Abstract
Recent epigenomic studies have identified significant differences between developmental stages and cell types. While the importance of epigenetic regulation has been increasingly recognized, it remains unclear how the global epigenetic patterns are established and maintained. Here I review a number of recent studies with the emphasis on the role of the genomic sequence in shaping the epigenetic landscape. These studies strongly suggest that the sequence information is important not just for controlling target specificity but for orchestrating the diversity of epigenetic patterns among different cell types. The epigenome is maintained by the complex network of a large number of interactions. Integrative approaches are needed to gain insights into these networks.
Introduction
The genome provides a blueprint for gene regulation, but this blueprint is interpreted differently in different cell types. A partial explanation for these differences is the fact that the DNA sequences are packaged into chromatin, and that only a small portion of the genome is accessible in any cell type. DNA accessibility in a cell is highly controlled by the combined effects of multiple epigenetic marks including nucleosome positions, histone modifications, and DNA methylation [1].
The basic repeating unit of chromatin is the nucleosome, which consists of 147 base pairs of DNA wrapped around a core histone octamer in nearly two times [2]. The nucleosome is a formidable barrier for transcription factors (TF) binding, thereby serving as a repressor for transcription. The N-terminal histone tails can be covalently modified at numerous locations in different ways. The combinatorial pattern not only serves as a landmark for open or closed chromatin but recruit specific regulatory proteins [3, 4]. Epigenetic information can also be carried by the genomic DNA itself, where certain cytosine nucleotides are methylated at the C-5 position [5]. While promoter DNA methylation has been long recognized to be associated with gene silencing, genome-wide studies have identified new functions associated with coding region methylation [6, 7].
During the past decade a large amount of epigenomic data has been generated (recently reviewed by [8-10]), providing high-resolution maps of the entire epigenome which can be compared to gain functional insights. Significant differences have been identified between cells under different growth conditions or at developmental stages. What is more clinically relevant is the identification of global epigenetic aberrations in cancer and a number of other diseases [11, 12], suggesting dysfunctional epigenetic regulation may be an important step to these diseases.
Despite these exciting discoveries, we are still far from understanding the mechanisms that regulate the genome-wide epigenetic patterns. A number of fundamental questions remain unresolved. How are the chromatin regulators and DNA methytransferases recruited to the chromatin? How is the global epigenetic pattern stably maintained after initial establishment? What are the factors that cause epigenetic aberrations during the development of a certain disease? Is it possible to restore normal epigenetic patterns by clinical intervention? Answers to these questions are needed both for understanding the function of epigenetic mechanisms and for developing effective approaches for disease treatment.
It has been increasingly recognized that epigenomic patterns are regulated by the complex, dynamic interactions among multiple classes of factors, including chromatin modifiers, DNA binding proteins, noncoding RNAs (ncRNA), and signaling molecules. In this review, I will focus on the role of DNA sequence in mediating these interactions. This is a central question that has been extensively investigated yet remains incompletely understood. More comprehensive discussions can be found in several recent reviews [13-16].
Computation methods for predictions of epigenomic patterns
Sequence analysis has traditionally been used to predict TF binding sites, which often target specific short DNA sequence patterns called motifs (reviewed by [17]). However, most chromatin regulators either do not or weakly interact with DNA. While interactions with certain TFs may play an important role in recruitment, such interactions often involve many TFs. As a result, an epigenetic pattern is often associated with many sequence features each only making a minor contribution. A recent trend in computational biology is to develop effective predictive models by integrating multiple weak features, as described below.
A commonly used approach is supervised learning [18]. The availability of genome-wide data makes it possible to select sample sequences from either positive or negative regions. In order to build a predictive model, it is important to convert these sequences to numerical values. If the relevant sequence features are known, their occurring frequencies can be used as numerical predictors. However, in most cases discriminative sequence features are not known a priori. To address this problem, various classes of sequence features have been explored, including TF motifs [19-21], word counts [22], repetitive elements [19, 21, 23], DNA structural parameters [24], and periodic patterns [25]. Table 1 shows an incomplete list of DNA sequence features and their associated factors.
Table 1.
A list of common sequence features and known associated recruiting factors.
| Sequence Features | Binding Factors |
|---|---|
| CpG | CXXC domain containing factors; transcription factors; core histone; methylation sensitive binding factors |
| Poly A:T | core histone |
| TF motifs | transcription factors |
| repetitive elements | RNAi machineries |
| boundary elements | insulator proteins; transcription factors |
| uncharacterized sequences |
noncoding RNA |
Due to the large number of potentially useful sequence features (it is not uncommon to have more than 100 features), one important concern is overfitting – failure of a complex model that is trained based on a small sample to generate useful predictions. This risk can be reduced by using variable selection techniques. This can be done either in the traditional linear regression framework, by using principal component analysis, stepwise regression, penalized regression [26], or through machine learning approaches such as decision trees [27] and support vector machines [28]. Finally, prediction accuracy can be improved by ensemble-based prediction models such as boosting [29] and Bayesian additive regression trees (BART) [30]. These methods have been applied to prediction of nucleosome positioning [22, 24, 25], histone modifications [20, 31, 32], and DNA methylation [19-21, 33]. The performance of different methods in predicting protein-DNA interactions has been compared, with the results in favor of boosting and BART [34].
An alternative approach is to estimate the position specific sequence pattern directly as an extension of traditional motif analysis methods [35]. In this approach, the input sequences are first aligned, and then the probability of observing a certain nucleotide at a specific position is empirically estimated and further normalized. This approach has been applied to predict nucleosome positioning. Accurate predictions can be achieved when complemented by word frequency features [36, 37].
Another approach for nucleosome positioning prediction is based on biophysical properties [38-40]. Viewing DNA as a flexible polymer, the sequence dependent, free energy associated with a specific geometric configuration can be calculated. Since the nucleosome crystal structure has been experimentally identified [41], the most favorable sequence composition can be identified by minimizing the free energy [38-40]. While these models do not require genome-wide data for model training, the prediction accuracy is surprisingly competitive. However, the utility of this approach to other epigenetic marks is limited due to the lack of of high-resolution protein structure information.
The basic output of each method is a propensity score for an input sequence fragment; this can be extended to predict genome-wide epigenetic patterns by scanning with sliding windows. Additional constraints, such as the steric hindrance effect of neighboring nucleosomes, can be further incorporated, for example, by using a hidden Markov model [35, 42]. An extension of this approach has been applied to predict multiple factor binding profiles [43, 44].
Quantitative evaluation of the role of DNA sequences in regulating epigenomic patterns
The computational methods described above have been applied to quantitatively evaluate the association between DNA and epigenomic patterns. As discussed below, while each epigenetic mark involves a distinct set of sequence features and recruiting factors, the underlying principle is strikingly similar.
Nucleosome positioning
The nucleosome is the fundamental unit for packaging DNA. Its highly conserved three dimensional structure was identified by Richmond and colleagues [41], showing more than 120 direct histone-DNA interactions unevenly distributed across the nucleosome surface. Each individual interaction is rather weak, allowing the flexibility to package almost the entire genome. A long-standing question is to what extent the genome-wide nucleosome positioning is dictated by such variations.
The development of genomic technologies and computational methodologies has greatly advanced our understanding of the role of DNA sequence in global regulation of nucleosome positioning. The best studied model system is S. cerevisiae. Here high-resolution, genome-wide mapping of nucleosome positions has been obtained by crosslinking nucleosome with DNA, followed by MNase digestion and microarray hybridization or high-throughput sequencing [24, 42, 45-47]. A number of computational methods have been developed to predict nucleosome positions from the genomic sequence [22, 24, 25, 35-39, 48, 49]. While the prediction accuracy is significantly better than random guess, it is highly variable across the genome (see [50] for a review). While the nucleosome-free region (NFR) and +1 nucleosome position at the 5′ end of genes can be well predicted, the accuracy in other regions is poorer. One significant limitation is that while the in vivo nucleosome positions display regular phasing surrounding NFRs, predicted nucleosome positions do not have this pattern.
To test whether the DNA sequences indeed play a causal role, several groups have carried out experimental validations by deleting certain sequences that are predicted to be associated with nucleosome positions followed by comparison of the nucleosome positions in wild-type and mutants [51, 52]. The nucleosome positions in the mutants are indeed altered, but the degree of alteration gradually increases with respect to the deletion size [51], suggesting that nucleosome positions are mediated by the cumulative effect of many weak histone-DNA interactions.
Two independent studies have used a similar approach to investigate the direct role of DNA sequences in nucleosome positioning [37, 53]. In both studies, the yeast genome was extracted from the nucleus and reassembled into nucleosomes in a cell-free media. The in vitro assembled nuclesome positioning pattern was then detected by using next generation sequencing. Both studies have found that the in vitro and in vivo nucleosome occupancy patterns are highly correlated; notably, the DNA sequence of most yeast promoters intrinsically disfavor nucleosome assembly. On the other hand, the center positions of the nucleosomes vary significantly between the two conditions. The nucleosomes assembled in vitro are more delocalized [53], suggesting that the DNA sequence plays an important but insufficient role for genome-wide nucleosome positioning.
One striking feature of in vivo nucleosome positioning pattern is the regular phasing of nucleosomes around NFR [42], but such a pattern cannot be reproduced by in vitro nucleosome assembly. This difference can be explained by a simple model, called statistical positioning, proposed by Kornberg and Stryer more than twenty years ago [54]. This model assumes that nucleosome positions are bounded by a fixed barrier but distributed at random elsewhere. If averaged over all possible configurations, similar to averaging over a large cell population, the resulting pattern appears to be regular. Despite the elegance of the above model, recent experimental studies have uncovered important additional complexity [55]. In one study [56], the investigators reconstituted nucleosomes in a whole-cell-extract media supplemented by ATP and were able to recapitulate the in vivo-like nucleosome positioning pattern. Notably, the spacing between neighboring nucleosomes is not strongly dependent upon the local nucleosome density, as predicted by the statistical positioning model. These findings strongly suggest that in vivo nucleosome positions are not simply formed by statistical positioning but result from active packing by ATP-dependent remodeling complexes.
There is a great interest in to what extent the DNA sequence is conserved across different species. Genome-wide nucleosome positions have been mapped in a number of organisms, including S. pombe [57], C. elegans [58], D. melanogaster [59], A. thaliana [60], and human [61]. All these species share a similar pattern, that is, most promoters contain an NFR flanked by regular nucleosome arrays on each side. This is striking, considering that the promoter sequences have diverged significantly during evolution. For example, while the GC-content is typically low for a yeast promoter, it is significantly higher in mammalian promoters, which often harbor a CpG island. Indeed, computational studies have predicted human promoters to be encoded for high nucleosome occupancy [62]. One explanation of this counter-intuitive result is that most human genes are tissue-specific therefore are silenced by default and overridden by transcriptional regulators in a cell type specific manner [62]. This view is supported by in vitro nucleosome positioning data, which show no depletion of nucleosomes at CpG islands [63]. Similar to yeast, the in vitro data do not show regular phasing. In addition, the CpG islands seem to affect the function of the SWI/SNF remodeling complexes, which are required for induction of only those genes harboring CpG island promoters [64].
Histone modification
Investigations of the regulatory mechanisms for histone modification patterns are complicated by the fact that there are a large number of modifications each having its own global distributions [65]. Furthermore, genome-wide studies have been limited to those histone marks for which high-quality antibodies are available [66]. An important role of the DNA sequence has been recognized [16], but it is also possible that certain modifications are regulated mainly through sequence independent mechanisms.
There are three histone marks whose recruiting mechanisms have been systematically investigated: H3K4me3, H3K27me3, and H3K9me3. While H3K4me3 is an active mark that is typically present at the transcription start sites of active genes, both H3K27me3 and H3K9me3 marks are associated with gene silencing. Despite their opposite functions, H3K4me3 and H3K27me3 colocalize at certain genomic regions called bivalent domains [67], which play an important role in development by marking genes that are poised for activation during cell differentiation. The H3K9me3 mark is a typically associated with heterochromatin but can also be found at certain euchromatic regions.
The genome-wide distribution of H3K4me3 can be predicted with high accuracy from the DNA sequence in different cell types [31]. There is a high degree of overlap between H3K4me3 targets and CpG islands [67], which is at least in part due to selective binding of the Setd1/MLL complex member Cfp1, which is a CXXC domain containing protein, to non-methylated CpG [68]. Depletion of Cfp1 substantially reduces H3K4me3 recruitment at the CpG islands. Furthermore, H3K4me3 recruitment in ES cells is also mediated by interaction between Wdr5, another component in Setd1/MLL complex and Oct4 [69].
While the H3K4me3 distribution is mainly characterized by sharp peaks (<1kb), the distribution of H3K27me3 is more complex. It not only has peaks at selected promoters but also spans over broad domains. The global distribution of H3K27me3 highly colocalizes with PcG complexes, which contain the catalytic unit for H3K27 methylation. The core PcG complexes are highly evolutionarily conserved. Sequence-based models have been developed to predict PcG target genes in mammalian ES cells [32, 70], and many of target genes can be predicted based on the CpG density alone. An intriguing hypothesis is that PcG complexes target high CpG density regions by default [71]. A direct relationship between high CpG density and PcG recruitment has been experimentally verified by transgenic assays [71]. On the other hand, it is likely that the recruitment can be further modulated by TFs and ncRNAs. Incorporating TF motifs, such as Myc and E2F, in a computational model can substantially improve the accuracy for PcG target predictions [32, 70].
An important question is what DNA elements are required for PcG recruitment [72, 73], and this question cannot be simply answered by genome-wide location studies. Such DNA elements are referred to as Polycomb response elements (PRE). The PREs are well-characterized in Drosophila, containing binding sites for a number of TFs, such as Pho and the GAGA factor. Sequence-based computational models can predict PREs locations with high accuracy [74, 75]. Importantly, in one study the investigators followed up by experimentally validation using transgenic assays and found that 29 out of 43 predicted PREs can be experimentally validated [74]. In contrast, only few PREs have been experimentally identified in mammals [71, 76, 77]. There is growing evidence that the recruiting mechanism for mammals is fundamentally different; in particular, ncRNAs seem to play an important role in genome-wide recruitment of PcG complexes [78, 79].
Genome-wide analysis of the H3K9me3 is more difficult compared to the two marks discussed above, in part due to its strong association with regions which cannot be uniquely mapped to the reference genome. The distribution over mappable regions is much more diffusive compared to the two marks discussed above. The prediction accuracy for the H3K9me3 distribution is also lower [31], suggesting that the role of DNA sequence information is weaker. However, these observations cannot exclude the possibility that the DNA sequence still plays an important role at the recruitment step, and once recruited, the histone mark spreads along the chromosome in a sequence independent manner. Supporting this hypothesis, it has been shown that the RNA transcribed from repetitive DNA elements can promote heterochromatin formation through an RNAi dependent pathway [80, 81]. Deletion of these repetitive elements can abandon both heterochromatin formation and H3K9me3 recruitment [80]. While these experiments were done in S. pombe, such a mechanism seems to be conserved among eukaryotes [82]. In mammals, H3K9me3 appears to be more enriched in satellites than other classes of repetitive sequences [83].
In addition to the three histone marks discussed above, the DNA sequence is likely to play an important role in regulating chromatin states in a general sense. Recently, computational models have been developed to identify chromatin states from combinatorial histone modification patterns [84-87]. One study has investigated the link between chromatin states and DNA sequence patterns and found that a number of TF motifs are enriched with the enhancer chromatin state [88]. Furthermore, computational methods have been developed to predict p300 binding sites from DNA sequence patterns with good accuracy [89]. Interestingly, the role of DNA sequence for regulating the pattern of H3K4me1, a mark for enhancer regions, seems to be highly cell-type specific, and is strongest in ES cells [90].
DNA methylation
In vertebrate animals, DNA methylation almost exclusively occurs in the context of CpG dinucleotides [6]. Genome-wide DNA methylation level is highly associated with the CpG density and can be predicted with high-accuracy [19, 20]. While CpG islands are normally unmethylated, a subset may become hypermethylated in cancer, which may silence tumor suppressor genes thereby playing an important role in tumorgenesis [11]. Several groups have developed algorithms to predict hypermethylated CpG islands with moderate accuracy despite being statistically significant [19-21, 23]. In addition to CpG density, several TF motifs, including Sp1, NRF1, and YY1, have also been found to be methylation resistant, and motifs such as GAGA have been found to be methylation prone [91]. Most of these motifs are also associated with PcG targets [32, 70], which is not surprising since hypermethylated genes in cancer significantly overlap with PcG targets in ES cells [92]. In general, DNA methylation and chromatin state patterns are closely related, but a detailed discussion is beyond the scope of this review. Also, repetitive sequences, such as the Alu elements, have been found to be enriched with DNA methylation [19, 93]. In addition, a motif sequence associated with the G-quadruplex structure has been shown to be associated with hypomethylation [94]. In this study, the investigators also found that hypomethylated G-quadruplex sequence motif is enriched at the DNA breakpoints in many cancer types [94], suggesting a link between epigenetic pattern and genome stability.
With the rapid technological development, recent studies have been able to profile DNA methylome genome-wide [6, 95-97]. As a result, a more dynamic picture has emerged: hyper- and hypo-methylation are no longer viewed as isolated events but likely the results of dynamic boundary shift near CpG island shores [95, 96]. While it is clear that such transitions are mediated by the CpG density, it will be interesting to investigate whether there are additional DNA elements that are involved in the regulation.
A causal role of DNA elements in regulating DNA methylation pattern has been recently experimentally validated by using a transgenic approach [98]. These authors inserted selected promoter DNA sequences into the beta-globin locus in mouse embryonic stem cells and measured the DNA methylation level by using bisulfite sequencing. They found that the DNA methylation pattern at the inserted sites is quite similar to the endogenous sites, suggesting that the DNA sequence is sufficient for maintenance of DNA methylation pattern. Furthermore, they found that CpG density alone is not sufficient for maintenance of DNA methylation pattern, but it also requires combination of TF motifs.
Insights into recruitment factors
Computational sequence analyses are useful not only to predict regions targeted by a specific epigenetic factor but also to generate hypotheses regarding the recruiting factors, which can be validated in the laboratory. Here we briefly discuss the role of three general classes of recruiting factors.
Transcription factors
Since chromatin regulators often cannot directly bind DNA, it has been long speculated that their recruitment may be mediated by interactions with TFs. The sequence analysis results discussed above strongly support this hypothesis. In addition, recent proteomic studies have identified extensive interactions between TFs and chromatin modifiers [99, 100]. The impact of TF activities on epigenome organization is probably best illustrated by cell-state reprogramming. An adult cell can be reprogrammed to an induced pluripotent stem (iPS) cell by ectopic expression of four TFs: Oct4, Nanog, Klf4, and Myc [101]. The reprogramming process is accompanied by dramatic epigenetic changes. Most significantly, the DNA methylation marks at the pluripotent regulators are erased and bivalent domains are reestablished. All four reprogramming factors interact with chromatin modifiers. Specifically, Oct4 interacts with the SWI/SNF complex member Wdr5 [69]; Nanog interacts with the histone deacetylase NuRD and SWI/SNF complex [102], Klf4 can interact with the histone acetyltransferase p300 [103]; and most strikingly, Myc, can interact with many chromatin factors such as histone acetylases (GCN5, p300), chromatin-remodeling complexes, histone deacetylases, and histone demethylases. It remains unclear to what extent the global epigenetic changes are directly caused by these TF-chromatin interactions.
Computational studies of various epigenomic datasets have suggested that, in general, the contribution of each individual TF in maintenance of the epigenetic patterns is typically not significant. For example, for nucleosome positioning, there are at least three TF motifs that are enriched in nucleosomal sequences: ABF1, REB1, and STB2 [24]. For DNA methylation, a number of TF motifs such as SP1, NRF1, and YY1, are enriched with unmethylated regions [91]. However, in both cases the identified motifs alone can only explain a small fraction of the variation observed in the experimental data.
General factors
A surprising result from computational studies is that the genome-wide association between DNA sequences and epigenetic patterns is mainly contributed by a few features that are traditionally viewed as degenerative, whereas the sophisticated models described above often lead to quantitative but not qualitative improvement. For example, while many methods have been developed to predict nucleosome positions, it seems that the overall accuracy is only moderately higher than a simple model using the G+C density as the single predictor [62]. Similarly, for DNA methylation, the genome-wide pattern is most strongly correlated with the CpG density, whereas including other features such as TF motifs and repetitive sequences, only adds moderate prediction power [19-21]. As discussed above, the CpG density is also strongly associated with histone modifications such as H3K4me3 and H3K27me3.
Interestingly, eukaryotic cells have developed various mechanisms to recognize these general features. For nucleosome positioning, the G+C content can directly affect the nucleosome-DNA binding affinity. For different histone modifications, the sequence signals seem to be mediated by different factors. The recruitment of H3K4me3 is mediated by the CXXC domain-containing protein Cfp1 [68], which recognizes unmethylated CpG. The association between PcG complexes and CpG islands may be mediated by interaction with Jarid2 [104-107], which binds DNA with bias toward G+C rich sequences [104]. It has been proposed that the PcG complexes target CpG islands by default, and this default recruitment is antagonized by the simultaneous recruitment of activators by specific TF motifs [60]. Furthermore, cross-talk between these machineries is likely to play an important role in establishing the overall epigenetic pattern. For example, for DNA methylation, the preference seems to be dictated by interaction with histone modifying complexes such as G9a and PcG proteins [14, 108].
Noncoding RNA
NcRNAs are an emerging class of potentially important regulators [109, 110]. Two types of ncRNAs, long noncoding RNAs (lincRNA) and short RNAs, have been implicated in chromatin recruitment. Many short RNAs are involved in the RNAi pathway, which plays an important role in heterochromatin formation. An unrelated class of short RNAs have been found to be transcribed from the 5′ end of PcG target genes [111], and these short RNAs form a stem-loop structure that is able to physically interact with PRC2. In addition, thousands of lincRNAs have been detected in mammalian cells [112]. About 20% of these transcripts can physically interact with PRC2 [113]. While the function of most lincRNAs remains uncharacterized, a few examples, such as Xist and HOTAIR, have been investigated intensively. Xist is the major regulator for X-inactivation and able to recruit PcG complexes to the parent locus, to which it remains tethered [79]. The 5′ end of Xist contains a RepA domain which can interact with PRC2 [79]. The ability to interact with both PRC2 and DNA makes Xist act as a linker between them. HOTAIR also physically associates with PRC2 [78], but its target region is located on a different chromosome, and the underlying mechanism remains unclear.
A major challenge to further investigate the role of ncRNAs is to understand their targeting mechanism. As discussed above, while some ncRNAs act in cis, others target distant regions that may be located on a different chromosome. Several intriguing models for trans-action have been recently proposed [113], including 1) forming a RNA-DNA triplex; 2) serving as a link by interaction with both chromatin complexes and TFs; 3) inducing specificity of a chromatin remodeling protein through allosteric modification; and 4) mediating long-range chromatin interactions.
DNA sequence associated with overall epigenetic variability
While epigenetic patterns are cell type specific, the variability is not uniform across the genome. A growing body of literature suggests that the genome-wide distribution of epigenetic variability is closely associated with the underlying DNA sequence. In the following I briefly review some recent results.
Nucleosome positioning
In yeast, most promoters are associated with NFRs independent of the transcription status. These promoters are typically TATA-less [36, 47, 114]. In contrast, the nucleosome positions in the TATA-box containing promoters are dynamic depending on the growth conditions. Under an inducible condition, nucleosomes can be evicted by SWI/SNF, thereby making the DNA regulatory elements accessible for TF binding followed by gene activation [115]. In human and mouse, it has been shown that eviction of nucleosomes containing the H3K4me2 marks are generally associated with active enhancers [61, 116]. This relationship is likely to be valid in a wide range of species rather than mammal specific. Interestingly, the insulator protein CTCF targets DNA sequences with intrinsically high nucleosome occupancy [25, 117]. Once bound, CTCF displaces neighboring nucleosomes, thereby creating a regular nucleosome array [117, 118].
Experimental techniques have also been developed to measure the histone turnover rate [119, 120]. These investigators labeled ancestral and newly synthesized histones with different tags and then measured the enrichment ratio of two tags on a genome-scale. They found that, in cells that are arrested in the G1 stage of the cell-cycle, the histone turnover rate is higher in promoter regions and chromatin boundary elements [119]. In dividing cells, the ancestral histones are redistributed as a result of DNA replication. Surprisingly, the ancestral histones are not reincorporated at the same location but can be displaced within a window of ~400bp which results in accumulation of ancestral histones at 5′ end of genes [120].
Histone modification
Changes of the histone modification pattern in promoter regions are directly related to gene regulation. For both H3K4me3 and H3K27me3, the degree of cell type specific variability is highly associated with its CpG density. In particular, High CpG promoters (HCP) are highly associated with H3K4me3, regardless the transcription activities [68, 121]. Dynamic changes are mostly associated with low CpG promoters (LCP). The variability of H3K27me3 is also associated with CpG content [32, 70]. During cell differentiation, the loss of focal peaks is compensated by expansion of large-scale domains. As a result, the overall occupancy is nearly preserved [9, 122]. In addition to the CpG density, a handful of TF motifs (such as E2F, ZF5, SP1, and MYC) can also contribute to the overall variability [32].
The overall variability of promoter histone modification patterns is surprisingly low [9, 88, 118]. Many genes that are expressed only in certain cell types are already associated with open chromatin at the promoter regions. On the other hand, the patterns are much more dynamic at enhancers, and such variation is highly correlated with gene activities. Enhancers are often enriched with highly conserved noncoding elements and contain clusters of TF motif sites [123].
DNA methylation
The variability of DNA methylation is also highly associated with the CpG density. During normal cell differentiation, dynamic changes are most concentrated on LCPs, while most HCPs are unmethylated across different cell types [124]. Indeed, the hypermethylated HCPs also tend to have lower CpG content than the unmethylated ones. Recent studies have shown that this relationship also holds on the genome-scale [6, 7, 68, 125-127]. Large-scale aberrant DNA methylation patterns have been long recognized in cancer and may play an important step toward tumorgenesis [11, 128]. A recent study has identified significant overlap between regions that are differentially methylated among tumors and those among different normal tissues, suggesting a developmental origin of these aberrations [126, 129]. Interestingly, these studies have also observed large-scale domains of high variability [6, 95]. While it is unclear how these domains are established, it is possible that certain sequence features, such as the CTCF motif sites, may play an important role in maintenance of the boundaries.
Discussions
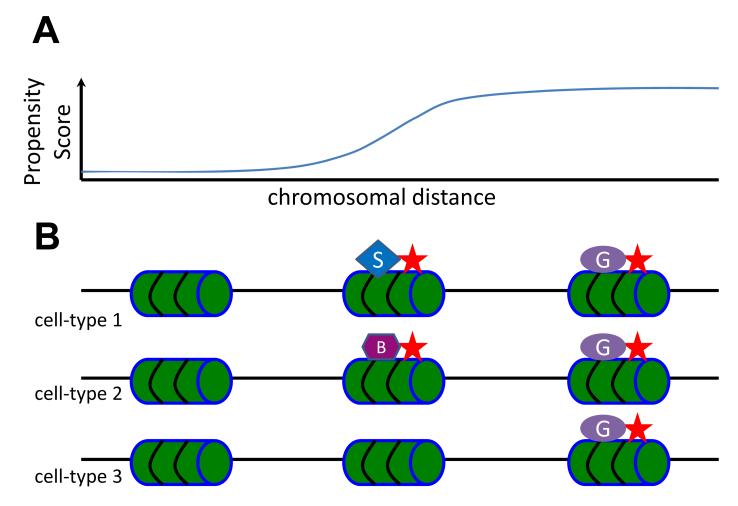
What has emerged from these studies is a consensus that the overall epigenetic pattern is not independent of, but closely associated with the genomic sequence (Figure 1). The DNA sequence seems to play at least three important roles: 1) Generic features, such as the G+C content and CpG density, may be used to shape the skeleton of the epigenomic patterns, by facilitating various protein-DNA interactions mostly involving a small set of general factors; 2) Specific features, such as TF motifs, can recruit TFs, ncRNAs, and other factors in a cell type specific manner, but the immediate impact of each factor may be limited to a specific biological pathway or cell type; and 3) the DNA sequence information may also be used orchestrate the overall epigenetic variability, such that certain regions are more plastic than others. One likely scenario is that, while the general factors described above may play a major role in initializing the epigenetic patterns at an early developmental stage, cell type specific factors, such as TFs and ncRNAs, may play a more important role later on by mediating the divergence of epigenomic patterns across different cell types.
Figure 1.
A model for the role of the genomic sequence in guiding genome-wide epigenetic patterns. (A) The intrinsic association between an epigenetic mark and a genome locus is sequence-dependent and can be quantified by a propensity score. (B) An epigenetic mark (represented by the stars) is constitutively recruited to regions with high propensity scores, mediated by interaction with general factors (represented by the ovals) that target distinct sequence features. These features are usually degenerative and are the main determinants of the propensity scores. On the opposite end, this mark is excluded from regions with low propensity score. In the middle range, the epigenetic pattern is highly variable among different cell type. Occupancy can be enhanced or inhibited due to interactions with many cell type specific factors (represented by other shapes).
While I have mainly focused on the DNA sequence, it is important to recognize that this is only one component of the complex network of diverse interactions that regulates global epigenetic patterns. For example, once initialized, the epigenetic marks may serve as a platform that can be used to recruit additional marks, thereby serving as a self-enhancing feedback loop, which may also lead to significant spatial expansion, resulting large-scale domains spanning hundreds of kilobases [6, 9, 67, 130]. On the other hand, different epigenetic marks compete through cross-talk between chromatin readers, effectors, and erasers, and between these machineries and DNA sequence binding factors [131]. Long-range chromatin interactions may play an important role in maintenance of domain boundaries [132]. All in all, the genome-wide epigenetic pattern should not be viewed as a simple equilibrium but a steady state resulting from the delicate balance of these complex interactions. An important gap in our current knowledge is how distinct combinatorial epigenetic patterns are established, which is not clear even for promoter regions [133-135]. We are just beginning to understand this complex network of these interactions (see [13-15] for more comprehensive reviews). Future investigations will benefit from integration of diverse data-types [9].
Systems biology studies have shown the expression level of many genes are not precisely controlled but undergo significant stochastic fluctuation [136]. It has been theorized that phenotypic diversity is evolutionarily beneficial for robustness against environmental perturbations [129, 137-139]. It has been increasingly recognized that epigenetic regulation plays an important role in the maintenance of gene expression diversity. It is very interesting that the degree of epigenetic variability is associated with certain DNA sequence signatures, providing a simple mechanism for inheritance. As suggested by several recent studies [139, 140], elucidating the connection between DNA sequence in epigenetic variability will provide mechanistic insights into the evolutionary history of phenotypic diversity.
Acknowledgement
I apologize for omission of relevant references due to space restrictions. I thank Drs. Franziska Michor and Shirley Liu for helpful discussions. This research was supported by an HSPH Career Incubator Award and the NIH grant 1R01HG005085-01A2.
References
- 1.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128(4):669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98(3):285–94. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 4.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 5.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 6.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu Rev Biochem. 2009;78:245–71. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010;11(7):476–86. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 11.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8(4):253–62. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136(2):200–6. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 15.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145(6):835–50. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moazed D. Mechanisms for the inheritance of chromatin states. Cell. 2011;146(4):510–8. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji H, et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26(11):1293–300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hastie T, Tibishirani R, Friedman J. The elements of statistical learning. Springer; 2001. p. 533. [Google Scholar]
- 19.Das R, et al. Computational prediction of methylation status in human genomic sequences. Proc Natl Acad Sci U S A. 2006;103(28):10713–6. doi: 10.1073/pnas.0602949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bock C, et al. CpG island methylation in human lymphocytes is highly correlated with DNA sequence, repeats, and predicted DNA structure. PLoS Genet. 2006;2(3):e26. doi: 10.1371/journal.pgen.0020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goh L, et al. Genomic sweeping for hypermethylated genes. Bioinformatics. 2007;23(3):281–8. doi: 10.1093/bioinformatics/btl620. [DOI] [PubMed] [Google Scholar]
- 22.Peckham HE, et al. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17(8):1170–7. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang F, et al. Predicting methylation status of CpG islands in the human brain. Bioinformatics. 2006;22(18):2204–9. doi: 10.1093/bioinformatics/btl377. [DOI] [PubMed] [Google Scholar]
- 24.Lee W, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39(10):1235–44. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 25.Yuan GC, Liu JS. Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput Biol. 2008;4(1):e13. doi: 10.1371/journal.pcbi.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tibshirani R. Regression Shrinkage and Selection via the Lasso. Journal of the Royal Statistical Society. Series B (Methodological) 1996;58(1):267–288. [Google Scholar]
- 27.Breiman L. Classification and regression trees. x. Wadsworth International Group; Belmont, Calif.: 1984. p. 358. (The Wadsworth statistics/probability series). [Google Scholar]
- 28.Cortes C, Vapnik V. Support-vector networks. Machine Learning. 1995;20:273–297. [Google Scholar]
- 29.Freund Y. Boosting a weak learning algorithm by majority. Information and Computation. 1995;121(2):256–285. [Google Scholar]
- 30.Chipman H, George E, McCulloch R. Bayesian ensemble learning. In: Scholkopf B, editor. Neural information processing systems. MIT Press; 2007. [Google Scholar]
- 31.Yuan GC. Targeted recruitment of histone modifications in humans predicted by genomic sequences. J Comput Biol. 2009;16(2):341–55. doi: 10.1089/cmb.2008.18TT. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Shao Z, Yuan GC. Prediction of Polycomb target genes in mouse embryonic stem cells. Genomics. 2010;96(1):17–26. doi: 10.1016/j.ygeno.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 33.Feltus FA, et al. DNA motifs associated with aberrant CpG island methylation. Genomics. 2006;87(5):572–9. doi: 10.1016/j.ygeno.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Q, Liu JS. Extracting sequence features to predict protein-DNA interactions: a comparative study. Nucleic Acids Res. 2008;36(12):4137–48. doi: 10.1093/nar/gkn361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442(7104):772–8. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Field Y, et al. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4(11):e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458(7236):362–6. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miele V, et al. DNA physical properties determine nucleosome occupancy from yeast to fly. Nucleic Acids Res. 2008;36(11):3746–56. doi: 10.1093/nar/gkn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morozov AV, et al. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Res. 2009;37(14):4707–22. doi: 10.1093/nar/gkp475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Locke G, et al. High-throughput sequencing reveals a simple model of nucleosome energetics. Proc Natl Acad Sci U S A. 2010;107(49):20998–1003. doi: 10.1073/pnas.1003838107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luger K, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 42.Yuan GC, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309(5734):626–30. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 43.Wasson T, Hartemink AJ. An ensemble model of competitive multi-factor binding of the genome. Genome Res. 2009;19(11):2101–12. doi: 10.1101/gr.093450.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raveh-Sadka T, Levo M, Segal E. Incorporating nucleosomes into thermodynamic models of transcription regulation. Genome Res. 2009;19(8):1480–96. doi: 10.1101/gr.088260.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Albert I, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446(7135):572–6. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 46.Mavrich TN, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18(7):1073–83. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shivaswamy S, et al. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6(3):e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ioshikhes IP, et al. Nucleosome positions predicted through comparative genomics. Nat Genet. 2006;38(10):1210–5. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- 49.Gupta S, et al. Predicting human nucleosome occupancy from primary sequence. PLoS Comput Biol. 2008;4(8):e1000134. doi: 10.1371/journal.pcbi.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10(3):161–72. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell. 2005;18(6):735–48. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 52.Wippo CJ, et al. Differential cofactor requirements for histone eviction from two nucleosomes at the yeast PHO84 promoter are determined by intrinsic nucleosome stability. Mol Cell Biol. 2009;29(11):2960–81. doi: 10.1128/MCB.01054-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, et al. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol. 2009;16(8):847–52. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kornberg RD, Stryer L. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 1988;16(14A):6677–90. doi: 10.1093/nar/16.14.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sadeh R, Allis CD. Genome-wide “re”-modeling of nucleosome positions. Cell. 2011;147(2):263–6. doi: 10.1016/j.cell.2011.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Z, et al. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science. 2011;332(6032):977–80. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lantermann AB, et al. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17(2):251–7. doi: 10.1038/nsmb.1741. [DOI] [PubMed] [Google Scholar]
- 58.Valouev A, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18(7):1051–63. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mavrich TN, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453(7193):358–62. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chodavarapu RK, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466(7304):388–92. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132(5):887–98. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tillo D, et al. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One. 2010;5(2):e9129. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valouev A, et al. Determinants of nucleosome organization in primary human cells. Nature. 2011;474(7352):516–20. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramirez-Carrozzi VR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138(1):114–28. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 66.Egelhofer TA, et al. An assessment of histone-modification antibody quality. Nature structural & molecular biology. 2011;18(1):91–3. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 68.Thomson JP, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464(7291):1082–6. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ang YS, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145(2):183–97. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ku M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4(10):e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mendenhall EM, et al. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6(12):e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 73.Schuettengruber B, et al. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009;7(1):e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ringrose L, et al. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev Cell. 2003;5(5):759–71. doi: 10.1016/s1534-5807(03)00337-x. [DOI] [PubMed] [Google Scholar]
- 75.Fiedler T, Rehmsmeier M. jPREdictor: a versatile tool for the prediction of cis-regulatory elements. Nucleic Acids Res. 2006;34(Web Server issue):W546–50. doi: 10.1093/nar/gkl250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sing A, et al. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009;138(5):885–97. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 77.Woo CJ, et al. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140(1):99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao J, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297(5588):1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 81.Verdel A, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303(5658):672–6. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Djupedal I, Ekwall K. Epigenetics: heterochromatin meets RNAi. Cell research. 2009;19(3):282–95. doi: 10.1038/cr.2009.13. [DOI] [PubMed] [Google Scholar]
- 83.Martens JH, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. Embo J. 2005;24(4):800–12. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nature biotechnology. 2010;28(8):817–25. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput Biol. 2009;5(11):e1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Larson JL, Yuan GC. Epigenetic domains found in mouse embryonic stem cells via a hidden Markov model. BMC Bioinformatics. 2010;11:557. doi: 10.1186/1471-2105-11-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471(7339):480–5. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–9. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee D, Karchin R, Beer MA. Discriminative prediction of mammalian enhancers from DNA sequence. Genome Res. 2011;21(12):2167–80. doi: 10.1101/gr.121905.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pinello L, et al. A motif-independent metric for DNA sequence specificity. BMC Bioinformatics. 2011;12(1):408. doi: 10.1186/1471-2105-12-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gebhard C, et al. General transcription factor binding at CpG islands in normal cells correlates with resistance to de novo DNA methylation in cancer cells. Cancer Res. 2010;70(4):1398–407. doi: 10.1158/0008-5472.CAN-09-3406. [DOI] [PubMed] [Google Scholar]
- 92.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39(2):157–8. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 93.Edwards JR, et al. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 2010;20(7):972–80. doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.De S, Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nature structural & molecular biology. 2011;18(8):950–5. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43(8):768–75. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Molaro A, et al. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146(6):1029–41. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Berman BP, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2011 doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lienert F, et al. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43(11):1091–7. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- 99.Kim J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–24. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ravasi T, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140(5):744–52. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 102.Wang J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444(7117):364–8. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 103.Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276(37):34355–8. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 104.Li G, et al. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24(4):368–80. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pasini D, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010 doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 106.Peng JC, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shen X, et al. Jumonji modulates Polycomb activity and self-renewal versus differentiation of stem cells. 2009. submitted. [DOI] [PMC free article] [PubMed]
- 108.Vire E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 109.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23(16):1831–42. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–61. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 111.Kanhere A, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38(5):675–88. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr Opin Genet Dev. 2010;20(2):142–8. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tirosh I, Berman J, Barkai N. The pattern and evolution of yeast promoter bendability. Trends Genet. 2007;23(7):318–21. doi: 10.1016/j.tig.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 115.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–73. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 116.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42(4):343–7. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fu Y, et al. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4(7):e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19(1):24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dion MF, et al. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315(5817):1405–8. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 120.Radman-Livaja M, et al. Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol. 2011;9(6):e1001075. doi: 10.1371/journal.pbio.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vastenhouw NL, et al. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464(7290):922–6. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mohn F, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30(6):755–66. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 123.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19(6):541–9. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39(4):457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 125.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008 doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ji H, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010 doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Feinberg AP. Genome-scale approaches to the epigenetics of common human disease. Virchows Arch. 2010;456(1):13–21. doi: 10.1007/s00428-009-0847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Feinberg AP, Irizarry RA. Evolution in health and medicine Sackler colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1757–64. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wen B, et al. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41(2):246–50. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–8. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 132.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Weishaupt H, Sigvardsson M, Attema JL. Epigenetic chromatin states uniquely define the developmental plasticity of murine hematopoietic stem cells. Blood. 2010;115(2):247–56. doi: 10.1182/blood-2009-07-235176. [DOI] [PubMed] [Google Scholar]
- 135.Ucar D, Hu Q, Tan K. Combinatorial chromatin modification patterns in the human genome revealed by subspace clustering. Nucleic Acids Res. 2011;39(10):4063–75. doi: 10.1093/nar/gkr016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Raser JM, O’Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309(5743):2010–3. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wolf DM, Vazirani VV, Arkin AP. Diversity in times of adversity: probabilistic strategies in microbial survival games. J Theor Biol. 2005;234(2):227–53. doi: 10.1016/j.jtbi.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 138.Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell. 2007;128(4):655–68. doi: 10.1016/j.cell.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 139.Tirosh I, Barkai N, Verstrepen KJ. Promoter architecture and the evolvability of gene expression. J Biol. 2009;8(11):95. doi: 10.1186/jbiol204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Field Y, et al. Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat Genet. 2009;41(4):438–45. doi: 10.1038/ng.324. [DOI] [PMC free article] [PubMed] [Google Scholar]