SUMMARY
Kinase inhibitors have limited success in cancer treatment because tumors circumvent their action. Using a quantitative proteomics approach, we assessed kinome activity in response to MEK inhibition in triple negative breast cancer (TNBC) cells and genetically engineered mice (GEMMs). MEK inhibition caused acute ERK activity loss, resulting in rapid c-Myc degradation that induced expression and activation of several receptor tyrosine kinases (RTKs). RNAi knockdown of ERK or c-Myc mimicked RTK induction by MEK inhibitors, whereas prevention of proteasomal c-Myc degradation blocked kinome reprogramming. MEK inhibitor-induced RTK stimulation overcame MEK2 but not MEK1 inhibition, reactivating ERK and producing drug resistance. The C3Tag GEMM for TNBC similarly induced RTKs in response to MEK inhibition. The inhibitor-induced RTK profile suggested a kinase inhibitor combination therapy that produced GEMM tumor apoptosis and regression where single agents were ineffective. This approach defines mechanisms of drug resistance, allowing rational design of combination therapies for cancer.
INTRODUCTION
Kinase-targeted cancer therapies can fail when tumor cells circumvent the action of a single agent, facilitating therapeutic resistance. Acquired or selected mutations can decrease affinity for kinase inhibitors, but resistance also develops through alternate routes of kinase pathway activation. For example, RTK upregulation has been observed following targeted inhibition of selective kinases (Chandarlapaty et al., 2011; Johannessen et al., 2010; Nazarian et al., 2010; Villanueva et al., 2010); this kinome reprogramming circumvents inhibition of proto-oncogenic kinases. Alternatively, genomic loss of PTPN12 phosphatase expression similarly causes activation of multiple tyrosine kinases (Sun et al., 2011). Thus, dynamic and system-wide changes in multiple kinases can occur in tumor cells following pharmacological or progressive genetic perturbations. An understanding of these kinome responses and the mechanisms by which they occur will be key in determining how to abrogate therapeutic resistance. With over 130 kinase-specific inhibitors currently in Phase 1-3 clinical trials, developing combination therapies relevant for molecularly-defined cancer subtypes is a highly tractable goal. However, rational design of kinase inhibitor combinations requires an overall knowledge of kinome activity and response, not just a simple measure of an inhibitor’s effect on one or two kinase pathway components. Currently, there is no optimal discovery mechanism to define the entire kinome and its dynamic activity. Such a technique could globally assess tumor kinome response to small molecule inhibitors and suggest more effective combination therapies.
To meet this challenge, we developed a chemical proteomics approach using multiplexed kinase inhibitor beads and mass spectrometry (MIB/MS) to define and quantitate the activity and drug responsiveness of a significant percentage (50-60%) of the expressed kinome. We applied this technique to triple negative breast cancer cell lines, pre-clinical tumor models and human tumors. Analysis of patient TNBC showed activated RAF-MEK1/2-ERK1/2 signaling, supporting MEK as a target in TNBC. Pharmacologic MEK inhibition in TNBC cell lines and GEMM tumors resulted in rapid kinome reprogramming through the induced expression and activation of multiple Tyr and Ser/Thr kinases that bypassed the initial MEK-ERK inhibition. Alterations in virtually every Tyr and Ser/Thr kinase family were observed. The mechanism of this kinome reprogramming involved the proteolytic degradation of c-Myc following MEK1 and MEK2 inhibition which resulted in increased expression and activity of RTKs. MIB/MS analysis showed that reprogrammed kinase activation overcame MEK2 (but not MEK1) inhibition leading to therapeutic resistance. The MEK inhibitor kinome response signature allowed us to predict and test the efficacy of a novel small molecule kinase inhibitor combination. The combination synergistically inhibited TNBC cell line proliferation and caused apoptosis and tumor regression in the C3Tag GEMM of basal-like/claudin-low TNBC.
RESULTS
Kinome profiling of TNBC
TNBC clinical trials of single kinase inhibitors have largely failed, consistent with drug-induced activation of alternative survival signaling pathways. Figure 1A outlines our strategy to interrogate kinome dynamics with the goal of defining endpoints leading to rational design of combination therapies. RNA-seq defined the transcript-level expressed kinome and affinity capture of endogenous kinases followed by quantitative mass spectrometry measured kinome activity profiles in tumors and cells. The proteomic assessment was used to define the kinome response to targeted inhibition of kinases. RNAi tested growth and survival functions of the kinases activated in response to inhibitors, and the cumulative results were used to rationally predict kinase inhibitor combinations to test in models of TNBC.
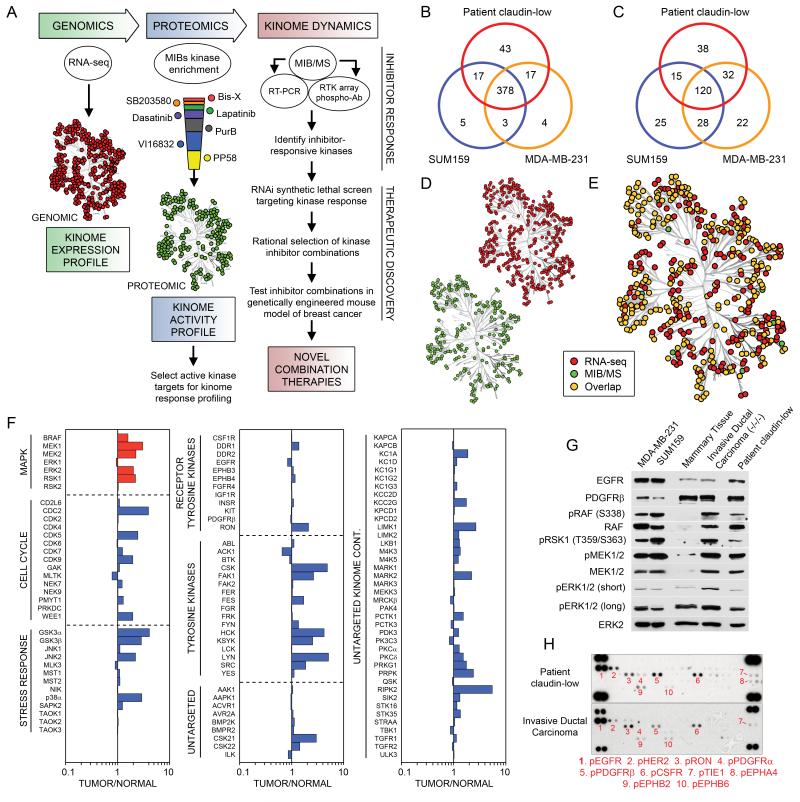
Figure 1. Kinome profiling of TNBC reveals elevated ERK signaling.
(A) Experimental strategy for the rational design of kinase inhibitor combination therapies. To define kinome inhibitor response signatures, expression profiling is integrated with kinase affinity capture and MS quantitative assessment of the activation state of the kinome. RNAi is used to analyze kinase function in survival response to inhibitors.
(B) Venn diagram shows number of expressed kinases defined by RNA-seq across patient TNBC and MDA-MB-231 and SUM159 cell lines. See Table S1 for normalized read count transcript expression profiles.
(C) Venn diagram shows number of kinases captured and identified by MIB-based proteomics across patient-sample TNBC and MDA-MB-231 and SUM159 cell lines. MIB/MS captures 50-60% of the expressed kinome as defined by RNA-seq. See Table S2 for MS identifications of protein kinases.
(D) Distribution and (E) overlap of expressed and MIB-bound kinases across patient-sample TNBC and MDA-MB-231 and SUM159 cell lines.
(F) RAF-MEK-ERK pathway activated in patient TNBC tumors. Quantitative comparison of patient TNBC to matched uninvolved mammary tissue using MIB/MS. The line graphs show iTRAQ determined quantitative changes in MIB binding as a ratio of tumor/uninvolved. Ratio <1 denotes decreased MIB binding and >1 increased MIB binding of kinase in tumor versus control tissue.
(G) Immunoblotting confirms an activated RAF-MEK-ERK pathway in TNBC cell lines and TNBC patient samples.
(H) RTK array analysis of patient TNBC tumors reveals multiple Tyr phosphorylated RTKs, including EGFR and PDGFRβ.
RNA-seq defined the kinome transcript expression profile of a patient’s claudin-low breast tumor and two claudin-low TNBC lines, SUM159 and MDA-MB-231. Greater than 400 of the 518 human protein kinases were expressed in the claudin-low human TNBC tumor and cell lines (Figure 1B). Approximately 10% of the kinases expressed in the claudin-low patient tumor were unique compared to the claudin-low cell lines, undoubtedly due to the tumor’s complex cellular composition (Table S1). The majority of expressed kinases are common between tumor and claudin-low cell lines, suggesting that interrogating the cellular kinome response to inhibitors will be relevant to patient tumors.
Profiling kinase activity in tumors and cell lines was carried out using Multiplexed Inhibitor Beads (MIBs), which consist of mixtures of Sepharose beads with covalently immobilized, linker adapted, kinase inhibitors of moderate selectivity for different kinases (Bisindoylmaleimide-X, SB203580, Dasatinib and Lapatinib) and relatively broad pan-kinase inhibitors (VI16832, Purvalanol B, and PP58) (Figure S1A) (Daub et al., 2008). Kinase capture is reproducible and is a function of kinase expression, the affinity of kinases for the immobilized inhibitors, and the activation state of the kinase (Bantscheff et al., 2007). Acute changes in activation-dependent binding were demonstrated by the increased binding of MAPK pathway kinases in EGF-stimulated cells and the increased retention of Tyr kinases from cells treated with the Tyr phosphatase inhibitor pervanadate (Figures S1B and C). Our data showed that MIBs capture the majority of the expressed kinome estimated by RNA-seq and detect altered kinome activity profiles in response to stimulus or clinical kinase inhibitors.
Using MIBs and mass spectrometry (MIB/MS), we have cumulatively sequence identified more than 320 kinases from cell lines and tumors (Table S2). MIB/MS profiling of an invasive ductal carcinoma breast tumor and two claudin-low cell lines identified approximately 50-60% of the expressed kinome (Figures 1C-E and Tables S1 and S3). Kinases from all major kinome subfamilies were captured, with a large percentage representing the untargeted kinome (Fedorov et al., 2010). iTRAQ labeling of digested MIB elutions allowed quantitative profiling of kinases in patient invasive ductal carcinoma compared to adjacent uninvolved mammary tissue (Figure 1F). Of the kinases detected, there was a general increase in MIB binding of tumor kinases, suggesting escalated kinome activity in the tumor compared to uninvolved mammary tissue. For example, the RAF-MEK-ERK pathway is increased in MIB binding in the tumor relative to control tissue, consistent with ERK activity being a driver for TNBC proliferation. Immunoblots confirmed the activation of RAF-MEK-ERK signaling in the patient invasive ductal carcinoma (Figure 1G). RTK arrays further revealed Tyr phosphorylated RTKs in two human tumors, which showed phosphorylation of EGFR, HER2, PDGFRβ, CSF1R, RON and EPHB2 (Figure 1H). Although our data pointed to the potential importance of Tyr phosphorylated EGFR and PDGFRβ in patient TNBC, clinical trials targeting these RTKs have largely failed (Bianchi G, 2009; Finn et al., 2009). The failure of single agent RTK inhibitors in TNBC is consistent with drug-induced activation of multiple kinases or compensatory tumor kinome responses. Since many expressed RTKs drive ERK activation, we profiled claudin-low breast cancer cells after MEK inhibition (e.g. AZD6244 currently in clinical trials), to determine if dynamic kinome reprogramming occurs. Our goal was to define kinome alterations that would suggest a more effective, rationally designed combination therapy.
Reprogramming the kinome in response to MEK inhibition
MEK inhibitors AZD6244 or U0126 inhibited growth of SUM159 (Figure 2A) and MDA-MB-231 cells (Figure S2A). ERK remained inhibited after 4h of MEK inhibitor treatment, while MEK phosphorylation was enhanced (Figure 2B). Inhibitor treatment for 24h resulted in reactivation of ERK, demonstrating both lines overcame the initial MEK inhibition (Figures 2B and S2B). Phosphoproteomic analysis revealed loss of ERK-mediated feedback regulation of both BRAF and MEK1 (Figure 2C). Reduced phosphorylation of negative feedback sites on BRAF and MEK1 indicate escape from the suppressive feedback regulation on the ERK pathway (Ritt et al., 2010). Analysis of MIB isolated protein kinases identified 52 peptides with decreased and 59 peptides with increased phosphorylation, while the phosphorylation status of 365 phosphopeptides was unchanged after MEK inhibition (Figure S2C and Table S3). The majority of these phosphorylation sites were Ser, Thr and Pro-directed Ser/Thr sites, but pTyr changes were also included, suggesting a broad change in kinome activity in response to AZD6244.
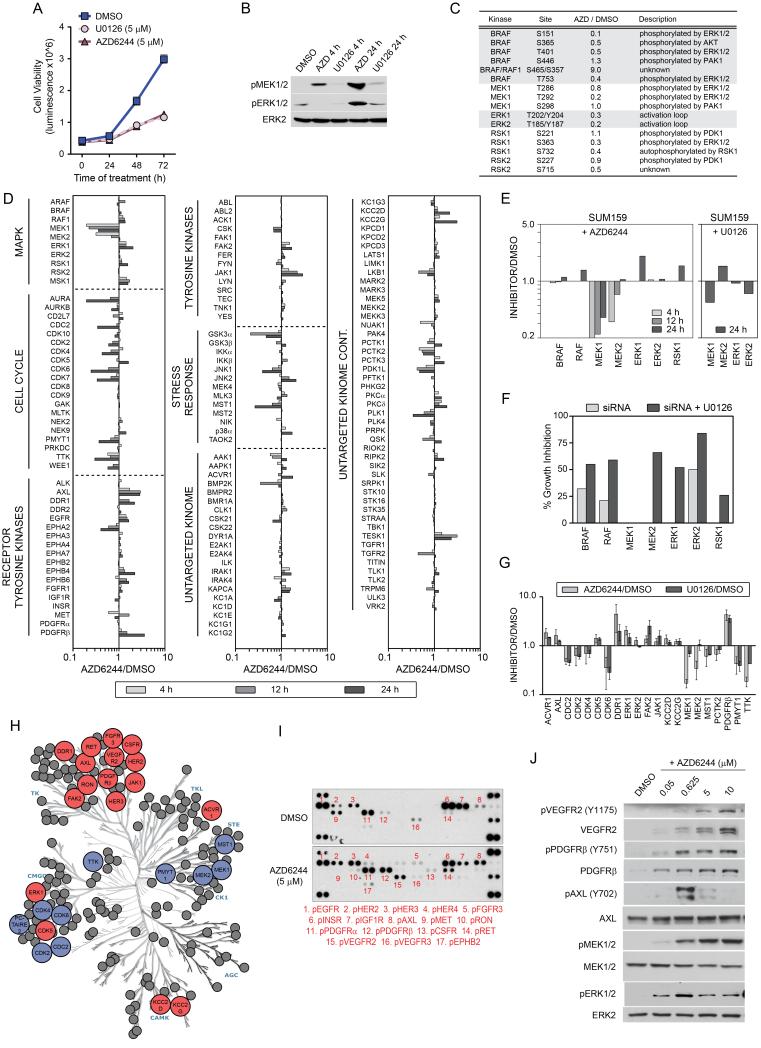
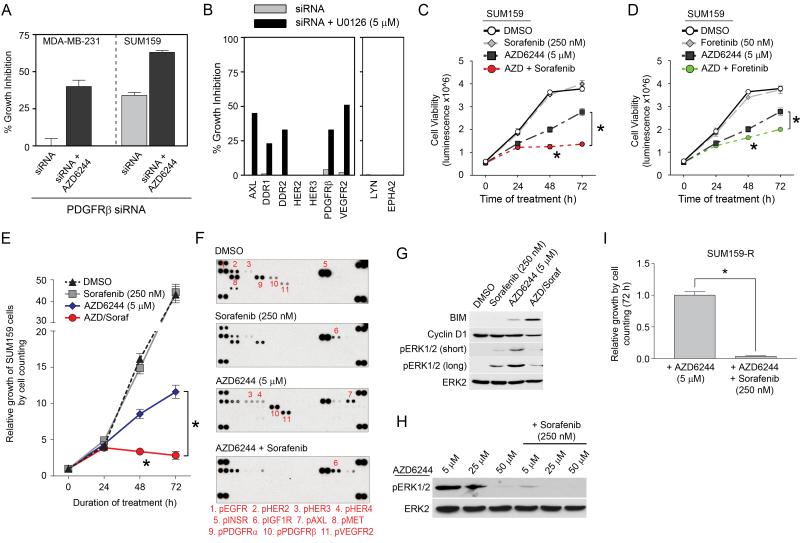
Figure 2. Reprogramming of the kinome in response to MEK inhibition.
(A) Growth inhibition of SUM159 cells in response to AZD6244 or U0126. Triplicate experiments + SD.
(B) Reactivation of MEK and ERK in the continued presence of 5 μM AZD6244 shown by western blot.
(C) Loss of ERK regulated feedback of the RAF-MEK-ERK pathway and downstream signaling. SUM159 cells were treated with 5 μM AZD6244 for 12h and kinome phosphorylation analyzed by MIB/MS.
(D) Activation and repression of the kinome in response to 5 μM AZD6244 in SUM159 cells. Line graphs show iTRAQ-determined quantitative changes in MIB binding as a ratio of AZD6244/DMSO. Ratio <1 denotes decreased and >1 denotes increased MIB binding of kinases in treated versus control cells.
(E) MEK2 and ERK1 escape AZD6244 inhibition. MIB/MS binding profile of RAF-MEK-ERK from SUM159 cells treated with 5 μM AZD6244 for 4, 12 and 24h or 5 μM U0126 for 24h.
(F) MEK2 and ERK1 promote survival following MEK inhibition. siRNA knockdown of MAPK signaling components in SUM159 cells shows loss of MEK2, but not MEK1, inhibits growth in the presence of U0126.
(G) Kinome response signature for MEK inhibition in SUM159 cells. Triplicate MIB/MS runs of SILAC-labeled SUM159 cells ± 5 μM AZD6244 or U0126 relative to DMSO. Error bars represent mean + SD where kinases are significant at FDR of 0.05.
(H) Kinome map of AZD6244 response (blue: inhibited, red: induced) as determined by MIB/MS and RTK arrays.
(I) Increased Tyr phosphorylation of RTKs in response to MEK inhibition. SUM159 cells were treated with 5 μM AZD6244 for 24h and analyzed by RTK array.
(J) Dose-dependent RTK reprogramming in response to AZD6244. Dose-dependent induction of RTK expression and activity in 24h-treated SUM159 cells was determined by western blot.
We next used MIB/MS to profile the SUM159 kinome response after exposure to AZD6244 (Figure 2D). MEK inhibition resulted in time-dependent MIB binding changes for more than 140 kinases, including cell cycle regulatory kinases, MAPK pathway kinases, RTKs, cytosolic Tyr kinases and other Ser/Thr kinases. Figure 2E highlights the MIB binding dynamics for MAPK component kinases during the time course of MEK inhibitor response in SUM159 cells. At 4h of AZD6244 treatment both MEK1 and MEK2 are inhibited, as measured by loss of MIB binding. However, while MEK1 binding remains largely inhibited, MEK2 binding to MIBs increases at 12h of treatment and by 24h was similar to control cells, indicating a return of MEK2 activity. In parallel to restored MEK2 binding to MIBs, RAF1 and ERK1 binding to MIBs increases over the time course of AZD6244 treatment, correlating with activation of these kinases. We used RNAi for each kinase in the MAPK pathway to determine if knockdown had a differential growth affect in response to MEK inhibition (Figure 2F). RNAi knockdown shows that loss of MEK2 and ERK1 inhibited SUM159 cell growth in the presence of MEK inhibitor, whereas MEK1 knockdown did not enhance growth inhibition. Taken together, these data indicate that MEK2 and ERK1 can escape from inhibition by AZD6244, suggesting a critical role for MEK2/ERK1 in SUM159 growth and survival during AZD6244 treatment.
Figures 2G and H show a 21-kinase signature defining a reprogrammed kinome in response to MEK inhibitors. This signature shows a loss of cyclin dependent kinases, consistent with growth inhibition, and increased ERK binding to MIBs indicating escape from MEK inhibition. RTKs including AXL, DDR1 and PDGFRβ, cytosolic Tyr kinases FAK2 and JAK1, and the Ser kinase ACVR1 all showed increased MIB binding. While MDA-MB-231 cells have a somewhat less robust kinome response to AZD6244, they displayed a significant kinome reprogramming that included a strong increase in PDGFRβ binding to MIBs (Figure S2D).
RTK arrays confirm the increased Tyr phosphorylation of multiple RTKs, including PDGFRβ and AXL in response to MEK inhibition (Figures 2I and S2E). In SUM159 cells AZD6244 also significantly increased Tyr phosphorylation of VEGFR2 and RET. The AZD6244 response of SUM159 cells is dose-dependent (Figure 2J), as PDGFRβ and VEGFR2 show increased RTK phosphorylation and expression with increasing AZD6244. These results demonstrate that a significant number of kinases were induced in response to MEK inhibition. Relevant to the changes in the kinome to MEK inhibition, Figure S2F lists the 40 highest expressed kinase transcripts of a patient claudin-low tumor. Of these 40 kinases, 14 (24%) were dynamically regulated in SUM159 and/or MDA-MB-231 cells in response to AZD6244, suggesting patient tumors could have a similar kinome reprogramming in response to targeted kinase inhibition.
MEK inhibition deregulates transcription, expression and activation of RTKs
Figures 3A and S3A define the time course of kinome reprogramming to AZD6244 in SUM159 and MDA-MB-231 cells. MEK and ERK were rapidly inhibited, allowing accumulation of MKP3, the MAPK phosphatase that inactivates ERK (Sandrine Marchetti, 2005). Increased MKP3 expression combined with AZD6244 to strongly suppress ERK activity, but MKP3 protein was lost as MAPK pathway activity returned. Over time, VEGFR2, PDGFRβ and DDR1 expression was increased with AZD6244 treatment, as was the phosphorylation of HER3 and AXL. qRT-PCR analysis of SUM159 (Figure 3B) and MDA-MB-231 (Figure S3B) cells treated with AZD6244 demonstrated elevated RNA levels for several of these RTKs, including DDR1/2, PDGFRβ, VEGFR2 (SUM159 only) and HER2/3. Analysis of cytokine RNA expression showed EGF, Gas6, PDGFB and PDGFD induction, indicating the establishment of autocrine/paracrine loops for RTK activation (Figures 3C and S3C). RTK arrays further showed a time dependent increase in Tyr phosphorylation of PDGFRβ, VEGFR2 and HER2/3 (DDR1/2 are not on the array) (Figure 3D). PDGFRβ, whose RNA and protein expression was induced in response to AZD6244, was phosphorylated at Tyr 751, 857 and 1009; sites required for PDGFRβ activation and recruitment of PI3K and PLCγ (Figure 3E).
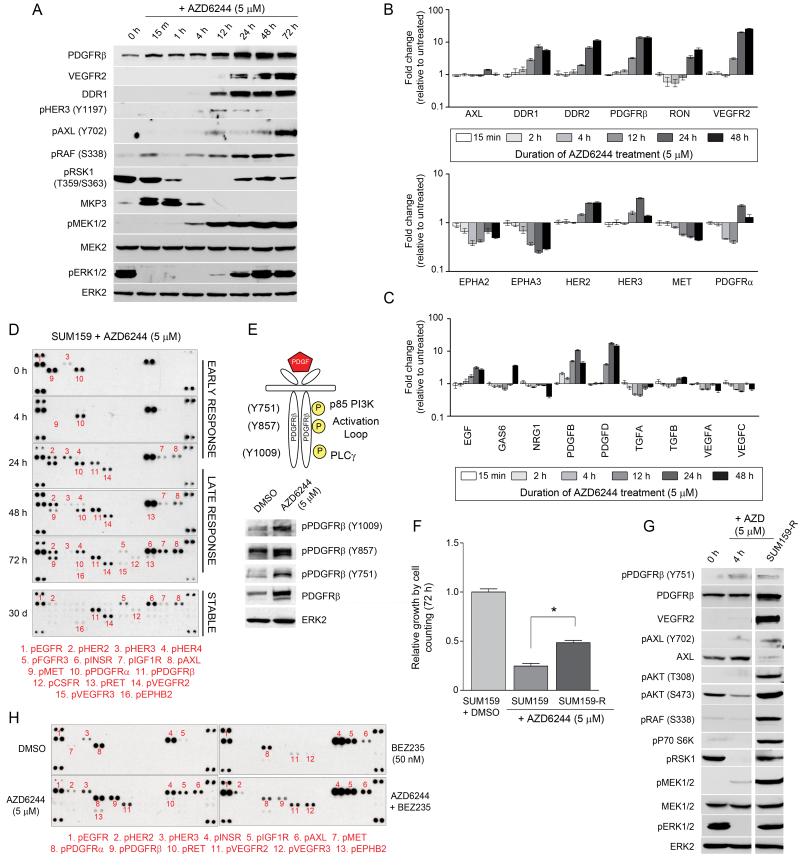
Figure 3. AZD6244-induced kinome reprogramming is target specific and involves rapid, stable transcriptional upregulation of RTKs and cytokines.
(A) AZD6244 treatment initially causes ERK inhibition and accumulation of MKP3. With prolonged AZD6244 treatment, increased RTK expression and downstream survival signaling coincides with the reactivation of RAF-MEK-ERK signaling.
(B) Time-dependent increase in RTK and (C) cytokine gene expression in SUM159 cells after AZD6244 treatment, determined by qRT-PCR.
(D) Prolonged treatment of SUM159 cells with AZD6244 leads to stable upregulation of RTKs. SUM159 cells were treated with DMSO or AZD6244 for 4-72h or 30d and RTK tyrosine phosphorylation determined by RTK antibody arrays.
(E) Treatment with AZD6244 enhances phosphorylation of PDGFRβ at multiple sites, including the activation loop.
(F) Generation of AZD6244-resistant SUM159 cells following stable treatment with AZD6244. Increased cell growth of SUM159-R cells compared to SUM159 cells treated with AZD6244 for 72h determined by cell counts (*p-value<0.001).
(G) Maintenance of RTK reprogramming in SUM159-R cells accompanied by increased survival signaling. Kinome reprogramming in SUM159 cells treated with DMSO or AZD6244 for 4h was compared to SUM159-R cells by western blot.
(H) AZD6244 and BEZ235 are target-specific in their reprogramming of kinome response. BEZ235 induces a kinase response different from AZD6244 in SUM159 cells, despite similar growth inhibition.
Error bars represent triplicate experiments ± S.D.
See also Figure S3.
After 30d of continuous exposure to AZD6244, SUM159 cells have become significantly resistant to MEK inhibitor-induced growth arrest (Figure 3F). Expression of cyclins A2 and B1 have recovered, consistent with increased proliferation (Figure S3D). The AZD6244-resistant cells (SUM159-R; grown continuously in 5 μM AZD6244) continue to have a reprogrammed kinome where PDGFRβ and VEGFR2 exhibited both increased expression and Tyr phosphorylation, and AXL showed increased Tyr phosphorylation (Figures 3D and G). Activation of these RTKs is accompanied by increases in phosphorylated AKT, RAF, p70 S6 kinase, MEK, ERK and RSK1, showing that the cells overcame MEK inhibition by RTK activation of the ERK, AKT and mTOR pathways (Figure 3G).
These findings indicate that targeted MEK inhibition significantly alters the activity of multiple kinases. It was therefore important to determine if the changes in kinase activity were specific for MEK inhibition or a function of growth arrest. BEZ235 is a dual PI3K/mTOR inhibitor that strongly inhibits SUM159 cell growth (Figure S3E). BEZ235 inhibited p70 S6 kinase activity consistent with mTOR inhibition but had no effect on the ERK pathway (Figure S3F). We compared the SUM159 kinome responses to BEZ235 and AZD6244 to determine if kinome reprogramming was target-specific or a function of growth arrest. Whereas AZD6244 induced PDGFRβ, VEGFR2 and AXL phosphorylation, BEZ235 treatment did not change the RTK phosphorylation profile except for increased phosphorylation of INSR, IGF1R and AXL (Figure 3H). MIB/MS confirmed that AZD6244 altered the kinome differently from BEZ235, indicating that drug-induced kinome reprogramming is target-specific (Figure S3G).
MEK-ERK inhibition induces c-Myc degradation leading to RTK reprogramming
ERK phosphorylates the transcription factor c-Myc at Ser62 and stabilizes c-Myc protein by preventing its proteasomal degradation (Sears R, 2000). Treatment of both SUM159 and MDA-MB-231 cells with AZD6244 caused rapid loss of c-Myc protein and mRNA (Figures 4A, S4A and 4B). This AZD6244-mediated repression of c-Myc protein and transcript, along with reduced phosphorylation of c-Myc at Ser62 (Figures 4B, S4A-C), resulted in decreased Myc-Max heterodimerization that is required for Myc transcriptional regulation (Figure 4C) (Marampon F, 2006; Wierstra and Alves, 2008). Despite partial recovery of MEK-ERK activation after 24-72h, total c-Myc expression remained repressed in the continued presence of AZD6244 (Figures 4A-C and S4A-C).
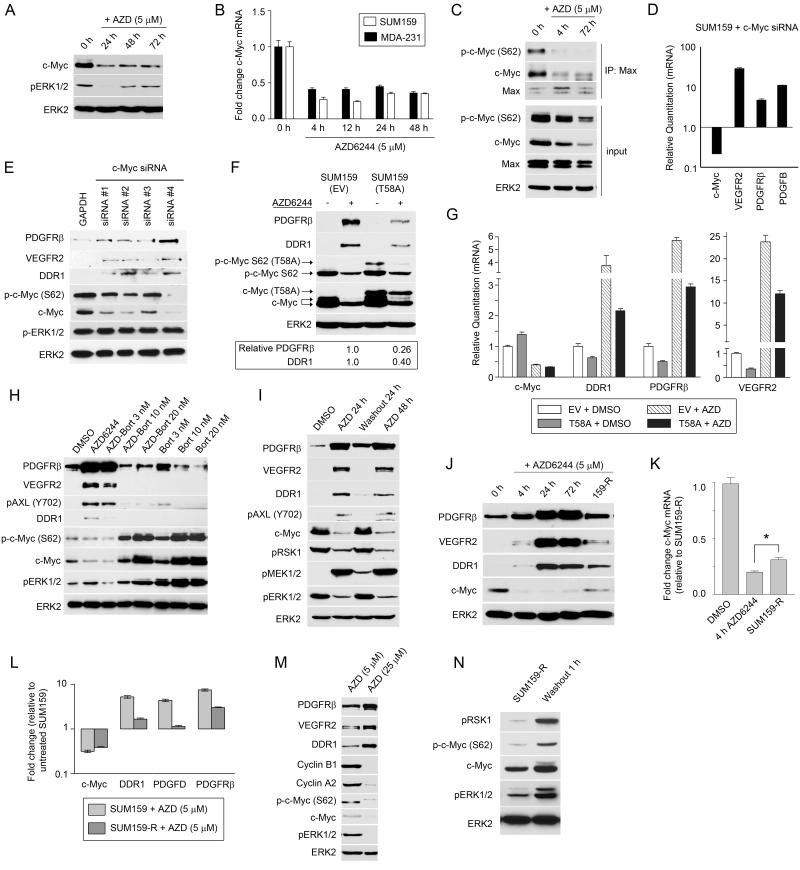
Figure 4. AZD6244–mediated loss of ERK1/2 causes rapid degradation of c-Myc, destabilization of Myc-Max complexes and promotion of RTK expression.
(A) Loss of ERK1/2 activity following AZD6244 treatment promotes c-Myc degradation. SUM159 cells were treated with AZD6244 and monitored by western blot.
(B) Stable suppression of c-Myc RNA levels following AZD6244 treatment. MDA-MB-231 and SUM159 cells were treated with AZD6244 and c-Myc gene expression determined by qRT-PCR.
(C) Disruption of Myc-Max complexes following AZD6244 treatment. SUM159 cells were treated with AZD6244 for 0, 4 and 72h. Nuclear extracts were immunoprecipitated with anti-Max antibodies and immunoblotted for Myc and Max.
(D) RNAi knockdown of c-Myc using pooled siRNA for 72h in SUM159 cells upregulates gene expression of PDGFRβ, VEGFR2 and PDGFB as determined by qRT-PCR.
(E) RNAi knockdown of c-Myc upregulates PDGFRβ, VEGFR2 and DDR1 expression. SUM159 cells were transfected with deconvolved c-Myc or GAPDH siRNA for 48h and kinome reprogramming determined by western blot.
(F) Retroviral expression of non-degradable c-Myc(T58A) in SUM159 cells suppresses RTK reprogramming after 24h treatment with 5 μM AZD6244, as shown by western blot and quantified by densitometry.
(G) Transcript levels of DDR1, PDGFRβ and VEGFR2 are reduced by the expression of c-Myc(T58A) in the presence and absence of 5 μM AZD6244. SUM159 cells were treated for 24h and analyzed by qRT-PCR.
(H) Stabilization of c-Myc protein levels by bortezomib prevents AZD6244-mediated kinome reprogramming. SUM159 cells were treated with AZD6244 (5 μM) or bortezomib alone or in combination for 24h. Bortezomib blocked the AZD6244-dependent induction of RTKs as shown by western blot.
(I) Removal of AZD6244 results in stabilization of c-Myc protein and reversal of RTK reprogramming. AZD6244 was removed from the media of SUM159 cells after 24h of treatment and cells were grown without AZD6244 for a further 24h.
(J) c-Myc protein levels partially return in SUM159-R cells, while AZD6244-mediated RTK reprogramming is reduced but still maintained. SUM159 cells were treated with AZD6244 and RTK and c-Myc levels compared to SUM159-R cells by western blot.
(K) Increased c-Myc RNA levels in SUM159-R cells relative to AZD6244 treated SUM159 cells. SUM159 cells were treated with DMSO or 5 μM AZD6244 for 4h and c-Myc gene expression compared to SUM159-R cells using qRT-PCR (*p-value<0.001).
(L) AZD6244-induced RTK expression is maintained at reduced levels in SUM159-R cells. qRT-PCR was used to compare gene expression in SUM159 cells treated with AZD6244 for 24h or SUM159-R cells relative to DMSO-treated cells.
(M) c-Myc stabilized by RTK-mediated ERK activation in SUM159-R cells. RTK reprogramming and c-Myc levels were determined by western blot comparing SUM159-R treated with AZD6244 for 24h.
(N) RTK-mediated reactivation of ERK is incomplete in the continued presence of AZD6244. AZD6244 (5 μM) was removed from media of SUM159-R cells for 1h and ERK1/2 phosphorylation of c-Myc and RSK1 determined by western blot.
Error bars represent triplicate experiments ± S.D.
See also Figure S4.
c-Myc binds the promoter of human PDGFRβ (Figure S4D) to repress PDGFRβ expression (Oster et al., 2000). To define the role of c-Myc loss in the AZD6244 reprogramming response, we applied RNAi to knockdown expression of c-Myc; the effect was analogous to the reprogrammed RTK and cytokine response seen with AZD6244 treatment (Figures 4D, 4E and S4E). Similar to the AZD6244 response, knockdown of c-Myc induced expression of PDGFRβ, VEGFR2 and PDGFB, and increased Tyr phosphorylation of PDGFRβ, VEGFR2, HER3 and AXL. RNAi knockdown of ERK1/2 confirmed that ERK inhibition was the primary signal inducing loss of c-Myc mRNA expression in the AZD6244 reprogramming of the kinome. Dual ERK1/2 knockdown resulted in reduced c-Myc and increased PDGFRβ expression (Figure S4F). Thus, reprogramming of RTKs in response to AZD6244 occurs by loss of ERK-mediated stabilization of c-Myc and the subsequent transcriptional derepression of RTKs and cytokines that are negatively regulated by c-Myc. BEZ235 inhibition of mTOR and PI3K inhibited cell growth but did not change ERK activity, c-Myc expression (Figure S4G) or RTK reprogramming (Figure S3G), confirming the specificity of MEK-ERK in controlling c-Myc levels.
Proteasomal degradation of c-Myc lacking Ser62 phosphorylation triggers AZD6244-induced kinome reprogramming. Expression of a non-degradable c-Myc(T58A) mutant in SUM159 cells significantly blocked AZD6244-mediated induction of PDGFRβ, DDR1 and VEGFR2 (Figures 4F and G). GSK3β promotes c-Myc degradation, and inhibition of GSK3β stabilized c-Myc protein to repress the induction of PDGFRβ (Figure S4H). Similarly, treatment of SUM159 or SUM159-R cells with the proteasome inhibitor bortezomib prevented AZD6244-mediated c-Myc degradation, blocked c-Myc mRNA repression, and inhibited the induction of PDGFRβ, DDR1 and VEGFR2 (Figures 4H, S4I-M). Washout of AZD6244 from SUM159 or SUM159-R cells led to increased ERK activity, stabilization of c-Myc expression and subsequent loss of RTK reprogramming (Figures 4I, S4N-P). Thus, stabilizing c-Myc protein levels prevented the onset of RTK reprogramming to AZD6244 and reversed the reprogramming in SUM159-R cells. Taken together, these findings show that AZD6244-induced c-Myc proteasomal degradation is responsible for kinome reprogramming and RTK upregulation.
In SUM159-R cells c-Myc protein, mRNA levels and Myc-Max heterodimers have partially returned due to reactivated ERK stabilizing c-Myc (Figure 4J, 4K and S4Q). This correlates with SUM159-R cells having an increased growth rate compared to cells acutely treated with MEK inhibitor (Figure 3F). The level of c-Myc protein, however, is insufficient to completely repress RTK expression, which remains elevated compared to control cells but at lower levels than cells treated with AZD6244 for 4-72h (Figures 4K and L). A 5-fold increase in AZD6244 concentration inhibited ERK activation in SUM159-R cells (Figure 4M) because the higher dose of MEK inhibitor more effectively prevented RTK-stimulated reactivation of MEK-ERK signaling. As expected, the resulting loss of phospho-c-Myc S62 and total c-Myc protein led to a corresponding increase in RTK expression in SUM159-R cells. Notably, the return of phospho-ERK in the continued presence of AZD6244 was insufficient to completely reverse RTK reprogramming, suggesting ERK may not be fully reactivated. This was shown by measuring the phosphorylation of two ERK substrates, RSK1 and c-Myc, after only 1h of AZD6244 washout from SUM159-R cells (Figure 4N). Phosphorylation of both substrates and phospho-ERK was increased, demonstrating further activation of ERK shortly after the removal of MEK inhibitor. Thus, the combination of persistent c-Myc transcriptional repression and partial MEK-ERK reactivation allows the maintenance of RTK reprogramming, leading to MEK inhibitor resistance.
RTK reprogramming rescues cells from AZD6244-induced growth arrest
RNAi knockdown of PDGFRβ in SUM159 cells enhanced growth inhibition by AZD6244 (Figure 5A), indicating the induction of RTK signaling was critical for growth and survival of cells inhibited by AZD6244. To test the role of additional RTKs in the rescue response of cells to MEK inhibition, we performed siRNA knockdown of RTKs found to be transcriptionally induced and/or Tyr phosphorylated in response to U0126 in SUM159 (Figure 5B) and MDA-MB-231 cells (Figure S5A). As controls we used siRNA to knockdown BRAF, RAF1 and ERK1/2; knockdown of each pathway member enhanced growth arrest observed with MEK inhibition (Figures S5A and B). Knockdown of PI3K and AKT produced a greater growth arrest response in SUM159 than MDA-MB-231 cells, consistent with mutant PI3K being a driver in SUM159 cells. siRNA knockdown of LYN and EPHA2 had no effect on the growth of either cell type in the presence or absence of MEK inhibitor (LYN and EPHA2 show no change or loss of MIB binding in response to MEK inhibitor). While knockdown of HER2 or HER3 had little effect in SUM159 and MDA-MB-231 cells, knockdown of AXL, DDR1, DDR2, PDGFRβ and VEGFR2 each resulted in a strong synthetic lethal-like effect in the presence of U0126 (Figures 5B, S5A, S5C and S5D). Thus, loss of MEK-ERK signaling causes induction of multiple RTKs, each contributing to the subversion of MEK inhibition.
Figure 5. RTK inhibition synergizes to enhance AZD6244-induced growth arrest.
(A) RNAi knockdown of PDGFRβ enhances AZD6244-induced growth arrest. PDGFRβ knockdown was performed in the presence or absence of 1.25 μM AZD6244 in MDA-MB-231 and SUM159 for 96h and cell growth monitored using Cell-Titer Glo.
(B) RNAi knockdown of MEK inhibitor-responsive RTKs in SUM159 cells synergizes with U0126 to inhibit growth. Cell proliferation was determined at 96h of treatment by Cell-Titer Glo.
(C) Cotreatment with AZD6244 and sorafenib synergizes in cell growth inhibition of SUM159 cells, as determined by Cell-Titer Glo.
(D) Cotreatment of SUM159 cells with AZD6244 and foretinib enhances growth inhibition, as determined by Cell-Titer Glo.
(E) Cell counting confirms synergistic growth inhibition of combined AZD6244 and sorafenib treatment in SUM159 cells.
(F) Sorafenib inhibits AZD6244-mediated activation of RTKs. SUM159 cells were treated as indicated for 72h and RTK Tyr phosphorylation determined by RTK antibody arrays.
(G) Cotreatment of SUM159 cells with AZD6244 and sorafenib enhances inhibition of ERK activity and primes cells for apoptosis. SUM159 cells were treated with for 72h and BIM, cyclin D1 expression and ERK activity determined by western blot.
(H) AZD6244 activation of ERK1/2 requires RTK and MEK activity. Inhibition of ERK activity in SUM159-R cells occurs after treatment with 50 μM AZD6244 or cotreatment of 5 μM AZD6244 with 250 nM sorafenib.
(I) SUM159-R cells require AZD6244-induced RTK activity for drug resistance. SUM159-R cells treated with 250 nM sorafenib are growth arrested over 72h, as determined by cell counts.
*p-value<0.001; Error bars represent triplicate experiments ± S.D.
AZD6244 in combination with RTK inhibitors
Our results suggested RTK inhibitors in combination with AZD6244 could block the growth-promoting activity of the reprogrammed kinome. Given the repertoire of AZD6244-activated RTKs, we tested sorafenib and foretinib alone or in combination with AZD6244 for their ability to inhibit cell growth (Figures 5C, 5D, S5E and S5F, see Table S4 for targeted kinases by each inhibitor). Whereas the two RTK inhibitors were ineffective as single agents, both were synergistic in inhibiting cell growth in combination with AZD6244, with sorafenib being most effective. Cell counting assays reinforced the strong synergistic growth arrest of SUM159 cells with the AZD6244/sorafenib combination (Figure 5E); RTK arrays validated that sorafenib inhibited Tyr phosphorylation of multiple RTKs induced by AZD6244 (Figures 5F and S5G). The combination of AZD6244/sorafenib enhanced the inhibition of ERK1/2, decreased cyclin D1 levels and increased expression of the pro-apoptotic Bim protein compared to AZD6244 alone, indicating the cells were primed for apoptosis (Figures 5G and S5H).
Sorafenib inhibits PDGFRα and β, VEGFR2 and DDR1/2, but is also an inhibitor of BRAF and RAF. Therefore, we assayed the action of different RAF inhibitors in combination with AZD6244 to determine if the effect of sorafenib could be mimicked by other BRAF/RAF inhibitors (Figures S5I and J). The RAF inhibitor SB590885 inhibited MDA-MB-231 proliferation in combination with AZD6244 to a similar extent as sorafenib, consistent with the role of BRAF activation downstream of the observed RTK reprogramming as a driver of the proliferation. RAF inhibitors, but not sorafenib, in combination with AZD6244 actually stimulated the growth of SUM159 cells (Figure S5I), consistent with the known activation of wild-type RAF signaling by both PLX4720 and SB590885. At 250-500 nM, only sorafenib synergistically inhibited growth of SUM159 cells in combination with AZD6244. Thus, sorafenib in combination with AZD6244 inhibits growth of SUM159 cells more effectively than BRAF inhibitors by cotargeting induced RTKs.
SUM159-R cells that have become resistant to AZD6244 rely on RTK-driven reactivation of ERK for drug resistance. If a 10-fold higher dose of AZD6244 is used, ERK activity can be inhibited (Figure 5H). At the 5 μM of AZD6244 that was used to develop SUM159-R cells, the addition of sorafenib inhibited ERK activity and cell growth (Figure 5I), confirming that AZD6244-induced activation of upstream RTKs drives ERK reactivation. In SUM159-R cells the combination of low doses of AZD6244 and sorafenib was similarly effective as high dose AZD6244 at inhibiting ERK activation and cell growth (Figures 5H and I).
Kinome reprogramming in the C3Tag TNBC GEMM
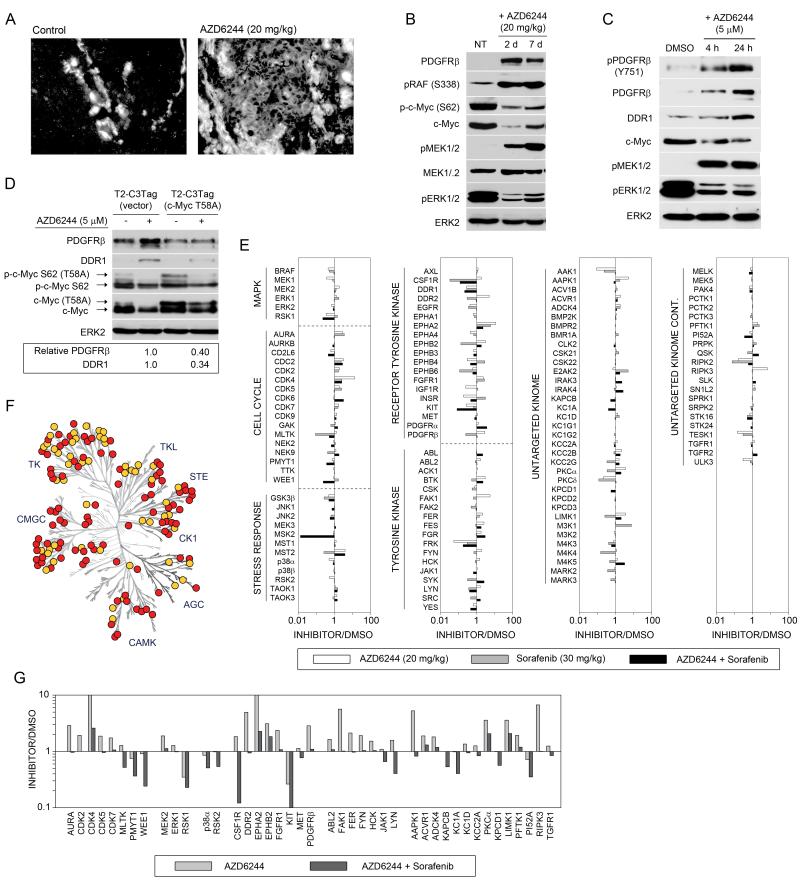
The genetically engineered C3Tag mouse model has a gene expression signature similar to human TNBC. To define AZD6244-mediated kinome reprogramming in vivo, we harvested tumor tissue before or after oral delivery of AZD6244. Figures 6A and S6 show increased expression of PDGFRβ in response to AZD6244 in both the tumor cells and stroma of C3Tag tumors, demonstrating in vivo induction of PDGFRβ. Rapid degradation of c-Myc protein and induction of PDGFRβ was observed in 2d and 7d AZD6244-treated tumors, consistent with loss of c-Myc repression of RTK expression (Figure 6B). A C3Tag-derived breast cancer cell line (T2-C3Tag) isolated from the GEMM tumor responded to AZD6244 with upregulation of PDGFRβ and DDR1, confirming the tumor cell response to MEK inhibitor (Figure 6C). Expression of non-degradable c-Myc(T58A) in T2-C3Tag cells prevented the induction of PDGFRβ and DDR1, further indicating that proteasomal degradation of c-Myc is responsible for RTK reprogramming in C3Tag tumor cells (Figure 6D).
Figure 6. AZD6244-mediated kinome reprogramming in C3Tag mouse model of TNBC.
(A) PDGFRβ is induced in C3Tag tumors after 2d AZD6244 treatment, as shown by anti-PDGFRβ immunofluorescence.
(B) AZD6244 treatment of C3Tag mice for 2 and 7d causes c-Myc degradation and induced PDGFRβ expression, as shown by western blot.
(C) Tumor-derived C3Tag cell line shows AZD6244-mediated c-Myc loss and induction of RTKs. T2-C3Tag cells were treated with AZD6244 and RTK reprogramming determined by western blot.
(D) Expression of c-Myc(T58A) in T2-C3Tag cells suppresses AZD6244-mediated RTK reprogramming. T2-C3Tag cells stably expressing vector or human c-Myc(T58A) were treated with AZD6244 for 24h and analyzed by western blot.
(E) MIB/MS profile of C3Tag tumors in response to AZD6244 for 28d, sorafenib for 26d, or combined AZD6244 and sorafenib for 26d show distinct kinome responses. The line graphs show iTRAQ-determined quantitative changes in MIB binding as a ratio of inhibitor/untreated. Ratio <1 denotes decreased MIB binding and >1 increased MIB binding of kinases in treated versus control tumors.
(F) AZD6244 plus sorafenib inhibits AZD6244-induced kinome response in C3Tag tumors, as identified by MIB/MS. Changes in MIB binding (>1.5 fold) of AZD6244-responsive kinases following cotreatment with sorafenib are shown in yellow.
(G) Cotreatment of C3Tag mice with AZD6244 and sorafenib prevents upregulation of kinases observed by AZD6244 treatment alone, including CDKs and PDGFRβ. These kinases were selected from (E) with a >1.5-fold reduction in MIB binding between AZD6244 treatment alone and cotreatment with AZD6244 and sorafenib.
See also Figure S6.
Profiling kinome response to targeted combination therapies in the C3Tag mouse model of TNBC
MIB/MS was then used to define the kinome response profile of C3Tag tumors from mice treated with AZD6244, sorafenib or the combination of AZD6244 and sorafenib (Figure 6E). The MIB/MS signatures of tumors continuously treated with AZD6244 or sorafenib share some overlap but exhibit significant differences, demonstrating drug selective reprogramming of the kinome. AZD6244-treated tumors have upregulation of RTKs PDGFRβ, DDR2 and CSF1R, as well as a number of tyrosine kinases similar to the AZD6244 response in human TNBC cell lines. Importantly, the escape of MEK2 and ERK1 from AZD6244 inhibition was recapitulated in MIB/MS profiles of AZD6244-treated C3Tag tumors. Sorafenib-treated tumors showed decreased MIB binding of the previously reported sorafenib targets: BRAF, PDGFRβ, CSF1R, DDR1, DDR2, KIT, MLTK and FRK (Karaman et al., 2008). Both AZD6244- and sorafenib-treated tumors showed increased MIB-binding of cyclin-dependent kinases, indicating the tumors have circumvented the action of the single agents to reenter cell cycle progression. MIB/MS profiling of tumors treated with the combination of AZD6244 and sorafenib showed reduced MIB-binding of kinases activated by AZD6244 treatment (Figure 6F). Sorafenib inhibited AZD6244-mediated activation of RTKs PDGFRβ, DDR2 and CSF1R, as well as a number of intracellular Tyr kinases, including JAK1. RTK-driven activation of MEK2-ERK1 was inhibited by sorafenib in tumors and loss of cyclin-dependent kinase binding to MIBs was also observed, consistent with the combination of AZD6244 and sorafenib arresting tumor growth (Figure 6G).
AZD6244 plus sorafenib causes tumor regression
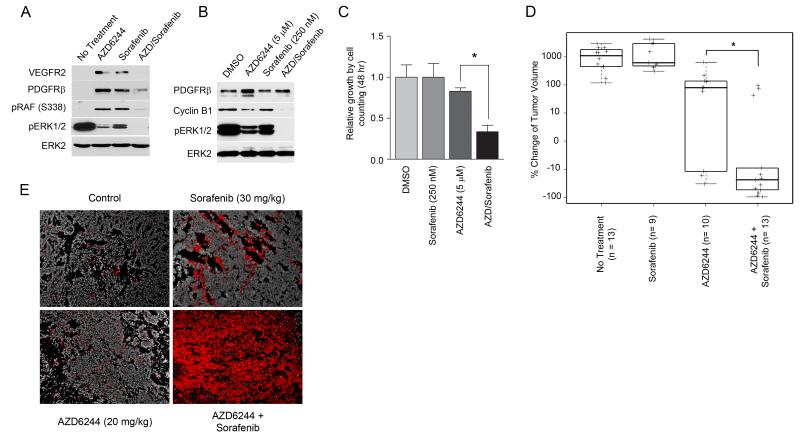
After only 2d of AZD6244 or sorafenib treatment, the expression of VEGFR2 and PDFGRβ was increased along with increased phosphorylation of RAF at Ser338, demonstrating RAF activation (Figure 7A). The combination of AZD6244 and sorafenib reduced VEGFR2 and PDGFRβ expression, suppressed RAF activation and synergistically inhibited reactivated ERK. Figure 7B shows the combination of AZD6244 and sorafenib blocked ERK activation and induction of PDGFRβ in the T2-C3Tag cell line. Cyclin B1 levels are also reduced by the combination therapy, and a strong growth arrest was observed in cells cotreated with AZD6244 and sorafenib, indicating that AZD6244 sensitizes cells to sorafenib treatment (Figure 7C).
Figure 7. Combination of AZD6244 and sorafenib causes apoptosis and tumor regression in C3Tag TNBC mouse model.
(A) AZD6244 (20 mg/kg) or sorafenib (30 mg/kg) fed in chow results in ERK activation after 2d of treatment in C3Tag GEMM, while cotreatment with AZD6244 and sorafenib inhibits RTK-mediated ERK activation. RTK reprogramming was monitored in tumors treated with AZD6244 and/or sorafenib relative to untreated tumors by western blot.
(B) Sorafenib inhibits AZD6244-dependent reactivation of ERK, promoting c-Myc degradation and loss of cyclin B1 expression in T2-C3Tag cells. T2-C3Tag cells were treated for 24h and analyzed by western blot.
(C) AZD6244 and sorafenib synergize to inhibit cell growth in C3Tag cell line. T2-C3Tag cells were treated with AZD6244 and sorafenib, alone or in combination, and cell growth determined by cell counting (*p-value<0.001; quadruplicate experiments).
(D) Cotreatment of C3Tag mice with AZD6244 and sorafenib for 21d causes significant tumor regression compared to AZD6244 alone. C3Tag mice were treated with AZD6244 (20 mg/kg), sorafenib (30 mg/kg) or the combination of AZD6244 and sorafenib and compared to untreated tumors. Percent change in tumor volume of drug treated relative to untreated is shown (* Wilcoxon p-value=0.007).
(E) Increased apoptosis of C3Tag mouse tumors following cotreatment with AZD6244 and sorafenib. Apoptosis in C3Tag tumors treated for 2d was determined by TUNEL staining (shown in red; DAPI is grayscale).
See also Figure S7.
Our findings showed that the combination of AZD6244 and sorafenib was significantly more effective in inhibiting ERK activation in 2d treated C3Tag mice and the C3Tag tumor cell line. Therefore, C3Tag mice were allowed to develop tumors and then treated for 21d with AZD6244 and/or sorafenib (Figures 7D and S7A). Sorafenib treatment alone had no effect on tumor progression, whereas 30% of the AZD6244-treated mice showed some tumor regression. In contrast, 77% of mice treated with AZD6244 and sorafenib had tumor regression, demonstrating a significantly greater effect of the combination therapy versus AZD6244 alone. TUNEL assays of the tumors showed that the combination of AZD6244 and sorafenib induced a strong apoptotic response in only 2d of treatment, in stark contrast with single drug treatment (Figures 7E and S7B).
DISCUSSION
We describe a novel approach to study the reprogramming of protein kinase networks “en masse”. Our methods allowed the isolation and analysis of protein kinases from cells and tumors with 50-60% of the expressed kinome assayed in a single mass spectrometry run. Profiling MIB binding of kinases is a highly sensitive method to simultaneously monitor activation and inhibition of numerous kinases. This profiling technique allows interrogation of kinases known by sequence but which have been understudied due to lack of biologic or phenotypic knowledge or reagent availability. An example of the latter is the ability to distinguish changes in MEK1 and MEK2.
This technique identified a kinome response signature to the selective MEK1/2 kinase inhibitor AZD6244. The only defined substrates for MEK are ERK1 and 2, yet we observed changes in activity of kinases in every subfamily of the kinome in response to MEK inhibition. Kinome assessment showed a time-dependent reprogramming that involved an early loss of ERK feedback regulation of RAF and MEK, as well as increased MKP3 protein stability. The increased expression of MKP3 functions to enhance ERK inactivation. In contrast, the loss of RAF and MEK feedback inhibition would allow upstream activation of the pathway. The time-dependent change in MIB binding of specific RTKs such as PDGFRβ and DDR1 was readily detected and provided the critical experimental observation that MEK inhibition was driving the expression and activation of multiple RTKs, each of which are capable of stimulating the RAF-MEK-ERK pathway. Importantly, we identified c-Myc degradation as a key mechanism mediating kinome reprogramming; preventing proteasomal degradation of c-Myc inhibited the reprogramming response. RNAi knockdown of ERK or c-Myc recapitulated the MEK inhibitor-induced expression and Tyr phosphorylation of several RTKs, demonstrating ERK regulation of c-Myc stability is critical in controlling the expression and activation of specific kinases. The fact that multiple RTKs are activated in response to MEK inhibition demonstrates the difficulty in using single kinase inhibitors to arrest tumor progression.
In addition to c-Myc, inhibition of AKT and mTOR also causes kinome reprogramming in different breast cancer subtypes (Chandarlapaty et al., 2011; Rodrik-Outmezguine et al., 2011). Whereas c-Myc functions as a repressor of PDGFRβ, DDR1/2 and VEGFR2 expression in claudin-low breast cancer, AKT has been shown to negatively regulate FOXO-dependent expression of HER3, IGF1R and INSR in several breast cancer cell lines. Inhibition of mTOR kinase activity leads to AKT inhibition and subsequent RTK reprogramming (Rodrik-Outmezguine et al., 2011). Differential kinome reprogramming is seen not simply with targeting the MEK-ERK and AKT pathways but with tyrosine kinase inhibitors as well. HER3 upregulation was shown to play a major role in lapatinib resistance (Garrett, 2011) and in lung cancer MET amplification leads to gefitinib resistance (Engelman et al., 2007).
Analysis of the ERK pathway in cells treated with AZD6244 showed a time-dependent rescue of BRAF/RAF, MEK2, ERK1 and RSK1 binding to MIBs. We demonstrated that MIB binding of these kinases is a function of their activation. The time course of recovery parallels that of AZD6244-induced RTK expression. The C3Tag tumor shows a comparable increase in MEK2 and ERK1 binding after AZD6244 treatment, mimicking the reprogramming response observed in SUM159 cells. Published work with a similar MEK inhibitor, GSK1120212, which binds to the MEK allosteric regulatory site (as does AZD6244) provides insight into how MEK2 escapes inhibition (Gilmartin et al., 2011). MEK phosphorylated at the activation loop serines has a 20-fold lower affinity for GSK1120212 than nonphosphorylated MEK, effectively alleviating allosteric site inhibition of MEK. Because ERK activity is increasing over time, MEK1 would be feedback phosphorylated at its negative regulatory site Thr292, preventing MEK1 reactivation even in the setting of RTK reprogramming; MEK2, however, lacks this regulatory site and selectively escapes inhibition. This suggests a unique paradigm of activation of an upstream signaling pathway increasing the IC50 of an inhibitor for a target kinase.
In many tumor types Tyr kinases are molecular drivers of transformation and also play a major role in resistance to therapy. Claudin-low SUM159 cells and the C3Tag breast cancer GEMM were remarkably similar in response to AZD6244, with induction and activation of PDGFRβ, VEGFR2, CSFR1, DDR1/2 and AXL. The claudin-low MDA-MB-231 cell line was somewhat less responsive, but still showed the induction of PDGFRβ, DDR1 and DDR2 and activation of AXL with AZD6244 treatment. RNAi knockdown of the different RTKs indicated that each kinase contributed to the survival response of SUM159 and MDA-MB-231 cells. Given the repertoire of RTKs whose expression and activity is induced with AZD6244 treatment, we predicted that the combination therapy of sorafenib and AZD6244 would “broaden” the kinase targeting sufficiently to produce significant therapeutic benefit. The combination therapy increased apoptosis and tumor regression significantly compared to either drug alone in the C3Tag TBNC GEMM.
We identified AZD6244-induced RTKs (and Ser/Thr kinases) using a combination of MIB/MS and immunoblotting of cell lines and C3Tag tumors. We created a signature of therapeutic resistance allowing a rational prediction of combinatorial therapies. This approach can be extended to human tumors using so-called “window trials” in which a patient is treated with a targeted agent prior to surgery and their tumor analyzed at excision for kinome-resistance signatures. Importantly, we have shown that the kinome response is unique for inhibitors targeting different kinases and the response of different tumor types to a common inhibitor may also vary. Thus, this systems kinome approach can be applied to help define patterns of resistance for a variety of drugs and biopsy-accessible tumor types.
EXPERIMENTAL PROCEDURES
Cell culture
MDA-MB-231 and T2-C3Tag cells were grown in DMEM/F12 supplemented with 10% FBS. SUM159 cells were grown in DMEM/F12 supplemented with 5% FBS, 1 μg/ml hydrocortisone and 5 μg/ml insulin. SUM159-R cells were continually grown in the presence of 5 μM AZD6244. For SILAC labeling, cells were grown for five doublings in arginine- and lysine-depleted media (as above) supplemented with either unlabeled L-arginine (42 mg/L) and L-lysine (71 mg/L) or equimolar amounts of [13C6,15N4]arginine (Arg10) and [13C6]lysine (Lys6) (Cambridge Isotope Laboratories). Proliferation was quantified by Cell-Titer Glo Luminescent Cell Viability Assay (Promega). Media containing kinase inhibitors was replaced daily.
MIB chromatography
Cells and tumors were harvested as previously described (Oppermann et al., 2009). Briefly, lysates were brought to 1 M NaCl and passed through columns of inhibitor-conjugated beads (Bisindoylmaleimide-X, SB203580, Lapatinib, Dasatinib, Purvalanol B, VI16832, PP58) to isolate protein kinases from the lysates (see Supplemental Procedures for MIBs preparation). Kinase-bound inhibitor beads were washed with high- and low-salt buffers and 0.1% SDS before elution in 0.5% SDS solution in high heat. Proteins were purified using chloroform/methanol extraction, resuspended in 50 mM ammonium bicarbonate (pH 8) or 50 mM HEPES (pH 8) for SILAC or iTRAQ respectively. Samples were digested overnight at 37°C with sequencing grade modified trypsin (Promega). iTRAQ labeling of digested peptides was carried out using iTRAQ 4-plex reagent (AB SCIEX) for 2h at room temperature in the dark. Peptides were fractionated with Mini SCX Spin Columns and cleaned with PepClean C18 Spin Columns (Thermo Scientific) before separation by a Tempo LC MALDI Spotting System.
MS analysis
MS and MS/MS data were acquired with a MALDI TOF/TOF 5800 (ABSCIEX) and analyzed by ProteinPilot Software Version 3.0 (ABSCIEX) with a UniProtKB/Swiss-Prot database. Proteins were accepted when ≥1 unique peptide was identified at 99% confidence. Peptide quantitation by ProteinPilot was performed using the Pro Group Algorithm and quant ratios are bias-corrected. All MIB/MS kinome response profiles can be found in Table S5. MIB/MS analysis with cell lines was done in 2-3 independent experiments. A set of 3 independent experiments using SILAC labeled SUM159 cells treated with AZD6244 or DMSO was used to assess statistical significance and reproducibility of MIB/MS (Supplementary Procedures; Table S6).
Western blotting and RTK arrays
Western blotting and RTK array analyses were performed as previously described (Amin et al., 2010). Detailed methods and antibodies are provided in Supplemental Procedures.
qRT-PCR
RNA was isolated from cell lines or murine tumors using the RNeasy® Plus Mini Kit (Qiagen). qRT-PCR was performed on diluted cDNA with the Applied Biosystems 7500 Fast Real-Time PCR System and inventoried TaqMan® Gene Expression Assays.
In vivo tumorigenesis experiments
Animal handling and procedures were approved by the UNC Institutional Animal Care and Use Committee and followed the NIH guidelines. Male C3Tag mice were bred with wildtype females to produce experimental offspring. Treatment began the same day a palpable mass was found. Drugs were incorporated into chow and food was provided ab libitum. Tumor size was assessed twice weekly by caliper measurements of tumor areas ((width)2 × length))/2 for 21d. Percent change of tumor volume was calculated using (Final volume – Initial Volume)/Initial Volume and graphed using R (http://www.r-project.org/). Tumors at harvest were halved and either snap-frozen in liquid nitrogen and stored at −80°C or placed in neutral buffered 10% formalin solution.
Human breast tissue procurement
All human breast tissue was obtained from the Tissue Procurement Facility in compliance with the laws and institutional guidelines as approved by the UNC IRB committee. Clinical specimens were phenotyped by gene expression analysis in the lab of CMP.
Supplementary Material
Highlights.
Inhibition of the MEK-ERK pathway rapidly reprograms kinome activity in tumors.
c-Myc degradation induces expression and activation of receptor tyrosine kinases.
Receptor tyrosine kinase activation overcomes MEK inhibition causing resistance.
Kinome inhibitor response profiling rationally predicts combination therapies.
ACKNOWLEDGEMENTS
This work was supported by NIH grants GM30324 and DK37871, NCI Breast SPORE CA58223 and the University Cancer Research Fund. NVJ and MW are supported by NIH training grants GM007040 and CA71341. JD was supported by CIHR postdoctoral fellowship. The authors also acknowledge support by the High-throughput Sequencing and Proteomics Cores. Kinome illustration reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com).
Footnotes
SUPPLEMENTARY DATA Supplemental data includes Supplemental Procedures, 6 tables and 7 figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Amin DN, Sergina N, Ahuja D, McMahon M, Blair JA, Wang D, Hann B, Koch KM, Shokat KM, Moasser MM. Resiliency and Vulnerability in the HER2-HER3 Tumorigenic Driver. Science Translational Medicine. 2010;2:16ra17. doi: 10.1126/scitranslmed.3000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotech. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- Bianchi G, L.S., Zamagni C, Salvagni S, Raab G, Siena S, Laferriere N, Peña C, Lathia C, Bergamini L, Gianni L. Phase II multicenter, uncontrolled trial of sorafenib in patients with metastatic breast cancer. Anticancer Drugs. 2009;20:616–624. [PubMed] [Google Scholar]
- Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Körner R, Greff Z, Kéri G, Stemmann O, Mann M. Kinase-Selective Enrichment Enables Quantitative Phosphoproteomics of the Kinome across the Cell Cycle. Molecular Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Fedorov O, Muller S, Knapp S. The (un)targeted cancer kinome. Nat Chem Biol. 2010;6:166–169. doi: 10.1038/nchembio.297. [DOI] [PubMed] [Google Scholar]
- Finn RS, Press MF, Dering J, Arbushites M, Koehler M, Oliva C, Williams LS, Di Leo A. Estrogen Receptor, Progesterone Receptor, Human Epidermal Growth Factor Receptor 2 (HER2), and Epidermal Growth Factor Receptor Expression and Benefit From Lapatinib in a Randomized Trial of Paclitaxel With Lapatinib or Placebo As First-Line Treatment in HER2-Negative or Unknown Metastatic Breast Cancer. Journal of Clinical Oncology. 2009;27:3908–3915. doi: 10.1200/JCO.2008.18.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett J, Olivares MG, Rinehart C, Granja-Ingram ND, Sanchez V, Chakrabarty A, Dave B, Cook RS, Pao W, McKinely E, Manning HC, Chang J, Arteaga CL. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. PNAS. 2011;1:6. doi: 10.1073/pnas.1016140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin AG, Bleam MR, Groy A, Moss KG, Minthorn EA, Kulkarni SG, Rominger CM, Erskine S, Fisher KE, Yang J, et al. GSK1120212 (JTP-74057) Is an Inhibitor of MEK Activity and Activation with Favorable Pharmacokinetic Properties for Sustained In Vivo Pathway Inhibition. Clinical Cancer Research. 2011;17:989–1000. doi: 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotech. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- Marampon F, C.C., Zani BM. Down-regulation of c-Myc following MEK/ERK inhibition halts the expression of malignant phenotype in rhabdomyosarcoma and in non muscle-derived human tumors. Mol Cancer. 2006;5:2–17. doi: 10.1186/1476-4598-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, Mann M, Daub H. Large-scale proteomics analysis of the human kinome. Molecular & cellular proteomics : MCP. 2009;8:1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster SK, Marhin WW, Asker C, Facchini LM, Dion PA, Funa K, Post M, Sedivy JM, Penn LZ. Myc Is an Essential Negative Regulator of Platelet-Derived Growth Factor Beta Receptor Expression. Mol Cell Biol. 2000;20:6768–6778. doi: 10.1128/mcb.20.18.6768-6778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritt DA, Monson DM, Specht SI, Morrison DK. Impact of Feedback Phosphorylation and Raf Heterodimerization on Normal and Mutant B-Raf Signaling. Mol Cell Biol. 2010;30:806–819. doi: 10.1128/MCB.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, Baselga J, Guichard S, Rosen N. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer discovery. 2011;1:248–259. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrine Marchetti CG, Jean-Claude Chambard, Thomas Touboul, Danièle Roux, Jacques Pouysségur, Gilles Pagès. Extracellular Signal-Regulated Kinases Phosphorylate Mitogen-Activated Protein Kinase Phosphatase 3/DUSP6 at Serines 159 and 197, Two Sites Critical for Its Proteasomal Degradation. Molecular and Cellular Biology. 2005;25:854–864. doi: 10.1128/MCB.25.2.854-864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, N.F., Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes and Development. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Aceto N, Meerbrey KL, Kessler JD, Zhou C, Migliaccio I, Nguyen DX, Pavlova NN, Botero M, Huang J, et al. Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell. 2011;144:703–718. doi: 10.1016/j.cell.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, Wubbenhorst B, Xu X, Gimotty PA, Kee D, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierstra I, Alves J. The c-myc Promoter: Still MysterY and Challenge. In: George FVW, George K, editors. Advances in Cancer Research. Academic Press; 2008. pp. 113–333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.