Abstract
The ecology and physiology of ectomycorrhizal (EcM) symbiosis with conifer trees are well documented. In comparison, however, very little is known about the molecular regulation of these associations. In an earlier study, we identified three EcM-regulated Pinus expressed sequence tags (EST), two of which were identified as homologous to the Medicago truncatula nodulin MtN21. The third EST was a homologue to the receptor-like kinase Clavata1. We have characterized the expression patterns of these genes and of auxin- and mycorrhiza-regulated genes after induction with indole-3-butyric acid in Pinus sylvestris and in a time course experiment during ectomycorrhizal initiation with the co-inoculation of 2,3,5-triiodobenzoic acid, an auxin transport inhibitor. Our results suggest that different P. sylvestris nodulin homologues are associated with diverse processes in the root. The results also suggest a potential role of the Clv1-like gene in lateral root initiation by the ectomycorrhizal fungus.
Electronic supplementary material
The online version of this article (doi:10.1007/s00572-011-0402-2) contains supplementary material, which is available to authorized users.
Keywords: Auxin, IBA, TIBA, Pinus sylvestris, Laccaria bicolor, Ectomycorrhiza, Symbiosis, MtN21, Clavata1-like
Introduction
Symbiotic associations between tree roots and ectomycorrhizal fungi play important roles in boreal forests. Ectomycorrhizal fungi enhance the growth and fitness of trees by providing them with mineral and organic nutrients from the soil and by protecting the roots from pathogenic organisms in exchange for carbohydrates and organic compounds resulting from photosynthesis (Read 1991). In order to understand what characterizes the interaction between ectomycorrhizal (EcM) fungi and trees, a number of transcriptome studies has been carried out (Voiblet et al. 2001; Podila et al. 2002; Le Quere et al. 2005; Heller et al. 2008). Studies comparing the host responses to symbiotic, pathogenic, and saprotrophic interactions (Adomas et al. 2008) are also available. These studies have indicated that the number of mycorrhiza-specific genes in both partners may be relatively small. A characteristic of EcM interactions, which may explain these observations, is the morphogenetic differentiation in the plant root with the formation of a symbiotic organ in association with the fungal partner. As a result of colonization by EcM fungi, the tips of pine short roots may undergo dichotomous branching (Kaska et al. 1999). The molecular mechanisms regulating root development during interaction with the EcM fungus are still unclear (Felten et al. 2009, 2010). Phytohormones, especially auxins, play central role in EcM formation (Barker and Tagu 2000, and references therein; Jambois and Lapeyrie 2005; Felten et al. 2009). EcM fungi, e.g., Laccaria bicolor produce auxin (Ek et al. 1983) that can influence the root morphology and mycorrhization process (Gay et al. 1994). The interaction between auxin signaling and fungal volatile signals such as ethylene (Kaska et al. 1999; Splivallo et al. 2009; Felten et al. 2010) seems to be very important in this process.
In a previous study of the transcriptome in ectomycorrhizal development in Pinus sylvestris with L. bicolor, we have identified a set of expressed sequence tags (ESTs) representing Pine genes which were expressed at early stages in EcM formation (Heller et al. 2008). In the present study, we focused on three ESTs with a potential role in root morphogenesis. The first of these three ESTs showed similarity to Clavata1 receptor-like kinase (RLK) genes of Medicago truncatula and Arabidopsis (Clv1-like). Several genes with homology to Clavata1 RLK genes of Arabidopsis have been implicated in various types of organogenesis (Oka-Kira and Kawaguchi 2006; van Noorden et al. 2006). The remaining two are MtN21-like-a and MtN21-like-b clones showing similarity to an MtN21 nodulin-like gene of M. truncatula and to 5NG4, another member of the Pinus MtN21 gene family (Busov et al 2004). 5NG4 has been implicated in adventitious root formation (Busov et al 2004), suggesting that especially the MtN21-like-a and MtN21-like-b ESTs could be suitable marker genes for the onset of lateral root formation in the P. sylvestris/L. bicolor symbiosis. In the present work, we studied the expression of these candidate genes during rhizogenesis and EcM development. We also compared the transcription of the genes under indole-3-butyric acid (IBA) and auxin transport inhibitor treatments to elucidate the role of auxin and auxin signaling in their regulation.
Materials and methods
Sequences and phylogenetic analysis
The conserved regions in the amino acid sequences of Pt_MtN21-like (BG485740.1) and Pt_Clv1-like (BF517399.1) and orthologous protein sequences (Shiu et al. 2004; Schnabel et al. 2005; Morillo and Tax 2006) were aligned using ClustalW. Thereafter, neighbor-joining trees were constructed. All the sequences used in this study can be found in Electronic supplementary material S1. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 (Tamura et al. 2007). Statistical support was estimated with a bootstrap analysis of 1,000 replicates.
Biological material
Four fungal isolates were used in the study: L. bicolor (Maire) Orton 238A (Finlay et al. 1992) and Paxillus involutus Maj (Gafur et al. 2004), which are capable of forming EcM in vitro with pine and birch, respectively, Heterobasidion parviporum FS6 (Karlsson et al. 2007), and Phanerochete chrysosporium RP78 (Stewart et al. 2000). P. sylvestris (Scots pine) seeds from Svenska Skogsplantor (Saleby FP-45) were surface-sterilized with 33% hydrogen peroxide (H2O2) for 15 min, rinsed in several changes of distilled water, sown on 1% water agar Petri plates, and left to germinate under a photoperiod of 16 h light at a temperature of 18°C.
Preparation of the co-cultivation study
All fungi were cultivated on modified Melin-Norkrans (MMN) medium (Marx 1969) solidified with 1.5% agar. A 5-mm diameter agar plug was added to 15 mL of MMN medium in a 50-mL Falcon tube. After 14 days at 19°C, 14-day-old pine seedlings were transferred to tubes containing actively growing fungal colonies. Mock inoculations were prepared by transferring seedlings to 15 mL of MMN medium.
The co-cultivation experiment was incubated with a photoperiod of 16 h light at a temperature of 20°C. Three biological replicates were harvested at 0, 3, 6, 10 24, 48, and 72 h post initiation of the experiment (hpi), flash-frozen in liquid nitrogen, and stored at −80°C until processed. All manipulations in the co-cultivation experiment were performed under axenic conditions.
EcM, auxin treatment, and spatial analysis of gene expression in P. sylvestris
Two-week-old P. sylvestris seedlings were transferred under axenic conditions to a hydroponic box system and were grown for 14 days in MMN medium devoid of glucose. Systems were kept at 21°C under a 16-h photoperiod.
To quantify the basal expression of the candidate genes, 1-month-old seedlings were harvested with RNAlater (Ambion). To facilitate the infiltration of RNAlater into the tissues, a light vacuum was applied for 2 min; thereafter, seedlings were dissected in RNAlater and the fractions of root primordia, rot tips, hypocotyl, cotyledon, epicotyl, elongated root, intact shoot, and intact root were collected in two pooled representatives for each fraction.
For analysis of the effect of IBA on root morphogenesis and gene expression, P. sylvestris seedlings were treated with 100 μM IBA (Saveen Werner). Three biological replicates, each of 100 seedlings, were harvested after 2, 8, 16, and 24 hpi.
In the EcM experiment, EcM fungal inoculum (L. bicolor) was grown for 21 days in liquid Hagem medium (Stenlid 1985), washed with sterile-distilled water, and homogenized for 60 s in a sterile blender. The homogenized mycelium was then added to the box systems. For treatments with L. bicolor and the auxin transport inhibitor 2,3,5-triiodobenzoic acid (TIBA; Saveen Werner), 10 μM of TIBA was added to the box system in combination with L. bicolor. Control plants were grown with sterile MMN medium alone. Three biological replicates, each of 100 seedlings, were harvested for each treatment at 1, 5, 15, and 30 days post inoculation (dpi), flash-frozen in liquid nitrogen at harvest, and stored at −80°C until use.
In order to follow the physiological changes occurring after treatment with IBA, L. bicolor and the combination of L. bicolor and TIBA, the numbers of lateral roots, and the size of the main root were recorded for 100 seedlings per treatment over 30 days spanning the experiment and pictures were taken throughout the course of the experiment.
qRT-PCR analysis of gene transcription
Total RNA was extracted according to the methods of Chang et al. (1993) but samples less than 100 mg were homogenized directly in the extraction buffer. The final pellet was dissolved in RNAse free water. One microgram of total RNA was digested with deoxyribonuclease I (Sigma-Aldrich). The RNA samples were reverse-transcribed with MMLV reverse transcriptase (Invitrogen) or the MMLV-derived enzyme iScript (Bio-Rad). All specific primer pairs were designed using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) (Electronic supplementary material S2). The relative transcript abundance was detected using SYBR Green PCR Master Mix (Applied Biosystems) with 1 μL of template (concentration of template about 10 ng/μL) and 0.5 or 0.75 μM of each primer depending on the optimization results. The program used was as follows: initial denaturation for 1 min at 95°C, followed by a cycle of 15 s at 95°C, 1 min at 60°C, repeated 40 times with a 5 min final extension at 60°C. After the completion of the protocol, samples were subjected to a melting curve analysis.
Relative expression levels were calculated using REST software (Pfaffl et al. 2002; Vandesompele et al. 2002) using two internal reference genes: alpha-tubulin (Karlsson et al. 2007) and elongation factor 1a (Electronic supplementary material S2). The amount of product generated by qRT-PCR from control roots at each time point was used as a reference sample. In the spatial distribution experiment, the intact root and shoot were used as well to establish the relative expression levels among the fractions in the shoot and the root. The results were expressed as the fold change of messenger RNA level over the reference. The effect of time points and treatments within gene studies were tested by one-way ANOVA, and the results were confirmed with non-parametrical Wilcoxon test. The differences between the time points for every studied gene were determined by Tukey–Kramer test. The confidence level of all tests was set to 0.05. Data analysis was performed with STATISTICA 8.0 (StatSoft Inc. USA).
Results
In P. sylvestris, lateral roots emerge about 5 to 6 weeks following seed germination without the addition of any exogenous factor. We observed that 4-week-old seedlings inoculated for 15 days with a homogenate of L. bicolor displayed lateral root formation.
At 30 dpi, the roots treated with L. bicolor, L. bicolor and TIBA, or IBA displayed distinctive features. Extension growth of the main root axis was reduced in the presence of the EcM fungus, and the number of lateral roots increased compared to the control (Supplementary Material S4). At 30 dpi, these lateral roots displayed the typical dichotomous branching of EcM pine roots and possessed a mantle of fungal mycelium (Electronic supplementary material S4). When L. bicolor was inoculated together with the addition of 10 μM of TIBA, an auxin transport inhibitor, roots were shortened but no lateral roots could be observed even though the fungus was growing near the roots (Electronic supplementary material S4). P. sylvestris roots treated with IBA had excessive numbers of lateral roots, which were much shorter in length compared to control roots (Electronic supplementary material S4).
Expression of the auxin-induced genes GH3 and iaa88
The effect of IBA on the transcription levels of the genes coding for P. sylvestris GH3 and iaa88 genes led to 18- and 5-fold changes, respectively, when compared to the basal expression. Although the transcription levels were elevated throughout the experiment, they reached a significant peak at 8 h of treatment (p > 0.05; Table 1). In the presence of the EcM fungus, the patterns observed were very different. The transcript levels for GH3 were comparable to control at 5 dpi and decreased at 15 and 30 days (p < 0.05). Transcript levels for iaa88 decreased initially at day 1, and then gradually increased until day 15 (Fig. 2), after which they dropped to levels comparative to 1 dpi. Treatment with TIBA led to a transient increase of GH3 at day 1 of the interaction with L. bicolor, but did not affect the iaa88 transcription pattern (p > 0.05; Fig. 2).
Table 1.
Relative gene expression of Pinus sylvestris roots treated with indole-3-butyric acid (IBA)
| Gene | 2 h | 8 h | 16 h | 24 h |
|---|---|---|---|---|
| Fold change | Fold change | Fold change | Fold change | |
| GH3 | 4.2 (0.02)a | 17.7 (0.06)b | 13.3 (0.05)c | 9.1 (0.19)c |
| iaa88-like | 2.3 (0.04)a | 5.0 (0.18)b | 4.7 (0.11)bc | 2.5 (0.14)ac |
| MtN21-like-a | 1.7 (0.02)a | 1.1 (0.01)a | 1.6 (0.01)a | 0.3 (0.01)b |
| MtN21-like-b | 1.6 (0.02)a | 0.6 (0.01)b | 0.8 (0.03)b | 0.2 (0.01)c |
| 5NG4 | 1.0 (0.12)a | 1.6 (0.23)b | 2.5 (0.28)c | 1.4 (0.17)b |
| Clv1-like | 1.6 (0.02)a | 2.2 (0.00)b | 2.9 (0.07)b | 1.8 (0.01)ab |
Transcript levels were assessed by qPCR at 2, 8, 16, and 24 h of 100 μM IBA treatment. The numbers represent the mean fold change of transcript levels compared to control roots. Values within brackets represent the standard deviation. Different letters indicate significant difference between the time points for each analyzed gene (Tukey–Kramer test, p < 0.05)
Fig. 2.
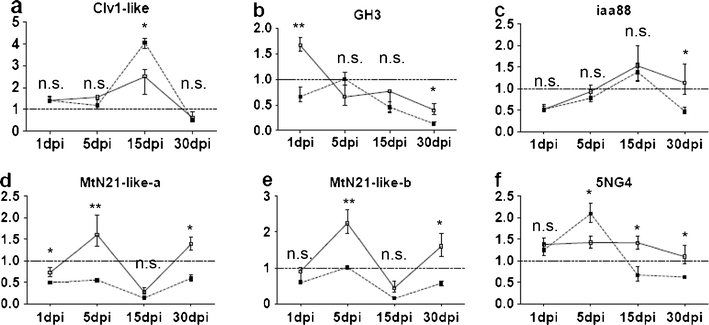
Relative gene expression of a Clv1-like, b GH3, c iaa88, d MtN21-like-a, e MtN21-like-b, and f 5NG4 in P. sylvestris roots during interaction with the EcM fungus, L. bicolor. The relative fold change of the genes was assessed by qPCR at 1, 5, 15, and 30 days post inoculation in the presence (full connectors) or absence (dashed connectors) of 2,3,5-triibenzoic acid (TIBA). The average of fold changes of three biological replicates are presented for every time point, and error bars demonstrate standard errors. Statistical difference between the treatments within every time point is shown as follows: n.s. not significant, *p < 0.05, **p < 0.01
Expression of Nodulin-like genes (MtN21-like-a/b and 5NG4)
The Pinus taeda MtN21-like-a and MtN21-like-b sequences showed a certain sequence overlap (seven amino acids, Supplementary Information S1 and S2) and they align within the same P. taeda EST contig (TC86303, DFCI Pinus Gene Indices; http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=pine). This indicates that they originated from the same P. taeda gene and consequently the PtMtN21-like gene is referred to as MtN21-like-a/b unless otherwise specified. The gene is a new member of the MtN21 gene family in P. taeda. Transcript levels of the 5NG4 and MtN21-like-a/b genes were higher in roots than the seedling as a whole in 1-month-old roots P. sylvestris seedlings (MtN21-like-a/b 6.6 times higher, p = 0.05; 5NG4 3.2 times higher, p = 0.066). There is also a higher accumulation of 5NG4 and MtN21-like-a/b transcripts in regions with protruding root primordia (45- and 85-fold increase, respectively) compared to the intact plants. Co-cultivation with a 21-day-old L. bicolor colony led to a pronounced increase in MtN21-like-a/b transcript levels (about 28 times the control) after 24 h of contact with the colony (p < 0.05; Fig. 1). P. involutus induced a small increase in MtN21-like-a/b transcript levels at 12 and 24 hpi. In contrast, the orthologous gene with similarity to Pt5NG4, responded to all fungal isolates with elevated transcript levels at 12 hpi and throughout the experiment (Fig. 1).
Fig. 1.
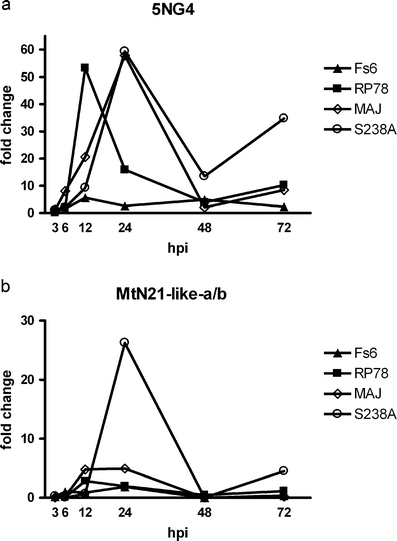
Gene expression measured by qPCR of the a 5NG4 and b MtN21-like-a/b genes in P. sylvestris roots during the interaction with 21-day-old growing colonies of H. annosum (FS6, triangles), P. chrysosporium (RP78, squares), P. involutus (MAJ, diamonds), and with L. bicolor (S238A, circles)
In the presence of homogenized L. bicolor, the expression of the MtN21-like-a/b gene was repressed compared to mock-inoculated controls (Fig. 2). The addition of TIBA resulted in an induction of the MtN21-like-a/b gene at 5 and 30 dpi compared to the treatment with L. bicolor alone (p < 0.05). The patterns of gene expression observed for the MtN21-like-a/b and 5NG4 differed after treatment with the plant growth regulator IBA (Table 1). MtN21-like-a/b showed a small upregulation at 2 h, and at 24 h, a downregulation was seen while 5NG4 showed a 2.5 fold upregulation at 16 h (p < 0.05). Transcript levels of 5NG4 seem to increase after 5 days of co-cultivation with homogenized L. bicolor and decreased slightly at 15 and 30 dpi compared to the mock-inoculated control. Treatment with TIBA inhibits this regulation (Fig. 2).
Expression of the Clavata-like gene (Clv1-like)
The Clv1-like gene appears to be a member of the LRR-XI superfamily of RLKs in plants. However, the sequence was positioned basally with respect to angiosperm LRR-XI (Electronic supplementary material S1). The gene was expressed at very low levels in P. sylvestris. In the experiment aiming to analyze the spatial distribution, the Clv1-like gene was close to the detection limit at about 10 copies at the start of the qPCR in all tissues except the root primordial, where 200–500 copies could be detected. The levels of Clv1-like gene expression remained under the detection limit in the co-cultivation experiment (data not shown). Treatment with IBA led to a relative increase in transcript level of the Clv1-like gene compared to the control at all time points during the 24-h monitoring period (p < 0.05; Table 1). In the presence of L. bicolor, the transcript levels of the Clv1-like gene rose to about 4-fold compared to the control from 5 to 15 dpi (Fig. 2). Addition of TIBA led to attenuated transcript accumulation levels at 15 dpi (p < 0.05; Fig. 2).
Discussion
We studied the expression of the candidate genes during rhizogenesis and EcM development under normal conditions as well as the influence of the auxin transport inhibitor TIBA. The application of auxin transport inhibitors has been reported to lead to local increase in auxin concentration in plant tissues through a blockage of the PIN1 cycling (Geldner et al. 2001). It has been shown that L. bicolor can induce lateral root formation in Populus tremula × Populus tremuloides and Arabidopsis thaliana even without physical contact. This induction can be completely inhibited by the auxin transport inhibitor 1-naphthylthalamic acid, showing that polar auxin transport is involved in lateral root formation in the early L. bicolor–plant interaction (Felten et al. 2009). We also observed an almost complete inhibition of L. bicolor-induced lateral root formation in our material after treatment with TIBA (Electronic supplementary material S4).
MtN21-like-a/b was identified through its upregulation during EcM formation (Heller et al. 2008), and just like the P. taeda gene 5NG4, it shared homology with the nodulin 21 gene from M. truncatula (Gamas et al. 1996). The two genes have a similar expression pattern in the untreated plant, with higher basal expression levels in roots than in shoots. However, in our system, the P. sylvestris homologues 5NG4 and MtN21-like-a/b did not respond similarly to fungal contact and colonization (Figs. 1 and 2). The 5NG4 gene is associated with adventitious root formation in P. taeda (Busov et al. 2004). Accordingly, the P. sylvestris 5NG4 transcript accumulated in root primordia. The P. taeda 5NG4 expression is induced by auxin in roots, stems, and hypocotyls (Busov et al. 2004) and the P. sylvestris homologue also responded to IBA treatment. Our results seem to emphasize the association of 5NG4 with local increases of auxin levels in developing tissues and a possible role in (adventitious) root formation as the gene responded to colonization by L. bicolor at 5 dpi, but application of TIBA inhibited the accumulation of 5NG4 transcripts at 5 dpi. The MtN21-a and MtN21-b clones correspond to a previously not studied member of the MtN21 family in Pinus. It has recently been shown that MtN21 homologues encode members of a transmembrane drug/metabolite exporter family and that the Arabidopsis mutant Walls are thin1 encoding an MtN21 exhibit defect cell elongation (Ranocha et al 2010). In Pinus, EcM colonization occurs primarily at short roots that have limited capacity to elongate, possibly because of peroxidase-mediated auxin degradation (Tarkka et al. 2001). We found that 24 h of IBA treatment and colonization with L. bicolor lead to lower steady-state levels of MtN21-like-a/b compared to untreated roots. Treatment with TIBA led to higher levels of MtN21-like-a/b expression compared to L. bicolor colonization alone. This indicates that inhibited auxin transport in roots may activate the expression of MtN21-like-a/b and that 5NG4 and MtN21-like-a/b are involved in different processes in the root, making MtN21-like-a/b and 5NG4 interesting candidates for more detailed functional and localization studies.
To validate the comparison of 5NG4 and MtN21-like-a/b expression patterns in response to L. bicolor and auxin treatment of P. sylvestris roots, the expression of two known auxin-regulated genes was also tested in our material. The upregulation of the auxin homeostasis gene GH3 in P. sylvestris in the presence of IBA and the gradual downregulation with L. bicolor colonization agree with the findings in Pinus pinaster where the transcript levels of Pp-GH3.16 decreased gradually in the presence of the EcM fungus Hebeloma cylindrosporum, but increased in auxin treatments (Reddy et al. 2006). Our results confirm that the expression of plant genes associated with EcM formation is similar between closely related plant species. This is further underlined by the observation on the expression of iaa88-like genes in EcM colonization. We could detect expression of a P. sylvestris iaa88-like sequence with primers designed on the basis of two homologous genes, Pp-iaa88 in P. pinaster and PTIAA2 in P. taeda (Charvet-Candela et al. 2002 ; Goldfarb et al. 2003). The Pp-iaa88 gene was reported to be highly expressed during fungal sheath formation (Charvet-Candela et al. 2002). Similarly, we found that the P. sylvestris iaa88-like sequence is upregulated at 15 dpi (Fig. 2), which also coincides with fungal sheath formation. TIBA application had no influence on the expression levels of P. sylvestris iaa88-like gene up to 15 dpi; however, it repressed the P. sylvestris gene significantly at 30 days, indicating that the induction of the P. sylvestris iaa88-like gene at 15 dpi is associated with the development of the symbiotic structures, supporting the concept of general EcM-associated transcription programs within the genus Pinus, possibly associated with organogenesis.
There are at least 38 ESTs with similarities to the LRR-XI superfamily of RLKs in the Pine EST databases. However, the expression pattern has been described for only one gene up to now (PsRLK, Avila et al. 2006). The LRR-XI RLK superfamily includes the well-known Clavata1 gene of Arabidopsis (Shiu and Bleecker 2001) and the M. truncatula SUNN gene involved in autoregulation of nodulation (Oka-Kira and Kawaguchi 2006). The Clv1-like amino acid sequence clusters basally among the LRR-XI RLK superfamily (Electronic supplementary material Fig. 1). In rhizobia symbiosis, SUNN controls the symbiosis, although it acts in the shoot (Oka-Kira and Kawaguchi 2006; van Noorden et al. 2006). On the basis of sequence similarity, it is tempting to suggest the possibility of an LRR-RLK-mediated autoregulatory mechanism in EcM formation as well, but the expression pattern of the Clv1-like gene suggests that the gene product is involved in other processes compared to other LRR-RLK members, which are active in aerial tissues. The Clv1-like gene showed higher expression levels in roots and root primordia compared to aerial tissues. The Clv1-like gene was induced by L. bicolor at 15 dpi in the EcM experiment, coinciding with lateral root and fungal sheath formation (Heller et al. 2008). The role of the Clv1-like gene in lateral root formation and fungal sheath formation deserves further exploration in future studies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Sequences used in the MtN21 and Clv1 neighbor-joining analyses (DOC 55 kb)
a Alignment with TC86303 (TC86303, DFCI Pinus Gene Indices) showing the seven amino acid sequence overlap between the Pinus taeda MtN21-like-a and MtN21-like-b ESTs indicating that they originate from the same P. taeda gene. Neighbor-joining trees of b the Pinus taeda MtN21-like-a/b (Ptnod21) and c Clv1-like (PtCLV1 orf) amino acid sequences. The analyses were conducted using MEGA version 4, statistical support was estimated with a bootstrap analysis of 1,000 replicates (JPEG 167 kb)
qPCR primers used in the study (XLS 18 kb)
Morphology of Pinus sylvestris roots after 30 days of treatment with a control, b Laccaria bicolor, c Laccaria bicolor and 10 μM of TIBA, and d 100 μM of IBA photographed in the dissecting microscope (JPEG 69 kb)
Acknowledgments
We thank Professor Anders Tunlid, Lund University, Lund, Sweden for the generous gift of P. involutus MAJ and Professor Jill Tuskan Gaskell, USDA Forest Service Forest Products Laboratory for the generous gift of P. chrysosporium RP78. This work was supported by grants from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS), the Swedish Science Research Council (VR), Carl Tryggers Stiftelse (CTS), and by the Swedish Organization for International Co-operation in Research and Higher Education (STINT).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Adomas A, Heller G, Olson Å, Osborne J, Karlsson M, Nahalkova J, Van Zyl L, Sederoff R, Stenlid J, Finlay R, Asiegbu FO. Comparative analysis of transcript abundance in Pinus sylvestris after challenge with a saprotrophic, pathogenic or mutualistic fungus. Tree Physiol. 2008;28:885–897. doi: 10.1093/treephys/28.6.885. [DOI] [PubMed] [Google Scholar]
- Avila C, Perez-Rodriguez J, Canovas FM. Molecular characterization of a receptor kinasae gene from pine (Pinus sylvestris L.) Planta. 2006;224:12–19. doi: 10.1007/s00425-005-0184-x. [DOI] [PubMed] [Google Scholar]
- Barker SJ, Tagu D. The roles of auxins and cytokinins in mycorrhizal symbioses. J Plant Growth Regul. 2000;19:144–154. doi: 10.1007/s003440000021. [DOI] [PubMed] [Google Scholar]
- Busov VB, Johannes E, Whetten RW, Sederoff RR, Spiker SL, Lanz-Garcia C, Goldfarb B. An auxin-inducible gene from loblolly pine (Pinus taeda L.) is differentially expressed in mature and juvenile-phase shoots and encodes a putative transmembrane protein. Planta. 2004;218:916–927. doi: 10.1007/s00425-003-1175-4. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. doi: 10.1007/BF02670468. [DOI] [Google Scholar]
- Charvet-Candela V, Hitchin S, Ernst D, Sandermann H, Marmeisse R, Gay G. Characterization of an Aux/IAA cDNA upregulated in Pinus pinaster roots in response to colonization by the ectomycorrhizal fungus Hebeloma cylindrosporum. New Phytol. 2002;154:769–777. doi: 10.1046/j.1469-8137.2002.00423.x. [DOI] [PubMed] [Google Scholar]
- Ek M, Ljungquist PO, Stenström E. Indole-3-acetic acid production by mycorrhizal fungi determined by gas chromatography–mass spectrometry. New Phytol. 1983;94:401–407. doi: 10.1111/j.1469-8137.1983.tb03454.x. [DOI] [Google Scholar]
- Felten J, Kohler A, Morin E, Bhalerao RP, Palme K, Martin F, Ditengou FA, Legué V. The ectomycorrhizal fungus Laccaria bicolor stimulates lateral root formation in poplar and Arabidopsis through auxin transport and signalling. Plant Physiol. 2009;151:1991–2005. doi: 10.1104/pp.109.147231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felten J, Legué V, Ditengou FA. Lateral root stimulation in the early interaction between Arabidopsis thaliana and the ectomycorrhizal fungus Laccaria bicolor. Is fungal auxin the trigger? Plant Signalling and Behavior. 2010;5(7):864–867. doi: 10.4161/psb.5.7.11896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay RD, Frostegård Å, Sonnerfelt A-M. Utilization of organic and inorganic nitrogen sources by ectomycorrhizal fungi of different successional stages grown in pure culture and in symbiosis with Pinus contorta (Dougl. ex Loud) New Phytol. 1992;120:105–115. doi: 10.1111/j.1469-8137.1992.tb01063.x. [DOI] [Google Scholar]
- Gamas P, De Carvalho NF, Lescure N, Cullimore JV. Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol Plant Microbe Interact. 1996;9:233–242. doi: 10.1094/MPMI-9-0233. [DOI] [PubMed] [Google Scholar]
- Gafur A, Schützendübel A, Langenfeld-Heyser R, Fritz E, Polle A. Compatible and Incompetent Paxillus involutus Isolates for Ectomycorrhiza Formation in vitro with Poplar (Populus×canescens) Differ in H2O2. Plant Biol. 2004;6:91–99. doi: 10.1055/s-2003-44718. [DOI] [PubMed] [Google Scholar]
- Gay G, Normand L, Marmeisse R, Sotta B, Debaud JC. Auxin overproducer mutants of Hebeloma cylindrosporum Romagnesi have increased mycorrhizal activity. New Phytol. 1994;128:645–657. doi: 10.1111/j.1469-8137.1994.tb04029.x. [DOI] [Google Scholar]
- Geldner N, Friml J, Stierhof Y-D, Jürgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413:425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- Goldfarb B, Lanz-Garcia C, Lian Z, Whetten R. Aux/IAA gene family is conserved in the gymnosperm, loblolly pine (Pinus taeda L.) Tree Physiol. 2003;23:1181–1192. doi: 10.1093/treephys/23.17.1181. [DOI] [PubMed] [Google Scholar]
- Heller G, Adomas A, Li G, Osborne J, van Zyl L, Sederoff R, Finlay RD, Stenlid J, Asiegbu FO. Transcriptional analysis of Pinus sylvestris roots challenged with the ectomycorrhizal fungus Laccaria bicolor. BMC Plant Biol. 2008;8:19. doi: 10.1186/1471-2229-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambois A, Lapeyrie F. Jasmonates, together with zeatin, induce hypaphorine accumulation by the ectomycorrhizal fungus Pisolithus microcarpus. Symbiosis. 2005;39:137–141. [Google Scholar]
- Karlsson M, Hietala AM, Kvaalen H, Solheim H, Olson Å, Stenlid J, Fossdal CG. Quantification of host and pathogen DNA and RNA transcripts in the interaction of Norway spruce with Heterobasidion annosum. Physiol Mol Plant Pathol. 2007;70:99–109. doi: 10.1016/j.pmpp.2007.07.006. [DOI] [Google Scholar]
- Kaska DD, Myllyla R, Cooper JB. Auxin transport inhibitors act through ethylene to regulate dichotomous branching of lateral root meristems in pine. New Phytol. 1999;142:49–58. doi: 10.1046/j.1469-8137.1999.00379.x. [DOI] [Google Scholar]
- Le Quere A, Wright DP, Soderstrom B, Tunlid A, Johansson T. Global patterns of gene regulation associated with the development of ectomycorrhiza between birch (Betula pendula Roth.) and Paxillus involutus (Batsch) fr. Mol Plant Microbe Interact. 2005;18:659–673. doi: 10.1094/MPMI-18-0659. [DOI] [PubMed] [Google Scholar]
- Marx D. The influence of ectotrophic fungi on the resistance of pine roots to pathogenic infection. Phytopathology. 1969;59:153–163. [PubMed] [Google Scholar]
- Morillo SA, Tax FE. Functional analysis of receptor-like kinases in monocots and dicots. Curr Opin Plant Biol. 2006;9:460–469. doi: 10.1016/j.pbi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Oka-Kira E, Kawaguchi M. Long-distance signaling to control root nodule number. Curr Opin Plant Biol. 2006;9:496–502. doi: 10.1016/j.pbi.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podila GK, Zheng J, Balasubramanian S, Sundaram S, Hiremath S, Brand JH, Hymes MJ. Fungal gene expression in early symbiotic interactions between Laccaria bicolor and red pine. Plant and Soil. 2002;244:117–128. doi: 10.1023/A:1020275330363. [DOI] [Google Scholar]
- Ranocha P, Denancé N, Vanholme R, Freydier A, Martinez Y, Hoffmann L, Köhler L, Pouzet C, Renou JP, Sundberg B, Boerjan W, Goffner D. Walls are thin 1 (WAT1), an Arabidopsis homolog of Medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers. Plant J. 2010;63:469–483. doi: 10.1111/j.1365-313X.2010.04256.x. [DOI] [PubMed] [Google Scholar]
- Read DJ. Mycorrhizas in ecosystems. Experientia. 1991;47:376–391. doi: 10.1007/BF01972080. [DOI] [Google Scholar]
- Reddy SM, Hitchin S, Melayah D, Pandey AK, Raffier C, Henderson J, Marmeisse R, Gay G. The auxin-inducible GH3 homologue Pp-GH3.16 is downregulated in Pinus pinaster root systems on ectomycorrhizal symbiosis establishment. New Phytol. 2006;170:391–400. doi: 10.1111/j.1469-8137.2006.01677.x. [DOI] [PubMed] [Google Scholar]
- Schnabel E, Journet EP, De Carvalho-Niebel F, Duc G, Frugoli J. The Medicago truncatula SUNN gene encodes a Clv1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol Biol. 2005;58:809–822. doi: 10.1007/s11103-005-8102-y. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci U S A. 2001;98(19):10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KFX, Li WH. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splivallo R, Fischer U, Göbel C, Feussner I, Karlovsky P. Truffles regulate plant root morphogenesis via the production of auxin and ethylene. Plant Physiol. 2009;150:2018–2029. doi: 10.1104/pp.109.141325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenlid J. Population structure of Heterobasidion parviporum as determined by somatic incompatibility, sexual incompatibility and isozyme patterns. Can J Bot. 1985;63:2268–2273. doi: 10.1139/b85-322. [DOI] [Google Scholar]
- Stewart P, Gaskell J, Cullen D. A homokaryotic derivative of a Phanerochaete chrysosporium strain and its use in genomic analysis of repetitive elements. Appl Environ Microbiol. 2000;66:1629–1633. doi: 10.1128/AEM.66.4.1629-1633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tarkka MT, Nyman TA, Kalkkinen N, Raudaskoski M. Scots pine expresses short-root-specific peroxidases during development. Eur J Biochem. 2001;268:86–92. doi: 10.1046/j.1432-1327.2001.01853.x. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):research0034.1–0034.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noorden G, Ross JJ, Reid JB, Rolfe BG, Mathesius U. Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol. 2006;140:1494–1506. doi: 10.1104/pp.105.075879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiblet C, Duplessis S, Encelot N, Martin F. Identification of symbiosis-regulated genes in Eucalyptus globules–Pisolithus tinctorius ectomycorrhiza by differential hybridization of arrayed cDNAs. Plant J. 2001;25:181–191. doi: 10.1046/j.1365-313x.2001.00953.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences used in the MtN21 and Clv1 neighbor-joining analyses (DOC 55 kb)
a Alignment with TC86303 (TC86303, DFCI Pinus Gene Indices) showing the seven amino acid sequence overlap between the Pinus taeda MtN21-like-a and MtN21-like-b ESTs indicating that they originate from the same P. taeda gene. Neighbor-joining trees of b the Pinus taeda MtN21-like-a/b (Ptnod21) and c Clv1-like (PtCLV1 orf) amino acid sequences. The analyses were conducted using MEGA version 4, statistical support was estimated with a bootstrap analysis of 1,000 replicates (JPEG 167 kb)
qPCR primers used in the study (XLS 18 kb)
Morphology of Pinus sylvestris roots after 30 days of treatment with a control, b Laccaria bicolor, c Laccaria bicolor and 10 μM of TIBA, and d 100 μM of IBA photographed in the dissecting microscope (JPEG 69 kb)


