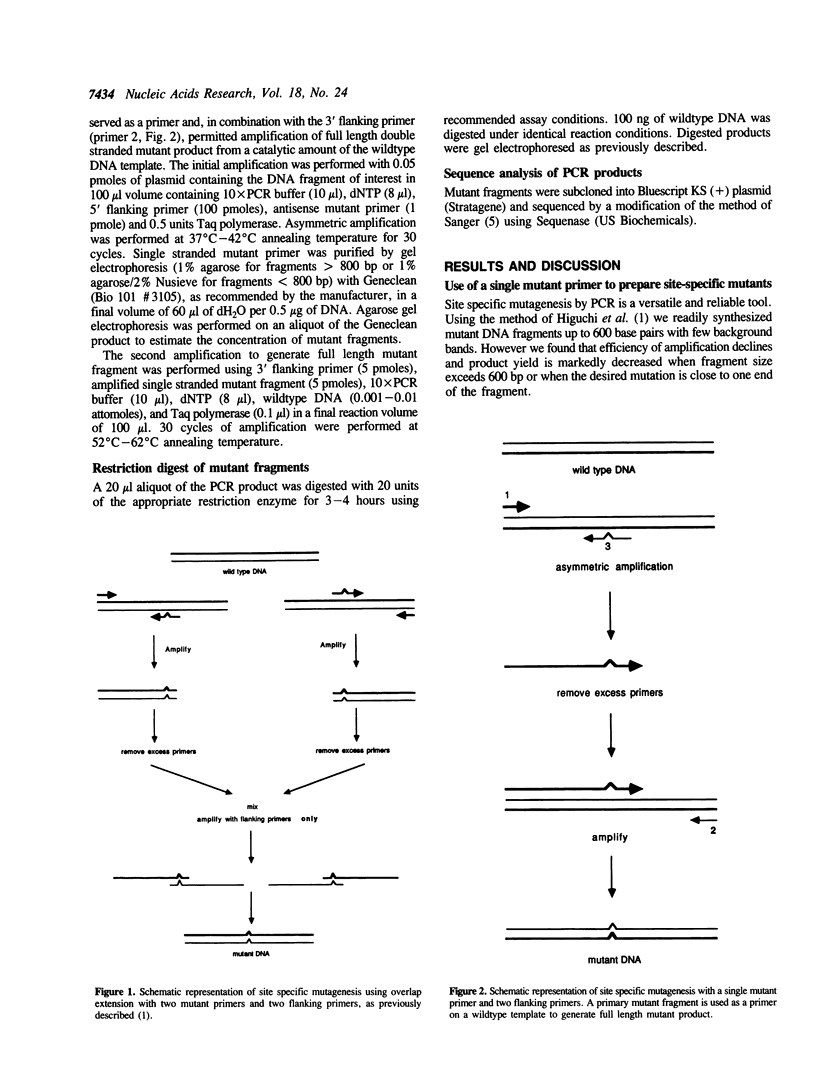
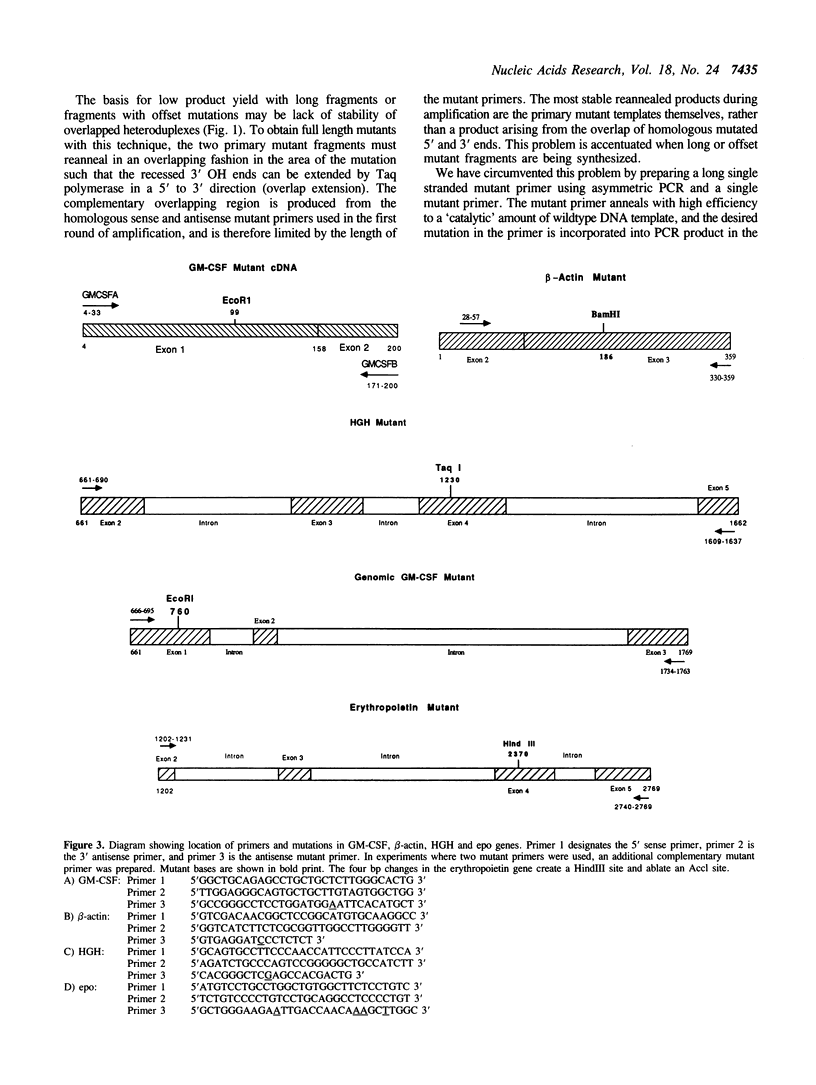
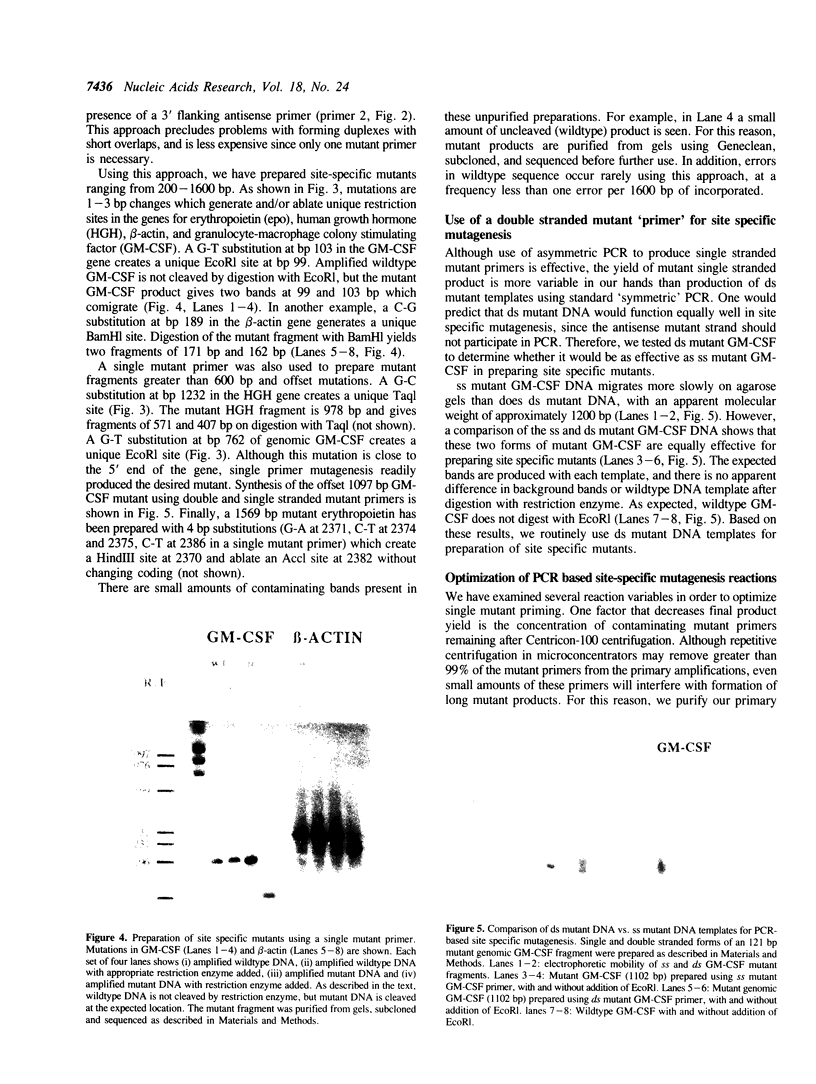
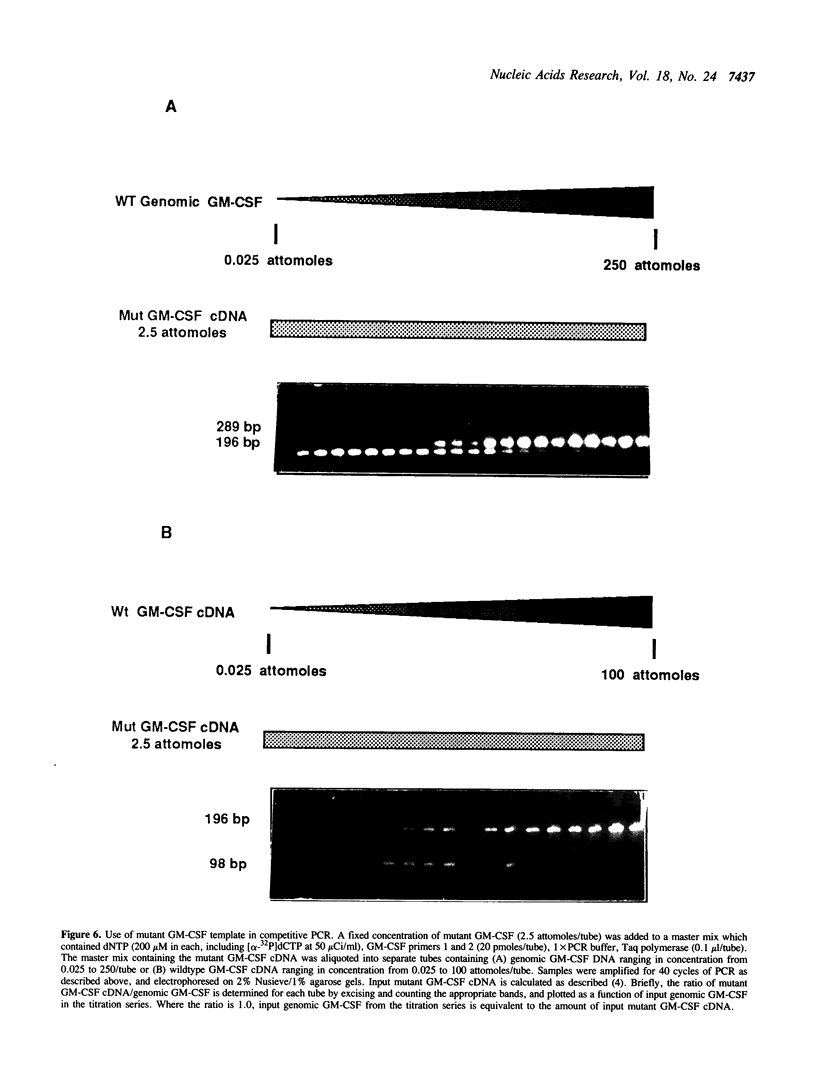
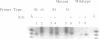Abstract
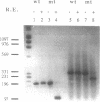
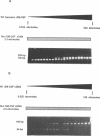
A method is described for preparing site-specific mutants using a polymerase chain reaction (PCR) based protocol. The protocol requires a single mutant primer, and has been used to introduce mutations into DNA fragments ranging in size from 200 bp to 1569 bp in length in the GM-CSF, beta-actin, human growth hormone and erythropoietin genes. Sequence analysis of PCR derived mutant fragments shows an error rate of less than one bp change per 1500 bp incorporated. Single base pair mutations have been introduced into these genes which create unique restriction sites. We demonstrate that these mutant templates may be used for competitive PCR to quantitate mRNA and DNA. This method thus offers a rapid means for producing competitive templates for use in quantitative PCR.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Higuchi R., Krummel B., Saiki R. K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988 Aug 11;16(15):7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]