Abstract
Protein solubility is a problem for many protein chemists, including structural biologists and developers of protein pharmaceuticals. Knowledge about how intrinsic factors influence solubility is limited due to the difficulty of obtaining quantitative solubility measurements. Solubility measurements in buffer alone are difficult to reproduce, because gels or supersaturated solutions often form, making it impossible to determine solubility values for many proteins. Protein precipitants can be used to obtain comparative solubility measurements and, in some cases, estimations of solubility in buffer alone. Protein precipitants fall into three broad classes: salts, long-chain polymers, and organic solvents. Here, we compare the use of representatives from two classes of precipitants, ammonium sulfate and polyethylene glycol 8000, by measuring the solubility of seven proteins. We find that increased negative surface charge correlates strongly with increased protein solubility and may be due to strong binding of water by the acidic amino acids. We also find that the solubility results obtained for the two different precipitants agree closely with each other, suggesting that the two precipitants probe similar properties that are relevant to solubility in buffer alone.
Introduction
Protein solubility is important to structural biologists (1), the pharmaceutical industry (2), and all scientists who work with protein in solution. Structural studies (1,3,4) and pharmaceutical applications (2,5,6) often require very-high-concentration protein samples. The solubility of a protein in aqueous solution varies from almost completely insoluble to hundreds of milligrams per milliliter. For instance, crambin has been reported to be completely insoluble in water (7), and serum albumins have solubilities of >500 mg/mL (8). Low protein solubility has also been implicated in a number of human diseases (9–12). The P23T mutation in human γD-crystallin shows a markedly decreased solubility and leads to childhood onset of cataracts (10). Therefore, understanding the factors that contribute to protein solubility is an important area of research.
Protein solubility is a thermodynamic parameter defined as the concentration of protein in a saturated solution that is in equilibrium with a solid phase, either crystalline or amorphous, under a given set of conditions (13,14). Solubility can be influenced by a number of extrinsic and intrinsic factors. Extrinsic factors that influence protein solubility include pH, ionic strength, temperature, and the presence of various solvent additives (3). Varying these extrinsic factors can lead to increased solubility (1,4); however, altering the solution conditions is not always appropriate or sufficient to increase protein solubility to the extent required. The intrinsic factors that influence protein solubility are defined primarily by the amino acids on the protein surface, but a detailed understanding of how one can alter the intrinsic properties of a protein to increase its solubility is lacking (1,4,15). Recently, our laboratory has taken steps toward elucidating the intrinsic factors that influence protein solubility, with the goal of developing a strategy for making mutations that increase protein solubility (16,17).
One of the reasons for the lack of understanding of intrinsic protein solubility is the difficulty of obtaining quantitative solubility measurements. Two methods used to measure protein solubility in aqueous solution are: 1), adding lyophilized protein to solvent; and 2), concentrating a protein solution by ultrafiltration. Both of these methods require that the concentration of protein in solution be increased until saturation is reached; however, this is often difficult to do, especially with very soluble proteins, because gel-like or supersaturated solutions may form, making it difficult to determine the solubility values accurately (15). When lyophilized protein is added to solvent, the variable water and salt content of the lyophilized powder is difficult to control and can have a significant effect on solubility measurements (15).
One way to avoid the difficulties of measuring protein solubility is to make use of an extraneous agent that lowers the solubility of a protein called a precipitant. Protein precipitants can be divided into three classes: salts, organic solvents, and long-chain polymers (18). These precipitants are used by crystallographers to achieve slow precipitation and crystal formation; however, they can also be used to induce amorphous precipitation by direct mixing with protein solutions (16,17,19–25). Common examples from each of these three classes of precipitants, respectively, are ammonium sulfate, isopropanol, and polyethylene glycol (PEG). The relationship between precipitant concentration to protein solubility is described by the following general expression (13,19):
| (1) |
where S is the measured solubility at a given concentration of precipitant, and β is the dependence of solubility on precipitant concentration for a given protein. The constant is the y-intercept of the solubility plot, and for PEG precipitations is equal to the logarithm of the protein activity. For dilute protein solutions, as the activity constant approaches one, Eq. 1 becomes
| (2) |
where S0 is the solubility in the absence of precipitant. Middaugh et al. (19) showed that for PEG precipitations, the linearity of Eq. 1 extends to zero precipitant for proteins whose solubility can be accurately measured in buffer alone. In this case, the constant portion of Eq. 2, Log S0, can be used as an estimate of solubility in the absence of precipitant. For salts, Eq. 1 only describes the salting-out region of the solubility plot. At low salt concentrations, salting-in is observed and the solubility is higher than in the absence of salt. Therefore, the constant obtained from salt precipitations represents a projection of the salting-out region onto the y axis. A relationship between this constant and protein solubility has not been established for salt precipitations and will be investigated in this study.
Each class of precipitants decreases protein solubility by a different mechanism. For salts, kosmotropic ions bind water more tightly than water binds itself (26) and the surface tension of the solution increases, effectively competing with the surface of the protein for water molecules for hydration. As less water becomes available to hydrate the protein surface, the protein molecules self-associate and precipitate (13). Organic solvents, such as alcohols, lower the dielectric constant of the solution. As the dielectric constant decreases, the solution becomes a poorer solvent for the protein. Consequently, the relative favorability of protein-protein interactions increases and the protein precipitates (18). By the same mechanism, organic solvents decrease the strength of hydrophobic interactions, leading to decreased protein stability (27). Special care must be taken to ensure that the protein remains folded under experimental conditions if organic solvents are used as precipitants. Long-chain polymers occupy more space in solution than does a protein of similar molecular mass and lower the solubility of a protein through an excluded volume mechanism (25,28), effectively crowding the protein out of solution. Due to the different mechanisms of action employed by each class of precipitants, the results obtained with one precipitant may not coincide with results obtained with another precipitant, and may relate differently to solubility in the absence of precipitant. Because of this limitation and the absence of a quantitative relationship between solubility in the absence and presence of precipitant, solubility measurements are best used in a comparative manner. Our purpose in this study was twofold: 1), to compare the solubility results obtained with two different classes of precipitants (i.e., salts (ammonium sulfate) and long-chain polymers (PEG-8000)) for a number of different proteins; and 2), to examine the molecular properties of the proteins used in this study with the hope of gaining a better understanding of the solubility results obtained and insight into the intrinsic factors that influence protein solubility.
Materials and Methods
For details about the materials and methods used in this work, see the Supporting Material.
Results
Proteins are folded under experimental conditions
For this study, we are interested in examining the solubility of folded proteins. The low solubility of the unfolded state stands as a challenge for those studying the denatured-state ensemble; however, we chose to focus on the solubility of the native state because of its relevance to crystallographers, protein chemists, and developers of protein pharmaceuticals (15). The precipitants used in this study are common crystallographic precipitants; if the protein is folded in solution, the precipitate is expected to be native protein (18,41). To confirm this, we used thermal unfolding experiments to examine the effect of ammonium sulfate and PEG-8000 on stability. Originally, this study was designed to include isopropanol as a precipitant. Thomas and Dill (27) investigated the mechanism by which alcohols destabilize proteins and found that it was complex and dependent on protein sequence and structure. They concluded that alcohols destabilize proteins mainly by weakening hydrophobic interactions. We found that the concentration of isopropanol required to achieve precipitation for many of the proteins in this study was great enough that a mixture of folded and unfolded protein would be present under experimental conditions (R. Kramer, C. Pace, and J. Scholtz, unpublished data), and therefore we excluded the class of organic solvents from this study. The use of isopropanol or other denaturing organic solvents for studying protein solubility should be reserved for proteins that remain folded in the presence of solvent concentrations necessary to induce precipitation. The temperatures at which precipitations by organic solvents are performed can be lowered to diminish the denaturing effects.
Figs. 1 and 2 show the change in observed melting temperature, Tm, as a function of ammonium sulfate and PEG-8000, respectively. For ammonium sulfate, the Tm increases for all of the proteins studied. This is to be expected because sulfate ions increase the surface tension of bulk water, and the folded state is favored due to the reduced water-protein interface as compared with the unfolded state (42,43). For example, the degree of stabilization at 1M ammonium sulfate varies from an increase in Tm of 5.2°C for human serum albumin to an increase of 14.3°C observed for α-lactalbumin. One might propose that the amount of surface buried and the degree of unfolding influence the level of stabilization for an individual protein; however, in this situation, that does not appear to be the case. The protein with the greatest degree of stabilization by ammonium sulfate is α-lactalbumin. It forms a molten globule upon unfolding (44), and likely buries less surface area than proteins that more fully unfold. This differential stabilization by ammonium sulfate deserves further investigation than can be accomplished here. For the purposes of this study, it is sufficient to say that all of the proteins used here are stabilized by ammonium sulfate and are folded under experimental conditions. Furthermore, it has been shown that upon precipitation by ammonium sulfate, the protein in both solution and solid phase remains folded (41).
Figure 1.
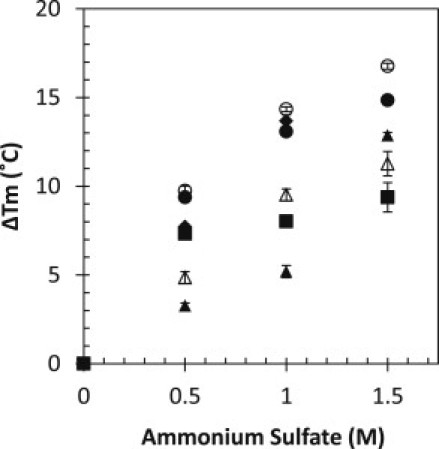
Change in Tm as a function of ammonium sulfate concentration for RNase Sa (solid diamonds), α-chymotrypsin (solid circles), lysozyme (open triangles), human serum albumin (solid triangles), ovalbumin (solid squares), and α-lactalbumin (open circles). Thermal denaturation was followed by circular dichroism (see Supporting Material for details).
Figure 2.
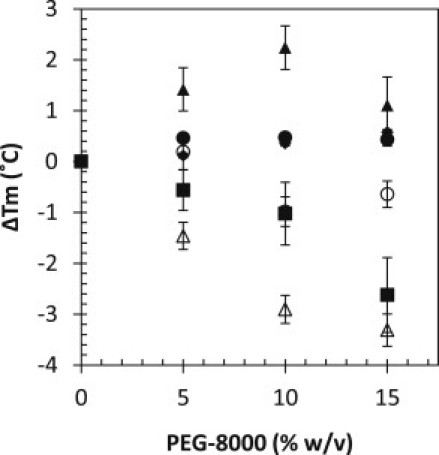
Change in Tm as a function of PEG for RNase Sa (solid diamonds), α-chymotrypsin (solid circles), lysozyme (open triangles), human serum albumin (solid triangles), ovalbumin (solid squares), and α-lactalbumin (open circles). Thermal denaturation was followed by circular dichroism (see Supporting Material for details).
In contrast to ammonium sulfate, high-molecular-mass PEGs are not expected to interact with or have a significant effect on protein stability (25,28); however, at high concentrations they may destabilize some proteins (14), and PEG molecules with a molecular mass of ≤6000 Da have been shown to destabilize some proteins (45). Fig. 2 shows that our data agree with those previous observations. The effect of PEG-8000 on stability is small relative to the effect of ammonium sulfate, although in the case of lysozyme and ovalbumin, we observe a slight decrease in stability. For example, the effect of PEG-8000 on Tm for all proteins ranges from an increase in Tm of 2.2°C for human serum albumin to a decrease in Tm of 2.9°C for lysozyme at 10% (w/v) PEG-8000. For the two proteins that show a decrease in stability, the decrease is not enough to cause a significant change in the population of unfolded protein present at room temperature. The Tm-values for lysozyme and ovalbumin in the absence of precipitant are 70.5°C and 70.8°C, respectively (data not shown), and the maximum concentration of PEG-8000 used is <15% (w/v) for these two proteins. As previously reported (46), fibrinogen exhibits a complicated multistate unfolding curve that is not amenable to this type of analysis; however, the curves clearly show that fibrinogen is folded under the conditions used in this study (data not shown). In conclusion, the proteins used in this study are folded under experimental conditions in the presence of either ammonium sulfate or PEG-8000.
Solubility measurements rapidly reach equilibrium
Because solubility values are defined at equilibrium, it is necessary to ensure that samples are allowed adequate time to equilibrate. For all proteins used in this study, samples were prepared in duplicate and were either centrifuged and quantified immediately or were allowed to sit for 24 h before centrifugation. Measurements obtained immediately and at 24 h yielded the same result (Table 1), signifying that protein precipitations reach equilibrium quickly after precipitation by either PEG-8000 or ammonium sulfate. This was seen for all protein and precipitant combinations tested in this study, and agrees with previously reported results for amorphous protein precipitation (16,47). Furthermore, we observed that precipitation could be reversed by the addition of buffer lacking precipitant (data not shown).
Table 1.
Dependence of solubility measurements on equilibration time
| Protein | Precipitant | Equilibration time | Solubility (mg/mL) |
|---|---|---|---|
| α-Chymotrypsin | 15% PEG-8000 | 10 min | 5.8 ± 0.5 |
| 24 h | 5.7 ± 0.2 | ||
| 2 M ammonium sulfate | 10 min | 31.9 ± 0.8 | |
| 24 h | 31.7 ± 0.8 | ||
| Lysozyme | 5% PEG-8000 | 10 min | 14.1 ± 0.2 |
| 24 h | 14.1 ± 0.1 | ||
| 1.5 M ammonium sulfate | 10 min | 18.2 ± 0.7 | |
| 24 h | 17.7 ± 0.4 | ||
| Serum albumin | 20% PEG-8000 | 10 min | 9.8 ± 0.2 |
| 24 h | 10.1 ± 0.2 | ||
| 2.5 M ammonium sulfate | 10 min | 8.9 ± 0.3 | |
| 24 h | 9.0 ± 0.6 | ||
| RNase Sa | 10% PEG-8000 | 10 min | 17.3 ± 0.6 |
| 24 h | 17.2 ± 0.2 | ||
| 1.5 M ammonium sulfate | 10 min | 8.9 ± 0.3 | |
| 24 h | 8.9 ± 0.4 | ||
| Ovalbumin | 7.5% PEG-8000 | 10 min | 22.3 ± 2.1 |
| 24 h | 22.2 ± 1.6 | ||
| 2 M ammonium sulfate | 10 min | 16.8 ± 0.8 | |
| 24 h | 16.7 ± 0.6 | ||
| α-Lactalbumin | 25% PEG-8000 | 10 min | 7.8 ± 1.2 |
| 24 h | 8.0 ± 0.7 | ||
| 1.5 M ammonium sulfate | 10 min | 22.2 ± 0.1 | |
| 24 h | 22.5 ± 1.3 | ||
| Fibrinogen | 2.5% PEG-8000 | 10 min | 4.5 ± 0.2 |
| 24 h | 4.3 ± 0.3 | ||
| 0.7 M ammonium sulfate | 10 min | 3.3 ± 0.2 | |
| 24 h | 3.4 ± 0.2 |
Experiments were performed in pH 7.0 5 mM citrate buffer.
Solubility curves in PEG
The solubility of seven proteins was determined as a function of PEG-8000. Fig. 3 shows the plot of log solubility as a function of PEG-8000 concentration. Equation 3 was fit to the data, and linear fit parameters are given in Table 2 (left side). The dependence of solubility on PEG-8000 concentration (β) varies over a 40-fold range from −0.01 for α-chymotrypsin to −0.42 for fibrinogen. As reported previously (19), PEG precipitation curves are linear over a wide range of PEG concentrations and can be extrapolated to zero concentration of PEG, yielding an estimate of solubility in the absence of precipitant from the y-intercept (log S0) of the fit. The relative solubility of the proteins is determined by the log S0-values, which vary between 0.9 for fibrinogen and 4.2 for α-lactalbumin. The log S0-values indicate that human serum albumin and α-lactalbumin are the most soluble of the proteins, which makes sense given that they are present at high concentrations in their respective biological environments (human plasma (48) and bovine milk (49), respectively).
Figure 3.
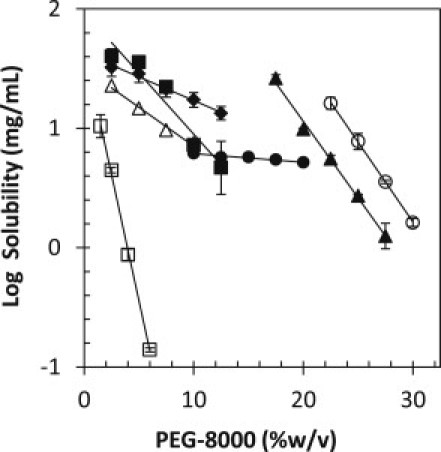
Solubility of several proteins in PEG-8000. The solubility of RNase Sa (solid diamonds), α-chymotrypsin (solid circles), lysozyme (open triangles), human serum albumin (solid triangles), ovalbumin (solid squares), α-lactalbumin (open circles), and fibrinogen (open squares) was measured at room temperature (25°C) and in pH 7.0 citrate. Equation 2 was fit to the data, and the fitted parameters can be found in Table 2.
Table 2.
Parameters obtained for PEG-8000 and ammonium sulfate solubility curves
| Protein‡ | PEG-8000†,§ |
Ammonium sulfate†,¶ |
||||
|---|---|---|---|---|---|---|
| Slope (β) | Intercept (log S0) | R2 | Slope (β) | Intercept (log S0∗) | R2 | |
| α-Chymotrypsin | −0.01 | 0.9 | 0.92 | −0.92 | 2.5 | 1.00 |
| Lysozyme | −0.07 | 1.5 | 0.99 | −1.49 | 3.5 | 0.99 |
| Human serum albumin | −0.13 | 3.6 | 0.99 | −2.68 | 7.5 | 0.97 |
| RNase Sa | −0.04 | 1.6 | 0.99 | −1.63 | 3.4 | 1.00 |
| α-Lactalbumin | −0.13 | 4.2 | 1.00 | −0.17 | 1.6 | 0.98 |
| Fibrinogen | −0.42 | 1.7 | 1.00 | −4.13 | 3.5 | 0.98 |
| Ovalbumin | −0.10 | 2.0 | 0.93 | −2.46 | 6.0 | 0.97 |
Equation 2 was fit to the data from Figs. 3 and 4, respectively.
Protein concentration is expressed in units of mg/mL.
PEG concentration is express in units of % w/v.
Ammonium sulfate concentration is expressed in units of M.
Solubility curves in ammonium sulfate
The solubility of the proteins was determined as a function of ammonium sulfate concentration. Fig. 4 shows the plot of log solubility versus ammonium sulfate concentration. The data were similarly fit by Eq. 3, and the best-fit values are given in Table 2 (right side). The linearity of a salting-out curve does not extend to the y axis due to salting-in at low concentrations of salt, so log S0∗ will be used in place of log S0 to signify that the y-intercept is projected from the salting-out region. The dependence of solubility on ammonium sulfate concentration (β) was found to be similar for all proteins except α-lactalbumin. For those six proteins, β varies only ∼4-fold from −0.92 for α-chymotrypsin to −4.13 for fibrinogen compared with β-values for PEG-8000, which vary over a 40-fold range. Clearly, the dependence of solubility on precipitant concentration is more variable for PEG-8000 than for ammonium sulfate. α-Lactalbumin is a clear outlier, with a β-value of −0.17 that is 13-fold lower than the average β-value and fivefold lower than the next-closest β-value of −0.92. This suggests that the ability of ammonium sulfate to precipitate α-lactalbumin is reduced in comparison with the other six proteins. The case of α-lactalbumin will be addressed further in the Discussion. Based on the log S0∗-values, human serum albumin is still predicted to have a high solubility, and α-chymotrypsin is still predicted to have a low solubility, as seen with the results for PEG-8000. Of interest, α-lactalbumin, which was predicted to have the highest solubility by PEG-8000, is now predicted to have the lowest solubility.
Figure 4.
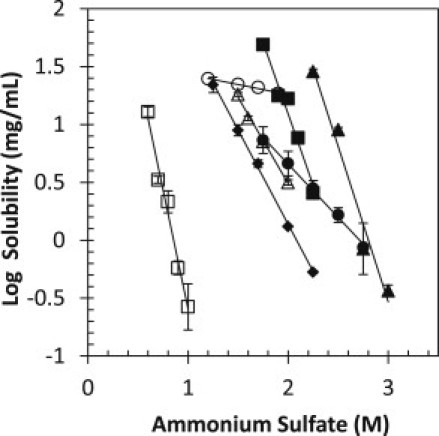
Solubility of several proteins in ammonium sulfate. The solubility of RNase Sa (solid diamonds), α-chymotrypsin (solid circles), lysozyme (open triangles), human serum albumin (solid triangles), ovalbumin (solid squares), α-lactalbumin (open circles), and fibrinogen (open squares) was measured at room temperature (25°C) and in pH 7.0 citrate. Equation 2 was fit to the data, and the fitted parameters can be found in Table 2.
Discussion
Comparing protein solubility in PEG-8000 and ammonium sulfate
To compare the solubility measured using PEG-8000 with that measured using ammonium sulfate, we evaluated the log S0-values from the two fits. Fig. 5 shows the plot of log S0∗ obtained with ammonium sulfate (log S0∗ (NH4)2SO4) versus log S0 obtained with PEG-8000 (log S0 PEG). A remarkably strong correlation between the solubility results for ammonium sulfate and those for PEG-8000 is seen. This suggests that log S0∗ (NH4)2SO4 is a parameter that is related to protein solubility in the absence of precipitant. Because log S0 PEG can be used to estimate solubility in the absence of buffer, the correlation of log S0 PEG with log S0∗ (NH4)2SO4 suggests that log S0 (NH4)2SO4 can be used qualitatively to determine differences in solubility.
Figure 5.
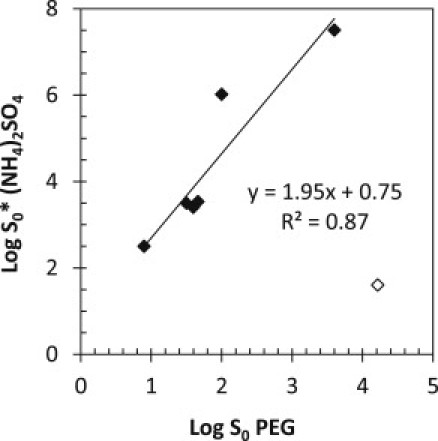
Comparison of the solubility data obtained in ammonium sulfate and PEG-8000. Log S0-values obtained from PEG-8000 precipitations are plotted against Log S0∗-values from ammonium sulfate precipitations for all of the proteins. The data correlate strongly, suggesting a relationship between solubility results obtained with PEG-8000 and ammonium sulfate. α-Lactalbumin is shown as an open diamond and is excluded from the fit.
The solubility of α-lactalbumin warrants further discussion. In the case of PEG precipitation, α-lactalbumin is predicted to have the highest solubility of the proteins used in this study. This is not surprising given that α-lactalbumin is present in high concentrations in bovine milk (49) and the fact that we are able to make stock concentrations of α-lactalbumin that are in excess of 100 mg/mL. α-Lactalbumin has a β-value in PEG-8000 that is intermediate of the slopes observed for the other proteins. In the case of ammonium sulfate, α-lactalbumin is predicted to have the lowest solubility among the proteins studied, and the slope observed with α-lactalbumin is a clear outlier. It is the smallest slope observed: 13-fold lower than the average slope, and fivefold lower than the next-closest slope in ammonium sulfate. This suggests that ammonium sulfate is not as effective as a precipitant for α-lactalbumin as it is for the other proteins. This may be due in part to the high surface charge on α-lactalbumin: two thirds of the exposed surface residues carry a charge at pH 7 (data not shown) and nearly a third of the accessible surface area is charged (see Table 3, column 9). We previously suggested that ammonium sulfate may underestimate the contribution of charged surface residues to protein solubility (16). This likely is related to the mechanism by which ammonium sulfate lowers protein solubility (i.e., increasing surface tension and competing for waters with the protein surface). The high level of charged surface area on α-lactalbumin (roughly equal amounts positive and negative) likely affects the ability of ammonium sulfate to act as a precipitant. The kosmotropic carboxylates on the protein surface compete strongly for water molecules with the sulfate ions, and the chaotropic amino and guanidino groups may lower the water surface tension at the protein water interface, partially opposing the effect of ammonium sulfate. Due to the unique nature of the salting-out curve of α-lactalbumin, the ammonium sulfate data were fit without α-lactalbumin in subsequent correlations.
Table 3.
Protein properties and surface properties used for correlations
| Protein | Molecular mass (kDa) | Amino acids | pI∗ | Charge∗ | Absolute charge∗ | Fraction of ASA† |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nonpolar | Polar | Charged | Positive | Negative | ||||||
| A-chymotrypsin | 25.2 | 241 | 8 | 3.1 | 3.1 | 0.52 | 0.48 | 0.18 | 0.12 | 0.06 |
| Lysozyme | 14.3 | 129 | 10 | 7.9 | 7.9 | 0.48 | 0.52 | 0.23 | 0.18 | 0.05 |
| Human serum albumin | 66.5 | 585 | 6 | −12.2 | 12.2 | 0.59 | 0.41 | 0.27 | 0.12 | 0.14 |
| RNaseSa | 10.5 | 96 | 3.5 | −6.6 | 6.6 | 0.56 | 0.44 | 0.14 | 0.06 | 0.08 |
| α-Lacalbumin | 14.2 | 123 | 5 | −6.6 | 6.6 | 0.50 | 0.50 | 0.30 | 0.14 | 0.15 |
| Fibrinogen | 160 | 1424 | 6.8 | −3.7 | 3.7 | 0.58 | 0.42 | 0.19 | 0.11 | 0.09 |
| Ovalbumin | 42.8 | 385 | 5.3 | −10.5 | 10.5 | 0.52 | 0.48 | 0.21 | 0.09 | 0.12 |
pI and charge were calculated at pH 7 using Protein Calculator (54).
Fractions of nonpolar, polar, charged, positively charged, and negatively charged were calculated as fractions of total accessible surface area.
In an attempt to determine the intrinsic factors that influence protein solubility, we looked at several intrinsic protein properties and examined them with respect to protein solubility by comparing them with log S0-values obtained in this study. We looked at fundamental properties of the protein, such as size (molecular mass) and net charge. We also looked at properties of the surface of the protein, including polarity and charge, by determining the fraction of the surface area of the protein that was polar, nonpolar, charged, negatively charged, or positively charged. By looking at the correlation of these properties with protein solubility, we were able to determine their relative importance for determining protein solubility.
Correlation of molecular mass and net charge with solubility measurements
To investigate the contribution of intrinsic factors to protein solubility (see Table 3, columns 1–6), we plotted log S0-values for PEG-8000 and ammonium sulfate versus molecular mass (Fig. 6, A and B), net charge (Fig. 6, C and D), and the absolute value of the net charge (Fig. 6, E and F). Linear fits were made to the data, and R2 values are given. Because the mechanism of PEG precipitation is related to the excluded volume, solubility may increase with protein size or molecular mass; however, no correlation with molecular mass was observed. In general, the solubility of a given protein is at a minimum near the isoelectric point (pI) and increases with the absolute value of the net charge (13,50). To assess whether net charge plays a role in determining the solubility of a group of proteins, we plotted the net charge and absolute value of net charge versus log S0. A weak correlation was observed in all four cases. In the case of the absolute value of net charge, a weak positive correlation was observed, suggesting that with an increasing positive or negative net charge, protein solubility increases. For net charge versus pH, a weak negative correlation was observed. This suggests that, on average for this set of proteins, negatively charged proteins are more soluble than positively charged proteins; however, more data points are required to determine whether this is true for a larger set of proteins.
Figure 6.
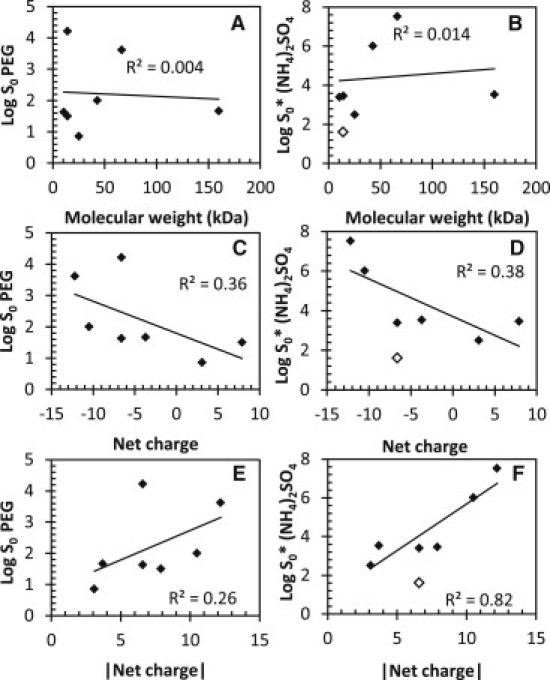
Correlation of molecular mass and net charge with PEG-8000 and ammonium sulfate solubility measurements. Log S0-values versus molecular mass (A and B), net charge (C and D), and absolute net charge (E and F) are shown. The lines and R2 values are from linear least-squares fits. α-Lactalbumin is shown as open diamonds and is excluded from the ammonium sulfate fits.
Correlation of the intrinsic properties of the accessible surface area with protein solubility
Because protein solubility is influenced largely by interactions between water and the protein surface, we investigated the correlation between solubility and the intrinsic properties of the surface of the proteins. The accessible surface areas (ASAs) of all atoms in the proteins were determined and the fractions that were polar, nonpolar, charged, positively charged, and negatively charged were calculated (see Table 3, columns 7–11). Fig. 7 depicts protein solubility as a function of fraction ASA that is polar or nonpolar for PEG-8000 (Fig. 7, A and B) and ammonium sulfate (Fig. 7, C and D). For PEG-8000, the correlation of the percentage of polar and nonpolar surface residues with protein solubility is very poor, although the correlation is positive for polar residues and negative for nonpolar residues, as might be predicted. This suggests that the surface polarity makes a minimal contribution to protein solubility in PEG-8000. For ammonium sulfate, a better correlation was observed, but the correlation with polar and nonpolar surface residues is negative and positive, respectively, which is the opposite of what we would have expected and what was observed in PEG-8000.
Figure 7.
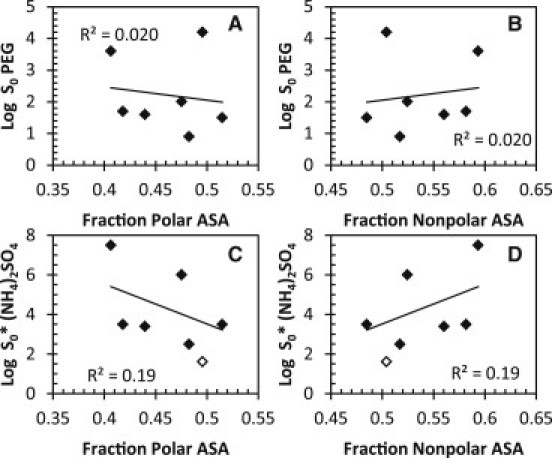
Correlation of fractions of polar and nonpolar ASAs with PEG-8000 and ammonium sulfate solubility measurements. The ASA for all atoms was calculated using PDB files and either pfis (31) or NACCESS (32). Carbon and sulfur atoms are considered nonpolar, and nitrogen and oxygen atoms are considered polar. Log S0-values versus fraction polar ASA (A and C) and fraction nonpolar ASA (B and D) are shown. The lines and R2 values are from linear least-squares fits. α-Lactalbumin is shown as open diamonds and is excluded from the ammonium sulfate fits.
The contribution of the ASA that is charged, positively charged, and negatively charged was evaluated (see Table 3, columns 9–11). Fig. 8 depicts the correlations of PEG-8000 and ammonium sulfate with the fraction of charged (Fig. 8, A and B), positively charged (Fig. 8, C and D), and negatively charged (Fig. 8, E and F) ASA. We find a strong correlation between solubility in PEG-8000 and fraction of charged ASA and a more moderate correlation for ammonium sulfate. A correlation between solubility and the fraction of positively charged ASA was not observed for either PEG-8000 or ammonium sulfate; however, a very strong correlation was observed between solubility and the fraction of negatively charged ASA in both PEG-8000 and ammonium sulfate. This strong correlation suggests that negatively charged surface area plays a significant role in determining protein solubility. This is supported by our previous findings that aspartic and glutamic acids contribute more favorably to protein solubility than do any of the other 18 amino acids (16). To understand the difference in contribution to protein solubility of negative versus positive charges, one needs to understand the properties of negatively and positively charged groups in proteins. Negatively charged groups in proteins include the kosmotropic carboxylate groups of aspartic acid and glutamic acid residues. Positively charged groups include the chaotropic amino and guanidino groups of lysine and arginine. In studies on ions in solution, Collins (26,51–53) described a Hofmeister series dependence for hydration of ions in solution. Collins showed that kosmotropes are highly hydrated and bind water more tightly than water binds itself, whereas chaotropes bind water more weakly than water binds itself and remain largely unhydrated in solution. Therefore, the differential contribution to the solubility of negative and positive groups on the protein surface appears to be due to the differential hydration of the carboxylates that bind water tightly and the amino and guanidino groups that bind water weakly.
Figure 8.
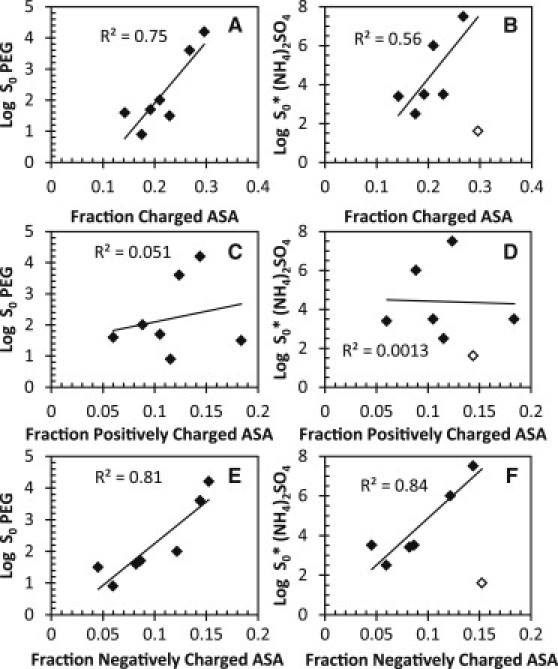
Correlation of the fraction of ASA that is charged (A and B), positively charged (C and D), and negatively charged (E and F) at pH 7.0 with PEG-8000 and ammonium sulfate solubility measurements. The ASA for all atoms was calculated using PDB files and either pfis (31) or NACCESS (32). The oxygen atoms glutamic acid and aspartic acid side chains and the C-terminus are considered negatively charged, and the nitrogen atoms from arginine and lysine side chains and the N-terminus are considered positively charged at pH 7. The lines and R2 values are from linear least-squares fits. α-Lactalbumin is shown as open diamonds and is excluded from the ammonium sulfate fits.
Conclusions
In this work, we found that negative surface charge had the strongest correlation with increased protein solubility, as measured by means of ammonium sulfate and PEG-8000 precipitation experiments. This is best explained by the strong water-binding properties of glutamic and aspartic acid (26). No correlation between solubility and positive surface charge was seen. We observed no correlation when we investigated the surface of the protein with respect to polarity. While comparing the two precipitants, we found that ammonium sulfate markedly increased protein stability for all proteins in this study, whereas PEG could have slight stabilizing or destabilizing effects. We found a remarkable correlation between the solubility results obtained with PEG-8000 and ammonium sulfate, even though they employ different mechanisms to decrease solubility. This suggests that solubility experiments using these two precipitants are probing similar intrinsic properties of the protein, making the choice between precipitants largely one of convenience. Because PEG precipitations can yield a quantitative estimate of solubility in buffer alone, and ammonium sulfate can only determine comparative solubility, PEG is a better choice of precipitant when absolute solubility values are of interest.
Acknowledgments
We thank Gerald Grimsley and Saul Trevino for critical readings of the manuscript.
This work was supported by Amgen, the Robert A. Welch Foundation (BE-1281), and the National Institutes of Health (Molecular Biophysics Training Grant T32GM65088 to R.K.). V.S. contributed to this work during an international rotation at Texas A&M University. N.M. contributed to this work as part of the Summer Research Experience for Undergraduates program funded by the National Science Foundation.
Supporting Material
References
- 1.Bagby S., Tong K.I., Ikura M., Thomas V.D., James L., Uli S. Methods in Enzymology. Academic Press; New York: 2001. Optimization of protein solubility and stability for protein nuclear magnetic resonance; pp. 20–41. [DOI] [PubMed] [Google Scholar]
- 2.Caldwell G.W., Ritchie D.M., Yan Z. The new pre-preclinical paradigm: compound optimization in early and late phase drug discovery. Curr. Top. Med. Chem. 2001;1:353–366. doi: 10.2174/1568026013394949. [DOI] [PubMed] [Google Scholar]
- 3.Riès-kautt M., Ducruix A. Inferences drawn from physicochemical studies of crystallogenesis and precrystalline state. In: Carter C.W. Jr., editor. Methods in Enzymology. Academic Press; New York: 1997. pp. 23–59. [DOI] [PubMed] [Google Scholar]
- 4.Schein C.H. Solubility and secretability. Curr. Opin. Biotechnol. 1993;4:456–461. doi: 10.1016/0958-1669(93)90012-l. [DOI] [PubMed] [Google Scholar]
- 5.Fowler S.B., Poon S., Zurdo J. Rational design of aggregation-resistant bioactive peptides: reengineering human calcitonin. Proc. Natl. Acad. Sci. USA. 2005;102:10105–10110. doi: 10.1073/pnas.0501215102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ricci M.S., Brems D.N. Common structural stability properties of 4-helical bundle cytokines: possible physiological and pharmaceutical consequences. Curr. Pharm. Des. 2004;10:3901–3911. doi: 10.2174/1381612043382611. [DOI] [PubMed] [Google Scholar]
- 7.Ahn H.C., Juranić N., Markley J.L. Three-dimensional structure of the water-insoluble protein crambin in dodecylphosphocholine micelles and its minimal solvent-exposed surface. J. Am. Chem. Soc. 2006;128:4398–4404. doi: 10.1021/ja057773d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters T. Academic Press; San Diego, CA: 1996. All About Albumin: Biochemistry, Genetics, and Medical Applications. [Google Scholar]
- 9.Evans P., Wyatt K., Slingsby C. The P23T cataract mutation causes loss of solubility of folded γD-crystallin. J. Mol. Biol. 2004;343:435–444. doi: 10.1016/j.jmb.2004.08.050. [DOI] [PubMed] [Google Scholar]
- 10.Pande A., Annunziata O., Pande J. Decrease in protein solubility and cataract formation caused by the Pro23 to Thr mutation in human γD-crystallin. Biochemistry. 2005;44:2491–2500. doi: 10.1021/bi0479611. [DOI] [PubMed] [Google Scholar]
- 11.Kim W., Hecht M.H. Generic hydrophobic residues are sufficient to promote aggregation of the Alzheimer's Aβ42 peptide. Proc. Natl. Acad. Sci. USA. 2006;103:15824–15829. doi: 10.1073/pnas.0605629103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaytor M.D., Warren S.T. Aberrant protein deposition and neurological disease. J. Biol. Chem. 1999;274:37507–37510. doi: 10.1074/jbc.274.53.37507. [DOI] [PubMed] [Google Scholar]
- 13.Cohn E.J., Edsall J.T. Hafner Publishing Company; New York: 1943. Proteins, Amino Acids, and Peptides. [Google Scholar]
- 14.Arakawa T., Timasheff S.N. Theory of protein solubility. Methods Enzymol. 1985;114:49–77. doi: 10.1016/0076-6879(85)14005-x. [DOI] [PubMed] [Google Scholar]
- 15.Middaugh C.R., Volkin D.B. Protein solubility. In: Ahern T.J., Manning M.C., editors. Stability of Protein Pharmaceuticals. Plenum Press; New York: 1992. pp. 109–134. [Google Scholar]
- 16.Trevino S.R., Scholtz J.M., Pace C.N. Amino acid contribution to protein solubility: Asp, Glu, and Ser contribute more favorably than the other hydrophilic amino acids in RNase Sa. J. Mol. Biol. 2007;366:449–460. doi: 10.1016/j.jmb.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trevino S.R., Scholtz J.M., Pace C.N. Measuring and increasing protein solubility. J. Pharm. Sci. 2008;97:4155–4166. doi: 10.1002/jps.21327. [DOI] [PubMed] [Google Scholar]
- 18.McPherson A. Introduction to protein crystallization. Methods. 2004;34:254–265. doi: 10.1016/j.ymeth.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 19.Middaugh C.R., Tisel W.A., Rosenberg A. Determination of the apparent thermodynamic activities of saturated protein solutions. J. Biol. Chem. 1979;254:367–370. [PubMed] [Google Scholar]
- 20.Shiau K.S., Chen T.L. Initial protein concentration effects on precipitation by salt. Biotechnol. Bioeng. 1997;53:202–206. doi: 10.1002/(SICI)1097-0290(19970120)53:2<202::AID-BIT10>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 21.Annunziata O., Payne A., Wang Y. Solubility of lysozyme in the presence of aqueous chloride salts: common-ion effect and its role on solubility and crystal thermodynamics. J. Am. Chem. Soc. 2008;130:13347–13352. doi: 10.1021/ja804304e. [DOI] [PubMed] [Google Scholar]
- 22.Przybycien T.M., Bailey J.E. Aggregation kinetics in salt-induced protein precipitation. AIChE J. 1989;35:1779–1790. [Google Scholar]
- 23.Saluja A., Crampton S., Gokarn Y.R. Anion binding mediated precipitation of a peptibody. Pharm. Res. 2009;26:152–160. doi: 10.1007/s11095-008-9722-0. [DOI] [PubMed] [Google Scholar]
- 24.Kumar V., Sharma V.K., Kalonia D.S. Effect of polyols on polyethylene glycol (PEG)-induced precipitation of proteins: Impact on solubility, stability and conformation. Int. J. Pharm. 2009;366:38–43. doi: 10.1016/j.ijpharm.2008.08.037. [DOI] [PubMed] [Google Scholar]
- 25.Atha D.H., Ingham K.C. Mechanism of precipitation of proteins by polyethylene glycols. Analysis in terms of excluded volume. J. Biol. Chem. 1981;256:12108–12117. [PubMed] [Google Scholar]
- 26.Collins K.D. Charge density-dependent strength of hydration and biological structure. Biophys. J. 1997;72:65–76. doi: 10.1016/S0006-3495(97)78647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas P.D., Dill K.A. Local and nonlocal interactions in globular proteins and mechanisms of alcohol denaturation. Protein Sci. 1993;2:2050–2065. doi: 10.1002/pro.5560021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arakawa T., Timasheff S.N. Mechanism of poly(ethylene glycol) interaction with proteins. Biochemistry. 1985;24:6756–6762. doi: 10.1021/bi00345a005. [DOI] [PubMed] [Google Scholar]
- 29.Pace C.N., Sholtz J.M. Measuring the conformational stability of a protein. In: Creighton T.E., editor. Protein Structure: A Practical Approach. IRL Press; Oxford: 1997. pp. 229–321. [Google Scholar]
- 30.Hebert E.J., Grimsley G.R., Pace C.N. Purification of ribonucleases Sa, Sa2, and Sa3 after expression in Escherichia coli. Protein Expr. Purif. 1997;11:162–168. doi: 10.1006/prep.1997.0776. [DOI] [PubMed] [Google Scholar]
- 31.Hebert E.J., Giletto A., Pace C.N. Contribution of a conserved asparagine to the conformational stability of ribonucleases Sa, Ba, and T1. Biochemistry. 1998;37:16192–16200. doi: 10.1021/bi9815243. [DOI] [PubMed] [Google Scholar]
- 32.Hubbard S.J., Thorton J.M. University College London; London, UK: 1993. NACCESS. Department of Biochemistry and Molecular Biology. [Google Scholar]
- 33.Lee B., Richards F.M. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 34.Kollman J.M., Pandi L., Doolittle R.F. Crystal structure of human fibrinogen. Biochemistry. 2009;48:3877–3886. doi: 10.1021/bi802205g. [DOI] [PubMed] [Google Scholar]
- 35.Bhattacharya A.A., Curry S., Franks N.P. Binding of the general anesthetics propofol and halothane to human serum albumin. High resolution crystal structures. J. Biol. Chem. 2000;275:38731–38738. doi: 10.1074/jbc.M005460200. [DOI] [PubMed] [Google Scholar]
- 36.LeBeau A.M., Singh P., Denmeade S.R. Prostate-specific antigen is a “chymotrypsin-like” serine protease with unique P1 substrate specificity. Biochemistry. 2009;48:3490–3496. doi: 10.1021/bi9001858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chrysina E.D., Brew K., Acharya K.R. Crystal structures of apo- and holo-bovine α-lactalbumin at 2. 2-Å resolution reveal an effect of calcium on inter-lobe interactions. J. Biol. Chem. 2000;275:37021–37029. doi: 10.1074/jbc.M004752200. [DOI] [PubMed] [Google Scholar]
- 38.Wang J., Dauter M., Dauter Z. Triclinic lysozyme at 0.65 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 2007;63:1254–1268. doi: 10.1107/S0907444907054224. [DOI] [PubMed] [Google Scholar]
- 39.Stein P.E., Leslie A.G.W., Carrell R.W. Crystal structure of uncleaved ovalbumin at 1.95 Å resolution. J. Mol. Biol. 1991;221:941–959. doi: 10.1016/0022-2836(91)80185-w. [DOI] [PubMed] [Google Scholar]
- 40.Sevcik J., Dauter Z., Wilson K.S. Ribonuclease from Streptomyces aureofaciens at atomic resolution. Acta Crystallogr. D Biol. Crystallogr. 1996;52:327–344. doi: 10.1107/S0907444995007669. [DOI] [PubMed] [Google Scholar]
- 41.Tobler S.A., Sherman N.E., Fernandez E.J. Tracking lysozyme unfolding during salt-induced precipitation with hydrogen exchange and mass spectrometry. Biotechnol. Bioeng. 2000-2001;71:194–207. doi: 10.1002/1097-0290(2000)71:3<194::aid-bit1009>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 42.Baldwin R.L. How Hofmeister ion interactions affect protein stability. Biophys. J. 1996;71:2056–2063. doi: 10.1016/S0006-3495(96)79404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Creighton T.E. W. H. Freeman; New York: 1993. Proteins: Structures and Molecular Properties. [Google Scholar]
- 44.Greene L.H., Grobler J.A., Brew K. Stability, activity and flexibility in α-lactalbumin. Protein Eng. 1999;12:581–587. doi: 10.1093/protein/12.7.581. [DOI] [PubMed] [Google Scholar]
- 45.Farruggia B., García G., Picó G. Destabilization of human serum albumin by polyethylene glycols studied by thermodynamical equilibrium and kinetic approaches. Int. J. Biol. Macromol. 1997;20:43–51. doi: 10.1016/s0141-8130(96)01150-6. [DOI] [PubMed] [Google Scholar]
- 46.Donovan J.W., Mihalyi E. Conformation of fibrinogen: calorimetric evidence for a three-nodular structure. Proc. Natl. Acad. Sci. USA. 1974;71:4125–4128. doi: 10.1073/pnas.71.10.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feher G., Kam Z. Nucleation and growth of protein crystals: general principles and assays. Methods Enzymol. 1985;114:77–112. doi: 10.1016/0076-6879(85)14006-1. [DOI] [PubMed] [Google Scholar]
- 48.Sugio S., Kashima A., Kobayashi K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 1999;12:439–446. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 49.Wickström E., Persson Waller K., Sternesjö A. Short communication: relationships between α-lactalbumin and quality traits in bulk milk. J. Dairy Sci. 2010;93:4577–4581. doi: 10.3168/jds.2009-2863. [DOI] [PubMed] [Google Scholar]
- 50.Tanford C. John Wiley and Sons; New York: 1961. Physical Chemistry of Macromolecules. [Google Scholar]
- 51.Collins K.D. Sticky ions in biological systems. Proc. Natl. Acad. Sci. USA. 1995;92:5553–5557. doi: 10.1073/pnas.92.12.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collins K.D. Ions from the Hofmeister series and osmolytes: effects on proteins in solution and in the crystallization process. Methods. 2004;34:300–311. doi: 10.1016/j.ymeth.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 53.Collins K.D., Neilson G.W., Enderby J.E. Ions in water: characterizing the forces that control chemical processes and biological structure. Biophys. Chem. 2007;128:95–104. doi: 10.1016/j.bpc.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 54.Protein Calculator. http://www.scripps.edu/∼cdputnam/protcalc.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


