Abstract
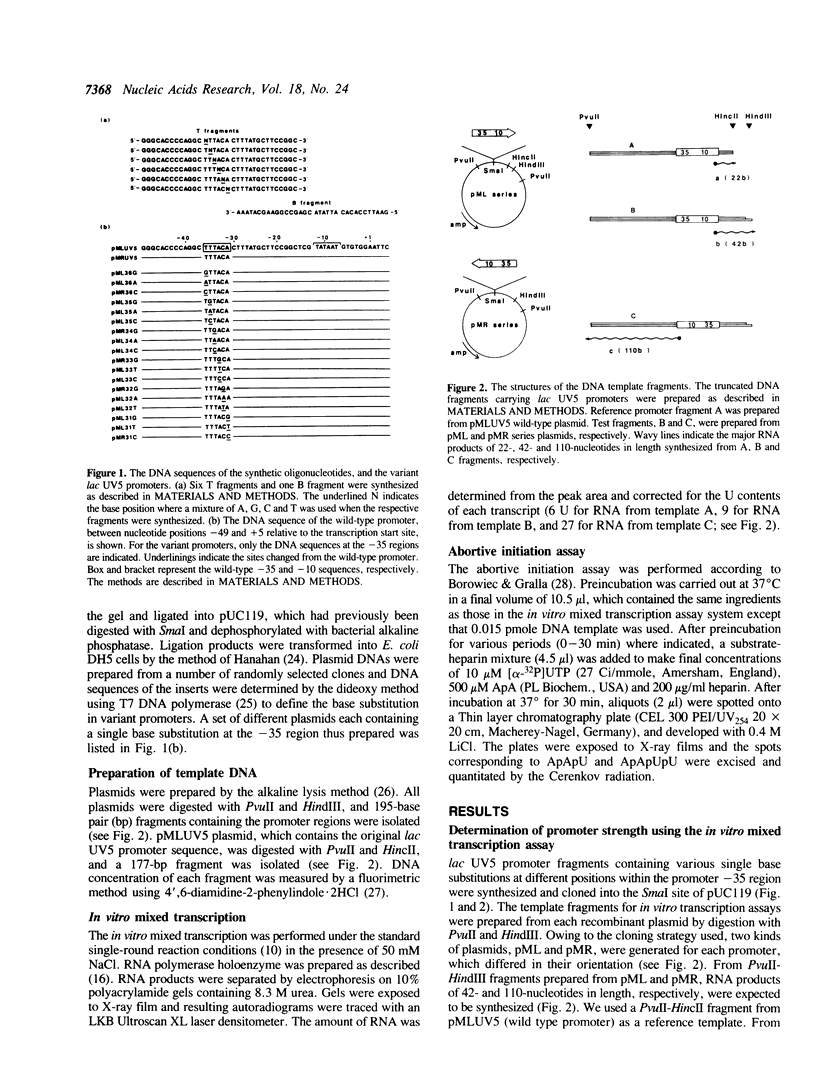
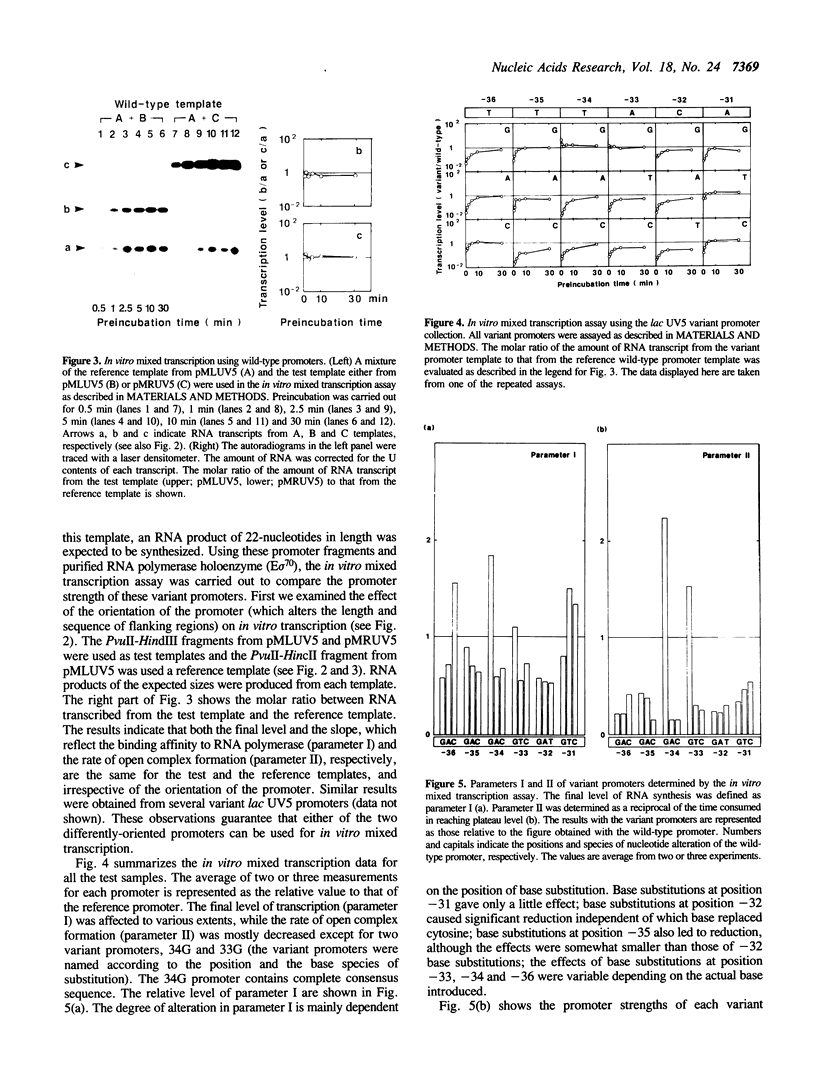
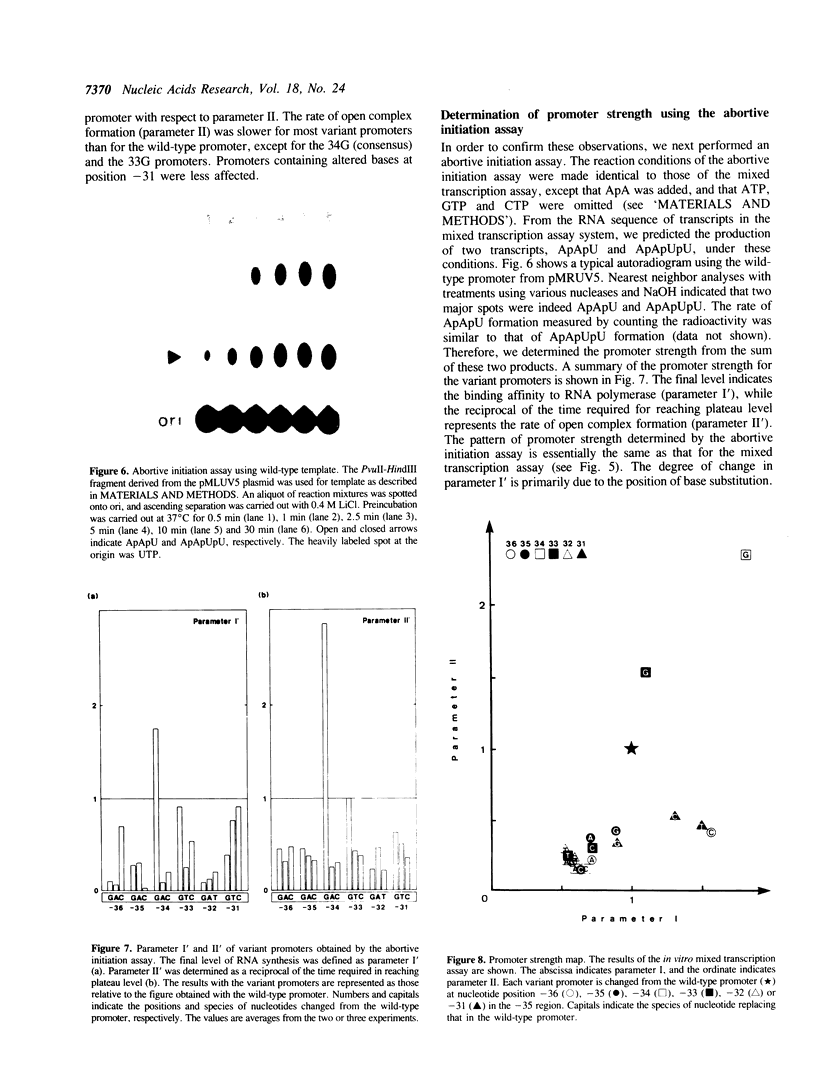
A set of 18 variant lac UV5 promoters was constructed, each carrying a single base substitution within the -35 region (nucleotide positions from -36 to -31 relative to the transcription start site). Using truncated DNA fragments carrying these variant promoters and purified Escherichia coli RNA polymerase holoenzyme, in vitro mixed transcription assays were performed to determine two parameters governing promoter strength: i.e., the binding affinity to RNA polymerase (parameter I) and the rate of open complex formation (parameter II). The following conclusions were drawn from the data presented: (1) Alteration in the promoter strength of variant promoters is dependent on both the position and base species of substitutions; (2) the consensus sequence (TTGACA) exhibits the highest values for both parameters; (3) base substitutions at nucleotide position -34 cause marked effect on both parameters; (4) cytosine at nucleotide position -32 can not be replaced with other nucleotides without significant reduction of the promoter strength; and (5) base substitution at nucleotide position -31 exerts only a little effect on parameter I. All these findings were confirmed by abortive initiation assays.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auble D. T., Allen T. L., deHaseth P. L. Promoter recognition by Escherichia coli RNA polymerase. Effects of substitutions in the spacer DNA separating the -10 and -35 regions. J Biol Chem. 1986 Aug 25;261(24):11202–11206. [PubMed] [Google Scholar]
- Ayers D. G., Auble D. T., deHaseth P. L. Promoter recognition by Escherichia coli RNA polymerase. Role of the spacer DNA in functional complex formation. J Mol Biol. 1989 Jun 20;207(4):749–756. doi: 10.1016/0022-2836(89)90241-6. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Gralla J. D. All three elements of the lac ps promoter mediate its transcriptional response to DNA supercoiling. J Mol Biol. 1987 May 5;195(1):89–97. doi: 10.1016/0022-2836(87)90329-9. [DOI] [PubMed] [Google Scholar]
- Buckle M., Buc H. Fine mapping of DNA single-stranded regions using base-specific chemical probes: study of an open complex formed between RNA polymerase and the lac UV5 promoter. Biochemistry. 1989 May 16;28(10):4388–4396. doi: 10.1021/bi00436a040. [DOI] [PubMed] [Google Scholar]
- Carpousis A. J., Gralla J. D. Interaction of RNA polymerase with lacUV5 promoter DNA during mRNA initiation and elongation. Footprinting, methylation, and rifampicin-sensitivity changes accompanying transcription initiation. J Mol Biol. 1985 May 25;183(2):165–177. doi: 10.1016/0022-2836(85)90210-4. [DOI] [PubMed] [Google Scholar]
- Gardella T., Moyle H., Susskind M. M. A mutant Escherichia coli sigma 70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol. 1989 Apr 20;206(4):579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Mechanism of activation of transcription initiation from the lambda PRM promoter. J Mol Biol. 1982 May 25;157(3):493–525. doi: 10.1016/0022-2836(82)90473-9. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Igarashi K., Fujita N., Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase: omega factor is responsible for the ppGpp sensitivity. Nucleic Acids Res. 1989 Nov 11;17(21):8755–8765. doi: 10.1093/nar/17.21.8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A. Promoter selectivity of prokaryotic RNA polymerases. Trends Genet. 1988 Oct;4(10):282–286. doi: 10.1016/0168-9525(88)90170-9. [DOI] [PubMed] [Google Scholar]
- Ishihama A. Transcription signals and factors in Escherichia coli. Adv Biophys. 1986;21:163–173. doi: 10.1016/0065-227x(86)90021-3. [DOI] [PubMed] [Google Scholar]
- Kajitani M., Ishihama A. Determination of the promoter strength in the mixed transcription system. II. Promoters of ribosomal RNA, ribosomal protein S1 and recA protein operons from Escherichia coli. Nucleic Acids Res. 1983 Jun 25;11(12):3873–3888. doi: 10.1093/nar/11.12.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani M., Ishihama A. Determination of the promoter strength in the mixed transcription system: promoters of lactose, tryptophan and ribosomal protein L10 operons from Escherichia coli. Nucleic Acids Res. 1983 Feb 11;11(3):671–686. doi: 10.1093/nar/11.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani M., Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase. Differential stringent control of the multiple promoters from ribosomal RNA and protein operons. J Biol Chem. 1984 Feb 10;259(3):1951–1957. [PubMed] [Google Scholar]
- Kapuściński J., Skoczylas B. Simple and rapid fluorimetric method for DNA microassay. Anal Biochem. 1977 Nov;83(1):252–257. doi: 10.1016/0003-2697(77)90533-4. [DOI] [PubMed] [Google Scholar]
- McClure W. R., Hawley D. K., Youderian P., Susskind M. M. DNA determinants of promoter selectivity in Escherichia coli. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):477–481. doi: 10.1101/sqb.1983.047.01.057. [DOI] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- McClure W. R. Rate-limiting steps in RNA chain initiation. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5634–5638. doi: 10.1073/pnas.77.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M. E., Hawley D. K., Entriken R., McClure W. R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Fujita N., Ishihama A. Expression of the leuX gene in Escherichia coli. Regulation at transcription and tRNA processing steps. J Mol Biol. 1987 Oct 20;197(4):659–670. doi: 10.1016/0022-2836(87)90472-4. [DOI] [PubMed] [Google Scholar]
- Nomura T., Fujita N., Ishihama A. Promoter selectivity of E. coli RNA polymerase: analysis of the promoter system of convergently-transcribed dnaQ-rnh genes. Nucleic Acids Res. 1985 Nov 11;13(21):7647–7661. doi: 10.1093/nar/13.21.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Fujita N., Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase: alteration by fMet-tRNAfMet. Nucleic Acids Res. 1986 Sep 11;14(17):6857–6870. doi: 10.1093/nar/14.17.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Shih M. C., Gussin G. N. Mutations affecting two different steps in transcription initiation at the phage lambda PRM promoter. Proc Natl Acad Sci U S A. 1983 Jan;80(2):496–500. doi: 10.1073/pnas.80.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Siegele D. A., Hu J. C., Walter W. A., Gross C. A. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989 Apr 20;206(4):591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- Stefano J. E., Gralla J. D. Mutation-induced changes in RNA polymerase-lac ps promoter interactions. J Biol Chem. 1982 Dec 10;257(23):13924–13929. [PubMed] [Google Scholar]
- Straney D. C., Crothers D. M. Comparison of the open complexes formed by RNA polymerase at the Escherichia coli lac UV5 promoter. J Mol Biol. 1987 Jan 20;193(2):279–292. doi: 10.1016/0022-2836(87)90219-1. [DOI] [PubMed] [Google Scholar]
- Szoke P. A., Allen T. L., deHaseth P. L. Promoter recognition by Escherichia coli RNA polymerase: effects of base substitutions in the -10 and -35 regions. Biochemistry. 1987 Sep 22;26(19):6188–6194. doi: 10.1021/bi00393a035. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana H., Ishihama A. Correlation between the rate of productive transcription initiation and the strand-melting property of Escherichia coli promoters. Nucleic Acids Res. 1985 Dec 20;13(24):9031–9042. doi: 10.1093/nar/13.24.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueshima R., Fujita N., Ishihama A. DNA supercoiling and temperature shift affect the promoter activity of the Escherichia coli rpoH gene encoding the heat-shock sigma subunit of RNA polymerase. Mol Gen Genet. 1989 Jan;215(2):185–189. doi: 10.1007/BF00339716. [DOI] [PubMed] [Google Scholar]
- Youderian P., Bouvier S., Susskind M. M. Sequence determinants of promoter activity. Cell. 1982 Oct;30(3):843–853. doi: 10.1016/0092-8674(82)90289-6. [DOI] [PubMed] [Google Scholar]
- Youderian P. Promoter strength: more is less. Trends Genet. 1988 Dec;4(12):327–328. doi: 10.1016/0168-9525(88)90049-2. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]





