Summary
The BLM helicase has been shown to maintain genome stability by preventing accumulation of aberrant recombination intermediates. We show here that the Saccharomyces cerevisiae BLM ortholog, Sgs1, plays an integral role in normal meiotic recombination, beyond its documented activity limiting aberrant recombination intermediates. In wild type meiosis, temporally and mechanistically distinct pathways produce crossover and noncrossover recombinants. Crossovers form late in meiosis I prophase, by polo kinase-triggered resolution of Holliday junction (HJ) intermediates. Noncrossovers form earlier, via processes that do not involve stable HJ intermediates. In contrast, sgs1 mutants abolish early noncrossover formation. Instead, both noncrossovers and crossovers form by late HJ intermediate resolution, using an alternate pathway requiring the overlapping activities of Mus81-Mms4, Yen1, and Slx1–Slx4, nucleases with minor roles in wild-type meiosis. We conclude that Sgs1 is a primary regulator of recombination pathway choice during meiosis, and suggest a similar function in the mitotic cell cycle.
Introduction
Homologous recombination is critical to the successful division of the diploid genome among haploid gametes during meiosis. The crossover (CO) products of recombination, visible at the chromosome level as chiasmata, provide stable connections between homologous chromosomes of different parental origin (homologs), and these connections are required for accurate homolog segregation at meiosis I, the first division of meiosis (Bascom-Slack et al., 1997)). In many organisms, early inter-homolog (IH) recombination intermediates create reversible contacts important for homolog association, alignment and pairing during meiosis I prophase (Bhalla and Dernburg, 2008). These early events occur in excess over COs, and many are resolved without exchange of flanking sequences as noncrossover (NCO) recombinants. Since excessive COs adversely affect homolog segregation (Koehler et al., 1996), it is of considerable interest to understand the mechanisms that distinguish CO and NCO recombination during meiosis.
Meiotic recombination is initiated by programmed double strand breaks (DSBs) produced by the Spo11 transesterase (Keeney, 2007). DSB ends are resected to produce 3’ overhangs, which initiate strand invasion of a homologous donor (Figure 1A). These initial strand invasion intermediates can be further processed in different ways, with different recombination product outcomes. For example, if a single DSB end, after priming DNA synthesis, is displaced and anneals with the other DSB end, a NCO is produced in a process called synthesis-dependent strand-annealing (SDSA, Figure S1A; Paques and Haber, 1999). Alternatively, stabilization of strand invasion intermediates, followed by capture of the second DSB end, can create a double Holliday junction joint molecule (dHJ-JM) intermediate (Schwacha and Kleckner, 1995; Szostak et al., 1983), which can be resolved by multiple mechanisms (Youds and Boulton, 2011). Coupled helicase and topoisomerase activities can disassemble dHJ-JMs, in a process called dissolution, to produce only NCOs (Figure S1B). dHJ-JMs can also be resolved by endonuclease cleavage of the two HJs to produce either a CO or a NCO, depending upon the relative orientation of the two cleavage events (Figure S1C).
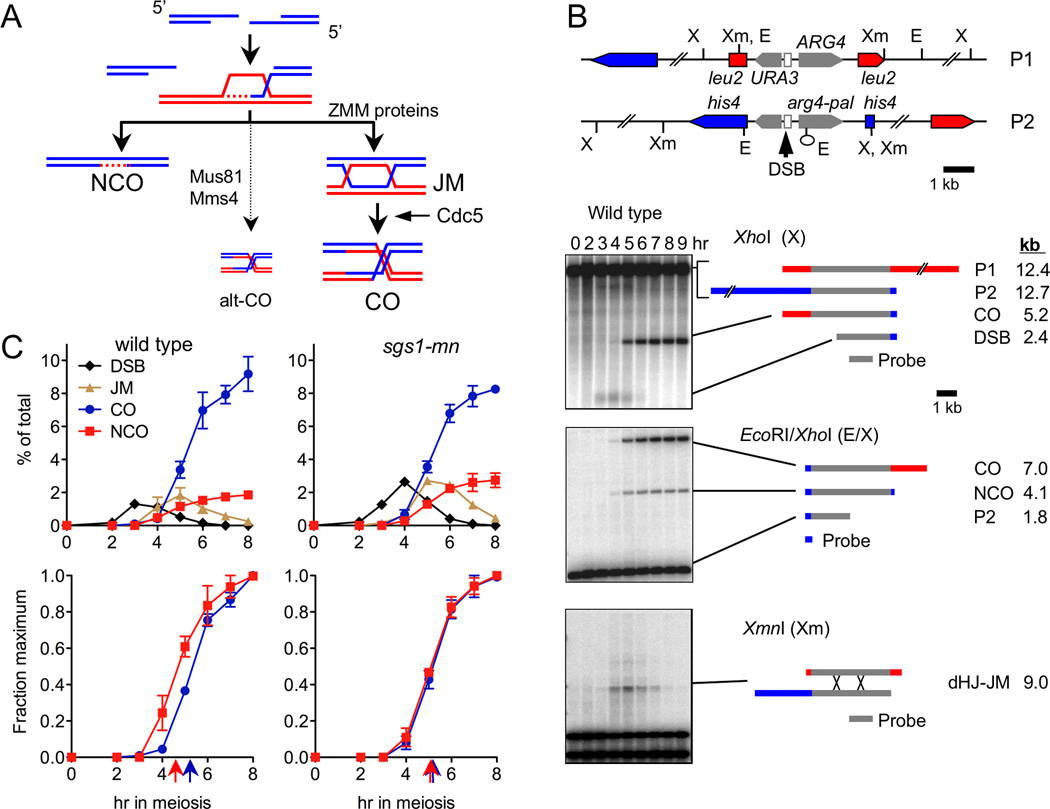
Figure 1. Sgs1 is involved in NCO formation.
(A) Early crossover decision model for meiotic recombination (Bishop and Zickler, 2004). A double-strand break (DSB) is resected to expose 3’-ended single-strand tails, which invade the homolog and initiate DNA synthesis, forming an early D-loop intermediate. D-loop disassembly (left) creates a noncrossover by annealing with the two DSB ends; early intermediate stabilization by synaptonemal complex components (ZMM proteins, right) leads to JM formation by capture of the other DSB end. At exit from pachytene, triggered by polo kinase Cdc5, JMs are resolved in a biased manner to produce crossovers. A minor fraction of crossovers are produced by an alternative, ZMM-independent mechanism that involves the Mus81-Mms4 nuclease (alt-CO, center).
(B) Recombination reporter system used to detect intermediates and products (Jessop et al., 2005). A 3.5 kb insert with URA3 and ARG4 genes (grey arrows) contains a strong meiotic DSB site (open box), and is inserted at LEU2 (red) on one chromosome III homolog and at HIS4 (blue) on the other. A short palindrome with an EcoRI site (lollipop) creates the arg4-pal allele. Restriction sites: Xm—XmnI; X—XhoI; E—EcoRI. XmnI digests probed with ARG4 sequences (grey bar) detect dHJ-JMs. XhoI digests probed with the same sequences detect DSBs and COs. EcoRI/XhoI double digests, probed with HIS4 sequences (blue bar), detect NCOs where arg4-pal is converted to ARG4 (full conversion shown), as well as a subset of COs. Representative Southern blots are shown.
(C) Recombination intermediates and products in wild type (MJL2984) and sgs1-mn (MJL3166). Top—DSB (black), JM (tan), CO (blue), and NCO (red) signals from southern blots. Bottom—COs and NCOs, expressed as a fraction of maximum levels. Arrows indicate times of half-maxima. Values are from two independent experiments; error bars indicate S.E.M.
In budding yeast, where meiotic recombination has been best characterized at the molecular level, NCOs and COs have been shown to form by distinct mechanisms (Bishop and Zickler, 2004). NCOs and JMs appear at the same time in meiosis, but COs appear only later, when JMs resolve, indicating that only COs are formed by the resolution of stable JMs (Allers and Lichten, 2001). In addition, several classes of mutants confer meiotic CO and JM defects without reducing NCOs. Mutants lacking the yeast polo kinase, Cdc5, or Ndt80, a transcription factor that drives meiotic Cdc5 expression (Chu and Herskowitz, 1998), show normal NCO formation but markedly reduced JM resolution and CO production, and ectopic Cdc5 expression in ndt80 mutants restores JM resolution and COs without additional NCO formation (Allers and Lichten, 2001; Clyne et al., 2003; Sourirajan and Lichten, 2008; Xu et al., 1995). A second class of mutants, lacking members of the ZMM (Zip1/2/3 Msh4/5 Mer3 Spo16/22) protein family, show diminished JMs and COs but normal NCO formation (Börner et al., 2004; Lynn et al., 2007). ZMM proteins are components of the synaptonemal complex (SC), a tripartite protein structure that pairs and aligns homologs at the pachytene stage of meiosis (Page and Hawley, 2003), and many ZMM proteins form foci at sites of IH recombination (Lynn et al., 2007). Mutants lacking MutL homologs Mlh1 or Mlh3, or lacking exonuclease I (Exo1), also show reduced COs, but available data suggest that NCOs and JMs are not similarly affected (Argueso et al., 2004; Khazanehdari and Borts, 2000; Wang et al., 1999; Zakharyevich et al., 2010; Zakharyevich et al., 2012). Taken together, these observations suggest that most meiotic NCOs are not derived from stable JMs, but instead are formed by SDSA or dissolution. COs, on the other hand, are produced by polo kinase-triggered, biased resolution of JMs that are stabilized by the ZMM proteins (Figure 1A, Figure S1C). In addition, a minor fraction of COs are produced by ZMM-independent processes (alt-CO in Figure 1A), as inferred from findings that COs are still present at reduced levels in zmm mutants (Argueso et al., 2004; de los Santos et al., 2003). However, the identity of factors and activities responsible for directing meiotic recombination events amongst these three pathways remains elusive.
One enzyme complex with the potential to regulate meiotic recombination pathway choice is the RecQ family helicase BLM (Sgs1 in budding yeast) and its partners topoisomerase III (Top3) and Rmi1, called BLAP75 in mammals (Bernstein et al., 2010). The BLM/Sgs1 complex has two in vitro activities that might promote NCOs at the expense of COs in vivo. First, BLM disassembles D-loops structures, analogous to early strand invasion intermediates, and thus could promote NCO formation by SDSA (Adams et al., 2003; Bachrati et al., 2006; McVey et al., 2004b; van Brabant et al., 2000). Second, the BLM/Sgs1 complex can drive dHJ dissolution in vitro, producing NCOs (Cejka et al., 2010; Wu and Hickson, 2003). Consistent with these in vitro activities, sgs1 mutants show increased mitotic JMs and COs (Bzymek et al., 2010; Ira et al., 2003), and mutants lacking BLM homologs show increased mitotic recombination in a variety of multicellular organisms (Bernstein et al., 2010). Thus, the BLM/Sgs1 complex appears to play an important role in regulating recombination during the mitotic cell cycle.
Evidence for a similar role in meiosis is limited, as sgs1 mutants produce COs and NCOs at near wild type levels (Jessop et al., 2006; Oh et al., 2007; Rockmill et al., 2003). JMs do accumulate at modestly elevated levels in sgs1 mutants, with particular increases in JMs involving sister chromatids and in JMs containing three or four chromosomes (multichromatid JMs, MC-JMs), but these JMs are resolved efficiently and with normal timing (Jessop et al., 2006; Oh et al., 2007). sgs1 mutation also partially suppresses the meiotic JM and CO defects of zmm mutants and the CO defects of mlh3 mutants (Jessop et al., 2006; Oh et al., 2007; Rockmill et al., 2003). These findings have been taken to indicate that Sgs1 acts as a recombination chaperone during meiosis, by disassembling aberrant recombination intermediates that form outside the context of the SC.
Similar uncertainty exists regarding the nucleases that participate in meiotic JM resolution. Biochemical studies have identified three structure-selective nucleases with potential JM-resolving activity (Schwartz and Heyer, 2011): XPF ortholog Mus81, which partners with Mms4 (Eme1 in some organisms); XPG ortholog Yen1(Gen1 in other organisms); and GIY-domain nuclease Slx1 and its partner Slx4 (BTBD12, HIM-18 and MUS312 in other organisms). Evidence for these nucleases being meiotic JM resolvases is incomplete and varied. Drosophila mutants lacking Slx4 homolog MUS312 show marked CO defects (Yildiz et al., 2002), but only minor defects are seen in analogous nematode and mouse mutants (Holloway et al., 2011; Saito et al., 2009). Fission yeast mus81 mutants show marked CO defects (Smith et al., 2003), but only minor meiotic CO defects are seen in budding yeast, Arabidopsis, Drosophila and mouse mus81 or mms4 mutants, primarily in ZMM-independent COs (Argueso et al., 2004; Berchowitz et al., 2007; de los Santos et al., 2003; Higgins et al., 2008; Holloway et al., 2008; Jessop and Lichten, 2008; Oh et al., 2008; Trowbridge et al., 2007). Budding yeast mus81 and mms4 mutants display limited meiotic chromosome segregation defects, but most JMs resolve, indicating that Mus81-Mms4 is required for timely resolution of a minor fraction of JMs (de los Santos et al., 2003; Jessop and Lichten, 2008; Matos et al., 2011; Oh et al., 2008). A recent study reported more severe segregation defects in mus81 yen1 double mutants, suggesting a greater JM resolution defect, but the extent of the resolution defect was not quantified (Matos et al., 2011). This study also showed that Mms4, Yen1, and Slx1 undergo programmed modification at the end of meiosis I prophase, with Mms4 and Slx1 being phosphorylated, and Yen1 being dephosphorylated, and that Cdc5 is the kinase that phosphorylates Mms4, thereby stimulating Mus81-Mms4 nuclease activity.
These findings identify Mus81-Mms4 as a Cdc5-stimulated nuclease that acts redundantly with other nucleases to resolve meiotic JMs, but also suggest that it acts mainly in secondary CO-forming processes. This suggestion is supported by the finding that Mus81-Mms4 is both necessary and sufficient to resolve many of the aberrant meiotic JMs that form in the absence of Sgs1 (Jessop and Lichten, 2008; Oh et al., 2008), indicating that Sgs1 and Mus81-Mms4 collaborate to prevent the accumulation of aberrant recombination intermediates that form outside of the primary pathways for meiotic recombination. However, because meiotic CO and NCO levels were not markedly altered in sgs1 single mutants, and because NCOs form at normal levels in sgs1 mus81 double mutants, it was not anticipated that Sgs1 would determine the outcome of the majority of meiotic recombination events.
We present here data indicating that, to the contrary, Sgs1 is a central regulator of most of the recombination events that occur during budding yeast meiosis. We show that, during normal meiosis, Sgs1 is responsible for directing meiotic recombination towards the alternate formation of either early NCOs or JMs, the latter being subsequently resolved as COs in a Mus81-Mms4, Yen1, and Slx1–Slx4 independent manner. In contrast, in sgs1 mutants, early NCO formation is abolished, and most meiotic recombination events form JMs that are later resolved as both COs and NCOs, by mechanisms that require Mus81-Mms4, Yen1, or Slx1–Slx4. Remarkably, Cdc5 promotes JM resolution under all circumstances, regardless of whether JMs are formed in the presence or absence of Sgs1, and regardless of whether resolution primarily produces CO, or both COs and NCOs.
Results
Sgs1 is required for early NCO formation during meiosis
NCOs form before COs during wild-type meiosis, consistent with NCO- and CO-forming processes diverging early in meiosis I prophase (Figure 1A; Allers and Lichten, 2001; Börner et al., 2004; Hunter and Kleckner, 2001). Using a recombination reporter (Jessop et al., 2005; Figure 1B), we asked if NCO and CO formation showed similar differential timing in sgs1 mutants. To avoid sgs1 mitotic growth defects, we used an sgs1 meiotic null (sgs1-mn) mutant (Jessop et al., 2006; Oh et al., 2008), in which SGS1 is transcribed from a CLB2 promoter that is active during the mitotic cell cycle and inactive during meiosis. As expected, NCOs preceded COs by about 45 min in wild-type cells. However, NCO formation was delayed in sgs1-mn, and COs and NCOs appeared at the same time (Figure 1C). Thus, Sgs1 is responsible for early NCO formation during wild-type meiosis. In addition, cotemporaneous formation of NCOs and COs in sgs1-mn suggests that, in the absence of Sgs1, NCOs and COs might be produced by resolution of a common JM precursor.
Sgs1-independent JMs resolve to form both COs and NCOs
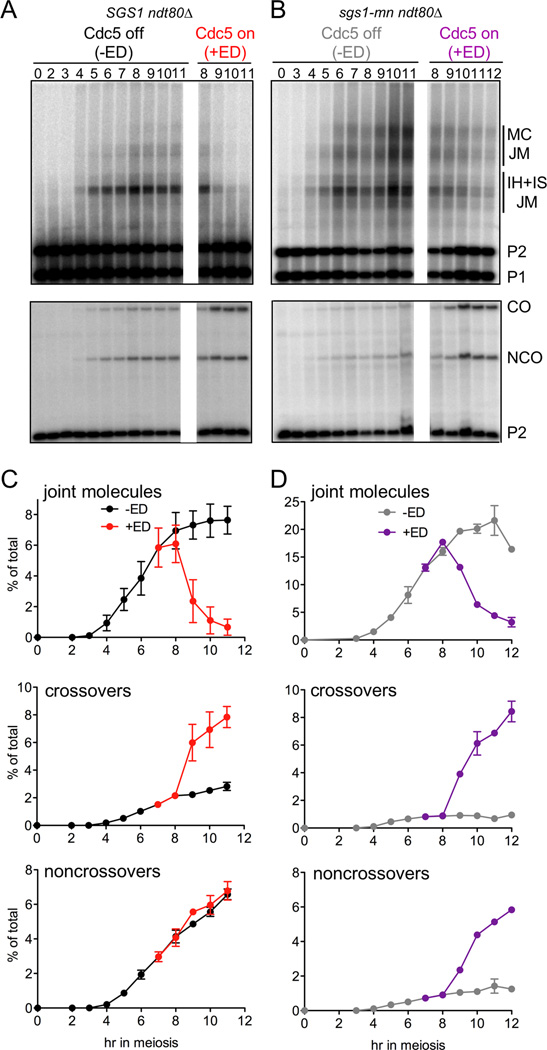
In wild type, JM resolution is triggered by the Cdc5 polo kinase, and the vast majority of JMs resolve as COs (Clyne et al., 2003; Sourirajan and Lichten, 2008). To test the hypothesis that JMs formed in sgs1-mn cells resolve as both NCOs and COs, we asked if resolution of these JMs requires Cdc5, and if so, what type of recombinants are produced. We used meiotic CDC5 expression-defective ndt80Δ mutants and a β-estradiol (ED)-inducible CDC5 allele (CDC5-IN; Sourirajan and Lichten, 2008) to allow controlled Cdc5 expression late in meiosis I prophase. In cells where Sgs1 was active (SGS1 ndt80Δ), NCOs formed normally, JMs accumulated, and COs were greatly reduced (Allers and Lichten, 2001; Figure 2A, C, −ED); these JMs resolve as COs without additional NCO formation when CDC5 is expressed (Sourirajan and Lichten, 2008; Figure 2A, C, +ED). In contrast, in sgs1-mn ndt80Δ cells, when Cdc5 was not expressed (Figure 2B, D, −ED), NCOs were greatly reduced, and JMs accumulated to levels roughly twice that seen in SGS1 ndt80Δ controls. This confirms that Sgs1 directs some meiotic recombination events towards NCOs and away from JM formation. Subsequent CDC5 expression triggered efficient JM resolution, but unlike in SGS1 ndt80Δ, both NCOs and COs were produced (Figure 2B, D, +ED). Thus, JMs that form in the absence of Sgs1 differ from JMs that form in wild type, in that the former resolve as both COs and NCOs, while the latter predominantly produce COs.
Figure 2. Polo kinase Cdc5 triggers JM resolution as COs and NCOs in sgs1-mn.
SGS1 ndt80Δ CDC5-IN cells (MJL3553) and sgs1-mn ndt80Δ CDC5-IN cells (MJL3557) were sporulated for 7h, and the culture was divided into two portions: uninduced (no β-estradiol added; CDC5 off; -ED), and induced (β-estradiol added to 1µM at 7h; CDC5 on; +ED).
(A, B) Southern blot detection of intermediates and products in SGS1 (A) and sgs1-mn (B). Top—XmnI digest to detect bimolecular interhomolog and intersister chromatid intermediates (IH + IS JM) and multichromatid JMs composed of 3 or 4 chromosomes (MC JM). Bottom—EcoRI/XhoI digest to detect CO and NCO recombinants. See Figure 1B for details.
(C, D) Frequencies of JMs (IH+IS+MC), COs, and NCOs, in SGS1 (C) and sgs1-mn (D), plotted as a percentage of total lane signal. Values are from two independent experiments; error bars indicate S.E.M.
Mus81-Mms4, Yen1 and Slx1–Slx4 resolve a minor fraction of JMs in wild-type meiosis but have redundant roles in resolving the majority of JMs that form in the absence of Sgs1
The different product spectra seen upon resolution of JMs formed in the absence or presence of Sgs1 suggests that these two classes of JMs are resolved by different nucleases. We therefore examined the contribution of three candidate HJ-resolving nucleases, Mus81-Mms4, Yen1, and Slx1–Slx4, to JM resolution and recombinant product formation in wild type and in sgs1-mn cells lacking one or more of these nucleases. We first examined wild type meiosis. All single nuclease mutants completed meiotic recombination and resolved the vast majority of JMs (Figure S2A). Thus, none of these putative resolvases are essential for JM resolution during wild-type meiosis.
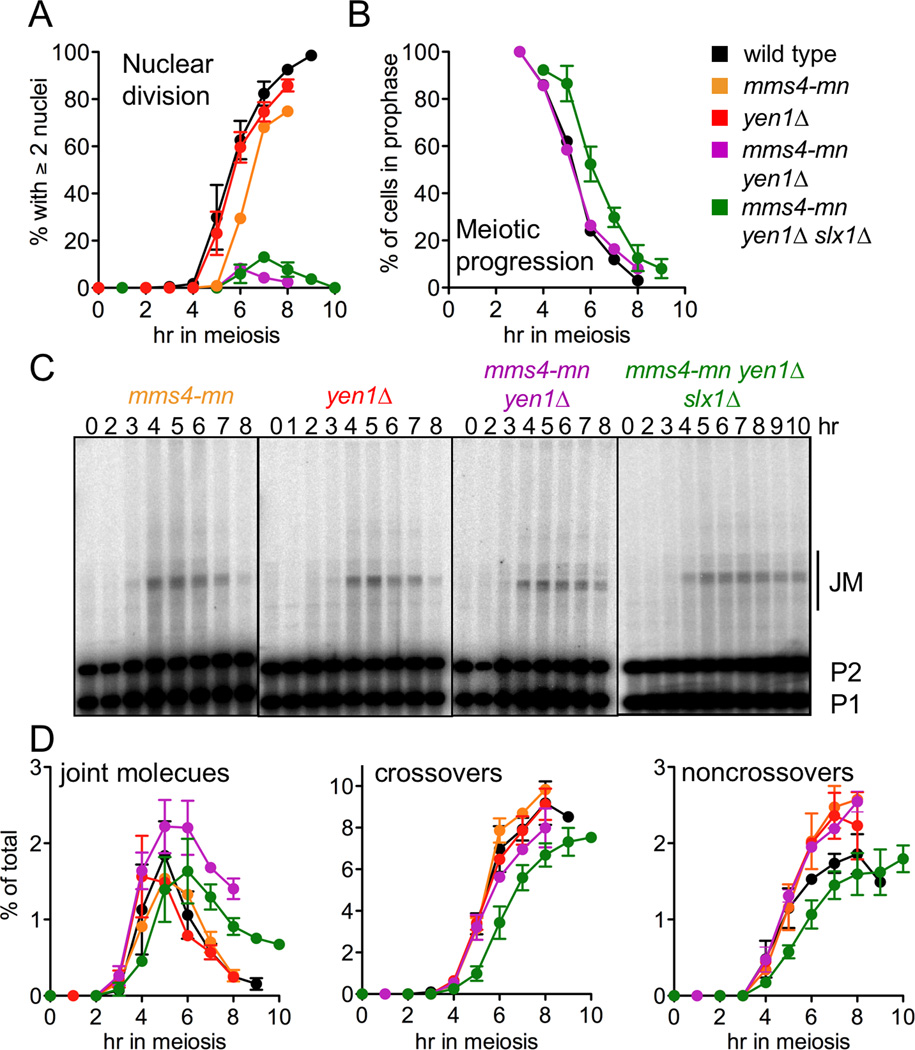
Previous studies had suggested redundant roles for Mus81-Mms4 and Yen1 during the mitotic cell cycle and during meiosis (Agmon et al., 2011; Blanco et al., 2010; Ho et al., 2010; Matos et al., 2011; Tay and Wu, 2010). We therefore examined meiotic recombination in mms4 yen1 double mutants, using a meiotic-null mms4-mn allele to avoid the marked growth defects seen in their combined absence (Agmon et al., 2011; L. J., E. K. and M. L., unpublished observations). As reported previously (Matos et al., 2011), mms4-mn yen1Δ mutant cells underwent meiotic catastrophe, failing to divide nuclei, even though spindle pole bodies separated and meiotic spindles assembled with normal timing (Figure 3A, B and data not shown). Instead, nuclei displayed transient nuclear stretching (Figure S2C and data not shown), a phenotype seen in cells that enter meiosis I with unresolved JMs (Jessop and Lichten, 2008; Oh et al., 2008). However, molecular analysis showed that only a minor fraction of JMs were not resolved (about 10–20% of total JMs formed, compare Figures 3D and 2C), with a corresponding modest decrease in COs (Figure 3C, D). These data indicate that Mus81-Mms4 and Yen1 are required for resolution of only a subset of JMs that form during normal meiosis. They also indicate that a small number of unresolved JMs is sufficient to block nuclear division. Consistent with this latter conclusion, a DSB/recombination-null spo11 mms4-mn yen1Δ mutant strain underwent efficient meiotic division (Figure S2B).
Figure 3. Mus81-Mms4, Yen1 and Slx1–Slx4 resolve a minor fraction of JMs in wild type meiosis.
(A) Fraction of cells undergoing the first meiotic nuclear division, scored as cells with two or more nuclei, including cells where 2 nuclei are connected by DNA bridges. Wild-type (black, MJL2984), yen1Δ (red, MJL3441), mms4-mn yen1Δ (purple, MJL3390) and mms4-mn yen1Δ slx1Δ (green, MJL3491) values are from two independent experiments; for mms4-mn (orange, MJL3172), a single experiment. Error bars indicate S.E.M.
(B) Meiotic progression. Cells with a single spindle pole body were scored as remaining in meiosis I prophase. Values for mms4-mn yen1Δ slx1Δ are from two experiments; error bars indicate S.E.M. Values for other strains are from a single experiment.
(C) Representative Southern blots used to detect JMs.
(D) Frequencies of JMs, COs, and NCOs, plotted as a percentage of total lane signal. JMs were quantified using XmnI digests; COs and NCOs were quantified using XhoI/EcoRI digests (see figure 1B). Values are from two independent experiments; error bars indicate S.E.M.
The finding that most JMs resolve in mms4-mn yen1Δ mutants prompted us to ask if the other candidate resolvase, Slx1–Slx4, acts redundantly with Yen1 or Mu81-Mms4. mms4-mn slx1Δ and yen1Δ slx1Δ strains displayed efficient nuclear division, JM resolution, and CO formation (Figure S2A), and mms4-mn yen1Δ slx1Δ mutants displayed no defects beyond those seen in mms4-mn yen1Δ (Figure 3C, D). Similar results were obtained when slx4Δ was used in place of slx1Δ (Figure S2A and data not shown). Therefore, activities other than Mus81-Mms4, Yen1 and Slx1–Slx4 must resolve the majority of JMs during wild-type meiosis.
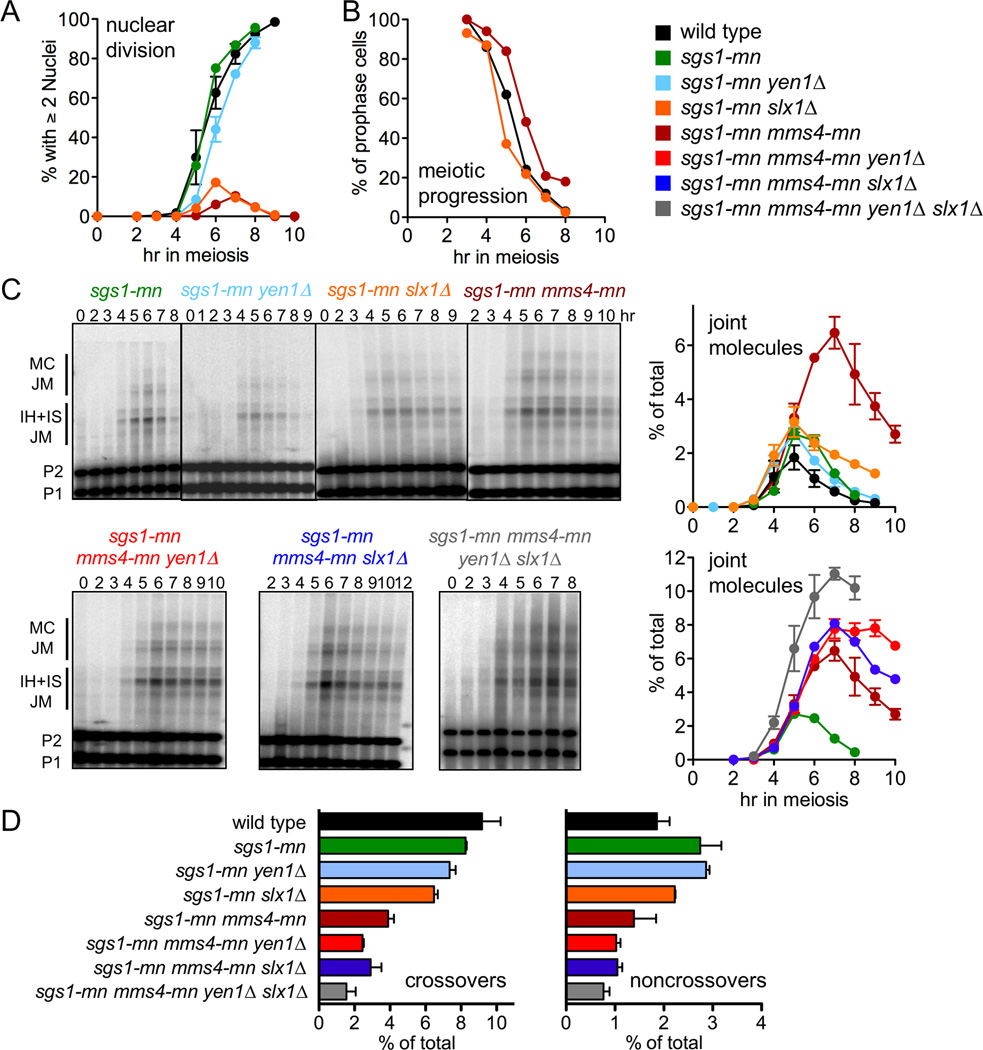
A very different picture emerged when resolution of JMs formed in sgs1 mutants was examined. Previous studies have shown that Mus81-Mms4 is required to resolve a substantial fraction of the JMs that form in the absence of Sgs1 (Jessop and Lichten, 2008; Oh et al., 2008). To ask if Yen1 and Slx1–Slx4 also resolved some of these JMs, we examined meiotic progression and monitored recombination in sgs1-mn mutants lacking one or more of these nucleases. Consistent with previous findings, sgs1-mn mms4-mn double mutants failed to separate nuclei at meiosis I; a similar phenotype was seen in sgs1-mn slx1Δ and in sgs1-mn slx4Δ (Figures 4A and S3A). All three strains accumulated unresolved JMs, consistent with a role for all three nucleases in resolving JMs that form in the absence of Sgs1 (Figures 4C and S3A). All three strains also displayed reductions in both COs and NCOs (Figures 4D and S3A), consistent with the conclusion that JMs that form in the absence of Sgs1 resolve as both COs and NCOs. Eliminating meiotic recombination restored nuclear division (Figure S3B; Jessop and Lichten, 2008), indicating that even the low level of unresolved JMs seen in sgs1-mn slx1Δ can completely block nuclear division. In contrast, sgs1-mn yen1Δ double mutants were similar to sgs1-mn YEN1 strains in terms of progression, JM resolution, and recombinant product formation (Figure 4). Multiple mutant analyses revealed limited redundancy between the three nucleases. sgs1-mn mutants lacking two of the three nucleases displayed greater levels of unresolved JMs and lower levels of COs and NCOs than did sgs1-mn strains lacking any single nuclease, and sgs1-mn mutants lacking all three nucleases (sgs1-mn mms4-mn yen1Δ slx1Δ) accumulated the greatest level of unresolved JMs and the lowest levels of CO and NCO recombinants (Figure 4, Figure S3A). Thus, when Sgs1 is absent, most meiotic JM resolution requires Mus81-Mms4, Yen1 and Slx1–Slx4.
Figure 4. Mus81-Mms4, Yen1 and Slx1–Slx4 have a major role in JM resolution during meiosis in the absence of Sgs1.
(A) Fraction of cells undergoing the first meiotic nuclear division, scored as cells with two or more nuclei, including cells where nuclei are connected by DNA bridges. Wild type (black, MJL2984), sgs1-mn (green, MJL3166) and sgs1-mn yen1Δ (light blue, MJL3363) values are from two independent experiments; error bars indicate S.E.M. Values for sgs1-mn slx1Δ (orange, MJL3467) and sgs1-mn mms4-mn (brown, MJL3171) are from a single experiment.
(B) Meiotic progression. Cells with a single spindle pole body were scored as remaining in meiosis I prophase. All values are from a single experiment.
(C) Left—representative Southern blots used to detect JMs. Additional strains are sgs1-mn mms4-mn yen1Δ (red, MJL3436), sgs1-mn mms4-mn slx1Δ (dark blue, MJL3544), and sgs1-mn mms4-mn yen1Δ slx1Δ (grey, MJL3582). Right—Total JM frequencies, plotted as percentage of total lane signal. Values are from two independent experiments; error bars indicate S.E.M.
(D) CO and NCO frequencies, 8 hr values, plotted as percent of total lane signal, from Southern blots of XhoI/EcoRI digests (see figure 1B). Values are from two independent experiments; error bars indicate S.E.M.
Taken together, these data indicate that a minor fraction of the JMs that form during wild type meiosis are resolved by the three candidate Holliday junction resolvases identified to date, Mus81-Mms4, Yen1 and Slx1–Slx4, while most of the JMs that form in the absence of Sgs1 are resolved by the combined activity of these three nucleases.
ZMM-independent JMs are resolved by Mus81-Mms4 and Yen1
Mus81-Mms4 is necessary for many of the residual genetic crossovers recovered from yeast zmm mutants (Argueso et al., 2004; de los Santos et al., 2003), suggesting that, like JMs that form in the absence of Sgs1, ZMM-independent JMs are resolved by Mus81-Mms4 and other resolvases. However, this suggestion has never been directly tested. We therefore examined the effect of mms4-mn yen1Δ on meiotic recombination in a msh4Δ background (Figure 5).
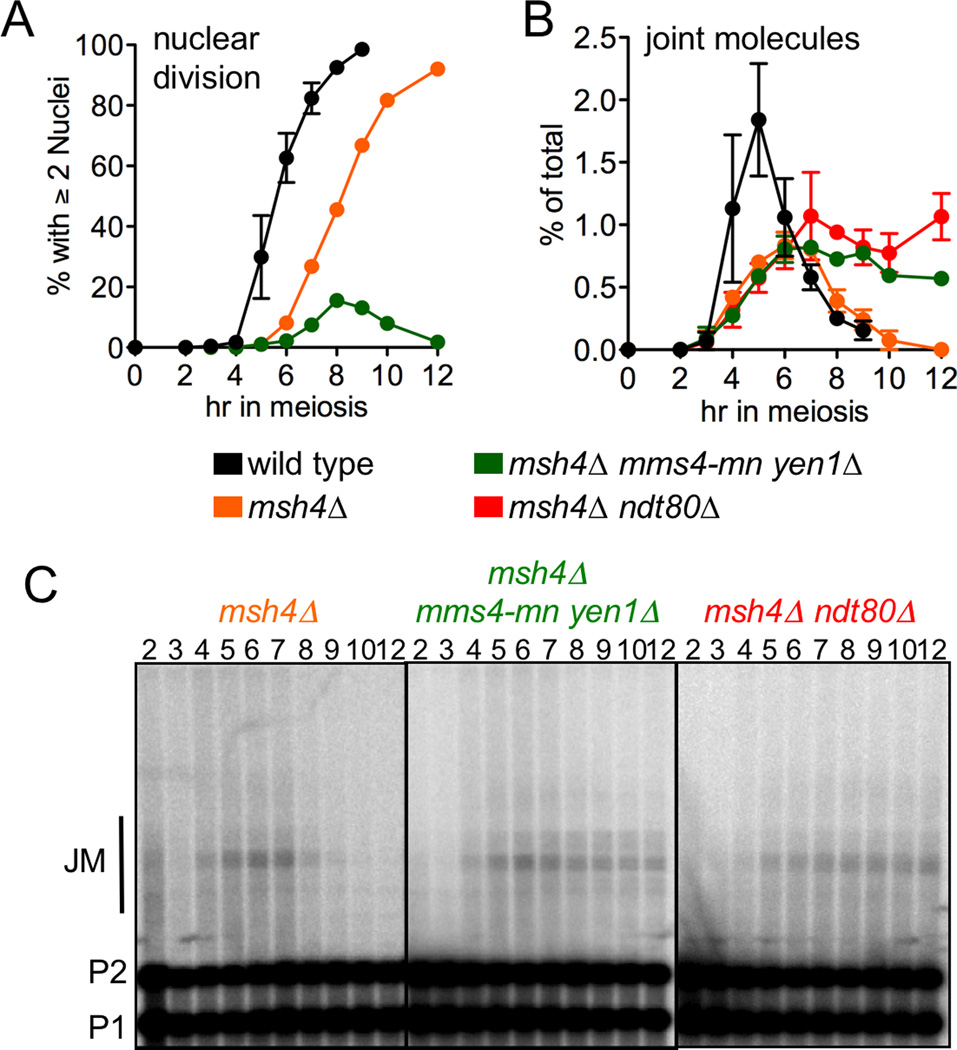
Figure 5. MSH4-independent JMs are not resolved in mms4-mn yen1Δ double mutants.
(A) Fraction of cells undergoing meiosis I nuclear division, scored as cells with two or more nuclei, including cells where 2 nuclei are by DNA bridges, in wild-type (black, MJL2984), msh4Δ (orange, MJL3020), and msh4Δ mms4-mn yen1Δ (green, MJL3489). Wild-type values are from two independent experiments; error bars indicate S.E.M. Other values are from a single experiment.
(B) JM frequencies, plotted as a percentage of total lane signal. Values are from two independent experiments; error bars indicate S.E.M.
(C) Representative Southern blots of XmnI digests used to detect JMs.
The vast majority of msh4Δ single mutant cells underwent nuclear division and distributed nuclear DNA among four spores (Figure 5 and data not shown). In contrast, msh4Δ mms4-mn yen1Δ triple mutants suffered nuclear division failure similar to that seen in mms4-mn yen1Δ, with a single unsegregated DNA mass that was excluded from spores (Figure 5A and data not shown). Nuclear division failure was accompanied by an accumulation of unresolved JMs, to levels similar to those seen in msh4Δ ndt80Δ mutants, where all JM resolution is blocked (Figure 5B, C). These data are consistent with the suggestion that most of the JMs formed in the absence of Msh4 are resolved by Mus81-Mms4 and Yen1.
Discussion
Sgs1 directs events towards resolvase-independent NCO formation and towards ZMM-dependent CO formation
We have examined contributions of the budding yeast BLM helicase homolog, Sgs1, and of the Mus81-Mms4, Yen1 and Slx1–Slx4 nucleases, to meiotic recombination intermediate metabolism and recombinant product formation. Sgs1 was identified as a regulator of meiotic recombination in previous studies (Jessop et al., 2006; Oh et al., 2007; Rockmill et al., 2003), but these had focused on Sgs1 activity in limiting intersister- and multichromatid-JMs during wild type meiosis, and in limiting all JM and CO formation in zmm mutants. In particular, the observation of similar NCO and CO levels in sgs1 mutants and in wild type seemed to indicate a limited role for Sgs1 in the majority of meiotic recombination events.
Our current findings indicate that, despite the numerical similarity in NCOs and COs, meiotic recombination differs in fundamental aspects in SGS1 and in sgs1-mn cells. Early, Cdc5-independent NCO formation does not occur in sgs1-mn mutants (Figure 1), and cumulative JM levels are roughly doubled relative to wild type (Figure 2). In addition, while most JMs that form in SGS1 cells are resolved as COs without contributions from Mus81-Mms4, Yen1, or Slx1–Slx4 (Figure 3), resolution of JMs that form in sgs1 mutants produces both COs and NCOs (Figure 2), and JM resolution is strongly dependent upon the Mus81-Mms4, Yen1 and Slx1–Slx4 resolvases (Figure 4). Similar observations are reported by Zakharyevich and coworkers (Zakharyevich et al., 2012).
On the basis of these and previous findings, we suggest that Sgs1 functions as a central regulator that impacts virtually all meiotic recombination (Figure 6). In the absence of Sgs1 activity, the majority of events form JMs in an unregulated manner and outside of the normal meiotic chromosomal context. Inter-sister and multichromatid JMs are frequently produced, in addition to biparental interhomolog JMs. Furthermore, these JMs are resolved in an unbiased manner to produce both COs and NCOs. In contrast, in wild type cells, Sgs1 directs about half of events towards NCO formation before they can form stable JMs, and directs most of the remaining events towards ZMM protein-associated, interhomolog biparental JMs, which undergo biased resolution as COs when Cdc5 triggers exit from pachytene.
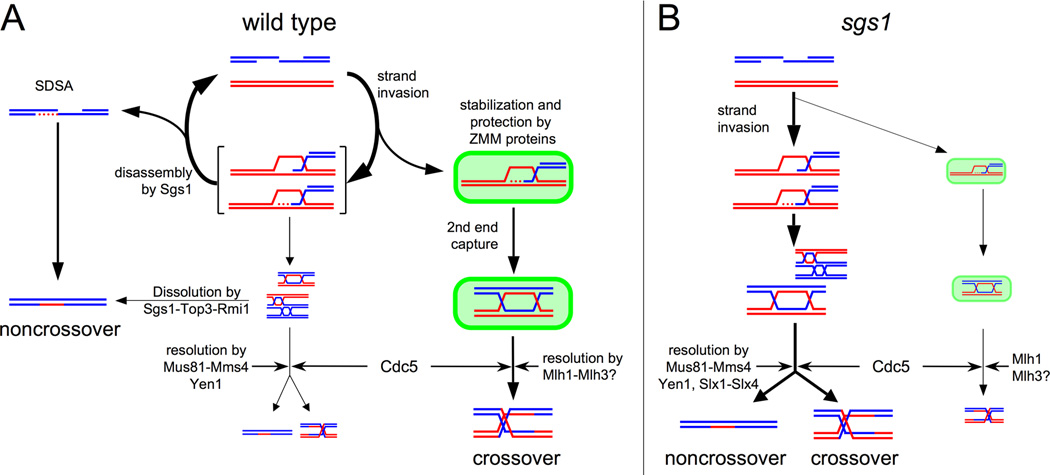
Figure 6. Model of how Sgs1 regulates meiotic recombination intermediate metabolism.
(A) During wild type meiosis, nascent recombination intermediates that contain branched DNA structures are disassembled by Sgs1 helicase. Disassembly of unprotected strand invasion intermediates can drive events towards strand annealing with the other DSB end to form NCOs (SDSA, left) or can return molecules to the broken state. Strand invasion events that are captured by ZMM proteins (right) are stabilized and protected from Sgs1-mediated disassembly, allowing second end capture and dHJ-JM formation. Branched intermediates that escape Sgs1 helicase can form both dHJ and multichromatid JMs (center), which are further vulnerable to Sgs1/Top3/Rmi1-mediated dissolution to form NCOs. JMs that are protected by ZMM proteins are designated by Mlh1-Mlh3 to be resolved as crossovers in a Cdc5-triggered process; ZMM-independent JMs that escape Sgs1 disassembly and dissolution undergo Cdc5-triggered resolution by Mus81-Mms4 and Yen1 to form both NCOs and COs. Strand invasion events involving two sister chromatids also occur, but are not illustrated here.
(B) In sgs1 mutant cells, most strand invasion recombination intermediates proceed directly to form JMs (both dHJ and multichromatid) without ZMM protein involvement, with a consequent reduction in ZMM-associated JMs. All JMs persist until Cdc5 triggers JM resolution. Most JMs are resolved by Mus81-Mms4, Yen1, and Slx1–Slx4 to form both COs and NCOs, with a minor contribution from the ZMM-dependent, CO forming processes that dominate in wild type. It is possible that minor fraction of NCOs are still formed by SDSA; these are not illustrated here.
Both of these Sgs1 functions can be explained by suggesting that the BLM/Sgs1 helicase complex has the potential to disassemble all of the branched recombination intermediates that form during meiosis (Figure 6A). We suggest that the D-loop unwinding activity of BLM/Sgs1 (Figure S1A) disassembles most early interhomolog strand invasion intermediates before they can capture a second DSB end, maintaining DSBs in a state of dynamic instability between strand invasion and free DSB ends. Events can escape BLM/Sgs1-mediated disassembly when break ends anneal to form an unbranched NCO via SDSA (McMahill et al., 2007; McVey et al., 2004a). Alternatively, when strand invasion intermediates are captured by the ZMM proteins and thus protected from BLM/Sgs1, they are stabilized for enough time to allow second end-capture, forming JMs that are later resolved as COs. On occasion, ZMM-independent JMs can form when strand invasion intermediates capture a second end before BLM/Sgs1 disassembles them. Because the ZMM proteins do not protect these intermediates, they are substrates for BLM/Sgs1-Top3-Rmi1 complex-mediated dissolution to form NCOs (Figure S1B).
Dissolution has been identified as a prominent mechanism for JM resolution during the mitotic cell cycle (Dayani et al., 2011), and studies have identified marker segregation patterns in some tetrads that are consistent with dHJ dissolution (Gilbertson and Stahl, 1996; Martini et al., 2011). However, because dissolution produces only fully duplex NCOs (Figure S1B), it cannot direct events towards subsequent ZMM-dependent JM formation. We therefore believe that mechanisms such as D-loop disassembly must also be involved in regulating meiotic recombination intermediate metabolism. Current data do not distinguish between these two possible BLM/Sgs1 activities, and addressing this issue is an important goal for ongoing research.
Mus81-Mms4, Yen1, and Slx1–Slx4 are not the major JM resolvase in normal meiosis
Studies of repair and recombination in budding yeast have suggested redundant roles for Mus81-Mms4 and Yen1 in JM resolution during mitotic cell cycle (Agmon et al., 2011; Blanco et al., 2010; Ho et al., 2010; Tay and Wu, 2010), while the role of Slx1–Slx4 has not been fully evaluated. Our data, and the data of others, indicate that none of these nucleases, either singly or in combination, are the main JM-resolving activity during budding yeast meiosis, since most meiotic JMs still are resolved in mms4-mn yen1Δ slx1Δ triple mutants (Figures 3 and S2; Zakharyevich et al., 2012). It is likely that the limited number of JMs that remain unresolved in mms4-mn yen1Δ slx1Δ triple mutants represent ZMM-independent JMs that escape Sgs1-mediated disassembly, and that these JMs contribute to what was previously described as a ZMM-independent, Mus81-Mms4-dependent “alternative” recombination pathway (Argueso et al., 2004; de los Santos et al., 2003; Figure 1A). Consistent with this suggestion, unresolved JMs are seen at similar levels in msh4Δ ndt80Δ and in msh4Δ mms4-mn yen1Δ cells (Figure 5). While this alternative pathway is usually described as producing COs, data from sgs1 mutants make it likely that it also produces NCOs (see below).
Mus81-Mms4, Yen1, and Slx1–Slx4 resolve JMs that form in the absence of Sgs1
Unlike in wild-type meiosis, Mus81-Mms4, Yen1 and Slx1–Slx4 play important roles in resolving the meiotic JMs that form in the absence of Sgs1, since JMs persist at high levels in sgs1-mn mms4-mn yen1Δ slx1Δ cells (Figure 4; Zakharyevich et al., 2012). As in wild type, these JMs are resolved by Cdc5-dependent mechanisms (Figure 2). Our data thus provide in vivo confirmation of recent reports that the HJ-resolving activity of Mus81-Mms4 is activated by the Cdc5-catalyzed phosphorylation (Matos et al., 2011). In addition, unresolved JM levels increase as sgs1-mn mms4-mn is combined with yen1Δ, slx1Δ or slx4Δ, and a further increase is seen in sgs1-mn mms4-mn yen1Δ slx1Δ cells (Figure 4, Figure S3; Zakharyevich et al., 2012). These synthetic JM resolution defects confirm that both Yen1 and Slx1–Slx4 can resolve meiotic JMs in vivo, that Mus81-Mms4 and Yen1 act redundantly (Matos et al., 2011), and also suggest that, like Mus81-Mms4, Yen1 and Slx1–Slx4 are activated in a Cdc5-dependent manner during meiosis. Furthermore, the increase in unresolved JMs in sgs1-mn strains lacking one or more of these nucleases is associated with a reduction in both NCOs and COs (Figure 4), indicating that the three nucleases resolve dHJ-JMs in an unbiased manner. This is incompatible with a class of models (Whitby, 2005) in which Mus81-Mms4 or other nucleases cleave nascent recombination intermediates in a manner that generates only COs.
Concluding remarks
We have shown here that Sgs1, the budding yeast BLM helicase ortholog, controls meiotic recombination by preventing accumulation of unregulated JMs, which in the absence of Sgs1 comprise the default recombination intermediate. Sgs1 most likely does so, either alone or in complex with Top3 and Rmi1, by disassembling all unprotected branched DNA structures (Figure S1), thus channeling events both towards NCOs (which lack branched structures), and towards JMs that are protected by SC-associated ZMM proteins (Figure 6). These ZMM-protected JMs are later resolved, at the end of pachytene, by an as yet uncharacterized, Cdc5-activated resolvase. The nuclease(s) that resolve ZMM-dependent JMs remain to be identified. We suggest that ZMM-dependent JMs undergo biased resolution as COs because they reside in a structurally coordinated context that ensures cleavage of the two HJs in opposite orientations (Figure S1C). Mlh1, Mlh3, and Exo1, which are not required for JM formation or resolution but which are necessary for full CO formation, may provide this structural context, and possibly the HJ cleavage activity itself (Zakharyevich et al., 2010; Zakharyevich et al., 2012).
JMs that form in the absence of Sgs1, or that escape Sgs1 surveillance in wild type cells, also are resolved by Cdc5-triggered mechanisms, but these involve known nucleases (Mus81-Mms4, Yen1 and Slx1–Slx4) that cleave JMs in an unbiased manner, forming both COs and NCOs as predicted by the original DSBR model (Szostak et al., 1983). Our data indicate that this alternative mode of JM resolution acts in a minor fraction of the interhomolog recombination events that occur during wild-type meiosis. However, this alternative pathway contains sufficient JM resolution capacity to resolve most of the events that occur in the absence of Sgs1, thus revealing a remarkable robustness and flexibility in the budding yeast meiotic recombination program.
Studies in several other organisms, including mouse and Arabidopsis, have also suggested a dual contribution to crossover recombination, with ZMM-dependent processes being responsible for most crossovers, and with minor contributions from ZMM-independent processes involving nucleases such as Mus81-Mms4 (Berchowitz et al., 2007; Holloway et al., 2008). In other organisms, such as C. elegans, CO recombination is almost completely ZMM-dependent (Zetka, 2009). Finally, in organisms that lack ZMM protein homologs, such as S. pombe and Drosophila, CO recombination is almost completely dependent on structure-selective nucleases (Radford et al., 2005; Smith et al., 2003; Yildiz et al., 2002). We suggest that variation between organisms, in terms of dependence upon structure selective nucleases for JM resolution, reflects the relative efficiency with which helicases disassemble early recombination intermediates. It will be of considerable interest to test this suggestion in different organisms, to determine if requirements for alternative JM resolution activities become greater during meiosis in the absence of BLM helicase activity. It is important to note that, while Sgs1 appears to be the dominant helicase regulating meiotic recombination in budding yeast, other helicases have the potential to regulate meiotic recombination in multicellular organisms; these include the Srs2 homolog RTEL-1, the Fanconi anemia complementation group M (FANCM) protein, as well as other RecQ helicase homologs (Barber et al., 2008; Higgins et al., 2011; Whitby, 2010). Testing the impact of these helicases on meiotic recombination will be an important subject for future research.
Finally, our findings suggest a striking parallel between recombination that occurs during the mitotic cell cycle and the ZMM protein-independent recombination that occurs during meiosis, in that both types of recombination are regulated by Sgs1 to frequently form NCOs, rarely form JMs, and both use known structure-selective nucleases to resolve the JMs that do form in an unbiased manner (Figure S4). In this view, the recombination events that occur during meiosis proceed through a combination of two independent pathways. In one, the synaptonemal complex and associated proteins provide a structural context that stabilizes JMs and directs their resolution as COs, thus promoting homolog disjunction at the first meiotic division. In the second, additional interhomolog intermediates, needed to promote the earlier events of homolog pairing and synapsis, are disassembled or resolved by the same mechanisms that function during the mitotic cell cycle. Our data indicate that Sgs1 plays an important role in partitioning meiotic recombination between these coexisting processes.
Experimental Procedures
Yeast strains
Strains (Table S1) were derived from the haploid parents of MJL2984 (Jessop et al., 2005) by transformation or genetic crosses. Construction details are given in the legend to Table S1.
Sporulation
Yeast strains were grown in buffered liquid presporulation medium and shifted to sporulation medium (supplemented 1% potassium acetate) with vigorous aeration to induce meiosis, as described (Jessop et al., 2006). For experiments where CDC5 expression was induced, sporulation cultures were split at the indicated time, β-estradiol or vehicle were added as described (Sourirajan and Lichten, 2008), and aeration was continued.
DNA extraction and analysis
DNA was prepared using a CTAB extraction procedure that stabilizes joint molecule intermediates, and was analyzed on Southern blots of one-dimensional agarose gels as described, using electrophoresis conditions that stabilize joint molecules (Allers and Lichten, 2000, 2001; Jessop and Lichten, 2008). XhoI and XmnI digests, to score DSBs and JMs respectively, were probed with ARG4 coding sequences (+165 to +1413). XhoI/EcoRI double digests, to score NCO and CO recombinants, were probed with HIS4 coding sequences (+538 to +718).
Cytology
Nuclear morphology was scored by DAPI staining (Goyon and Lichten, 1993); cells with a stretched single nucleus or with more than one nucleus were scored as having initiated meiosis I. Progression through meiosis was monitored by scoring spindle pole body/spindle morphology, by immunostaining for β-tubulin as described (Jessop and Lichten, 2008); cells with a single monopolar spindle were scored as not having exited prophase I.
Highlights.
Sgs1 is required for normal noncrossover and crossover formation during meiosis
Noncrossovers and crossovers form via different pathways in wild-type.
Noncrossovers and crossovers form via a common pathway in sgs1 mutants.
Polo kinase Cdc5 triggers joint molecule resolution in both wild type and sgs1.
Supplementary Material
Acknowledgements
We thank Neil Hunter for communicating unpublished data, and Valérie Borde, Dhruba Chattoraj, Mathilde Grelon, Neil Hunter, Raphaël Mercier, Yikang Rong, and Denise Zickler for helpful discussions. This work was supported by the Intramural Research Program of the National Institutes of Health, through the Center for Cancer Research of the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams MD, McVey M, Sekelsky JJ. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–267. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- Agmon N, Yovel M, Harari Y, Liefshitz B, Kupiec M. The role of Holliday junction resolvases in the repair of spontaneous and induced DNA damage. Nucleic Acids Res. 2011;16:7009–7019. doi: 10.1093/nar/gkr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T, Lichten M. A method for preparing genomic DNA that restrains branch migration of Holliday junctions. Nucleic Acids Res. 2000;28:e6. doi: 10.1093/nar/28.2.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Argueso JL, Wanat J, Gemici Z, Alani E. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics. 2004;168:1805–1816. doi: 10.1534/genetics.104.032912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O'Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bascom-Slack CA, Ross LO, Dawson DS. Chiasmata, crossovers, and meiotic chromosome segregation. Adv. Genet. 1997;35:253–284. doi: 10.1016/s0065-2660(08)60452-6. [DOI] [PubMed] [Google Scholar]
- Berchowitz LE, Francis KE, Bey AL, Copenhaver GP. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 2007;3:e132. doi: 10.1371/journal.pgen.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N, Dernburg AF. Prelude to a division. Annu. Rev. Cell Dev. Biol. 2008;24:397–424. doi: 10.1146/annurev.cellbio.23.090506.123245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- Blanco MG, Matos J, Rass U, Ip SC, West SC. Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair. 2010;9:394–402. doi: 10.1016/j.dnarep.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Börner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464:937–941. doi: 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat. Struct. Mol. Biol. 2010;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, Nasmyth K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis. I. Nat. Cell Biol. 2003;5:480–485. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- Dayani Y, Simchen G, Lichten M. Meiotic recombination intermediates are resolved with minimal crossover formation during return-to-growth, an analogue of the mitotic cell cycle. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002083. e1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson LA, Stahl FW. A test of the double-strand break repair model for meiotic recombination in Saccharomyces cerevisiae. Genetics. 1996;144:27–41. doi: 10.1093/genetics/144.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C, Lichten M. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol. Cell. Biol. 1993;13:373–382. doi: 10.1128/mcb.13.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Buckling EF, Franklin FC, Jones GH. Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 2008;54:152–162. doi: 10.1111/j.1365-313X.2008.03403.x. [DOI] [PubMed] [Google Scholar]
- Higgins JD, Ferdous M, Osman K, Franklin FC. The RecQ helicase AtRECQ4A is required to remove inter-chromosomal telomeric connections that arise during meiotic recombination in Arabidopsis. Plant J. 2011;65:492–502. doi: 10.1111/j.1365-313X.2010.04438.x. [DOI] [PubMed] [Google Scholar]
- Ho CK, Mazon G, Lam AF, Symington LS. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol. Cell. 2010;40:988–1000. doi: 10.1016/j.molcel.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000186. e1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JK, Mohan S, Balmus G, Sun X, Modzelewski A, Borst PL, Freire R, Weiss RS, Cohen PE. Mammalian BTBD12 (SLX4) protects against genomic instability during mammalian spermatogenesis. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002094. e1002094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N, Kleckner N. The single-end invasion, an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, Allers T, Lichten M. Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics. 2005;169:1353–1367. doi: 10.1534/genetics.104.036509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol. Cell. 2008;31:313–323. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, Rockmill B, Roeder GS, Lichten M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of Sgs1. PLoS Genet. 2006;2:e155. doi: 10.1371/journal.pgen.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn. Stab. 2007;2:81–123. doi: 10.1007/7050_2007_026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazanehdari KA, Borts RH. EXO1 and MSH4 differentially affect crossing-over and segregation. Chromosoma. 2000;109:94–102. doi: 10.1007/s004120050416. [DOI] [PubMed] [Google Scholar]
- Koehler KE, Hawley RS, Sherman S, Hassold T. Recombination and nondisjunction in humans and flies. Hum. Mol. Genet. 1996;5:1495–1504. doi: 10.1093/hmg/5.supplement_1.1495. [DOI] [PubMed] [Google Scholar]
- Lynn A, Soucek R, Börner G. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 2007;15:591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- Martini E, Borde V, Legendre M, Audic S, Regnault B, Soubigou G, Dujon B, Llorente B. Genome-wide analysis of heteroduplex DNA in mismatch repair-deficient yeast cells reveals novel properties of meiotic recombination pathways. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002305. e1002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Blanco MG, Maslen S, Skehel JM, West SC. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell. 2011;147:158–172. doi: 10.1016/j.cell.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahill MS, Sham CW, Bishop DK. Synthesis-dependent strand annealing in meiosis. PLoS Biology. 2007;5:e299. doi: 10.1371/journal.pbio.0050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M, Adams M, Staeva-Vieira E, Sekelsky JJ. Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics. 2004a;167:699–705. doi: 10.1534/genetics.103.025411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M, LaRocque JR, Adams MD, Sekelsky JJ. Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion. Proc. Natl. Acad. Sci. U. S. A. 2004b;101:15694–15699. doi: 10.1073/pnas.0406157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY, Taylor AF, Smith GR, Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Taylor AF, Smith GR, Hunter N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol. Cell. 2008;31:324–336. doi: 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301:785–789. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford SJ, Goley E, Baxter K, McMahan S, Sekelsky J. Drosophila ERCC1 is required for a subset of MEI-9-dependent meiotic crossovers. Genetics. 2005;170:1737–1745. doi: 10.1534/genetics.104.036178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Fung JC, Branda SS, Roeder GS. The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr. Biol. 2003;13:1954–1962. doi: 10.1016/j.cub.2003.10.059. [DOI] [PubMed] [Google Scholar]
- Saito TT, Youds JL, Boulton SJ, Colaiacovo MP. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000735. e1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Schwartz EK, Heyer WD. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120:109–127. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GR, Boddy MN, Shanahan P, Russell P. Fission yeast Mus81· Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;165:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourirajan A, Lichten M. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 2008;22:2627–2632. doi: 10.1101/gad.1711408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Tay YD, Wu L. Overlapping roles for Yen1 and Mus81 in cellular Holliday junction processing. J. Biol. Chem. 2010;285:11427–11432. doi: 10.1074/jbc.M110.108399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge K, McKim K, Brill SJ, Sekelsky J. Synthetic lethality of Drosophila in the absence of the MUS81 endonuclease and the DmBlm helicase is associated with elevated apoptosis. Genetics. 2007;176:1993–2001. doi: 10.1534/genetics.106.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brabant AJ, Ye T, Sanz M, German IJ, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- Wang TF, Kleckner N, Hunter N. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13914–13919. doi: 10.1073/pnas.96.24.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby MC. Making crossovers during meiosis. Biochem. Soc. Trans. 2005;33:1451–1455. doi: 10.1042/BST0331451. [DOI] [PubMed] [Google Scholar]
- Whitby MC. The FANCM family of DNA helicases/translocases. DNA Repair. 2010;9:224–236. doi: 10.1016/j.dnarep.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz O, Majumder S, Kramer B, Sekelsky JJ. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol. Cell. 2002;10:1503–1509. doi: 10.1016/s1097-2765(02)00782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, Boulton SJ. The choice in meiosis - defining the factors that influence crossover or non-crossover formation. Cell Sci. 2011;124:501–513. doi: 10.1242/jcs.074427. [DOI] [PubMed] [Google Scholar]
- Zakharyevich K, Ma Y, Tang S, Hwang PY, Boiteux S, Hunter N. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol. Cell. 2010;40:1001–1015. doi: 10.1016/j.molcel.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, Hunter N. Joint molecule resolution during meiotic recombination. Cell. 2012 doi: 10.1016/j.cell.2012.03.023. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka M. Homologue pairing, recombination and segregation in Caenorhabditis elegans. Genome Dyn. 2009;5:43–55. doi: 10.1159/000166618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.