same adduct did not result in RMA. Interestingly, 4-HNE adduct reduction via sodium borohydride (NaBH4) treatment stabilized against the CID induced RMA. In a direct comparison of three forms of 4-HNE adducts, computational modeling revealed sizeable shifts in the shape and orientation of the lowest unoccupied molecular orbital (LUMO) density around the 4-HNE-Cys moiety. These findings demonstrate that ETD MS/MS analysis can be used to improve the detection of 4-HNE-protein modifications by preventing RMA reactions from occurring.
INTRODUCTION
Lipid peroxidation is known to occur during periods of sustained oxidative stress, most notably near lipid-rich cellular membranes.1 In particular, the peroxidation of linoleic and arachidonic acid generates numerous α,β-unsaturated reactive aldehydes, including 4-hydroxynonenal (4-HNE).1, 2 The highly reactive nature of this compound is imparted due to the bi-functional mechanism of adduction, forming both Michael addition and Schiff base adducts with nucleophilic amino acid side chains. The modification of proteins by 4-HNE can alter protein structure, stability and function.3-6 Multiple studies have characterized the underlying mechanism of 4-HNE protein adduction (Cys > His > Lys) and its impact on a host of cellular processes.3, 7-9
4-HNE has achieved prominence as a biomarker for a number of diseases, including early-stage detection of cancer, atherosclerosis and inflammation.10-13 While much is known regarding the biochemical impact of lipid peroxidation and the effects of pathophysiological concentrations of 4-HNE on cellular processes, little is known about the biochemical interactions of 4-HNE on an atomic scale. The ability of 4-HNE to modify amino acids and consequently alter protein function in vivo is an extremely complex process and presents a technically challenging area of investigation. Recently, comparison of electrophile/nucleophile interactions has provided significant insight into the reactivity of 4-HNE, specifically toward cysteine residues9; however, the complex amino acid microenvironment surrounding peptide residues as well as protein tertiary structure and solvent accessibility confound predictive measures. Used as an aid in detecting 4-HNE adducts, reductive stabilization via NaBH4 allows for the detection of more transient modifications. Successfully identifying protein carbonylation not only requires relatively stable adducts, but highly-sensitive MS instrumentation.14-16 Certain ionization and MS/MS conditions are known to cleave labile post-translational modifications (PTMs) before achieving peptide fragmentation, resulting in the neutral loss of the modification. The development of MS/MS techniques such as ETD has been critical in the identification of labile amino acid modifications such as phosphorylation. Specifically, the MS/MS-induced neutral loss of 4-HNE occurs through retro-Michael addition (RMA) reactions and has been characterized in a number of studies.17-20
In the present study, it was observed that the non-reduced 4-HNE-Cys280 adduct underwent a RMA reaction, resulting in the neutral loss of 4-HNE under CID conditions. Conversely, ETD fragmentation provided 4-HNE retention, preventing RMA during MS/MS fragmentation. NaBH4 reduction was employed to stabilize the 4-HNE-Cys280 adduct under CID conditions. Based on these observations, computational modeling was employed to provide a theoretical analysis of the role LUMO energies played in the CID/ETD fragmentation of 4-HNE under MS/MS conditions. Overall, these findings demonstrate the utility of ETD technology when applied to the detection of 4-HNE modified proteins.
EXPERIMENTAL PROCEDURES
Chemicals
All chemicals were obtained from Sigma-Aldrich (St. Louis, MO) unless specified otherwise. Solvents for LC-MS analysis and in-gel trypsin digestion were of HPLC grade purity or higher. Formic acid for LC-MS was obtained in 1 mL sealed glass vials from ThermoFisher Scientific (Waltham, MA). The active form of human recombinant SIRT3 was obtained from Cayman Chemical (Ann Arbor, MI). 4-HNE was prepared in our laboratory as previously described.21
Modification of SIRT3 with 4-HNE
Adduction of SIRT3 was achieved by incubating 1 μg (27 pmoles, 1.34 μM) of recombinant human protein with a 10-fold excess concentration of 4-HNE for 30 min at 37°C. Half of the samples were then subjected to reductive stabilization for 30 min at 25°C using a final concentration of 5 mM NaBH4 (stock concentration of 100 mM NaBH4 in 100 mM NaOH). Upon completion, samples were applied to SDS gel electrophoresis, coomassie stained for 10 min and then destained overnight. SIRT3 protein bands were excised from the gel and proteolysis was performed using a standard in-gel digestion procedure.21 Excised protein bands were destained in 50% acetonitrile (ACN), 50% 50 mM ammonium bicarbonate (NH4HCO3), dehydrated in 100% ACN and reduced for 45 min at 60°C with 10 mM dithiothreitol in 50 mM NH4HCO3. The gel pieces were then treated for 30 min in the dark at room temp in 40 mM iodoacetamide in 50 mM NH4HCO3, washed twice in 50 mM NH4HCO3 and resuspended in 25 μL 50 mM NH4HCO3 and 0.3 μg porcine-modified trypsin (Promega) and digested overnight at 37°C.
CID and ETD Mass Spectrometry
The digested samples (Control, 4-HNE and reduced 4-HNE SIRT3) were analyzed using nano liquid chromatography (EASY-nLC, Proxeon) at a flow rate of 300 nL/min with a gradient of 5 to 40% ACN (0.2% formic acid) over 40 min on C-18 Proxeon EASY-Column trapping (20 × 0.1 mm) and analytical columns (100 × 0.075 mm). The nLC was coupled to a nano-ESI source and Esquire HCT ion trap mass spectrometer (Bruker Daltonics, Inc., Billerica, MA). The instrument was operated using data-dependent CID MS/MS with a threshold of 30,000 TIC. Data analysis was performed using Mascot (v 2.1.04, www.matrixscience.com) and Esquire v5.2 data analysis package. BioTools (Bruker Daltonics) was used for MS/MS spectra annotation. Previous work identified a 4-HNE adduct on SIRT3 at Cys280; therefore, MS and MS/MS analysis was focused on the SIRT3 tryptic peptide containing Cys280, a 22-mer with the following sequence: CPVCTGVVKPDIVFFGEPLPQR.22 In order to obtain comparative CID and ETD fragmentation spectra, these same SIRT3 digests were analyzed using alternating CID/ETD MS/MS fragmentations on an amaZon-ETD ion trap mass spectrometer (Bruker Daltonics, Inc., Billerica, MA) coupled to a Dionex Ultimate3000 RSLCnano (Dionex, Sunnyvale, CA) using an LC gradient similar to that detailed above.
Quantum Mechanical Calculations
Computational modeling of the SIRT3 22-mer peptide containing 4-HNE-Cys280 was performed using Spartan (Wavefunction, Inc., Irvine, CA) and provides parameters such as the highest occupied molecular orbital (HOMO) and LUMO energies for each molecular species. The peptide species were assembled and minimized using Spartan. Computations were performed using Equilibrium Geometry at ground state with Hartree-Fock 3-21G in a vacuum, starting from MMFF geometry. Peptide geometry was initially optimized for the neutral molecule, basic sites were then protonated and the final minimum geometry was calculated using the +3 charge state to reflect the most abundant charge state observed during MS analysis.23, 24 The 4-HNE adduct was minimized in native, cyclic hemiacetal and reduced conformations. Due to the large mass of the model peptide (>2500 amu), a number of modifiers were used in obtaining a successful model. Specifically, the options maxpropweight=none, nosolvent and elcharge were used in order to complete the modeling.
RESULTS AND DISCUSSION
Cysteine residues are the most reactive nucleophilic amino acids towards 4-HNE, whereas 4-HNE adducts of histidine are more stable than either cysteine or lysine conjugates towards retro-Michael addition reactions.8, 17, 25, 26 Since oxo-cyclic equilibrium favors hemiacetal formation, the free aldehyde of the primary product may react with the 4-hydroxyl group to form the hemiacetal derivative.1, 27 Figure 1 presents the structures for both cyclic hemiacetal and reduced 4-HNE adducts. It remains unclear which 4-HNE structure occurs during ESI ionization and MS/MS fragmentation; therefore, both structures have been considered for theoretical analysis. Given the complex nature of 4-HNE chirality, resulting in at least 8 configurational isomers (Wakita et al.), and the inability to determine chiral selectivity of MS ion-trap fragmentations, these factors were not included in CID/ETD fragmentation analysis or the computational modeling.28 The following data present MS/MS fragmentations of native and NaBH4 reduced 4-HNE thiol adducts under CID/ETD conditions and resulting computational analysis.
Figure 1. HNE-cysteine adduct in linear, reduced and cyclic hemiacetal formation.
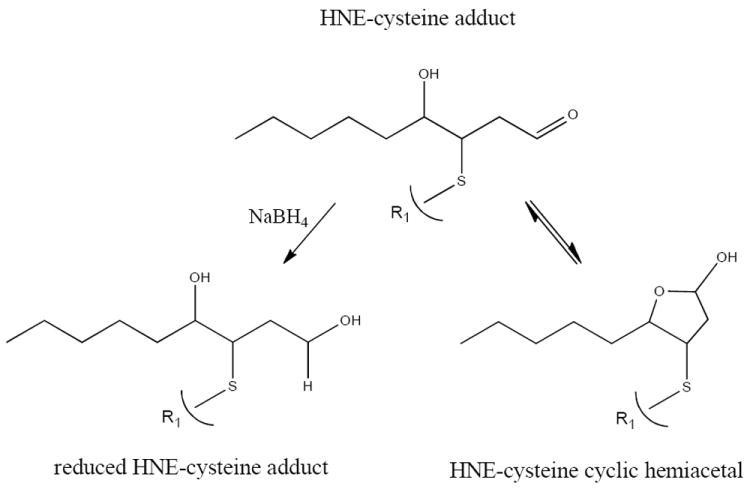
HNE is a bifunctionally reactive aldehyde that forms Michael-type adducts with cysteine residues. Michael additions are defined as the 1,4-addition of a doubly stabilized carbon nucleophile to an α,β-unsaturated carbonyl compound.36 NaBH4 reduction stabilizes Michael adducts by reducing the C=O bond to C-OH.
MS analysis of 4-HNE modified SIRT3 is presented in Figure 2A, where CID fragmentation yields primarily the +2 and +3 charged ions of the peptide without 4-HNE (1230.42 m/z and 820.62 m/z, respectively) representing the neutral loss of 4-HNE under CID. In contrast, the ETD spectrum in Figure 2A provides retention of the 4-HNE-Cys adduct during MS/MS. This comparison demonstrates the loss of a labile 4-HNE-Cys Michael adduct under CID fragmentation conditions, while ETD does not cleave 4-HNE from the peptide. Analysis of the CID data identified 27 unique fragmentation ions while the ETD data provided 41 unique identifications, yielding 52% greater sequence identity when ETD was utilized (BioTools spectra annotation, data not shown).
Figure 2.
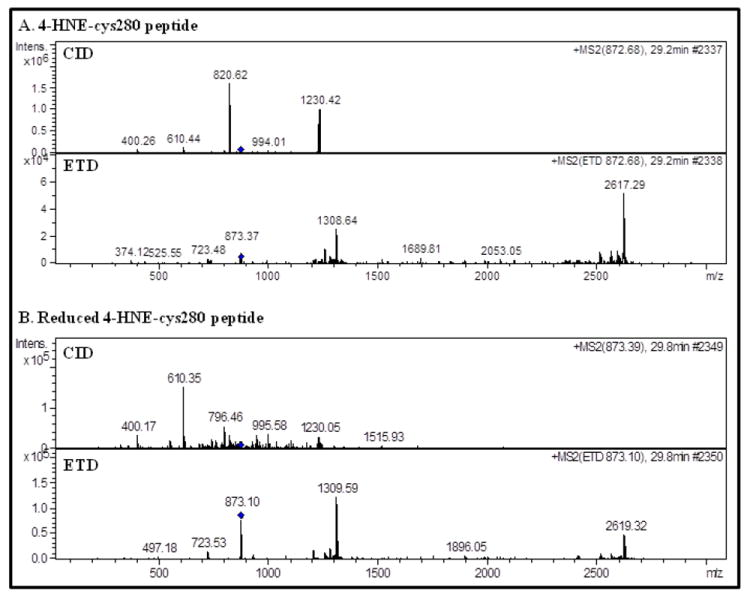
(A) Analysis of the 4-HNE adducted peptide reveals the CID neutral loss of 4-HNE from the parent ion (872.68 m/z) and the observed MS/MS ions representing the +2 and +3 charged peptide less the 4-HNE adduct, 1230.42 m/z and 820.62 m/z, respectively. ETD fragmentation did not result in the retro-Michael addition of the non-reduced 4-HNE adduct. (B) Reductive stabilization of 4-HNE provided sufficient adduct stability to prevent a retro-Michael addition reaction from occuring during CID conditions.
Contrary to the results obtained by CID of the 4-HNE adduct, CID analysis of the reduced 4-HNE adduct in Figure 2B demonstrates the advantages of stabilizing labile PTMs. Here, the neutral loss of 4-HNE does not occur, as the major neutral-loss peaks observed in Figure 2A are not present. The reduced 4-HNE peptide is also detected, and provides greater sequence coverage under ETD conditions. BioTools analysis of each spectra matched 22 ions following CID and 42 ions following ETD fragmentation, demonstrating an increase of 91% in peptide ion yield for ETD versus CID. This also reveals that despite stabilization by NaBH4 the overall ETD fragment ion yield is not improved for the non-reduced (41 ions identified) and reduced (42 ions identified) 4-HNE-peptides. Overall, MS/MS analysis demonstrates a clear advantage when utilizing sequential CID/ETD fragmentation in avoiding 4-HNE RMA reactions.
Given the recent studies by LoPachin et al. that intricately coupled computational modeling and quantum mechanics to the characterization of electrophile-nucleophile interactions,9, 29, 30 similar methods were employed to further explain the effects of adduct stability when the 4-HNE Michael-Cys280 adduct is fragmented by different mass spectrometry methods. Table 1 presents a number of quantum mechanical parameters comparing the three forms of 4-HNE. These results highlight a significant change in LUMO, where cyclization induces a 16% increase and NaBH4 reduction a 17% increase over the cyclized structure. The conversion of aldehyde to alcohol is known to provide stability to 4-HNE adducts.31 Here, the more negative LUMO of the non-reduced aldehyde may relate to the unstable nature of this particular 4-HNE Michael adduct. A report by Greaves et al. found a correlation between electron capture negative chemical ionization MS/MS, LUMO and calculated internal energies and provides support for this hypothesis.32 LoPachin et al. report that according to Frontier Orbital Theory, the most relevant frontier orbital for electrophiles is the LUMO, whereas the HOMO is most important for nucleophiles.9 Based on these studies, HOMO and LUMO were visualized using computational modeling. Examination of surface densities revealed no noticeable change in the size, shape and location of HOMO. Significant changes were observed in LUMO surface density and are presented in Figures 3A, 3B and 3C to provide a global view of LUMO surface fields as they reside along the 4-HNE-SIRT3 peptide for the 4-HNE, cyclized and reduced forms of the adduct. Remarkably, both cyclization and reduction significantly alter LUMO surface orientation away from the 4-HNE adduct and towards the more stable peptide backbone. This shift is magnified in Figures 3D-F. Visualizing the LUMO for each peptide-adduct species through surface density models supports the hypothesis that LUMO may play a role in predicting the stability of this PTM during MS/MS fragmentation. An important observation is that cyclization shifts the LUMO towards the peptide backbone, while NaBH4 reduction transfers the LUMO density completely onto the peptide backbone, removing the LUMO from the sulfur atom and the 4-HNE adduct. Indeed, the shift in LUMO towards the more stable peptide backbone is an interesting observation; however, precise mechanisms relating LUMO to MS-induced RMA reactions remain perplexing and require further investigation.
Table 1.
Calculated parameters for non-reduced, cyclic and reduced 4-HNE-Cys280 peptide.
| Peptide | Energy (au) | E (kJ/mol) | E HOMO (kJ/mol) | E LUMO (kJ/mol) |
|---|---|---|---|---|
| 4-HNE peptide | -9359 | -24573097 | -1108 | -236 |
| Cyclic 4-HNE peptide | -9359 | -24573314 | -1123 | -203 |
| Reduced 4-HNE peptide | -9360 | -24576100 | -1132 | -173 |
Figure 3.
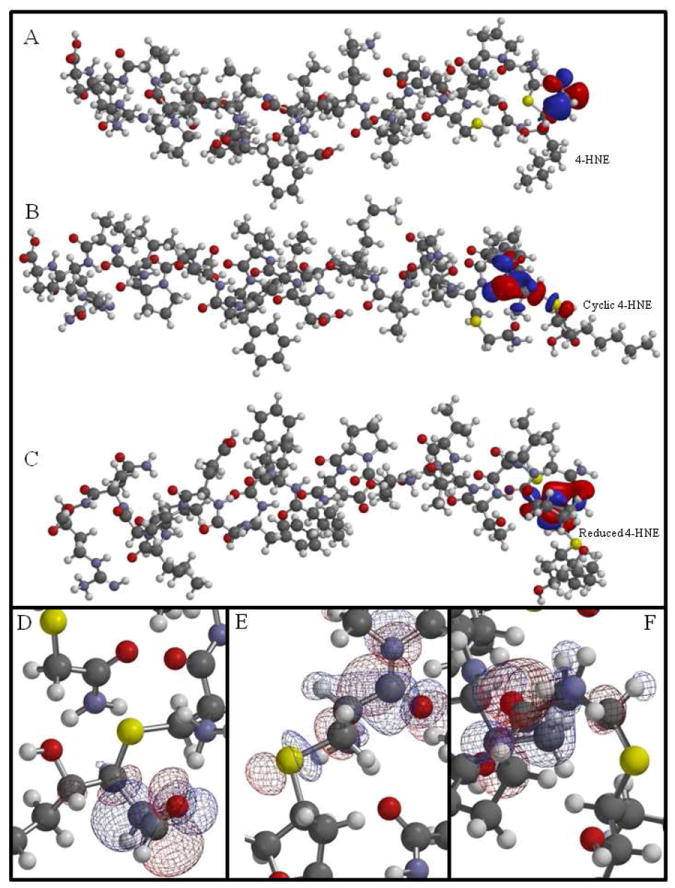
Molecular modeling illustrates the LUMO surface energy profile for the 4-HNE modified peptide containing Cys280 of SIRT3 (CHNEPVCCarbTGVVKPDIVFFGEPLPQR). Comparison of LUMO density surface maps for 4-HNE (A), cyclic hemiacetal (B) and reduced 4-HNE (C) adducts on Cys280. Analysis of the models finds the LUMO surface density located around C1-C3 of 4-HNE (D), around the thiol group at Cys280 (E) and surrounding the more stable peptide backbone of Cys280 of the reduced 4-HNE (F). This computational model presents a visual analysis of altered LUMO resulting from the increased stability of 4-HNE through cyclization (B, E) and reduction (C, F).
(Carb=carbamidomethylated) (Sulfur, yellow; oxygen, red; carbon, gray; nitrogen, purple; hydrogen, white)
The comparison of quantum mechanical analysis and molecular modeling to 4-HNE-peptide fragmentation by CID/ETD provides a unique approach to understanding protein carbonyl stability. Previous reports have examined the complementary nature of quantum mechanical parameters in developing therapeutics.23, 24 Reports by Rauniyar et al. detail MS techniques for studying 4-HNE, such as data-dependent neutral loss-driven MS3 acquisition and comparisons of CID to electron capture dissociation (ECD).33-35 Additionally, Wakita et al. demonstrate 2D-NMR analysis of the stereochemical configuration of a 4-HNE-thiol adduct which was enhanced through computational modeling of electrostatic surface potentials.28 This report represents the first analysis establishing a possible link between changes in E LUMO and LUMO surface density to the CID/ETD fragmentation characteristics of a 4-HNE adduct. It is likely, however, that numerous factors contribute to the stability, or lack thereof, of this 4-HNE adduct under MS/MS conditions and the observed neutral loss under CID fragmentation. Interestingly, ETD analysis did not initiate a RMA reaction for this 4-HNE-thiol adduct, similarly to NaBH4 reduction, and is a valuable tool for the direct analysis of unstable electrophilic adducts.
CONCLUSION
A host of in vivo studies have identified 4-HNE protein adducts as a direct result of sustained oxidative stress and persistent lipid peroxidation.11 Confirmed through immunohistochemical staining and Western blotting by anti-4-HNE antibodies, these adducts are proposed play a major role in disease states involving oxidative stress ranging from Alzheimer’s to liver disease. While the existence of these adducts remains unquestioned, the lack of adduct identification by mass spectrometry remains perplexing, as only a few in vivo adducts have been reported to date.6 The low-level abundance of these modifications remains a hurdle that is overcome through enrichment strategies. Assuredly, stability plays a crucial role in detecting these adducts and requires continued investigation to delineate the mechanistic roles of 4-HNE-protein adducts in cellular responses to sustained oxidative stress. As demonstrated here, the inclusion of ETD fragmentation will undoubtedly enhance the characterization of unstable electrophilic adducts.
Acknowledgments
We thank Bruker Daltonics (Billerica, MA.), Ben Gonzales, Anjali Alving and Matt Willetts for providing CID & ETD MS/MS. We appreciate the assistance of Phil Reigan and Donald Backos in the Skaggs School of Pharmacy Computational Biology Core.
FUNDING SOURCES
This work was supported, in part, by Grants NIH/NIAAA 5R37AA009300-15 (D.R.P.) and NIH/NIDDK 5R01DK074487-03 (D.R.P.).
ABBREVIATION
- 4-HNE
4-hydroxynonenal
- NaBH4
sodium borohydride
- PTM
post-translational modification
- LUMO
lowest unoccupied molecular orbital
- ETD
electron transfer dissociation
- CID
collision induced dissociation
- SIRT3
Sirtuin 3
References
- 1.Esterbauer H, Schaur RJ, Zollner H. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 2.Roede JR, Jones DP. Environ Mol Mutagen. 2010;51:380–390. doi: 10.1002/em.20553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Codreanu SG, Zhang B, Sobecki SM, Billheimer DD, Liebler DC. Mol Cell Proteomics. 2009;8:670–680. doi: 10.1074/mcp.M800070-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petersen DR, Doorn JA. Free Radic Biol Med. 2004;37:937–945. doi: 10.1016/j.freeradbiomed.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Smathers RL, Galligan JJ, Stewart BJ, Petersen DR. Chem Biol Interact. 2011 doi: 10.1016/j.cbi.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fritz KS, Petersen DR. Chem Res Toxicol. 2011;24:1411–1419. doi: 10.1021/tx200169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaur RJ. Mol Aspects Med. 2003;24:149–159. doi: 10.1016/s0098-2997(03)00009-8. [DOI] [PubMed] [Google Scholar]
- 8.Doorn JA, Petersen DR. Chem Res Toxicol. 2002;15:1445–1450. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 9.LoPachin RM, Gavin T, Petersen DR, Barber DS. Chem Res Toxicol. 2009;22:1499–1508. doi: 10.1021/tx900147g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poli G, Biasi F, Leonarduzzi G. Mol Aspects Med. 2008;29:67–71. doi: 10.1016/j.mam.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 11.Poli G, Schaur RJ, Siems WG, Leonarduzzi G. Med Res Rev. 2008;28:569–631. doi: 10.1002/med.20117. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhary P, Sharma R, Sharma A, Vatsyayan R, Yadav S, Singhal SS, Rauniyar N, Prokai L, Awasthi S, Awasthi YC. Biochemistry. 2010;49:6263–6275. doi: 10.1021/bi100517x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juric-Sekhar G, Zarkovic K, Waeg G, Cipak A, Zarkovic N. Tumori. 2009;95:762–768. doi: 10.1177/030089160909500620. [DOI] [PubMed] [Google Scholar]
- 14.Chung WG, Miranda CL, Maier CS. Electrophoresis. 2008;29:1317–1324. doi: 10.1002/elps.200700606. [DOI] [PubMed] [Google Scholar]
- 15.Carbone DL, Doorn JA, Kiebler Z, Petersen DR. Chem Res Toxicol. 2005;18:1324–1331. doi: 10.1021/tx050078z. [DOI] [PubMed] [Google Scholar]
- 16.Reed T, Perluigi M, Sultana R, Pierce WM, Klein JB, Turner DM, Coccia R, Markesbery WR, Butterfield DA. Neurobiol Dis. 2008;30:107–120. doi: 10.1016/j.nbd.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Guo J, Prokai L. J Proteomics. 2011;74:2360–2369. doi: 10.1016/j.jprot.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alary J, Fernandez Y, Debrauwer L, Perdu E, Gueraud F. Chem Res Toxicol. 2003;16:320–327. doi: 10.1021/tx025671k. [DOI] [PubMed] [Google Scholar]
- 19.Annangudi SP, Deng Y, Gu X, Zhang W, Crabb JW, Salomon RG. Chem Res Toxicol. 2008;21:1384–1395. doi: 10.1021/tx8000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rauniyar N, Prokai L. J Mass Spectrom. 2011;46:976–985. doi: 10.1002/jms.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roede JR, Carbone DL, Doorn JA, Kirichenko OV, Reigan P, Petersen DR. Chem Res Toxicol. 2008;21:2289–2299. doi: 10.1021/tx800244u. [DOI] [PubMed] [Google Scholar]
- 22.Fritz KS, Galligan JJ, Smathers RL, Roede JR, Shearn CT, Reigan P, Petersen DR. Chem Res Toxicol. 2011;24:651–662. doi: 10.1021/tx100355a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright P, Alex A, Nyaruwata T, Parsons T, Pullen F. Rapid Commun Mass Spectrom. 2010;24:1025–1031. doi: 10.1002/rcm.4458. [DOI] [PubMed] [Google Scholar]
- 24.Alex A, Harvey S, Parsons T, Pullen FS, Wright P, Riley JA. Rapid Commun Mass Spectrom. 2009;23:2619–2627. doi: 10.1002/rcm.4163. [DOI] [PubMed] [Google Scholar]
- 25.Lesgards JF, Frayne IR, Comte B, Busseuil D, Rheaume E, Tardif JC, Rosiers CD. Free Radic Biol Med. 2009;47:1375–1385. doi: 10.1016/j.freeradbiomed.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Drug Metab Rev. 2006;38:651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 27.Uchida K. Amino Acids. 2003;25:249–257. doi: 10.1007/s00726-003-0015-y. [DOI] [PubMed] [Google Scholar]
- 28.Wakita C, Maeshima T, Yamazaki A, Shibata T, Ito S, Akagawa M, Ojika M, Yodoi J, Uchida K. J Biol Chem. 2009;284:28810–28822. doi: 10.1074/jbc.M109.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopachin RM, Gavin T, Decaprio A, Barber DS. Chem Res Toxicol. 2011 doi: 10.1021/tx2003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martyniuk CJ, Fang B, Koomen JM, Gavin T, Zhang L, Barber DS, Lopachin RM. Chem Res Toxicol. 2011;24:2302–2311. doi: 10.1021/tx200437y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uchida K, Stadtman ER. Proc Natl Acad Sci U S A. 1992;89:5611–5615. doi: 10.1073/pnas.89.12.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greaves J, Harvey E, MacIntyre WG. Journal of the American Society for Mass Spectrometry. 1994;5:44–52. doi: 10.1016/1044-0305(94)85083-6. [DOI] [PubMed] [Google Scholar]
- 33.Rauniyar N, Stevens SM, Prokai-Tatrai K, Prokai L. Anal Chem. 2009;81:782–789. doi: 10.1021/ac802015m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rauniyar N, Prokai L. Proteomics. 2009;9:5188–5193. doi: 10.1002/pmic.200900116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rauniyar N, Stevens SM, Jr, Prokai L. Anal Bioanal Chem. 2007;389:1421–1428. doi: 10.1007/s00216-007-1534-2. [DOI] [PubMed] [Google Scholar]
- 36.Kohler EP, P H. Journal of the American Chemical Society. 1935;57:1316–1321. [Google Scholar]


