Abstract
Background & Aims
Legalon® SIL (SIL) is a chemically hydrophilized version of silibinin that has exhibited high antiviral effectiveness against hepatitis C virus (HCV). Its main mode of action (MOA) remains unclear, with contradicting in-vitro studies supporting either suppression of entry and cell-to-cell spread or suppression of viral RNA synthesis as the main MOA. We sought to provide new insight into SIL’s MOA in genotype-1/4 patients receiving intravenous SIL monotherapy for 7 days via mathematical modeling.
Methods
Changes in HCV RNA in 25 patients receiving 10, 15 or 20 mg/kg/day of SIL were analyzed and modeled using viral kinetic methods.
Results
In 15 patients virus declined in a biphasic manner, in which a sharp drop between days 0 and 2 was followed by a slower second phase of decline. In 10 patients the initial decline was weaker and virus declined in a single phase over the 7 day period. The blocking production effectiveness, ε, was dose dependent with mean ε=0.49 and 0.89 in the 10 or 15 mg/kg/day and 20 mg/kg/day dosing groups, respectively (P=0.020). The effectiveness of blocking viral infection, η, was estimated as 0.60 with no significant differences across dosing groups. For all patients the mean rate of viral-load decline measured between days 2 and 7 was high (0.3 logIU/mL/day), i.e., 4 fold higher than typically observed during the 2nd phase of (pegylated)-interferon-α±ribavirin treatment.
Conclusions
Modeling HCV kinetics in vivo suggests that SIL may block both viral infection and viral production/release with its main dose-dependent effect being blocking viral production/release.
Introduction
Chronic hepatitis C virus (HCV) infection has a worldwide prevalence of about 3% [1]. Achieving a long-term sustained virologic response (SVR), defined as undetectable serum HCV RNA 24 weeks after the end of antiviral treatment, is the most effective way to prevent disease progression permanently [2]. Treatment outcome with pegylated interferon (peg-IFN) and ribavirin (RBV) administered for 48 weeks, is correlated with HCV genotype and SVR is only achieved in approximately 50% of HCV genotype 1 patients, the most prevalent genotype in western countries [3]. Direct acting antiviral (DAA) agents constitute a new stage in HCV therapy and the combination of either of the two protease inhibitors (telaprevir or boceprevir), which recently have been approved, with peg-IFN plus RBV (PEG/RBV), should result in a dramatic improvement of SVR rates in both treatment-naive and treatment-experienced patients [4–7]. However, there is still a pressing need for alternative treatment strategies, as limitations associated with the PEG/RBV remain true (side effects and poor response) and new issues related to DAA have emerged (drug resistance).
Silymarin is a mixture of flavonolignans extracted from the milk thistle (Silybum marianum (L.) Gaertner) [8] that has been used since ancient times as a liver tonic [9]. Although the use of silymarin compounds (orally) is gaining popularity among HCV infected patients [10], the absence of well designed prospective studies on large populations precludes general conclusions on the effect of silymarin on serum transaminases, HCV RNA levels, or quality of life [9–12]. The major active component of silymarin is silibinin in a roughly 50:50 isomeric mixture of silibinin A and silibinin B. High doses of silibinin can be administered intravenously (IV) using its esterified modification silibinin-C-2′, 3-dihydrogen succinate, disodium salt (Legalon® SIL). Legalon® SIL (SIL) is currently authorized, in the name of Madaus GmbH (a company of the Rottapharm|Madaus Group), and marketed since the mid-1980s for the treatment of hepatic intoxication by Amanita phalloides mushrooms (“death cap”). Surprisingly, a strong antiviral effectiveness of SIL against HCV genotype 1 or 4 was shown in a pivotal study [13]. In the aftermath of this study, a case of SVR was reported in a patient treated with SIL monotherapy for one week, followed by PEG/RBV+SIL in the second week and continued with PEG/RBV for 15 weeks [14]. Interestingly, this patient achieved SVR in spite of having not responded to PEG/RBV previously, having the unfavorable T/T rs12979860 IL28B, being co-infected with HIV and receiving a short (16 week) duration of PEG/RBV therapy [15].
In spite of these encouraging clinical results, SIL’s mode of action (MOA) against HCV remains unknown, and results from in vitro experiments are controversial [16]. In particular, Ahmed-Belkacem et al. have experimentally shown that SIL mainly inhibits the HCV NS5B RNA-dependent RNA polymerase (RdRp) [17], while Wagoner et al. have presented results suggesting that SIL has a profound effect on HCV entry and cell-to-cell spread with only marginal suppression of HCV-RdRp activity [18, 19]. Other MOAs such as inhibition of T-cell inflammatory cytokines and hepatocyte NF-κB signaling have been suggested as well [18, 20].
While viral dynamic modeling has provided valuable insights by quantifying the effects of (peg)-IFN, ribavirin and HCV protease inhibitors and estimating its antiviral effectiveness in vivo (reviewed in [21]), it has not been used to analyze the effects of SIL treatment. Here, we analyzed and modeled HCV RNA kinetics from 25 patients, infected with HCV genotype 1 or 4, who were treated for 7 days with monotherapy of 10, 15 or 20 mg/kg/day of SIL. Using such an approach, we sought to provide new insights into SIL’s in vivo mode of action.
Patients & Methods
Patients and study protocol
All patients had failed prior treatment to full dose peg-IFN-α and ribavirin therapy. Treatment failure was defined by the lack of a >2 log-drop in viral load after 12 weeks of treatment and/or by not achieving SVR. The patients’ baseline characteristics are given in Table 1.
Table 1.
Baseline characteristics and empirical viral kinetic parameters
| Patient No. | Age [yr] | GT | Fibrosis stage | SIL Dose | V0 | ΔV0–1 | ΔV1–2 | ΔV0–2 | s2–7 | VK pattern* |
|---|---|---|---|---|---|---|---|---|---|---|
| 25 | 48 | 1b | 4 | 10 | 6.69 | 0.17 | 0.89 | 1.06 | 0.27 | B |
| 20 | 59 | 1a | 4 | 10 | 6.95 | 0.18 | 0.11 | 0.26 | 0.17 | M |
| 28 | 56 | 1a | 2 | 10 | 7.12 | 0.69 | 0.20 | 0.89 | 0.24 | B |
| Mean (SD) | 54 | 6.92 (0.22) | 0.35 (0.30) | 0.40 (0.43) | 0.75 (0.42) | 0.23 (0.05) | ||||
| 21 | 57 | 1b | 3 | 15 | 6.65 | 0.44 | 0.03 | 0.41 | 0.15 | B |
| 22 | 49 | 4 | 0 | 15 | 5.95 | 0.67 | 0.73 | 1.40 | 0.36 | B |
| 29 | 58 | 1a | 2 | 15 | 6.91 | 0.26 | 0.08 | 0.35 | 0.18 | M |
| 30 | 67 | 1a | 2 | 15 | 5.75 | 0.65 | 0.25 | 0.90 | 0.65 | M |
| 36 | 41 | 1b | 2 | 15 | 5.43 | 0.18 | 0.36 | 0.54 | 0.27 | M |
| Mean (SD) | 54 | 6.14 (0.62) | 0.44 (0.22) | 0.28 (0.28) | 0.72 (0.44) | 0.32 (0.20) | ||||
| 31 | 24 | 1b | 2 | 20 | 6.44 | 1.07 | 1.25 | 2.31 | 0.27 | B |
| 32 | 33 | 1a | n.a. | 20 | 6.98 | 1.15 | 1.04 | 2.19 | 0.50 | B |
| 33 | 48 | 1a | 1 | 20 | 6.04 | 1.31 | 0.42 | 1.73 | 0.33 | B |
| 34 | 60 | 1b | 4 | 20 | 6.50 | 0.57 | 0.39 | 0.96 | 0.18 | B |
| 35 | 49 | 1b | n.a. | 20 | 6.68 | 1.52 | 1.14 | 2.66 | 0.40 | B |
| 26 | 60 | 1a | 4 | 20 | 6.61 | 0.45 | 0.27 | 0.71 | 0.39 | M |
| 37 | 67 | 1b | 1 | 20 | 6.07 | 0.50 | 0.31 | 0.81 | 0.34 | M |
| 38 | 68 | 1b | 4 | 20 | 6.30 | 0.30 | 0.15 | 0.45 | 0.19 | M |
| 39 | 50 | 4 | n.a. | 20 | 5.77 | 0.39 | 0.65 | 1.04 | 0.27 | B |
| 40 | 69 | 1b | 3 | 20 | 6.32 | −0.32 | 0.53 | 0.21 | 0.19 | M |
| 41 | 50 | 1b | 4 | 20 | 6.27 | 0.63 | 0.74 | 1.36 | 0.24 | B |
| 43 | 30 | 1b | 2 | 20 | 5.65 | 0.59 | 0.41 | 1.00 | 0.26 | B |
| 44 | 55 | 1b | 2 | 20 | 6.25 | 0.30 | 0.26 | 0.56 | 0.26 | M |
| 45 | 52 | 1a | 2 | 20 | 7.66 | 0.30 | 0.75 | 1.06 | 0.35 | M |
| 46 | 68 | 4 | n.a. | 20 | 6.50 | 1.31 | 1.05 | 2.36 | 0.45 | B |
| 47 | 63 | 1 | 4 | 20 | 6.86 | 1.15 | 0.39 | 1.54 | 0.19 | B |
| 48 | 45 | 1a | n.a. | 20 | 6.32 | 1.12 | 0.49 | 1.61 | 0.45 | B |
| Mean (SD) | 52 | 6.42 (0.47) | 0.73 (0.49) | 0.60 (0.34) | 1.33 (0.73) | 0.31 (0.10) | ||||
| P-value& | 0.93 | 0.64 | 0.09 | 0.03 | 0.04 | 0.29 | 0.67$ | |||
VK, viral kinetic pattern: B= biphasic and M=monophasic; si–j, HCV decline slope between days i and j (logIU/mL/day); ΔVi–j, HCV decline between days i and j (logIU/mL); SIL dose (mg/kg/day); GT, HCV genotype; n.a., not available; V0, baseline viral load (logIU/mL); SD, standard deviation;
P-value to reject the null hypothesis that the distribution between 10+15 and 20 SIL dosing groups is the same (Mann-Whitney test and Fisher’s exact test, respectively).
Patients received 5 (N=3), 10 (N=3), 15 (N=5), 20 (N=17) mg/kg/day SIL infused over 4 hours for 7 days as monotherapy. During this period, HCV RNA was measured daily. The study protocol can be found in [13] (see “Protocol 2”). Compared to the initial study report [13], 8 patients in the 20 mg/kg/day silibinin were added to the present study (Table 1, patients numbered ≥ 40).
Data analysis
Patients receiving 5 mg/kg/day did not show a significant antiviral effect [13] and were excluded in our analysis. Further, given the small study populations and the fact that the viral responses of the 10 and 15 mg/kg/day dosing groups were largely comparable, we analyzed these groups together (termed 10+15 mg/kg/day dosing group).
Empirical analysis of the viral decline
Let ΔVi–j = log(Vi) − log(Vj) be the difference in log viral load between days i and j, and si–j the slope of the log viral decline between days i and j, calculated by linear regression.
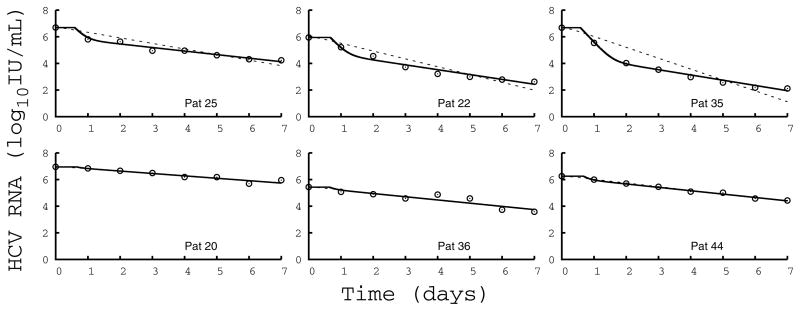
HCV RNA during therapy typically declines in a biphasic manner, where a rapid drop during the first 2 days of treatment, s0–2, is followed by a slower but sustained viral decline slope, s2–7 [21]; thus, during a typical two-phase decline the decline rate during the first phase is much larger than the second phase decline rate, i.e., s0–2≫s2–7. However, in some patients, viral declines during SIL treatment do not exhibit two clearly separated phases but rather decline in a single phase [16]. Since viral load was only measured daily, it was not possible to accurately determine s0–2 and we used max(ΔV0–1, ΔV1–2) as a proxy for the first phase decline slope s0–2. Then we defined a viral decline as monophasic if max(ΔV0–1, ΔV1–2) < 2s2–7, i.e., if the two typical phases of viral decline have slopes that differ by less than a factor of 2. As shown in Figure 1 and Table 1, in this case the two slopes (s0–2 and s2–7) cannot be distinguished empirically and only one single phase of viral decline is visible (termed monophasic).
Figure 1.
Fit of the model (Eq.1) to HCV RNA kinetics. Comparison of measured HCV RNA (circle) and best-fit prediction assuming SIL blocks viral production and viral infection (solid line) or blocks only infection (dotted line) in 6 representative patients dosed at (left) 10mg/kg/day, (center) 15mg/kg/day, and (right) 20 mg/kg/day. For each dosing group, one patient with a biphasic (upper row) and one with a monophasic (lower row) decline are shown. Parameter values used to generate the fits are given in Table S1.
Statistical analysis
Statistical comparisons were performed with SPSS V.18 (Chicago, IL). Continuous variables were compared using a 2-tailed Mann-Whitney U-test; to compare categorical variables we used a 2-tailed Fisher exact test. In all cases a p-value <0.05 was considered significant.
Viral kinetic modeling
The changes in viral load over time, V(t), were modeled using the standard model of HCV kinetics [22], which predicts that after therapy is initiated:
| Eq. (1) |
where V0 is the HCV RNA level at baseline, i.e., at t≤t0, ε and η are the drug’s effectiveness in blocking viral production/release and viral infection, respectively, and c and δ are the clearance rates of serum HCV RNA and HCV-infected cells, respectively, and t0 is the time after initiation of therapy that drug starts having an effect on viral load (see supplementary material).
Parameter identifiability, parameter estimation and model selection
The absence of frequent sampling during the first two days of treatment made it impossible to accurately estimate the viral clearance rate and therefore c was fixed at 6 day−1, consistent with values found in patients treated with IFN or PEG/RBV [22, 23]. The antiviral effectiveness in blocking infection, η, cannot be uniquely determined by fitting Eq. 1 to the viral load data [22] on a patient by patient basis but can be estimated using mixed-effect modeling (see supplementary material). Once parameters were estimated using Monolix 3.2© (http://software.monolix.org) [24], models were selected using the Akaike information criterion (AIC), which allows one to compare the ability of models with different numbers of parameters to fit experimental data [25]..
Results
Viral kinetic descriptive analysis
The drop in viral load levels during the first two days of treatment, ΔV0–2, was significantly associated with the dosing group, with mean ΔV0–2=0.73 logIU/ml vs 1.33 logIU/ml in the 10+15 mg/kg/day and in the 20 mg/kg/day dosing groups, respectively (P=0.048), and was highly variable among patients (Table 1). The subsequent slope of viral decline, s2–7, corresponded to rapid viral decay in all patients and was between 0.15 logIU/ml/day and 0.65 logIU/ml/day, and was not significantly correlated with the dosing group (mean s2–7=0.28 logIU/ml/day vs 0.31 logIU/ml/day in the 10+15 mg/kg/day and the 20 mg/kg/day, respectively, P=0.29). Interestingly, 40% (10/25) of the patients did not show the typical biphasic viral decline (Table 1), but rather exhibited a monophasic (see Methods) viral decline throughout the seven days of SIL monotherapy (Figure 1). This pattern was not dosing group dependent (as was seen in N=4/8 vs 6/17 patients in the 10+15 mg/kg/day and the 20 mg/kg/day dosing groups, respectively, P=0.67). Compared to patients with a biphasic viral decline, monophasic viral decline patients were associated with a significantly lower viral decline from baseline to day 2 (mean ΔV0–2=0.59 logIU/ml vs 1.50 logIU/ml in the monophasic and biphasic viral decline patients, respectively, P<10−4), but had a similar rate of second phase viral decline (mean s2–7= 0.30 logIU/ml/day in both groups).
Viral kinetic modeling
Next, we used the standard model of HCV infection and treatment (Eq. 1) to get a better understanding of the kinetics of viral decline and to infer SIL’s mode of action.
A model assuming that SIL only acts by blocking viral infection cannot reproduce a biphasic pattern of viral decline (see Methods and [22]), and therefore gave a poor description of the viral load data of patients with a biphasic viral decline (Figure 1, dotted lines in upper panels). The estimated treatment effectiveness in blocking infection was low (mean η=0.40), with no difference across dosing groups (P=0.25), and the estimate of the loss rate of infected cells was unrealistically high (mean δ=3.36 day−1), partially due to the poor fit of this model to the data.
A model assuming that SIL only acts by blocking viral production/release gave a very good description of the viral load data (see reduced AIC in Table 2), which clearly establishes that SIL’s main mode of action is blocking viral production/release rather than blocking viral infection. Using this model the effectiveness in blocking viral production/release was significantly associated with the dosing regimen (mean ε=0.69 vs 0.91 in the 10+15 mg/kg/day and in the 20 mg/kg/day dosing groups, respectively, P=0.031). The estimated loss rate of infected cells, although lower than in the model assuming no effect in blocking viral production (mean δ=0.91 day−1), remained larger than previous estimates [22, 23]. The rate of viral decline between day 2 and day 7, s2–7, is the net balance between the loss rate of infected cells and the rate of new cell infections. Contrary to biphasic viral decline patients, monophasic viral decline patients do not have their viral load levels rapidly fall by 1–2 logs, and thus a significant number of new cell infections can still occur. As s2–7 was similar in both groups, this can be either attributed to a higher δ in monophasic viral decline patients (mean δ=1.10 day−1 vs δ=0.83 day−1, respectively, P=0.002), or to the fact that SIL can also block new cell infections.
Table 2.
Mean viral kinetic parameters estimated using a population approach
| Model | t0 [day] | ε | δ [day−1] | η | AIC | ||
|---|---|---|---|---|---|---|---|
| 10+15 mg/kg/day dosing group | 20 mg/kg/day dosing group | ||||||
| Blocking infection only | Pop. parameters | 0** | 0 | 0 | 3.36 | 0.40* | 232.7 |
| SE | 1.30 | 0.072 | |||||
| Q1–Q3 | 2.0–5.81 | -- | |||||
| Blocking production only | Pop. parameters | 0.68 | 0.69 | 0.91 | 0.91 | 0 | 26.3 |
| SE | 0.036 | 0.13 | 0.034 | 0.076 | |||
| Q1–Q3 | 0.65–0.73 | 0.43–0.81 | 0.72–0.98 | 0.78–1.03 | |||
| Blocking production and infection | Pop. parameters | 0.66 | 0.49 | 0.89 | 0.77 | 0.60* | 16.7 |
| SE | 0.11 | 0.19 | 0.049 | 0.075 | 0.18 | ||
| Q1–Q3 | 0.63–0.70 | 0.24–0.80 | 0.66–0.98 | 0.65–0.90 | |||
Parameter estimated assuming no inter-individual variations; Pop. parameters, parameters estimated using mixed-effect modeling (see Methods);
t0 was fixed to zero; SE, standard error; Q1–Q3: first and third quartiles of the distribution of the individual a posteriori estimates (see Supp. Materials); t0, HCV delay before decline begins; ε, SIL effectiveness in blocking viral production/release; δ, HCV-infected cell loss rate; η; SIL effectiveness in blocking viral infection; AIC; Akaike information criterion. The model with the lowest AIC is considered the best.
Indeed assuming that SIL had an effect in blocking both viral infection and viral production/release yielded an improvement in the fit to the data (Table 2), suggesting that SIL may indeed have a significant effect in blocking infection (P=0.0016, likelihood ratio test). Further, the estimated value of δ was lower (mean 0.77 day−1) with no differences between biphasic and monophasic viral decline patients (P=0.15).. A precise estimate of the distribution of the effectiveness in blocking viral infection in the sample population, η, could not be obtained and thus we assumed this parameter to be the same for all patients. The best-fit to the data was obtained with η=0.60 (CI95%=[0.24;0.96]). Compared to the model assuming no effectiveness in blocking viral infection (i.e., η=0), the estimates of the treatment effectiveness in blocking viral production remained largely unchanged (mean ε=0.49 and 0.89 in the 10+15 mg/kg/day and in the 20 mg/kg/day dosing groups, respectively, P=0.020).
Accounting for pharmacokinetic related changes in treatment effectiveness
One could argue that the use of a model that assumes constant drug effectiveness over time, such as Eq. 1, is incorrect and makes the parameter estimates unreliable. To test this, we used data from 7 patients treated with 20 mg/kg/day for whom both the viral load and the SIL A concentration were frequently measured. Using a model that allows the treatment effectiveness in blocking viral production or viral infection to change according to the drug concentration [27, 28], we could estimate the viral kinetic parameters accounting for pharmacokinetic/pharmacodynamic effects. We found that the parameter estimates arrived at using this method and the constant effectiveness model were very close for all 7 patients (see supplemental material).
Discussion
Viral kinetics during IFN based therapy and protease inhibitor therapy is typically characterized by a rapid drop in HCV RNA level during the first 2 days of treatment, followed by a slower but sustained rate of viral decline afterwards [21]. Patterns of viral kinetics during SIL monotherapy were substantially different, with 40% (N=10/25) of patients showing a single phase of viral decline, consistent with other SIL studies [14, 29]. In all patients, the rate of viral load decline measured between days 2 and 7 was high with a mean value of 0.30 log IU/mL/day. Assuming this decline would be sustained beyond day 7, this value translates into a decline of 2.1 log IU/mL/week, which is about four times more rapid than what has typically been observed with PEG/RBV in HCV genotype 1 treatment-naive patients (c.f. [30, 31]).
Using the standard model of viral kinetics, we showed that SIL’s major mode of action is blocking viral production, which supports Ahmed-Belkacem et al.’s in vitro findings [17]. The effectiveness in blocking viral production/release was significantly higher in the 20 mg/kg/day than in the 10+15 mg/kg/day dosing group (mean ε=0.89 vs 0.49, P=0.02), and was nearly as high as values found in naïve patients treated with high daily doses of IFN [22]. Assuming that SIL also blocks viral infection gave a statistically significant improvement to the model’s ability to fit the patient viral load data (P=0.0016). We estimated SIL’s effectiveness in blocking infection to be moderate with η=0.60 in the population sample, however this estimate should be taken with caution as the confidence interval was rather large (CI95%=[0.24;0.96]). Therefore our in vivo analysis supports, in part, the in vitro findings of Wagoner at al. [18, 19], which suggested SIL may have an effect on HCV entry and cell-to-cell spread, and thus may have played a role in the prevention of HCV infection after liver transplantation [32, 33]. However, this effect is moderate; using our estimated parameters and simulating the viral decline assuming SIL only blocks production/release reduces the mean viral decline at the end of the 7 day treatment period of 0.53 log10 (mean decline of 2.15 log10 vs 2.68 log10).
The origin of the monophasic viral decline observed in 40% of the patients irrespective of the dose of drug received remains to be understood. It is related to having a weak first phase decline, i.e. relatively low value of ε, such that subsequent viral declines continue at approximately the same rate. We recently reported a similar monophasic decline pattern in some patients treated with mericitabine (RG7128), a potent NS5B polymerase inhibitor. For the case of mericitabine, which needs to be phosphorylated intracellularly, we showed that a monophasic decline could arise from a progressive increase in the drug effectiveness over time [34]. However accounting for the change in SIL A concentration over time did not substantially affect the parameter estimates provided by our model assuming constant effectiveness (Table S1). This supports the hypothesis that monophasic viral decline with SIL is not related to pharmacokinetic issues. However, our analysis was done only using SIL A concentrations in a limited number of patients, and further PK/PD analysis including both SIL A and SIL B need to be performed to confirm this hypothesis. Furthermore, such analyses will still be limited by the fact that drug concentrations are measured in the serum and not within liver cells.
Whatever the pattern of viral decline, all patients treated with SIL exhibited a rapid viral decline between days 2 and 7. The standard model of viral kinetics attributes this to a high loss rate of infected cells, δ, estimated here to be 0.77 day−1. Interestingly, this estimate is much more rapid that what is typically found during IFN-based treatment [22, 23], but is on the order of values found with telaprevir-based therapy [35]. Recently, Guedj and Perelson [35] showed that the large second phase slopes observed during telaprevir therapy were correlated with a high effectiveness in blocking viral production (ε). This example with SIL shows that one can have a high second phase slope without having an extremely high value of ε. Thus high drug effectiveness may be sufficient to generate a high second phase slope but may not be necessary.
The origin of the high δ seen with SIL remains to be elucidated. One possibility is that the high value of δ is due to an immunomodulatory effect of SIL [9], but no increase in aminotransferase (ALT) levels, a marker of cell necrosis, was reported during the dosing period [13]. Another possibility suggested by other viral dynamics models is that the second phase of HCV RNA decline is enhanced by loss of intracellular viral RNA [36] or blocking hepatocyte proliferation [37].
In summary, viral kinetics analyses show that SIL may block both viral infection and viral production/release with its main dose-dependent effect being blocking viral production/release. Further, SIL induces a high rate of viral decline between days 2 and 7 in all patients, which is about 4-fold higher than typically observed during (peg)-IFN±RBV treatment. Together, these results suggest that SIL could be a valuable candidate for combination with direct antiviral agents against HCV. HCV kinetic studies with SIL monotherapy that include frequent viral load sampling at the beginning of treatment along with frequent ALT measurements are needed to further understand SIL’s modes of action against HCV.
Supplementary Material
Acknowledgments
This work was performed under the auspices of the U.S. Department of Energy under contract DE-AC52-06NA25396, and supported by NIH grants RR006555, P20-RR018754, AI065256 and AI028433, and by the University of Illinois Walter Payton Liver Center GUILD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Health Organization. Hepatitis C Fact sheet No 164. Revised June 2011. http://www.who.int/mediacentre/factsheets/fs164/en/index.html.
- 2.Cardoso AC, Moucari R, Figueiredo-Mendes C, Ripault MP, Giuily N, Castelnau C, et al. Impact of peginterferon and ribavirin therapy on hepatocellular carcinoma: incidence and survival in hepatitis C patients with advanced fibrosis. J Hepatol. 2010;52:652–657. doi: 10.1016/j.jhep.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 3.Awad T, Thorlund K, Hauser G, Mabrouk M, Gluud C. Peginterferon alpha-2a is associated with higher sustained virological response than peginterferon alfa-2b in chronic hepatitis C: a systematic review of randomized trials. Hepatology. 2010;51:1176–1184. doi: 10.1002/hep.23504. [DOI] [PubMed] [Google Scholar]
- 4.McHutchison JG, Everson GT, Gordon SC, Jacobson IM, Sulkowski M, Kauffman R, et al. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360:1827–1838. doi: 10.1056/NEJMoa0806104. [DOI] [PubMed] [Google Scholar]
- 5.McHutchison J, Manns M, Muir A, Terrault N, Jacobson I, Afdhal N, et al. Telaprevir for previously treated chronic HCV infection. N Engl J Med. 2010;362:1292. doi: 10.1056/NEJMoa0908014. [DOI] [PubMed] [Google Scholar]
- 6.Bacon BR, Gordon SC, Lawitz E, Marcellin P, Vierling JM, Zeuzem S, et al. Boceprevir for previously treated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1207–1217. doi: 10.1056/NEJMoa1009482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, et al. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kroll DJ, Shaw HS, Oberlies NH. Milk thistle nomenclature: why it matters in cancer research and pharmacokinetic studies. Integr Cancer Ther. 2007;6:110. doi: 10.1177/1534735407301825. [DOI] [PubMed] [Google Scholar]
- 9.Mayer K, Myers R, Lee S. Silymarin treatment of viral hepatitis: a systematic review. J Viral Hepat. 2005;12:559–567. doi: 10.1111/j.1365-2893.2005.00636.x. [DOI] [PubMed] [Google Scholar]
- 10.Seeff LB, Curto TM, Szabo G, Everson GT, Bonkovsky HL, Dienstag JL, et al. Herbal product use by persons enrolled in the hepatitis C antiviral long term treatment against cirrhosis (HALT C) Trial. Hepatology. 2008;47:605–612. doi: 10.1002/hep.22044. [DOI] [PubMed] [Google Scholar]
- 11.Lawrence V Center SAE-bP, Veterans Evidence-based Research D, Center I, Research USAfH, Quality. Milk thistle: effects on liver disease and cirrhosis and clinical adverse effects. Agency for Healthcare Research and Quality, US Dept. of Health and Human Services; 2000. [Google Scholar]
- 12.Hawke RL, Schrieber SJ, Soule TA, Wen Z, Smith PC, Reddy KR, et al. Silymarin ascending multiple oral dosing phase I study in noncirrhotic patients with chronic hepatitis C. J Clin Pharmacol. 2010;50:434. doi: 10.1177/0091270009347475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferenci P, Scherzer TM, Kerschner H, Rutter K, Beinhardt S, Hofer H, et al. Silibinin Is a potent antiviral agent in patients with chronic hepatitis C not responding to pegylated interferon/ribavirin Therapy. Gastroenterology. 2008;135:1561–1567. doi: 10.1053/j.gastro.2008.07.072. [DOI] [PubMed] [Google Scholar]
- 14.Payer B, Reiberger T, Rutter K, Beinhardt S, Staettermayer A, Peck-Radosavljevic M, et al. Successful HCV eradication and inhibition of HIV replication by intravenous silibinin in an HIV-HCV coinfected patient. J Clin Virol. 2010;49:131–133. doi: 10.1016/j.jcv.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Dahari H, Guedj J, Perelson AS, Layden TJ. Hepatitis C Viral Kinetics in the Era of Direct Acting Antiviral Agents and Interleukin-28B. Current Hepatitis Reports. 2011:1–14. doi: 10.1007/s11901-011-0101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahari H, Guedj J, Perelson AS. Silibinin mode of action(s) against HCV: A controversy yet to be resolved. Hepatology. 2011;54:749. doi: 10.1002/hep.24310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmed-Belkacem A, Ahnou N, Barbotte L, Wychowski C, Pallier C, Brillet R, et al. Silibinin and related compounds are direct inhibitors of hepatitis C virus RNA-dependent RNA polymerase. Gastroenterology. 2010;138:1112–1122. doi: 10.1053/j.gastro.2009.11.053. [DOI] [PubMed] [Google Scholar]
- 18.Wagoner J, Negash A, Kane OJ, Martinez LE, Nahmias Y, Bourne N, et al. Multiple effects of silymarin on the hepatitis C virus lifecycle. Hepatology. 2010;51:1912–1921. doi: 10.1002/hep.23587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagoner J, Morishima C, Graf TN, Oberlies NH, Teissier E, Pécheur EI, et al. Differential In vitro effects of intravenous versus oral formulations of silibinin on the HCV life cycle and inflammation. PLoS ONE. 2011;6:e16464. doi: 10.1371/journal.pone.0016464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polyak SJ, Morishima C, Shuhart MC, Wang CC, Liu Y, Lee DYW. Inhibition of T-cell inflammatory cytokines, hepatocyte NF-[kappa] B signaling, and HCV infection by standardized silymarin. Gastroenterology. 2007;132:1925–1936. doi: 10.1053/j.gastro.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 21.Guedj J, Rong L, Dahari H, Perelson A. A perspective on modelling hepatitis C virus infection. J Viral Hepat. 2010;17:825–833. doi: 10.1111/j.1365-2893.2010.01348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 23.Adiwijaya B, Hare B, Caron P, Randle J, Neumann A, Reesink H, et al. Rapid decrease of wild-type hepatitis C virus on telaprevir treatment. Antivir Ther. 2009;14:591–595. [PubMed] [Google Scholar]
- 24.Kuhn E, Lavielle M. Maximum likelihood estimation in nonlinear mixed effects models. Comput Stat Data Anal. 2005;49:1020–1038. [Google Scholar]
- 25.Akaike H. A new look at the statistical model identification. IEEE Trans Automatic Control. 1974;19:716–723. [Google Scholar]
- 26.Dixit NM, Layden-Almer JE, Layden TJ, Perelson AS. Modelling how ribavirin improves interferon response rates in hepatitis C virus infection. Nature. 2004;432:922–924. doi: 10.1038/nature03153. [DOI] [PubMed] [Google Scholar]
- 27.Talal AH, Ribeiro RM, Powers KA, Grace M, Cullen C, Hussain M, et al. Pharmacodynamics of PEG-IFN alpha differentiate HIV/HCV coinfected sustained virological responders from nonresponders. Hepatology. 2006;43:943–953. doi: 10.1002/hep.21136. [DOI] [PubMed] [Google Scholar]
- 28.Dahari H, Affonso de Araujo ES, Haagmans BL, Layden TJ, Cotler SJ, Barone AA, et al. Pharmacodynamics of PEG-IFN-[alpha]-2a in HIV/HCV co-infected patients: Implications for treatment outcomes. J Hepatol. 2010;53:460–467. doi: 10.1016/j.jhep.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biermer M, Berg T. Rapid suppression of hepatitis C viremia induced by intravenous silibinin plus ribavirin. Gastroenterology. 2009;137:390–391. doi: 10.1053/j.gastro.2009.02.087. [DOI] [PubMed] [Google Scholar]
- 30.Bochud PY, Biber S, Negro F, Haagmans BL, Soulier A, Ferrari C, et al. IL28B polymorphisms significantly correlate with IFN antiviral effectiveness already on first day of pegylated interferon-a/ribavirin therapy of chronic HCV infection. Journal of Hepatology. in press. [Google Scholar]
- 31.Lindh M, Arnholm B, Eilard A, Färkkilä M, Hellstrand K, Lagging M, et al. Hepatitis C treatment response kinetics and impact of baseline predictors. J Viral Hepat. 2011;18:400–407. doi: 10.1111/j.1365-2893.2010.01323.x. [DOI] [PubMed] [Google Scholar]
- 32.Neumann UP, Biermer M, Eurich D, Neuhaus P, Berg T. Successful prevention of hepatitis C virus (HCV) liver graft reinfection by silibinin mono-therapy. J Hepatol. 2010;52:951–952. doi: 10.1016/j.jhep.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Beinhardt S, Rasoul-Rockenschaub S, Scherzer TM, Ferenci P. Silibinin monotherapy prevents graft infection after orthotopic liver transplantation in a patient with chronic hepatitis C. J Hepatol. 2011;54:591–592. doi: 10.1016/j.jhep.2010.09.009. author reply 592–593. [DOI] [PubMed] [Google Scholar]
- 34.Guedj J, Dahari H, Shudo E, Smith PF, Perelson AS. HCV viral kinetics with the nucleoside polymerase inhibitor RG7128 (mericitabine) Hepatology. 2011 doi: 10.1002/hep.24788. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guedj J, Perelson AS. Second-phase hepatitis C virus RNA decline during telaprevir-based therapy increases with drug effectiveness: implications for treatment duration. Hepatology. 2011;53:1801–1808. doi: 10.1002/hep.24272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guedj J, Neumann AU. Understanding hepatitis C viral dynamics with direct-acting antiviral agents due to the interplay between intracellular replication and cellular infection dynamics. J Theor Biol. 2010;267:330–340. doi: 10.1016/j.jtbi.2010.08.036. [DOI] [PubMed] [Google Scholar]
- 37.Dahari H, Shudo E, Cotler SJ, Layden TJ, Perelson AS. Modelling hepatitis C virus kinetics: the relationship between the infected cell loss rate and the final slope of viral decay. Antivir Ther. 2009;14:459–464. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.