Abstract
Understanding the process by which pancreatic beta-cells acquire their ‘fate’ is critical to the development of in vitro directed differentiation protocols for cell replacement therapies for diabetics. To date, these efforts are hampered by a paucity of markers that distinguish pancreatic endocrine cells at different stages of differentiation. Here, we identify EphB3 as a novel pro-endocrine marker and use its expression to track delaminating islet lineages. First, we provide a detailed developmental expression profile for EphB3 and other EphB family members in the embryonic pancreas. We demonstrate that EphB3 transiently marks endocrine cells as they delaminate from the pancreatic epithelium, prior to their differentiation. Using a Tet-inducible EphB3rtTA-lacZ reporter line, we show that short-term pulse-labeled EphB3+ cells co-express Pdx1, Nkx6.1, Ngn3 and Synaptophysin, but not insulin, glucagon or other endocrine hormones. Prolonged labeling tracks EphB3+ cells from their exit from the epithelium to their differentiation. These studies demonstrate that pro-endocrine cell differentiation during late gestation, from delamination to maturation, takes approximately two days. Together, these data introduce EphB3 as a new biomarker to identify beta-cells at a critical step during their step-wise differentiation and define the timeframe of endocrine differentiation.
Keywords: EphB3, pancreas, endocrine, delamination, epithelium, islet, lineage tracing
INTRODUCTION
Understanding the step-wise differentiation of pancreatic beta-cells is essential to diabetes research, as efforts to hone cell replacement therapy for diabetics have increased. In vitro directed-differentiation of naïve cells, such as ES or iPS cells, towards glucose-responsive, insulin-producing beta-cells have garnered much attention and been relatively successful (D’Amour et al., 2006). To date, however, these laboratory-derived cells have required implantation into mice for full maturation and functionality. The reasons for this required in vivo incubation are unknown, but it has been suggested that either in vitro generated cells diverged in their developmental trajectory early during the course of their differentiation or that their maturation required additional steps. To date, it has been difficult to detect or influence putative divergence in fate due in part to a dearth of useful markers that could track their step-wise differentiation. One effort to increase our understanding of these steps was the concerted effort by several groups, including the Beta Cell Biology Consortium (BCBC), to develop a large array of antibodies for delineating those steps (Hald et al., 2011). However, there remains a critical need to identify additional molecular landmarks and regulators of beta-cell differentiation, to provide greater resolution in understanding cell fate choices that endocrine progenitors make as they generate functional endocrine cells.
In recent decades, there have been intense efforts to identify the intrinsic regulators of pancreatic cell lineages. Endocrine specification and differentiation depends on the orchestrated action of cascades of transcription factors, which are dynamically expressed during pancreatic development. Pdx1, Ptf1a, Sox9 and Ngn3, for instance, are all transcription factors essential to endocrine cell differentiation, which display varied and transient patterns of expression in cells of the developing pancreatic bud. While all four factors are expressed throughout the early pancreatic epithelium, their expression diverge as endocrine cells differentiate: Pdx1 becomes restricted to β-cells, δ-cells, and PP (pancreatic polypeptide) cells at late gestation (Ahlgren et al., 1997; Guz et al., 1995); Ptf1a becomes first restricted to multipotent progenitors at epithelial branch termini, then to committed exocrine cells (Masui et al., 2007); Sox9 is extinguished as endocrine cells differentiate but continues to label tubular epithelium (ducts) into adulthood (Kopp et al., 2011); and Ngn3 is widespread in the early pancreatic epithelium (Villasenor, 2008), but later exhibits a highly transient and spatiotemporally restricted expression in all committed endocrine cell types, turning off as cells exit the epithelium (Gradwohl et al., 2000; Johansson et al., 2007). The timing of expression and interplay of these transcription factors is essential to determine endocrine fate. Forced co-expression of three transcription factors, pdx1 ngn3 and mafA, for instance, results in the transdifferentiation of exocrine cells into beta-cells (Zhou et al., 2008). Unveiling the stepwise roles of these powerful regulators is therefore of great importance to understand the molecular blueprint that underlies the beta-cell fate.
However, while many such intrinsic molecular regulators have been intensely studied (see excellent reviews (Madsen et al., 1997; Oliver-Krasinski and Stoffers, 2008; Pan and Wright, 2011; Sander and German, 1997; Wilson et al., 2003), relatively little is known about the extrinsic cell-cell signals that drive expression of these intrinsic factors although some inroads are being made (Apelqvist et al., 1999; Cleaver and MacDonald, 2009; Heller et al., 2002; Stafford and Prince, 2002). Recently, signaling by ephrin ligands to Eph receptor tyrosine kinases was shown to be required for proper pancreatic branching morphogenesis (Villasenor et al., 2010) as well as for optimal insulin secretion in the adult mouse (Konstantinova et al., 2007). Previous expression analysis had shown that EphBs and ephrinBs were expressed in the early pancreas (van Eyll et al., 2006; Villasenor et al., 2010).
Here, we extend original observations and provide a comprehensive spatiotemporal comparison of the different members of the EphB/ephrin family with a focus on the receptor EphB3. We use a Tet-inducible EphB3-rtTA BAC transgenic reporter line to identify and track the dynamics of EphB3 expressing cells along their developmental journey. We find that EphB3 is expressed in pro-endocrine cells in a tightly regulated manner, initiating just prior to their exit from the epithelium and becoming extinguished as they differentiate. Pulse-chase lineage labeling studies demonstrate that EphB3-expressing cells give rise to islet endocrine cells. Furthermore, these experiments show that endocrine differentiation, following the ‘secondary transition’, occurs in the span of approximately two days. To our knowledge, EphB3 is the first biomarker of delaminating endocrine cells, adding to the limited toolkit of endocrine markers currently available.
RESULTS
Eph/ephrinB expression analysis during pancreatic development
To characterize and compare Eph/ephrinB gene expression during pancreas development and endocrine differentiation, we carried out whole mount in situ hybridization and β-galactosidase (β-gal) staining of transgenic murine embryonic guts throughout mid- to late gestation. During embryonic development, endocrine cells emerge during two ‘waves’ of differentiation. Here, we show three representative developmental time frames: E9.5 (bud initiation), E10.5 (first wave endocrine cells) and E13.5 (second wave endocrine cells).
ephrinB ligands
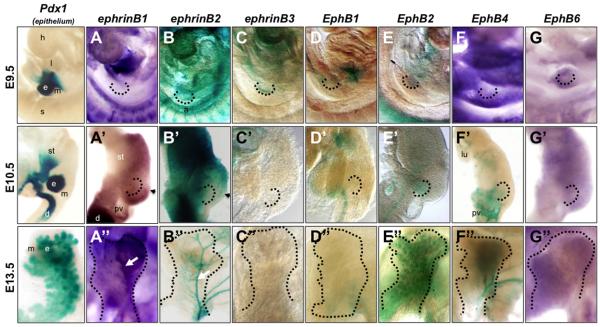
Whole mount in situ hybridization for ephrinB1 (efnb1) showed that starting around E10.5, efnb1 was expressed in the mesoderm surrounding the pancreatic bud epithelium (Fig. 1A,A’). Efnb1 expression showed an anterior-posterior gradient, with highest expression in the posterior mesoderm. By E13.5, efnb1 expression remained strong in the peripheral mesenchyme (Supp. Fig. S1A), with a noticeable enrichment in the pancreatic ‘ridge’ and the ‘right’ side of the pancreatic bud (when viewed laterally) (Fig. 1A”, white arrow) (Villasenor et al., 2010). Whole mount β-gal staining of ephrinB2lacZ embryos (Cowan et al., 2004; Dravis et al., 2004) and sections showed that efnb2 was expressed in the gut mesoderm at E9.0-9.5 (Fig. 1B,B’ and Supp. Fig. S1B), although it became enriched in the anterior bud mesoderm/mesenchyme at E10.5. At later stages, its expression became restricted to blood vessels as previously reported (Fig. 1B” and Supp. Fig. S1B’)(Wang et al., 1998). We note the interesting and relatively mutually exclusive and complementary pattern of efnb1 and efnb2 mesenchymal expression along the anteroposterior axis at E10.5 (compare Fig. 1A’ and 1B’, arrowheads). EphrinB3lacZ β-gal staining showed that efnb3 was briefly expressed in the pancreatic epithelium of the E9.5 bud, but was quickly extinguished in that tissue at later developmental stages (Fig. 1C,C” and Supp. Fig. S1C). Unlike the expression of efnb1 and efnb2, efnb3 was absent from the mesenchyme at all timepoints examined.
Figure 1. Expression of ephrin ligands and Eph receptors during pancreatic development.
Whole mount in situ hybridization and whole mount β-galactosidase staining of ephrinB ligands and EphB receptors. First column: β-galactosidase staining of Pdx1-lacZ embryonic midgut and pancreas at stages indicated; epithelium stains blue. In situ hybridization of ephrinB1 (A,A”); EphB4 (F); and EphB6 (G,G”). Whole mount β-galactosidase staining of ephrinB2 (B,B”); ephrinB3 (C,C”); EphB1 (D,D”); EphB2 (E,E”); and EphB4 (F’,F”) at stages indicated. E9.5 embryos in (A-G) are facing forward. E10.5 dissected gut tubes in (A’-G’) have anterior up and dorsal to the right. E13.5 Dorsal pancreatic buds in (A”-G”) have stomach in the background and are shown in lateral view (proximal at bottom, distal at top). The pancreatic endoderm is delineated by dotted black lines. Arrowheads in A’ and B’ show boundary between reciprocal expression domains of ephrinB2 and ephrinB1. White arrows point to ridge (A”) or to main blood vessel (B”). a, aorta; d, duodenum; e, pancreatic epithelium; h, heart; l, liver; lu, lung; m, mesoderm/mesenchyme; pv, portal vein; s, somites; st, stomach.
EphB receptors
β-gal staining of EphB1lacZ (Williams et al., 2003) embryonic guts revealed that the EphB1 receptor was expressed in the duodenum and foregut epithelium, but not in the pancreas at any developmental stage examined (Fig. 1D,D”). By contrast, β-gal staining of EphB2lacZ midguts (Henkemeyer et al., 1996) showed expression in the pancreatic epithelium throughout development (Fig. 1E,E”). At early developmental stages (E9.5-E10.5), EphB2 expression was absent in much of the foregut, but was clearly detectable in the pancreatic bud epithelium (Supp. Fig. S1D). By E13.5, EphB2 was detectable throughout the pancreatic epithelium, but enriched at branch tips (Supp. Fig. S1D’). At later developmental stages (data not shown), EphB2 became restricted to exocrine tissue and was further enriched at the apical membrane of acinar cell clusters. β-gal staining of early EphB4lacZ embryos (E9.5-E10.5) (Gerety et al., 1999), by contrast, showed that EphB4 was expressed in vessels throughout the mesoderm and in the portal vein posterior to the E10.5 dorsal bud (Fig. 1F,F’ and Supp. Fig. S1F), with later enrichment in veins as expected (Fig. 1F”) (Swift and Weinstein, 2009; Wang et al., 1998). Whole mount in situ hybridization showed that EphB6 was weakly and diffusely expressed throughout the pancreatic mesenchyme at all stages assayed (Fig. 1G,G” and Supp. Fig. S1G).
EphB3 is expressed in a subpopulation of cells in the early, stratified pancreatic epithelium
To characterize EphB3 expression, an inducible Tet-On transgenic mouse was generated by crossing BAC-Tg-EphB3rtTA-containing mice to a TRE-lacZ line (Ludwig et al., 2004) (Fig. 2A). The BAC-Tg-EphB3rtTA line expresses the reverse tetracycline-controller transactivator (rtTA) under the control of the EphB3 promoter (refer to methods), while the TRE-lacZ line expresses the lacZ reporter under the tetracycline inducible promoter (TRE). Doxycycline (Dox) induction leads to transcription of the lacZ open reading frame and production of β-gal in EphB3-expressing cells.
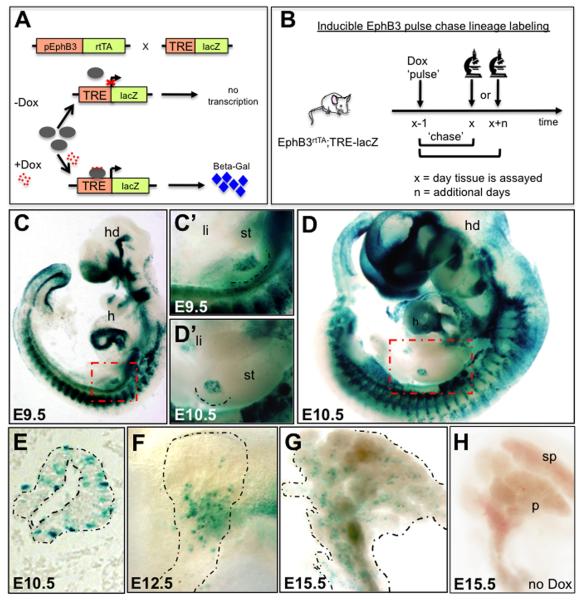
Figure 2. EphB3rtTA is expressed in the pancreatic anlage.
(A) Diagram showing the generation of BAC-Tg-EphB3rtTA-lacZ males. (B) Schematic exemplifying the strategy for EphB3 lineage tracing, either short or long term. (C,D) Whole mount β-galactosidase staining of E9.5 and E10.5 embryos (C’,D’ show close ups of gut tube in C,D, red dotted boxes). (E,F) Whole mount β-galactosidase staining of E12.5 and E15.5 pancreata. Distal is up, proximal is down. (G) Sagittal section of E10.5 pancreatic bud showing expression of EphB3 in scattered cells. (H) E15.5 uninduced pancreas showing no leakage of β-gal expression. Broken lines delineate pancreas. h, heart; hd, head.
We ‘Dox-induced’ double transgenic mice, here referred to simply as EphB3rtTA- lacZ, with a ‘pulse’ lasting approximately 20 hours and assayed for β-gal expression (Fig. 2B). Studies using similar tools have previously shown that Dox was systemically present for approximately 2 days and β-gal, once expressed, perdured for over 6 days (Ludwig et al., 2004). Therefore, cells labeled and assayed after 1 day would represent new EphB3 ‘expressors’, whereas those assayed 3 days following Dox labeling would comprise both downstream lineages, as well as newly emerging ‘expressors’. Beyond 5 days, these ‘older’ labeled cells would remain detectable (having persistent β-gal), but Dox would be systemically cleared and newly emerging progenitors would no longer be labeled.
Whole mount β-gal staining of E9.5 and E10.5 embryos revealed strong EphB3 expression in somites, telencephalon, primitive left ventricle of the heart, nasal epithelium, branchial arches and pancreas (Fig. 2C-D’). At later stages, this transgenic line also displayed expression in numerous other tissues, faithfully reporting EphB3 expression in tissues where EphB3 is known to be required for proper morphogenesis, such as in hindgut and foregut septations, and also in the developing palate (Dravis et al., 2004) and data not shown). Expression in the early gut tube was primarily restricted to the pancreatic endoderm and absent in the mesoderm (Fig. 2C’,D’). Sectioning of the E10.5 bud revealed EphB3 expression in a subpopulation of cells within the stratified pancreatic epithelium (Fig. 2E). At later stages, during the second wave of endocrine differentiation (E12.5 and E15.5), EphB3 expression was observed in scattered cells within the central core of the dorsal pancreas (Fig. 2F,G and Supp. Fig. S1E), suggesting that EphB3+ cells were pro-endocrine cells. No leakage of β-gal was detected in the absence of Dox (Fig. 2H).
To identify the identity of EphB3+, we performed a series of pulse-chase lineage labeling studies to trace descendants of EphB3+ cells at different timepoints. The different stages at which we induced with Dox and assayed β–gal expression throughout this paper are summarized in a schematic in Supp. Figure S2. Immunofluorescent stainings of EphB3rtTA-lacZ pancreata, using an anti-β-gal antibody, were performed to identify the EphB3+ cell lineages during the first wave of endocrine differentiation (primary transition). Given that Pdx1 is a marker of the pancreatic progenitor epithelium at early budding stages, including nascent endocrine cells therein (Guz et al., 1995), we analyzed its expression in the EphB3rtTA-lacZ pancreas and found that EphB3 marked a subset of Pdx1+ cells at E9.5 (Fig. 3A). Co-stainings of E-cadherin and β–gal at E10.5 confirmed that EphB3+ cells were indeed epithelial in nature (Fig. 3B), and were located both in peripheral ‘cap’ cells and in cells within the internal stratified epithelium.
Figure 3. Pulse-chase lineage tracing: First transition induction and assay after 1 day (at E9.5 or E10.5).
Immunostaining of anti-β-gal (green) and pancreatic markers (red) show that EphB3 is expressed in the early pancreatic epithelium. (A) Transverse section of E9.5 EphB3rtTA-lacZ pancreatic bud. EphB3+ cells (green) and Pdx-1+ cells (red). (B-F) Sagittal sections of E10.5 pancreata. EphB3 expression (green) and (B) E-cadherin, (C) Sox9, (D) Nkx6.1, (E) glucagon (red) and (F) Ngn3. Arrows indicate overlap. Asterisk indicate lack of overlap in panel C.
To determine whether EphB3+ cells represented endocrine progenitors during the first transition, as their distribution in whole mounts suggested (Fig. 2), we compared EphB3 expression to that of pro-endocrine markers, Sox9, Nkx6.1 and Ngn3 (Gradwohl et al., 2000; Sander et al., 2000; Seymour et al., 2007). Interestingly, we assayed co-expression of EphB3 with Sox9 and did not detect overlap (Fig. 3C). By contrast, 92% of EphB3+ cells expressed Nkx6.1, and 10% of EphB3+ cells expressed Ngn3 (Fig. 3C,E,F). These data suggest that a proportion of pro-endocrine cells initiated EphB3 expression during endocrine commitment, but after divergence from ductal fate. This was further evident in co-stains of EphB3 and glucagon, where a subset of first transition endocrine glucagon-expressing cells co-expressed EphB3 (Fig. 3D). In all cases, except for E-cadherin, the observed overlaps were partial, suggesting that either a proportion of endocrine cells had already downregulated EphB3 at the time of Dox induction (those that had already extinguished EphB3 transcription along their path to differentiation), or Dox induction is not fully penetrant and does not label all EphB3 expressing cells, or that EphB3 marks a distinct subset of endocrine cells.
EphB3 marks delaminating endocrine cells during the secondary transition
During the second wave of endocrine differentiation (E12.5-E15.5), EphB3+ cells appeared studded along pancreatic ducts (Fig. 4A). To identify the immediate progeny of these cells, we Dox-induced EphB3rtTA-lacZ transgenic embryos at E14.5 and co-stained for β-gal and various pancreatic markers one day later at E15.5 (Fig. 4 and Supp. Fig. S2).
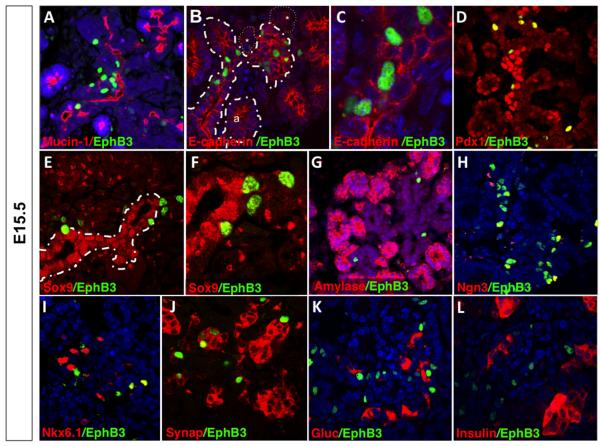
Figure 4. Pulse-chase lineage tracing: Second transition induction and assay after 1 day (at E15.5).
Immunostaining of anti-β-gal (green) and pancreatic markers (red) show that EphB3 is expressed in delaminating pro-endocrine cells. Sagittal sections of E15.5 EphB3rtTA-lacZ pancreata. EphB3+ cells (green) and (A) Mucin+, (B,C) E-cadherin+; (D) Pdx1+; (E,F) Sox9+; (G) amylase+; (H) Ngn3+; (I) synaptophysin+ (Synap); (J) Nkx6.1+; (K) glucagon+ (Gluc); or (L) insulin+ (Ins) cells (red). Pancreatic epithelium is outlined by stippled lines; endocrine clusters in B, *.
Mucin-1 (Muc1) (Fig. 4A) and E-cadherin (Fig. 4B,C) immunostainings confirmed that EphB3+ cells were located either in the pre-ductal cords or in close proximity to them (Fig. 4B,C), indicating that EphB3 marks a subpopulation of epithelial cells which may represent delaminating pro-endocrine cells. In addition, we found that most EphB3+ cells (92%) also expressed Pdx1 (Fig. 4D). To further assess whether EphB3+ cells might represent pro-endocrine cells, we assayed expression of the Sox9 transcription factor. Sox9 is expressed in endocrine progenitors within ductal epithelium, prior to their delamination, but becomes extinguished as endocrine cells exit the tubules (Seymour et al., 2007). Interestingly, we found that about 95% of EphB3+ cells expressed low to undetectable levels of Sox9 (Fig. 4E,F). EphB3+ cells seem to be exiting the epithelium suggesting that they represent ‘delaminating’ cells that have already acquired an endocrine fate.
To test this hypothesis, we performed co-staining with both exocrine and proendocrine markers. As expected, EphB3+ cells were excluded from pre-acini florets and did not co-express exocrine markers, such as amylase (Fig. 4G). Instead, EphB3 was expressed in 80% of Nkx6.1+ cells and 43% of Ngn3+ cells (Fig. 4H,I). These data indicated that EphB3 is expressed in committed endocrine progenitors, as Ngn3 is the universal pro-endocrine transcription factor and Nkx6.1 at this stage is restricted to β-cells.
We next asked whether those EphB3+ cells that did not express Ngn3 represented newly delaminated, differentiating endocrine cells. To assess later stages of endocrine differentiation, expression of synaptophysin was analyzed, as it marks all endocrine lineages as they begin to differentiate (Ray MacDonald personal communication; (Redecker et al., 1991). Most EphB3+ cells co-expressed synaptophysin (approximately 74%) (Fig. 4J). By contrast, EphB3+ cells did not express the endocrine hormones insulin, glucagon, somatostatin, or ghrelin (Fig. 4K,L and data not shown). Pulse-chase labeling for 1 day at a later embryonic stage (E18.5) similarly revealed lack of co-expression of EphB3+ cells and mature insulin or glucagon hormone markers (Supp. Fig. S3). Together, these results suggest that EphB3 expressing cells represent a transient state, starting with pro-endocrine cells as they prepare to delaminate, throughout the process of delamination, to the onset of cytodifferentiation.
EphB3+ cells contribute to pancreatic islets
Given β-gal stability (which remains detectable past 6 days) and slow systemic clearance rates of Dox (which remains in fetal and maternal tissues for 2 days (Ludwig et al., 2004)), we reasoned that labeled cells would retain β-gal beyond the immediate timeframe of Tet induction, allowing us to follow the fate of EphB3+ cells for an extended, but not indefinite, window of time. In other words, our technique labels cells that express EphB3 and also cells that once expressed EphB3. Since endocrine cells undergo only limited replication following Ngn3 expression and their exit from the epithelium (Miyatsuka et al., 2010), our brief ‘pulse labeling’ followed by a ‘chase’ of Dox withdrawal prior to immunostaining would provide a confirmation of the endocrine fate of EphB3+ cells, provided the time frame to final differentiation was not too lengthy and β-gal remained stable.
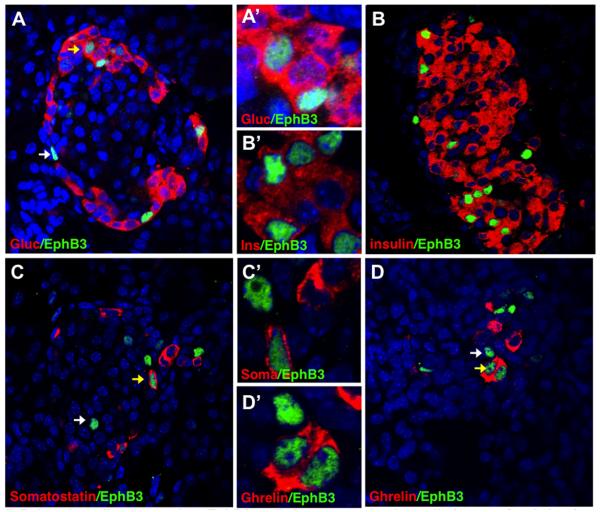
A Dox pulse was performed at E14.5 and pancreatic tissue was analyzed 8 days later at P4 (instead of the next day) (Supp. Fig. S2). Following this long ‘chase’, immunostainings with endocrine hormones showed that indeed β-gal activity was still present and EphB3+ cells became endocrine cells, located within islets. In addition, EphB3+ cells were found to contribute to different endocrine populations including glucagon (Fig. 5A,A’), insulin (Fig. 5B,B’), somatostatin (Fig. 5C,C’), and ghrelin expressing cells (Fig. 5D,D’).
Figure 5. Pulse-chase lineage tracing: EphB3+ cells become endocrine cells (assay after 8 days).
Dox pulse administered at E14.5 and pancreas assayed at P4. Immunostainings of postnatal EphB3rtTA-lacZ pancreata with anti-β-gal (green) and pancreatic markers (red) show that, following an 8 day chase, EphB3+ cells express (green): (A,A’) glucagon, (B,B’) insulin, (C,C’) somatostain (Sst), or (D,D’) ghrelin (red). Yellow arrows point to overlaps, while white arrows point to EphB3+ cells that do not overlap.
EphB3 a biomarker of delaminating endocrine cells
In order to track the timeframe of endocrine differentiation in the embryonic pancreas, and follow the progression of an endocrine progenitor from delamination to differentiation, we assayed Dox-pulsed pancreata at different timepoints and used different lengths of ‘chasing’ (Supp. Fig. S2). While ‘pulse’ labeling for 20 hours (and assaying after 1 day chase) allowed us to ‘catch’ a temporally narrow population of proendocrine cells as they delaminated, we reasoned that chasing for a longer timeframes (1, 2 or 3) would reveal multiple populations as they continuously exited the epithelium. The stability of β-gal would again allow detection of different sequential cell types along the developmental path taken by the earlier delaminating EphB3+ cells within that expanded population.
We Dox pulsed at E14.5 for 20 hours, and then assayed pancreatic tissue either 1 day later (E15.5); 2 days later (E17.5); or 3 days later (E18.5). As noted before, we observed that after 1 day of tracing, EphB3+ cells did not express insulin or glucagon (Fig. 6A,B). However, they were in close vicinity to endocrine cells, suggesting that they were in the process of aggregating into islets (Fig. 6A’,B’). When we assayed 2 days following the Dox pulse, some EphB3+ cells could be seen expressing glucagon (Fig. 6C,C’) and insulin (Fig. 6D,D’). These results showed that endocrine differentiation, from delamination to hormone expression, takes approximately 48 hours.
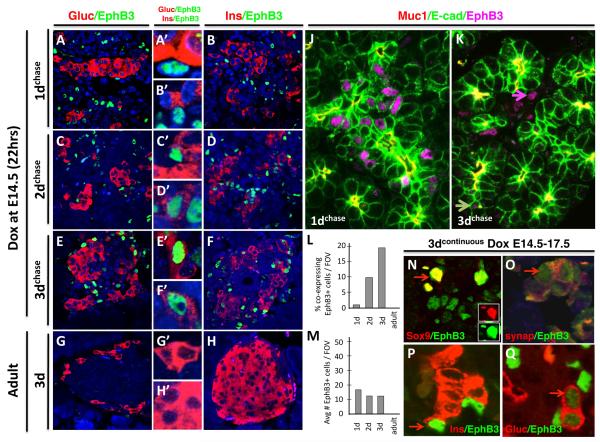
Figure 6. Timeframe of endocrine differentiation of EphB3+ cells: 2 days.
Dox pulse is administered at E14.5 for 20 hours, and pancreas is assayed 1 day later (E15.5) (A-B’); 2 days later (E17.5) (C-D’); or 3 days later (E18.5) (E-F’). Alternatively, Dox is administered continuously for 3 days (G-H’). (A-H) Panels show EphB3 co-expression with endocrine hormones: EphB3+ cells (green) and glucagon+ or insulin+ cells (red) are shown in adjacent columns (close ups in center column). Note lack of co-expression of EphB3 and hormones after 1 day pulse-chase, but a few cells co-express after 2 days, and more cells label after 3 days (A,B). Immunostains show pre-ductal epithelium and EphB3+ cells after 1 day of induction (J) and after 2 days (K): Muc1 marks apical side of ductal epithelium (red), E-cadherin marks epithelial cells (green) and EphB3+ (magenta). Graphs show the number of EphB3+ cells co-expressing hormonal markers (insulin and glucagon) (L) and total number of endocrine versus EphB3+ cells per field of view (FOV) (M), at different timepoints post-induction. (N-Q) Stainings of 3 day continuous Dox induction of EphB3rtTA-lacZ: EphB3+ (green) and (N) Sox9+, (O) synapthophysin+; (P) insulin+; or (Q) glucagon+ (red).
By the third day following Dox induction, increasing numbers of EphB3+ cells expressed glucagon (Fig. 6E,E’) and insulin (Fig. 6F,F’) appeared to have escaped the epithelial tubules. Muc-1 and E-cadherin co-stains of 1d and 2d induced pancreata confirmed that these results, as 1d-pancreata showed EphB3+ cells emerging from epithelial tubules (Fig. 6J), while 2d-pancreata showed EphB3+ cells clustering outside ductal tubules (Fig. 6K, pink arrow), with only some remaining within the epithelium (Fig. 6J, white arrow). The progression of decreasing epithelial EphB3+ and increased co-expression of EphB3 with hormone markers over developmental time is graphically depicted in (Fig. 6M,L, respectively). Continuous Dox induction for 3 consecutive days showed co-expression of EphB3 with sox9 (ducts); synaptophysin (early endocrine aggregates) and glucagon/insulin (mature hormones) confirming the progression of EphB3+ cells from ducts to islets (Fig. 6N,O).
Finally, to address whether EphB3 expression was restricted to embryonic islet neogenesis, we carried out similar assays in 6wk female adults. We performed Dox pulse labeling for 20hrs and assayed islets either 1d (not shown) or 3 days later (Fig. 6G-H’). We found no EphB3+ cells in any adult islets, either large or small, suggesting that EphB3 might only be appreciably activated in endocrine progenitors during embryonic development. While we detected low levels of endogenous EphB3 expression in adult pancreas by qPCR (data not shown), we never could detect expression by immunofluorence using anti-β-gal antibodies.
Together, these data support the idea that EphB3+ cells represent committed endocrine progenitors that give rise to islet endocrine cells during prenatal development. Moreover, using inducible transient lineage tracing and pulse-chase timing of these inductions, we establish that the timeframe for endocrine cells to delaminate from the epithelium and acquire the endocrine fate is about approximately 48hrs.
DISCUSSION
In recent years, it has become increasingly clear that the level of resolution, at the molecular level, of the developmental steps taken by beta cells is obscured in large part by the lack of molecular markers available as ‘bioindicators’ of beta cells from their birth to maturation. In an effort to increase the spectrum of tools for studying step-wise differentiation of beta cells, we identified EphB3 as a novel bioindicator of delaminating endocrine cells and we determine the time it takes for an embryonic endocrine cell to differentiate.
Throughout pancreas development, EphB3 proved a useful biomarker for delaminating and differentiating endocrine cells. We show that EphB3 marks proendocrine cells during both the primary and the secondary transitions of pancreatic development. During the primary transition, EphB3 is expressed in a subset of cells scattered in the stratified epithelium, while during the secondary transition, EphB3 marks pro-endocrine cells from their delamination from the pre-ductal tubular epithelium to the onset of their differentiation.
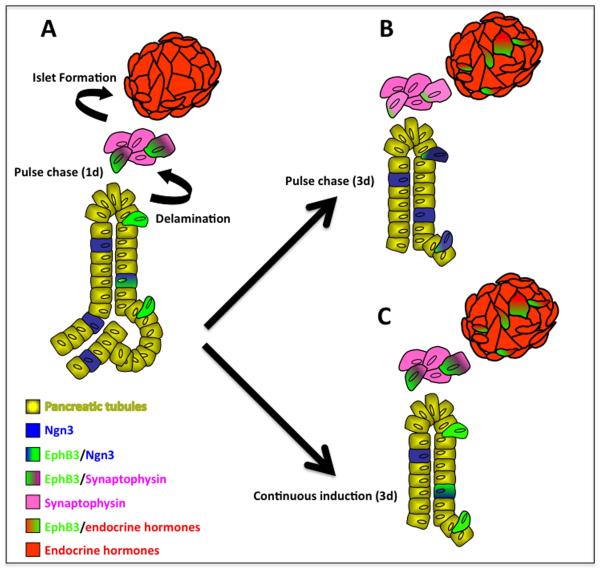
Because we used a transgenic transient labeling approach (not indelible), which depends on both Dox pulse induction and β-gal stability, we were able to determine the time progenitor cells took to differentiate. Pulse-chase lineage labeling using a Tetinducible approach, that ‘catches’ cells that express EphB3 at the time of induction, allowed us to dynamically identify the cell types expressing EphB3 in the developing pancreas. EphB3 expression was found in three subsets of pro-endocrine cells: a) cells within the pancreatic tubular epithelium, b) cells in the process of ‘delaminating’, and c) delaminated cells clustering into early islet aggregates. Short pulses of Dox followed by a short ‘chase’ period revealed EphB3 expression in pro-endocrine cells escaping the preductal epithelium, at the beginning of their developmental journey (Fig. 7A). By contrast, chasing for longer periods, such as 3-8 days, allowed us to follow them as they differentiated and to confirm their endocrine fate (Fig. 7B). After 2 days, cells that had expressed EphB3 could be found to give rise to mature endocrine cells located within the islet. Continuous Dox induction over a period of 3 days labeled cells at all stages during the progression of islet formation: from tubular epithelial cells, to delaminating, aggregating and finally to hormone-expressing cells within islets (Fig. 7C).
Figure 7. Model for EphB3-expressing cell contribution to islets.
Following the secondary transition, EphB3 is expressed in a continuum, including three key cell populations during the secondary transition: 1) pro-endocrine cells within the epithelium, 2) ‘delaminating’ pro-endocrine cells, and 3) pro-endocrine cells in early clusters and islets. (A) Short term labeling of EphB3+ cells shows that EphB3 is expressed in delaminating pro-endocrine cells. (B) Pulse-chase experiments determined that EphB3-expressing cells labeled at E14.5 begin to express endocrine hormones by E16.5 and contribute to islets. Newly emerging pro-endocrine cells are labeled due to residual systemic Dox (following a 20 hour pulse of Dox and 8 day chase, expression of EphB3 is restricted to endocrine cells, not shown here). (C) Continuous 3 day Dox induction of EphB3rtTA-lacZ mice labels all endocrine populations, during their step-wise differentiation, from their delamination to hormone expression: both newly emerging pro-endocrine cells and older maturing endocrine cells.
Importantly, our studies determine the developmental timing of pro-endocrine cell differentiation during pancreas development. Using different periods of chase, we determined that EphB3+ cells co-expressed hormonal markers starting the second day following removal of Dox, indicating that endocrine cells take approximately 48hrs from epithelial escape to maturation. Of interest, unlike the compressed timing of endocrine maturation observed during the first transition, when EphB3 cells are shown to co-express glucagon after 1 day, we found that following the secondary transition, EphB3 co-expression with hormonal markers began after 2 day following removal of Dox. This indicated that pro-endocrine cells from the first wave differentiated faster (~24hrs) than second wave endocrine cells (~48hrs).
We note that Eph-ephrin signaling has long been associated with fundamental cell behaviors that drive tissue morphogenesis, such as adhesion and repulsion (Dravis and Henkemeyer, 2011; Halloran and Wolman, 2006). Pro-endocrine cells tightly regulate adhesion properties during their delamination and subsequent aggregation into endocrine clusters and islets (Gouzi et al., 2011). We speculate pro-endocrine cells are likely influenced by Eph-ephrin mediated repulsion as they undergo EMT (epithelial-tomesenchymal transition) and delaminate from the pancreatic epithelium. This repulsive signaling is likely extinguished as pro-endocrine cells then adhere to each other and aggregate into islets. Expression of EphB3 mirrors these events, initiating during delamination and extinguishing prior to coalescence into endocrine clusters. While longer term pulse-chase lineage tracing shows that EphB3+ cells eventually give rise to islet endocrine cells, shorter term labeling never detects endocrine cells, suggesting EphB3 expression ceases prior to endocrine differentiation and islet formation.
Together, these results identify EphB3 as a new and useful marker of delaminating endocrine cells. In addition, we introduce BAC-Tg-EphB3rtTA mice as a new tool for analysis of both EphB3 embryonic expression and for elucidation of the developmental paths taken by EphB3 cells. We have used this transgenic mouse model to follow pancreatic endocrine cells from their specification within the progenitor epithelium to their eventual coalescence into islets during pancreatic development, however, it will also be useful for tracing fate of EphB3+ cells in other tissues. Studies such as ours will contribute to the growing molecular understanding of beta cell differentiation and advance efforts towards replacement therapies for diabetes.
EXPERIMENTAL PROCEDURES
BAC-Tg-EphB3rtTA mice
BAC-Tg-EphB3rtTA mice contain the second generation reverse tetracycline transactivator, rtTA-2sM2, knocked into the ORF of BAC EphB3 sequences (Dravis and Henkemeyer, 2011). Briefly, transgenic mice were generated as follows: BAC clone RP23-213m14 (GI: 61675810), a 208 kb BAC from a C57BL/6J background containing 42 kb upstream of the EphB3 start codon, was purchased from the BACPAC resources center at the Children’s Hospital Oakland. The cDNA for rtTA-2sM2 was targeted into the EphB3 ORF in rp23-213m14 using bacterial homologous recombination. Briefly, RP23-213m14 was first transformed into EL250 strain cells, and then targeted with a modified pL451 vector containing an Frt-sandwiched Neo/Kan cassette and the cDNA for rtTA-2sM2, flanked by 500 bp homology arms (the targeting vector is thus 500bpLHA-rtTA-2sM2-Frt/Neo/Kan/Frt-500bpRHA) guiding the cassette to the ORF of EphB3. Arabinose induction of flpE activity in the targeted BAC was then utilized to excise the Frt-Neo/Kan-Frt cassette, leaving rtTA-2s-M2 followed by a single copy of Frt in the EphB3 ORF. Proper targeting of the EphB3 ORF was confirmed by PCR and restriction endonuclease digests. Pronuclear injection of fertilized oocytes with this BACTg-EphB3-rtTA construct was performed by Transgenic Core facilities at the Department of Developmental Biology at UTSW.
Mice and embryo handling
The EphB3rtTA-lacZ, EphB1lacZ, EphB2lacZ, EphB4lacZ, ephrinB2lacZ and ephrinB3lacZ embryos were generated by crossing CD1 females to BAC-Tg-EphB3rtTA;TRE-lacZ males. EphB3rtTA-lacZ embryos were collected at different gestational stages and fixed in 4% paraformaldehyde (PFA)/PBS for 15 min, rinsed 3 times in 1X PBS, then stained overnight with lacZ staining solution (20 mM K4Fe(CN)6·3H2O, 20 mM K3Fe(CN)6, 2 mM MgCl2, 0.02% NP-40, 4μl of 100 mg/ml X-Gal). Embryos were viewed in a NeoLumar stereomicroscope (Zeiss) and photographed using a DP-70 camera (Olympus). Alternatively EphB3rtTA-lacZ embryos were fixed overnight in 4% paraformaldehyde (PFA)/PBS, washed, dehydrated to 70% ethanol and stored at −20°C for further processing.
Doxycycline Treatment
Animals were induced systemically for 20 hours by including doxycycline (Dox) in drinking water (150mg of Doxycycline in 250ml sucrose-sweetened water). For short-term lineage tracing, animals were induced for 20 hours and euthanized the next day. For long-term lineage tracing, animals were euthanized on the day indicated (i.e. day 2, day 3, etc). For continuous labeling, animals were induced with unrestricted access to Dox-drinking water for a period of approximately 72 hours (3 days) and then euthanized at mid-afternoon of the third day.
In situ hybridization
Experiments were performed as previously described (Villasenor et al., 2008). Probes used were: ephrinB1 (~800bp); EphB4 (4270bp, BE573518); and EphB6 (~2,067bp, BC018246) from Open Biosystems. In situs were carried out using a Biolane HTI automated incubation liquid handler (Holle & Huttner). Images were taken using a NeoLumar dissecting microscope (Zeiss) and a DP-70 camera (Olympus).
Histological Analysis
For Hematoxylin and Eosin (HE) staining, sections were de-waxed in xylene; rehydrated and stained with Hematoxylin (Fisher) for seven minutes. Then sections were submerged in an acid alcohol wash, washed in running water and stained with Eosin Y (Fisher) for 3 minutes. Sections were then mounted with Permount (Fisher).
Immunofluorescence studies
For paraffin sections, tissue was rehydrated via ethanol series; washed several times in 1X PBS; treated with R-Buffer A or R-Buffer B (Electron Microscopy Sciences) in a 2100 Retriever; blocked couple of hours with CAS-Block (Invitrogen); and incubated with primary antibody. The dilutions of primary antibodies used were: rabbit anti-β-gal (1:250 Cappel); chick anti-β-gal (1:2000 Abcam); rabbit anti-amylase (1:500 SIGMA); mouse anti-E-cadherin (1:200 INVITROGEN); rabbit anti-insulin (1:500 DakoCy or Linco); goat anti-ghrelin (Santa Cruz); guinea pig anti-glucagon (1:600 kindly provided by Raymond MacDonald); mouse anti-nkx6.1 (1:600 BCBC); mouse anti-ngn3 (1:500 BCBC and also 1:100 kindly provided by Dr. Guoqiang Gu); mouse anti-pdx1 (1:500 BCBC); goat anti-somatostatin (1:1000 Santa Cruz); rabbit anti-sox9 (1:600 Millipore). Secondary antibodies were either from Jackson ImmunoResearch Laboratories or from Invitrogen. Slides were counterstained with DAPI and mounted with ProLong Gold Antifade Reagent (Invitrogen). Images were acquired on a LSM510 META Zeiss confocal microscope. Quantifications: The values for the percentages of EphB3 co-expression with glucagon or insulin are sum of two independent insulin/glucagon experiments and the values for the total EphB3 are averages of two independent insulin/glucagon test. Quantified a minimium of 4 sections and maximum of 6 sections per animal. 1 day induction: n=2 animals for insulin and n=3 for glucagon. 2 day induction and 3 day induction: n=3 animals for insulin and n=3 animals for glucagon. Adult induction: n=2 animals for insulin and n=2 animals for glucagon. Ngn3 and Nkx6.1 were 5 sections per animal at N=2 at E10.5, 5 sections per animal with N=3 at E15.5.
Supplementary Material
Supplemental Figure 1. Expression of ephrin ligands and Eph receptors during pancreatic development (sections). Sections showing: in situ hybridization of ephrinB1 or EphB6 on pancreatic buds at E13.5 (A,G), and β-galactosidase stained sections of pancreata at stages indicated for ephrinB2 (B,B’), ephrinB3 (C), EphB2 (D,D’), EphB3 (E) and EphB4 (F). (A,B,B’,C,D’,E) Transverse sections; (D,F) sagittal sections. a, aorta; ar, artery; e, pancreatic epithelium; g, gut tube; m, mesoderm.
Supplemental Figure 2. Diagram of inducible EphB3rtTA;TRE-lacZ pulse-chase timecourses. Dox induction day, pulse-chase labeling times, and harvesting/assay days (scope) are shown in schematic representation. Corresponding figure panels are indicated (left). Contribution to pro-endocrine or endocrine lineages is summarized (right). Dox, doxycline.
Supplemental Figure 3. EphB3 is not co-expressed with mature hormone at late gestational stages following 1 day pulse induction. Immunostaining of anti-β-gal (green) and pancreatic markers (red) demonstrate that EphB3 is expressed in the late pancreatic epithelium, especially around ducts. Transverse section of E18.5 EphB3rtTA-lacZ pancreatic bud 1 day after Dox induction: EphB3+ cells (green) and (A) insulin+ cells (red) or (B) glucagon+ cells (red). d, ducts; i, islet.
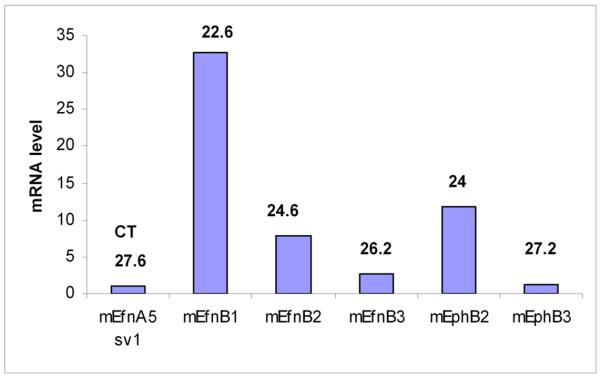
Figure 8. Eph/ephrinB are expressed in adult islets expression.
Q-PCR from mRna isolated from mice islets. Primers were validated with brabin mRNA. CT was calculated utilizing cyclophilin expression as baseline. Efn-ephrin.
ACKOWLEDGMENTS
We are grateful to Diana Chong for discussions and ephrinB2 stain at E13.5. We thank Guoqiang Gu for his kind gift of the Ngn3 antibody. We also thank Ray MacDonald, Thomas Wilkie, Rolf Brekken, Thomas Carroll, and their laboratories, as well as the whole Cleaver lab for invaluable discussions. This work was supported by grants JDRF Award 99-2007-472 and NIH R01 grant DK079862-01 to OC.
REFERENCES
- Ahlgren U, Pfaff SL, Jessell TM, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–60. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–81. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Cleaver O, MacDonald RJ. Developmental Molecular Biology of the Pancreas. In: Neoptolemos J, et al., editors. Handbook of Pancreatic Cancer. Springer; New York, NY: 2009. [Google Scholar]
- Cowan CA, Yokoyama N, Saxena A, Chumley MJ, Silvany RE, Baker LA, Srivastava D, Henkemeyer M. Ephrin-B2 reverse signaling is required for axon pathfinding and cardiac valve formation but not early vascular development. Dev Biol. 2004;271:263–71. doi: 10.1016/j.ydbio.2004.03.026. [DOI] [PubMed] [Google Scholar]
- D’Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, Moorman MA, Kroon E, Carpenter MK, Baetge EE. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- Dravis C, Henkemeyer M. Ephrin-B reverse signaling controls septation events at the embryonic midline through separate tyrosine phosphorylation-independent signaling avenues. Developmental biology. 2011;355:138–51. doi: 10.1016/j.ydbio.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravis C, Yokoyama N, Chumley MJ, Cowan CA, Silvany RE, Shay J, Baker LA, Henkemeyer M. Bidirectional signaling mediated by ephrin-B2 and EphB2 controls urorectal development. Dev Biol. 2004;271:272–90. doi: 10.1016/j.ydbio.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Gerety SS, Wang HU, Chen ZF, Anderson DJ. Symmetrical mutant phenotypes of the receptor EphB4 and its specific transmembrane ligand ephrin-B2 in cardiovascular development. Mol Cell. 1999;4:403–14. doi: 10.1016/s1097-2765(00)80342-1. [DOI] [PubMed] [Google Scholar]
- Gouzi M, Kim YH, Katsumoto K, Johansson K, Grapin-Botton A. Neurogenin3 initiates stepwise delamination of differentiating endocrine cells during pancreas development. Developmental dynamics : an official publication of the American Association of Anatomists. 2011;240:589–604. doi: 10.1002/dvdy.22544. [DOI] [PubMed] [Google Scholar]
- Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–11. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guz Y, Montminy MR, Stein R, Leonard J, Gamer LW, Wright CV, Teitelman G. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development. 1995;121:11–8. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- Hald J, Galbo T, Rescan C, Radzikowski L, Sprinkel AE, Heimberg H, Ahnfelt-Ronne J, Jensen J, Scharfmann R, Gradwohl G, Kaestner KH, Stoeckert C, Jr., Jensen JN, Madsen OD. Pancreatic islet and progenitor cell surface markers with cell sorting potential. Diabetologia. 2011 doi: 10.1007/s00125-011-2295-1. [DOI] [PubMed] [Google Scholar]
- Halloran MC, Wolman MA. Repulsion or adhesion: receptors make the call. Current opinion in cell biology. 2006;18:533–40. doi: 10.1016/j.ceb.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Heller RS, Dichmann DS, Jensen J, Miller C, Wong G, Madsen OD, Serup P. Expression patterns of Wnts, Frizzleds, sFRPs, and misexpression in transgenic mice suggesting a role for Wnts in pancreas and foregut pattern formation. Dev Dyn. 2002;225:260–70. doi: 10.1002/dvdy.10157. [DOI] [PubMed] [Google Scholar]
- Henkemeyer M, Orioli D, Henderson JT, Saxton TM, Roder J, Pawson T, Klein R. Nuk controls pathfinding of commissural axons in the mammalian central nervous system. Cell. 1996;86:35–46. doi: 10.1016/s0092-8674(00)80075-6. [DOI] [PubMed] [Google Scholar]
- Johansson KA, Dursun U, Jordan N, Gu G, Beermann F, Gradwohl G, Grapin-Botton A. Temporal control of neurogenin3 activity in pancreas progenitors reveals competence windows for the generation of different endocrine cell types. Dev Cell. 2007;12:457–65. doi: 10.1016/j.devcel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Konstantinova I, Nikolova G, Ohara-Imaizumi M, Meda P, Kucera T, Zarbalis K, Wurst W, Nagamatsu S, Lammert E. EphA-Ephrin-A-mediated beta cell communication regulates insulin secretion from pancreatic islets. Cell. 2007;129:359–70. doi: 10.1016/j.cell.2007.02.044. [DOI] [PubMed] [Google Scholar]
- Kopp JL, Dubois CL, Schaffer AE, Hao E, Shih HP, Seymour PA, Ma J, Sander M. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development. 2011;138:653–65. doi: 10.1242/dev.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Schlierf B, Schardt A, Nave KA, Wegner M. Sox10-rtTA mouse line for tetracycline-inducible expression of transgenes in neural crest cells and oligodendrocytes. Genesis. 2004;40:171–5. doi: 10.1002/gene.20083. [DOI] [PubMed] [Google Scholar]
- Madsen OD, Jensen J, Petersen HV, Pedersen EE, Oster A, Andersen FG, Jorgensen MC, Jensen PB, Larsson LI, Serup P. Transcription factors contributing to the pancreatic beta-cell phenotype. Horm Metab Res. 1997;29:265–70. doi: 10.1055/s-2007-979035. [DOI] [PubMed] [Google Scholar]
- Masui T, Long Q, Beres TM, Magnuson MA, MacDonald RJ. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev. 2007;21:2629–43. doi: 10.1101/gad.1575207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyatsuka T, Kosaka Y, Kim H, German MS. Neurogenin3 inhibits proliferation in endocrine progenitors by inducing Cdkn1a. Proc Natl Acad Sci U S A. 2010;108:185–90. doi: 10.1073/pnas.1004842108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Krasinski JM, Stoffers DA. On the origin of the beta cell. Genes Dev. 2008;22:1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240:530–65. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- Redecker P, Jorns A, Jahn R, Grube D. Synaptophysin immunoreactivity in the mammalian endocrine pancreas. Cell and tissue research. 1991;264:461–7. doi: 10.1007/BF00319036. [DOI] [PubMed] [Google Scholar]
- Sander M, German MS. The beta cell transcription factors and development of the pancreas. J Mol Med. 1997;75:327–40. doi: 10.1007/s001090050118. [DOI] [PubMed] [Google Scholar]
- Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, Schwitzgebel V, Hayes-Jordan A, German M. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127:5533–40. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- Seymour PA, Freude KK, Tran MN, Mayes EE, Jensen J, Kist R, Scherer G, Sander M. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci U S A. 2007;104:1865–70. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford D, Prince VE. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr Biol. 2002;12:1215–20. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- Swift MR, Weinstein BM. Arterial-venous specification during development. Circ Res. 2009;104:576–88. doi: 10.1161/CIRCRESAHA.108.188805. [DOI] [PubMed] [Google Scholar]
- van Eyll JM, Passante L, Pierreux CE, Lemaigre FP, Vanderhaeghen P, Rousseau GG. Eph receptors and their ephrin ligands are expressed in developing mouse pancreas. Gene Expr Patterns. 2006;6:353–9. doi: 10.1016/j.modgep.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Villasenor A, Chong DC, Cleaver O. Biphasic Ngn3 expression in the developing pancreas. Dev Dyn. 2008;237:3270–3279. doi: 10.1002/dvdy.21740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villasenor A, Chong DC, Henkemeyer M, Cleaver O. Epithelial dynamics of pancreatic branching morphogenesis. Development. 2010;137:4295–305. doi: 10.1242/dev.052993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HU, Chen ZF, Anderson DJ. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell. 1998;93:741–53. doi: 10.1016/s0092-8674(00)81436-1. [DOI] [PubMed] [Google Scholar]
- Williams SE, Mann F, Erskine L, Sakurai T, Wei S, Rossi DJ, Gale NW, Holt CE, Mason CA, Henkemeyer M. Ephrin-B2 and EphB1 mediate retinal axon divergence at the optic chiasm. Neuron. 2003;39:919–35. doi: 10.1016/j.neuron.2003.08.017. [DOI] [PubMed] [Google Scholar]
- Wilson ME, Scheel D, German MS. Gene expression cascades in pancreatic development. Mech Dev. 2003;120:65–80. doi: 10.1016/s0925-4773(02)00333-7. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–32. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Expression of ephrin ligands and Eph receptors during pancreatic development (sections). Sections showing: in situ hybridization of ephrinB1 or EphB6 on pancreatic buds at E13.5 (A,G), and β-galactosidase stained sections of pancreata at stages indicated for ephrinB2 (B,B’), ephrinB3 (C), EphB2 (D,D’), EphB3 (E) and EphB4 (F). (A,B,B’,C,D’,E) Transverse sections; (D,F) sagittal sections. a, aorta; ar, artery; e, pancreatic epithelium; g, gut tube; m, mesoderm.
Supplemental Figure 2. Diagram of inducible EphB3rtTA;TRE-lacZ pulse-chase timecourses. Dox induction day, pulse-chase labeling times, and harvesting/assay days (scope) are shown in schematic representation. Corresponding figure panels are indicated (left). Contribution to pro-endocrine or endocrine lineages is summarized (right). Dox, doxycline.
Supplemental Figure 3. EphB3 is not co-expressed with mature hormone at late gestational stages following 1 day pulse induction. Immunostaining of anti-β-gal (green) and pancreatic markers (red) demonstrate that EphB3 is expressed in the late pancreatic epithelium, especially around ducts. Transverse section of E18.5 EphB3rtTA-lacZ pancreatic bud 1 day after Dox induction: EphB3+ cells (green) and (A) insulin+ cells (red) or (B) glucagon+ cells (red). d, ducts; i, islet.