Abstract
Matriptase proteolytic activity must be tightly controlled for normal placental development, epidermal function, and epithelial integrity. Although hepatocyte growth factor activator inhibitor-1 (HAI-1) represents the predominant endogenous inhibitor for matriptase and the protein molar ratio of HAI-1 to matriptase is determined to be >10 in epithelial cells and the majority of carcinoma cells, an inverse HAI-1-to-matriptase ratio is seen in some ovarian and hematopoietic cancer cells. In the current study, cells with insufficient HAI-1 are investigated for the mechanisms through which the activity of matriptase is regulated. When matriptase activation is robustly induced in these cells, activated matriptase rapidly forms two complexes of 100- and 140-kDa in addition to the canonical 120-kDa matriptase-HAI-1 complex already described. Both 100- and 140-kDa complexes contain two-chain, cleaved matriptase but are devoid of gelatinolytic activity. Further biochemical characterization shows that the 140-kDa complex is a matriptase homodimer and that the 100-kDa complexes appear to contain reversible, tight binding serine protease inhibitor(s). The formation of the 140-kDa matriptase dimer is strongly associated with matriptase activation, and its levels are inversely correlated with the ratio of HAI-1 to matriptase. Given these observations and the likelihood that autoactivation requires the interaction of two matriptase molecules, it seems plausible that this activated matriptase homodimer may represent a matriptase autoactivation intermediate and that its accumulation may serve as a mechanism to control matriptase activity when protease inhibitor levels are limiting. These data suggest that matriptase activity can be rapidly inhibited by HAI-1 and other HAI-1-like protease inhibitors and “locked” in an inactive autoactivation intermediate, all of which places matriptase under very tight control.
Keywords: hepatocyte growth factor activator inhibitor-1, autoactivation, protease inhibitor
matriptase, a type 2 transmembrane serine protease (33), plays an essential role in the formation and maintenance of the common architectural features found in all epithelial tissues. The protease also plays several of the more tissue-specific epithelial functions characteristic of specialized epithelial tissues (17). Matriptase activity is tightly regulated primarily via control of zymogen activation and the rapid suppression of proteolytic activity once activated (14). Increased matriptase expression and an imbalance of matriptase to its endogenous protease inhibitors are associated with human cancers (16, 37). A detailed understanding of the processes involved in matriptase activation and inhibition should provide important insights into its role in the development and progression of disease. Matriptase is synthesized as an inactive, single chain zymogen (1). Cleavage of the matriptase zymogen into the two-chain enzyme is thought to occur through an autoactivation mechanism that is believed to involve interaction between two matriptase zymogen molecules (25). Matriptase activation can be induced by several nonprotease factors, including sphingosine 1-phosphate in mammary epithelial cells (2), androgens in LNCaP prostate cancer cells (10), and suramin in several matriptase-expressing epithelial and carcinoma cells (12). Aspects of the extracellular environment, such as acidity or the presence of reactive oxygen species, can also stimulate cells to activate matriptase (11, 36). Once activated, matriptase must apparently immediately act on its substrates, since free-active matriptase has a very short half-life. This is because inactivation of the enzyme, through binding with its endogenous inhibitor hepatocyte growth factor activator inhibitor (HAI)-1, seems to occur almost simultaneously with the activation of the enzyme (12, 23). The unusually rapid nature of the events associated with matriptase activation and inhibition was clearly manifested in a study of matriptase-mediated activation of prostasin, a glycosylphosphatidylinositol-anchored serine protease. With the use of cultured human keratinocytes, the minute-scale kinetics of these events demonstrates that active matriptase is capable of activating prostasin in the face of the rapid HAI-1-mediated inhibition of active matriptase (5).
The unusual dynamics of this system, with matriptase zymogen activation being so tightly coupled with the inhibition of active matriptase by HAI-1, is consistent with the apparently indispensible roles of matriptase in epithelial integrity and function. Thus it is not surprising that even moderate interruption of the matriptase-HAI-1 partnership can tip the proteolysis balance and causes disease. For example, a partial shift in the matriptase:HAI-1 balance in the epidermis of matriptase transgenic mice results in a 100% lifetime incidence of skin tumors and significantly potentiates sensitivity to chemical carcinogenesis in these transgenic mice (18). When the balance of matriptase and HAI-1 is reestablished in this model by crossing the matriptase mice with HAI-1-overexpressing mice, the oncogenic potential of matriptase is completely suppressed. An imbalance between matriptase and HAI-1 has been observed in human prostate and colorectal cancer and is associated with disease progression (29, 39, 41). In spite of the imbalance between matriptase and HAI-1, most cancer cells, however, maintain significant levels of HAI-1 expression, suggesting that some level of HAI-1 expression is required by cells that express matriptase, which is consistent with other observations (4, 20, 31, 32). Several animal studies of targeted deletion of HAI-1 or HAI-1 mutation support the idea that HAI-1 is required for matriptase regulation. The defects in placenta development, epidermal barrier function, epithelial integrity, and the control of skin inflammation that are associated with HAI-1 deficiency can be reversed by simultaneous deletion of matriptase (4, 20, 31, 32). It seems plausible that in the absence of HAI-1 matriptase activity would become excessive and hazardous to cells. The absence of HAI-1 in matriptase-expressing cells is an event that would normally not occur due to the extremely high concordance of HAI-1 and matriptase in most cells (3).
There are, however, several interesting exceptions from the rule of thumb that cells that express matriptase also express HAI-1. In human ovarian cancer, particularly advanced tumors, a proportion of matriptase-positive cancer specimens appears to not express HAI-1 at measurable levels as assessed by immunoblot analyses or immunohistochemistry (22, 24). Macrophages, monocytes, and some hematopoietic cancer cells also express matriptase with no or limited levels of HAI-1 (3, 9, 35). These observations raise the interesting question as to how matriptase proteolytic activity is controlled in these cells. In the current study, we set out to investigate how matriptase activity is controlled in those cells that express very low levels of HAI-1 and have identified and characterized two novel matriptase-containing species; an activated matriptase homodimer and a matriptase complex likely with a HAI-1-like serine protease inhibitor. Both complexes are enzymatically inactive and may represent the products of alternative mechanisms that are used by HAI-1-deficient cells to control matriptase activity such that the free-active matriptase only exists for a very short period time, similar to the situation in cells with high levels of HAI-1.
MATERIALS AND METHODS
Chemicals and reagents.
Media, supplements, and buffers used in these experiments were purchased or prepared as previously described (35). Agarose-bound wheat germ agglutinin (WGA) was obtained from Vector Laboratories (Burlingame, CA).
Cell lines and culture conditions.
184 A1N4 immortalized human mammary epithelial cells were cultured in a 50:50 mixture of DMEM/Ham's F-12 medium (DMEM-F-12 50/50; Mediatech) supplemented with 0.5% FBS (Gemini Bio-Products), 5 μg/ml recombinant human insulin (rh-insulin; Invitrogen), 5 μg/ml hydrocortisone (Sigma), 10 ng/ml recombinant human epidermal growth factor (rhEGF; Promega), 100 U/ml penicillin, and 100 μg/ml streptomycin (1% pen-strep; Mediatech). T-47D cells were cultured in RPMI-1640 medium (Mediatech) supplemented with 10% FBS, 45 μg/ml rh-insulin, and 1% pen-strep. LNCaP, OVCAR-3, OV2008, RPMI 8226, and OPM2 cells were cultured in RPMI-1640 medium supplemented with 10% FBS and 1% pen-strep. All cells were maintained at 37°C in a humidified atmosphere with 5% CO2.
Monoclonal antibodies.
Human matriptase was detected using the monoclonal antibodies (mAbs) M32, 21–9, or M24. These three mAbs recognize both latent and activated matriptase, and the epitopes recognized by mAb M24 and 21–9 likely lie in the noncatalytic domains of matriptase (15, 36). M32 mAb recognizes the third LDL receptor class A domain of matriptase (25). The mouse mAb M69 recognizes an epitope present only on the activated form of the enzyme and so can specifically detect activated matriptase but does not recognize latent matriptase (1, 2). Human HAI-1 was detected using the HAI-1-specific mouse mAb M19 (13). An additional matriptase/ST14 antibody was purchased from Bethyl Laboratories (Montgomery, TX), which recognizes the serine protease domain of matriptase.
Immobilization of mAbs.
M19, M69, and 21–9 mAbs were covalently coupled to Sepharose 4B at 5 mg/ml gel following the manufacturer's instructions (GE Healthcare). Briefly, the mAbs were purified and dialyzed against the coupling buffer (0.1 M sodium bicarbonate containing 0.5 M sodium chloride) and then incubated overnight at 4°C with preactivated Sepharose beads. Uncoupled mAbs were removed by washing the beads with the coupling buffer, and any remaining coupling sites on beads were blocked using 1 M Tris buffer pH 7.4. The mAb-Sepharose beads were stored in PBS at 4°C.
Acid-driven matriptase activation.
Acid-induced matriptase activation using intact cells and cell-free extracts was carried out as described previously (11, 36), and the procedures are summarized in results(see Fig. 5A). Briefly, cells or the insoluble fractions of cell homogenates were exposed to pH 6.0 buffers for 20 min to induce matriptase activation. PBS containing 1 mM 5,5′-dithiobis-(2-nitrobenzoic acid) and 1% Triton X-100 (TX-100) was then used to prepare the lysates.
Fig. 5.
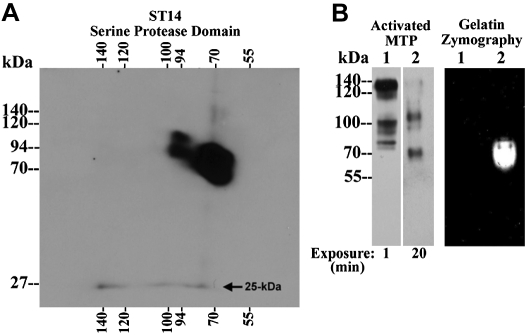
Matriptase complexes contain 2-chain, active matriptase but exhibit no gelatinolytic activity. A: diagonal gel electrophoresis analysis of matriptase complexes. Matriptase species were prepared using the cell-free activation method using extracts from OVCAR-3 cells (see Fig. 4A, lane 1, OVCAR-3). These matriptase species include 70-kDa matriptase zymogen, 94-kDa full length matriptase, 100-kDa matriptase complexes, 120-kDa matriptase-HAI-1 complex, and the 140-kDa complex. These matriptase species were resolved by 2-dimensional diagonal gel electrophoresis under boiled and reducing conditions and analyzed for the presence of 2-chain matriptase using a matriptase antibody directed against the serine protease domain of matriptase. The 25-kDa matriptase serine protease domain was indicated. B: Proteolytic activity analysis of matriptase complexes. Matriptase species from OVCAR-3 cells, prepared using the cell-free activation method (lanes 1) and from the shed fractions of RPMI 8226 cells, prepared using the intact-cell activation method (lanes 2), were analyzed by immunoblot for activated matriptase using the mAb M69 (left) and by gelatin zymography for the proteolytic activity (right). Differential exposure times to X-ray film were used for both samples in the immunoblot, as indicated.
Purification of matriptase complexes.
Samples were prepared using the method described in Acid-driven matriptase activation using cell-free extracts. Cell lysates prepared in this way from ∼600 culture dishes (150 mm) of 184 A1N4 cells were loaded onto a HAI-1 immunoaffinity column containing 1 ml mAb M19-Sepahrose (prepared as described above) at a flow rate of 5 ml/h to remove the 120-kDa matriptase-HAI-1 complex. The flow through was collected, and the resultant HAI-1 depleted fractions were then dialyzed against 20 mM Tris·HCl, pH 8.0. Insoluble debris was cleared by centrifugation, and the supernatant was loaded onto a DEAE-Sepharose FF column (2.5 × 20 cm; GE Healthcare) equilibrated with 20 mM Tris·HCl, pH 8.0. The column was washed with 10 column volumes of the equilibration buffer. Bound material was eluted with a linear gradient of 0–1 M NaCl in DEAE equilibration buffer using a total volume of 500 ml. Fractions (10 ml) were collected and assessed by immunoblotting with the matriptase mAb M24. Acid-activated cell lysates prepared from LNCaP cells (∼200 culture dishes) were loaded onto WGA columns (1.0 × 5 cm; Vector) equilibrated with PBS containing 1% TX-100. The columns were washed with four column volumes of equilibration buffer, and bound proteins were eluted with equilibration buffer containing 0.5 M N-acetylglucosamine (GlcNAc). The fractions containing matriptase complexes were pooled, and the 120-kDa matriptase-HAI-1 complex was removed by mAb M19-Sepahrose column as described above for the 184A1N4 cells. The HAI-1-depleted matriptase-containing fractions prepared in this way from 184 A1N4 or LNCaP cells were then further purified by sequential affinity chromatography using activated matriptase mAb M69 columns followed by total matriptase mAb 21–9 immunoaffinity columns. After being loaded, the columns were washed with 1% TX-100 in sodium phosphate-citric acid buffer, pH 6.0, and bound proteins then were eluted using 0.1 M glycine HCl, pH 2.4. Fractions were immediately neutralized using 2 M Trizma base.
Immunoblotting.
The protein concentration was determined using BCA protein assay reagents (Pierce, Rockford, IL) according to the manufacturer's protocol. Proteins were resolved on 7.5% gels by SDS-PAGE under nonreducing and nonboiled conditions (unless otherwise specified), transferred to Protran nitrocellulose membranes (Schleicher & Schuell, Keene, NH), and probed with the antibodies described above. Binding of the primary antibody was detected with the use of horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) and visualized using the Western Lightening Chemiluminescence Reagent Plus (Perkin-Elmer, Boston, MA).
Diagonal two-dimensional gel electrophoresis.
The various species of matriptase prepared from OVCAR-3 cells were first resolved by SDS-PAGE under nonreducing and nonboiled conditions. A vertical strip was then sliced from the gel, boiled in 1× SDS sample buffer containing 50 mM dithiothreitol, placed on the top of a new SDS-acrylamide gel, and electrophoresed in the second dimension. Protein bands were detected by Western blot using the matriptase/ST14 antibody that recognizes the serine protease domain.
Transient transfection assays.
A cDNA clone containing the full human HAI-1 coding sequence in the vector pcDNA 3.1 (25) was used in transient transfection studies with the empty pcDNA 3.1 as a control. Transient transfection of the HAI-1 construct into OVCAR-3 cells was accomplished using an Amaxa Nucleofector with Nucleofector kit VCA-1002 and program T-21. After nucleofection, the cells were immediately transferred and cultured in 6-well plates. Cells were harvested and analyzed two days after transfection.
Densitometry.
Images of the films from Western blot studies were generated using a Canon CanoScan 4400F scanner, and the intensity of the protein bands was determined using Image J software.
RESULTS
Matriptase activity is tightly controlled in most cells due to the high molar ratio of HAI-1 to matriptase.
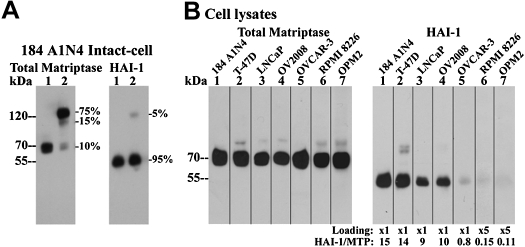
For most matriptase-expressing cells, the membrane-bound protease is detected by immunoblotting as a 70-kDa zymogen and HAI-1 as a 55-kDa species (Fig. 1A, lanes 1). To estimate the molar ratio of HAI-1 to matriptase at the protein level, zymogen activation of matriptase was induced by exposing the cells to pH 6.0 buffer for 20 min, a procedure we have previously described as the “intact-cell matriptase activation system” (36). This results in the vast majority of matriptase zymogen being converted into the active species and then almost immediately inhibited by binding to HAI-1 to form a 120-kDa matriptase-HAI-1 complex (Fig. 1A, lanes 2). Since the 120-kDa complex can be detected by both the matriptase mAb M24 and the HAI-1 mAb M19, the molar ratio of HAI-1 to matriptase can be estimated by comparing the ratio of HAI-1 in the 120-kDa complex to total HAI-1 with the ratio of matriptase in the 120-kDa complex to total matriptase. With the use of 184 A1N4 mammary epithelial cells as an example, HAI-1 in the 120-kDa complex represents only 5% of total HAI-1 (Fig. 1A, lane 2 HAI-1). In contrast, matriptase in the 120-kDa complex represents 75% of total matriptase. Since the amount of 120-kDa matriptase-HAI-1 complex is the same in both immunoblots, the molar ratio of HAI-1 to matriptase can be estimated to be ∼15 by dividing 75% by 5%. In addition to the 120-kDa matriptase-HAI-1 complex, a small portion (15%) of the matriptase was detected at 110-kDa. This band is made up of multiple complexes that are comprised of matriptase bound with a variety of other protease inhibitors, including antithrombin (6).
Fig. 1.
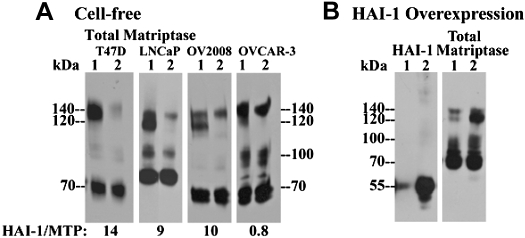
Ratios of hepatocyte growth factor activator inhibitor-1 (HAI-1) and matriptase protein levels relative to each other vary significantly among different cells. A: ratio of HAI-1 to matriptase in 184 A1N4 mammary epithelial cells. Cells were exposed to pH 6.0 buffer in the presence (lanes 1) or in the absence of 0.15 M sodium chloride at room temperature for 20 min to induce matriptase activation (lanes 2). Cells were lysed and analyzed by immunoblot for total matriptase using the mAb M24 and for HAI-1 using the mAb M19, as indicated. Ratios of individual matriptase species relative to total matriptase were determined from their protein band densities using a densitometer. Ratios of individual HAI-1 species relative to total HAI-1 were also determined, as indicated. B: ratios of HAI-1 relative to matriptase in a panel of matriptase-expressing cells. Matriptase levels were analyzed by immunoblot using matriptase mAb M24 among 7 different cells, and the loaded cell lysates were adjusted to yield similar levels of matriptase (total matriptase, lanes 1–7). HAI-1 levels of these 7 cells were further determined by immunoblot using HAI-1 mAb M19 against the normalized matriptase levels for 184 A1N4 (lanes 1), T-47D (lanes 2), LNCaP (lanes 3), OV2008 (lanes 4), and OVCAR-3 (lanes 5) and 5-fold matriptase levels for RPMI-8226 (lanes 6) and OPM2 (lanes 7) due to their extremely low levels of HAI-1. Band densities of matriptase and HAI-1 were determined by densitometer. Ratios of HAI-1 relative to matriptase for individual cells were determined by comparing to those of 184 A1N4 cells.
The ratio of HAI-1 to matriptase levels in 184 A1N4 cells was then used as the basis upon which to estimate the HAI-1-to-matriptase ratios in other matriptase-expressing cell lines in which the approach used in the 184 A1N4 cells could not be used for various reasons. This includes cells in which active matriptase is also inhibited by other protease inhibitors or forms homodimers (see below) or where matriptase zymogen is rapidly shed. The amount of lysate to be analyzed from seven different cells was first normalized with respect to the level of matriptase (Fig. 1B, total matriptase). HAI-1 levels were then compared among the seven cells lines (Fig. 1B, HAI-1). After these procedures, we estimate that the HAI-1-to-matriptase ratio in T47D, LNCaP, and OV2008 cells is ∼14:1, 9:1, and 10:1, respectively (Fig. 1B). In contrast, the ratio was found to be <1 in several cell lines, including OVCAR-3 ovarian cancer cells and RPMI 8226 and OPM2 multiple myeloma cells (Fig. 1B). The low expression of HAI-1 in the ovarian cancer and multiple myeloma cells appear to be consistent with observations made in clinical materials where loss of HAI-1 expression relative to matriptase levels has been documented in advanced human primary ovarian cancer (22, 24) and aggressive B-cell lymphomas and multiple myelomas (F. P. Chou and C.-Y. Lin, unpublished observations).
Activated matriptase is detected in three complexes in cells with HAI-1-to-matriptase ratios <1.
The low levels of HAI-1 expressed by these three cell lines allowed us to investigate alternative mechanisms for the control of matriptase in addition to inhibition by HAI-1. To do this, robust matriptase activation was induced and cell lysates were assayed for the appearance of various forms of activated matriptase (Fig. 2). In addition to 70-kDa latent matriptase, several matriptase complexes, including species at 140- and 120-kDa and an apparent doublet at 100-kDa were detected in the cell lysates of the three lines (Fig. 2, lanes 1, total MTP). These matriptase complexes appear to all contain activated matriptase as they were also detected by the mAb M69 (Fig. 2, lanes 1, activated MTP) that can specifically distinguish activated matriptase from matriptase zymogen (1, 2). The specificity of the mAb M69 for the cleaved (2-chain) form of matriptase was further confirmed in the current study. All of the matriptase species recognized by this mAb (see Fig. 5B, lane 1, activated MTP) could be dissociated by reducing agents, resulting in the release of the serine protease domain (see Fig. 5A). The 120-kDa species was shown to be a matriptase-HAI-1 complex, as expected, since it can be specifically immunodepleted by immobilized M19 HAI-1 mAb (Fig. 2, lanes 2). It is interesting to note that the activated matriptase mAb M69 apparently recognizes the 140-kDa complex better than the total matriptase mAb M24. Similarly the 100-kDa matriptase complexes were not detected or detected only at very low levels in the majority of high HAI-1-matriptase ratio cells, including 184 A1N4 cells (Fig. 1A). LNCaP prostate cancer cells are an interesting exception in which the 100-kDa complex was detected at high levels (see Fig. 6). These data suggest that cells with a low HAI-1-to-matriptase ratio may employ other mechanisms in addition to HAI-1 to control matriptase activity.
Fig. 2.
Fate of activated matriptase in HAI-1-deficient cells. 184 A1N4, OVCAR-3, RPMI 8226, and OPM2 cells were exposed to a pH 6.0 buffer for 20 min to induce matriptase activation. Cell lysates (lanes 1) were further incubated with HAI-1 mAb M19-Sepharose beads to immunodeplete the 120-kDa matriptase-HAI-1 complex (lanes 2). Matriptase (MTP) species of both samples were detected by immunoblot analysis using matriptase mAb M24 (lanes 1 in each cells) and activated matriptase mAb M69.
Fig. 6.
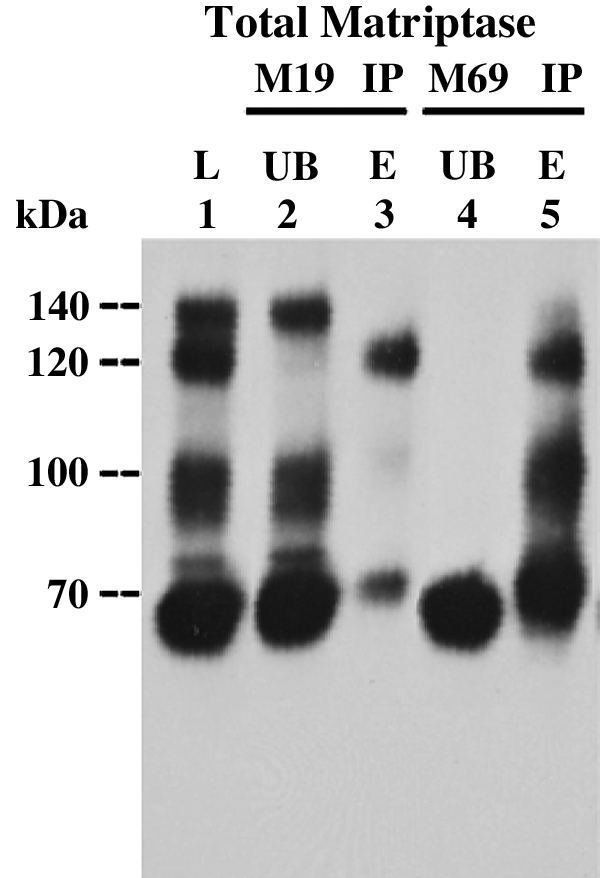
Stability and reversibility of matriptase complexes. LNCaP prostate cancer cell extracts were subjected to cell-free matriptase activation. Lysates (L, lane 1) were incubated with HAI-1 mAb M19-Sepharose beads (M19 IP) or activated matriptase mAb M69-Sepharose beads (M69 IP), respectively. After removal of the unbound fractions (UB, lane 2 and 4), the mAb-captured proteins were eluted by 0.1 M glycine buffer, pH 2.4 and immediately neutralized (lanes 3 and 5). Lysate, unbound fractions, and the eluted fractions (E) were analyzed for matriptase species by immunoblot using matriptase mAb M32.
Formation of the 140-kDa activated matriptase complex is a common event associated with matriptase activation and is reversely regulated by HAI-1.
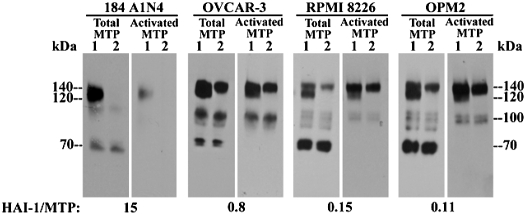
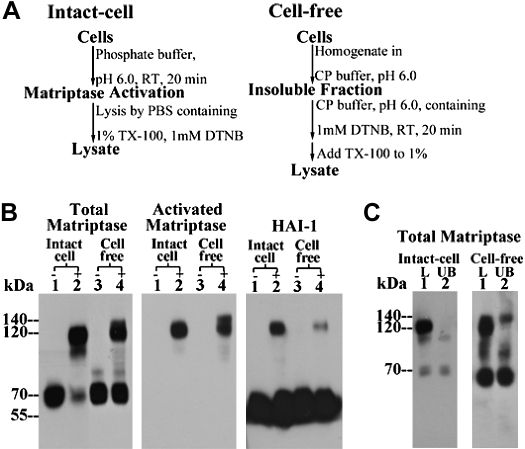
Although using the intact-cell matriptase activation system the 140-kDa complex was seen only in the cells with a low HAI-1-to-matriptase ratio, a complex of this size has frequently been observed when using a complementary matriptase activation system: the “in vitro, cell-free matriptase activation system” (11), regardless of the ratio of HAI-1 to matriptase in the cells used (Figs. 3 and 4). The cell-free matriptase activation system, similar to the intact-cell matriptase activation system, is also an acid-driven event. The procedures for both activation systems are outlined in Fig. 3A. Under regular culture conditions and at physiological pH, matriptase resides on the cell surface or inside the cell and is maintained in its zymogen form (36). In the in vitro, cell-free system, the cells are first homogenized by Dounce homogenizer and the internal matriptase residing in the perinuclear area of the cell is harvested along with the insoluble cell fractions by centrifugation. When these insoluble fractions, or intact cells, are exposed to pH 6.0 buffer, matriptase zymogen undergoes robust activation leading to the formation of activated matriptase complexes (Fig. 3B, comparing lanes 2 with lanes 1 or lanes 4 to lanes 3). With the use of the intact-cell system on 184 A1N4 cells, as described above, the vast majority of activated matriptase is in a HAI-1 complex that can be detected by a HAI-1 mAb in immunoblot analysis (Fig. 3B, lane 2, HAI-1) and immunodepleted using immobilized HAI-1 mAb M19 (Fig. 3C, comparing lanes 2 with lanes 1, intact cell). When using the cell-free activation system, however, in addition to the 120-kDa HAI-1 complex (Fig. 3B, lane 4 and Fig. 3C, lane 1, cell free), a 140-kDa complex, can be discerned, although not clearly, above the 120-kDa HAI-1 complex by immunoblot analysis using the total matriptase mAb M24 (Fig. 3B, lane 4, total matriptase). Both the 140- and 120-kDa complexes can, however, clearly be seen as two distinct protein bands when using the activated matriptase mAb M69 due to its different affinity for both complexes (Fig. 3B, lane 4, activated matriptase). The 140-kDa complex was not detected by the HAI-1 mAb M19 in immunoblot analysis (Fig. 3B, lane 4, HAI-1) and is not removed by HAI-1 immunodepletion (Fig. 3C, lane 2, cell free). The 140-kDa complex observed in the cell-free system, therefore, contains no HAI-1, consistent with its 140-kDa counterpart observed in the intact-cell activation system when using low HAI-1-to-matriptase ratio cells (Fig. 2). The appearance of the 140-kDa activated matriptase complex in the cell-free activation system is not unique to 184 A1N4 cells, having been seen with all cells tested. In Fig. 4, the 140-kDa matriptase complex can be clearly seen in extracts from LNCaP, OV2008, and OVCAR-3 cells (Fig. 4A, lanes 1). The presence of the 140-kDa complex can be further confirmed by immunodepletion of the 120-kDa complex (Fig. 4A, lanes 2). We were interested to note a loose inverse correlation between the level of the 140-kDa complex and the molar ratio of HAI-1 to matriptase was observed among these cells. This led us to further test the apparent inverse relationship between the ratio of HAI-1 to matriptase and the ratio of the 140-kDa complex to 120-kDa matriptase-HAI-1 complex by transiently transfecting OVCAR-3 cells with HAI-1 to increase the HAI-1-to-matriptase ratio. As can be seen, this intervention significantly reduced the ratio of the 140-kDa:120-kDa complex providing additional evidence for the relationship (Fig. 4B).
Fig. 3.
Formation of 140-kDa matriptase complex is induced in the cell-free matriptase activation system using 184 A1N4 cells that express high levels of HAI-1. A: flow chart of matriptase activation in the intact-cell and in vitro, cell-free systems. B: less robust matriptase activation in the cell-free activation system compared with that of intact-cell system. Living cells (lanes 1 and 2) or the insoluble fractions (lanes 3 and 4) of cell homogenates of 184 A1N4 mammary epithelial cells were exposed to pH 6.0 buffer alone (lanes 2 and 4, +) or in the presence of 150 mM NaCl as nonactivation control (lanes 1 and 3, −) at room temperature for 20 min. Lysates were prepared and analyzed by immunoblot for total matriptase using mAb M24, for activated matriptase using the mAb M69, and for HAI-1 using the mAb M19, as indicated. C: generation of the 140-kDa matriptase complex in 184 A1N4 cells by in vitro, cell-free matriptase activation method. 184 A1N4 cells were exposed to pH 6.0 buffer to induce matriptase activation in either intact-cell or cell-free settings, as indicated. Lysates were incubated with immobilized HAI-1 mAb M19 to deplete the 120-kDa matriptase-HAI-1 complex. Lysates (L, lane 1) and HAI-1-depleted fractions (UB, lane 2) were analyzed by immunoblot for matriptase species using total matriptase mAb M24. TX-100, Triton X-100; DNTB, 5,5-dithio-bis(2-nitrobenzoic acid); RT, room temperature.
Fig. 4.
Formation of the 140-kDa complex is a common event associated with cell-free matriptase activation and regulated by HAI-1 levels. A: 140-kDa matriptase complex in a panel of cells. Matriptase activation was induced in the cell-free setting using extracts from four different cells, as indicated. Lysates were incubated with HAI-1 mAb M19-Sepharose beads to deplete the 120-kDa matriptase-HAI-1 complex. Lysates (lanes 1) and HAI-1-depleted lysates (lanes 2) were analyzed by immunoblot for total matriptase using the mAb M24. Ratios of HAI-1 to matriptase for each cell line were also indicated. B: ratio of 140-kDa complex relative to 120-kDa matriptase-HAI-1 complex decreases with increased HAI-1 levels. HAI-1 was transiently overexpressed in OVCAR-3 cells. Matriptase activation was induced in the cell-free setting using the mock-transfected cells (lanes 1) and HAI-1-overexpressing cells (lanes 2). Lysates of both cells were analyzed for total matriptase using the mAb M24 and for HAI-1 using the mAb M19, as indicated.
The 140- and 100-kDa matriptase complexes contain two-chain matriptase but are enzymatically inactive.
The detection of the 140- and 100-kDa matriptase complexes by mAb M69 suggests that both complexes contain two-chain, activated matriptase, similar to that found in matriptase-HAI-1 complexes (1, 13). We tested this hypothesis by analyzing these matriptase complexes using two-dimensional, diagonal gel electrophoresis. Matriptase species were prepared from OVCAR3 cells by exposing cell homogenates, rather than the living cells, to the pH 6.0 buffer in a cell-free setting. In addition to the 70-kDa matriptase zymogen and the 94-kDa full-length matriptase (Fig. 4A, OVCAR-3), this preparation also contains the 140-, 120-, and 100-kDa activated matriptase complexes that can be detected by both the total matriptase mAb M24 (Fig. 4A, OVCAR-3) and the antibody specific for activated matriptase (Fig. 5B, left, lane 1, activated MTP). A matriptase species with a size slightly >70-kDa was also detected by the activated mAb (Fig. 5B, left, lane 1, activated MTP). As shown in Figs. 2, 3, and 4, the level of the 140-kDa complex relative to the 120-kDa matriptase-HAI-1 complex is significantly increased in this cell-free model, compared with what is observed using intact cells. This preparation, therefore, allows us to analyze the 140-kDa complex in the absence of significant levels of the 120-kDa matriptase-HAI-1 complex. The various matriptase species were analyzed by diagonal gel electrophoresis with first dimension under nonboiled and nonreducing conditions and second dimension under boiled and reducing conditions. Under this condition, the two-chain matriptase will be dissociated into a 45-kDa fragment of the noncatalytic domains and a 25-kDa fragment of the serine protease domain, as shown in our previous study (1). In contrast, matriptase zymogen will be detected as a signal-chain form. Immunoblot analysis of these matriptase species was performed using an antibody that recognizes the serine protease domain of matriptase. This analysis clearly shows that the 25-kDa matriptase serine protease domain was dissociated from the four matriptase species detected by the activated matriptase mAb (Fig. 5A). In contrast, matriptase zymogen and full-length matriptase were detected at 70- and 94-kDa, respectively, consistent with their single chain nature.
We next determined if these matriptase complexes are enzymatically active by gelatin zymography (Fig. 5B, right). The sample prepared from OVCAR-3 cells, containing the 140-, 120-, and 100-kDa activated matriptase complexes (Fig. 5B, left, lane 1, activated MTP), was analyzed by gelatin zymography (Fig. 5B, right, lane 1, gelatin zymography). This assay revealed that none of these matriptase complexes exhibits any detectable gelatinolytic activity. Interestingly, the two-chain matriptase species with a size slightly >70-kDa was also devoid of gelatinolytic activity. In contrast, a positive control consisting of free-active matriptase demonstrated strong gelatinolytic activity at 70-kDa, as expected (Fig. 5B, right, lane 2, gelatin zymography). The 70-kDa free-active matriptase was prepared from the shed fractions of RPMI-8226 multiple melanoma cells after exposed to a pH 6.0 buffer (6). The amount of active matriptase protein loaded on the gel (Fig. 5B, left, lane 2, activated MTP) was considerably less than the matriptase present in the 140- and 100-kDa activated matriptase complexes (Fig. 5B, left, lane 1, activated MTP), was determined by immunoblot using mAb M69 and employed different exposure times for the evaluation of both samples (Fig. 5B, left, activated MTP). These matriptase species were clearly seen after a 1 min exposure to the X-ray film. In contrast, it took an exposure of 20 min to detect the free-active matriptase. These data suggest that both the 140- and 100-kDa matriptase complexes are enzymatically inactive, as has previously been shown for the 120-kDa complex (1).
Differential acid-sensitive stability and reversibility of activated matriptase complexes.
One of the interesting biochemical features of the interaction between active matriptase and HAI-1 is the property of acid-sensitive, reversible complex formation (1), which can be demonstrated in an immunoprecipitation-immunoblotting analysis (Fig. 6). This acid-sensitive interaction is a common feature of the interaction between serine proteases and those serine protease inhibitors that employ a reversible tight binding mechanism to inhibit serine proteases (19). Lysates prepared from LNCaP cells, which contains the three matriptase complexes at similar levels (Fig. 6, lane 1), were used to this stability and reversibility test. The 120-kDa matriptase-HAI-1 complex (Fig. 6, lane 1) was first immunoprecipitated using immobilized HAI-1 mAb (Fig. 6, lane 2). The captured 120-kDa matriptase-HAI-1 complex was then eluted by a pH 2.4 buffer and immediately neutralized with Trizma base. After this procedure, the majority of the matriptase-HAI-1 complex regains its 120-kDa complex form with some limited dissociation resulting in a matriptase band detected as 70-kDa band (Fig. 6, lane 3). To test whether the 140- and 100-kDa matriptase complexes also exhibit the acid-sensitive and reversible interactions between their subunits exhibited by the 120-kDa complex, we captured all three activated matriptase complexes using immobilized M69 activated matriptase mAb (Fig. 6, lane 4). All three complexes were then eluted using a pH 2.4 buffer and immediately neutralized. After these procedures, the 100-kDa complexes and the 120-kDa complex are recapitulated (Fig. 6, lane 5). These data suggest that the 100-kDa species, like the matriptase-HAI-1 complex, is most likely comprised of matriptase bound with a serine protease inhibitor like HAI-1, in an acid labile, reversible complex. In contrast, the 140-kDa matriptase complex is not recovered after the immunoprecipitation, low pH elution, and neutralization procedures, and concurrent with the disappearance of this species, high levels of activated 70-kDa matriptase monomer are found in the sample (Fig. 6, lane 5).
The 140-kDa complex is a matriptase homodimer and the 100-kDa complexes contain matriptase and a HAI-like protease inhibitor.
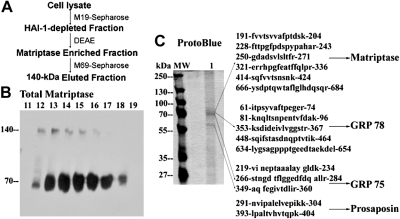
To identify the subunits present in both the 140- and 100-kDa matriptase complexes, we purified these complexes from two different cell lines subjected to in vitro, cell-free activation using slightly different methods. The first purification was carried out using lysates prepared from 184 A1N4 mammary epithelial cells, which contain the 140-kDa complex, the 120-kDa complex, and matriptase zymogen as seen in Fig. 3B, lane 4, total matriptase. While the level of the 140-kDa complex is very low in 184A1N4 cells, the absence of 100-kDa complex allowed us to purify the 140-kDa complex without contamination with the 100-kDa complex. A three-step purification scheme was employed to purify the 140-kDa species, as outlined in the purification scheme in Fig. 7A. Purification started with the removal of the 120-kDa complex using the HAI-1 mAb M19. The HAI-1-depleted fractions were then partially purified by DEAE ion exchange chromatography (Fig. 7B). Both the 140-kDa matriptase complex and the matriptase zymogen were eluted from DEAE at almost the same salt concentrations, suggesting that both matriptase species resemble one another with respect to their biochemical behavior on DEAE chromatography. The final step of purification was carried out using the M69 mAb immobilized on Sepharose, which can specifically capture the 140-kDa matriptase complex but not matriptase zymogen. Therefore, the 140-kDa complex can be purified without contamination of the 70-kDa matriptase zymogen. The proteins eluted from the M69 affinity column were analyzed by SDS-PAGE followed by ProtoBlue staining of the gel and found to contain two major protein bands at 70- and 60-kDa (Fig. 7C, lane 1). Proteomic protein identification of the proteins that make up the bands on the gel was conducted through the use of in-gel trypsin digestion, mass spectrometric analysis of the resultant tryptic fragments, and database research (Prottec, Norristown, PA). This analysis revealed that the 70-kDa protein band contained three proteins; matriptase, glucose-regulated protein 78, and glucose-regulated protein 75 (Fig. 7C). The 60-kDa protein band was identified as prosaposin. In spite of these data, however, using antibodies against the three proteins identified, we have been unable to generate any evidence that any of these proteins are in fact present in the 140-kDa matriptase complex. Therefore, these copurified proteins are not likely to be the matriptase binding partner in the 140-kDa complex. Given the apparent molecular mass of matriptase (70-kDa) combined with the lack of evidence that these copurified proteins are part of the 140-kDa complex, we came to the conclusion that the 140-kDa complex is most likely a homodimer of activated matriptase.
Fig. 7.
The 140-kDa complex is homodimer of activated matriptase. A: purification scheme for the 140-kDa matriptase complex. The 140-kDa matriptase complex was purified from 184 A1N4 lysates through a 3-step purification scheme. Complex preparation was first separated from the 120-kDa HAI-1 complex by HAI-1 M19-Sepharose, followed by DEAE and activated matriptase mAb M69 immunoaffinity chromatography. B: partial purification of 140-kDa matriptase complex by DEAE chromatography. HAI-1-depleted fractions were further purified by DEAE chromatography. Column fractions (11–19) were analyzed using total matriptase mAb M24. Both 140-kDa complex and latent matriptase were eluted in similar fractions 12–18. There was no matriptase-HAI-1 complex in these DEAE fractions. C: immunoaffinity purification and proteomic protein identification of 140-kDa matriptase complex. The 140-kDa matriptase complex-enriched DEAE fractions (from Fig. 7B) were then purified by immunoaffinity chromatography with activated matriptase mAb 69. Eluted proteins were resolved by SDS-PAGE under nonreducing and nonboiled condition and the gel stained with ProtoBlue (lane 1). The 70-kDa matriptase dissociated from the 140-kDa matriptase complex eluted as major proteins from immunoaffinity column and one 60-kDa minor protein was also seen in eluted fractions. Protein molecular weight markers (MW) are indicated. Proteomic protein identification of the subunits of the 140-kDa matriptase complex revealed that there were 14 tryptic fragments derived from the 70-kDa protein spots matched to 6 stretches of amino acid sequences of matriptase; 5 fragments matched to 5 stretches of amino acid sequences of GRP 78 and 3 fragments matched to 3 stretches of amino acid sequences of GRP 75. The 60-kDa spots yielded 2 fragments matched to 2 stretches of amino acid sequences of prosaposin. These tryptic fragments were presented by their amino acid sequences.
The 140-kDa matriptase complex was also purified from lysates prepared from LNCaP prostate cancer cells using a similar purification scheme in which immobilized WGA was used to replace DEAE chromatography. In addition to the 120-kDa matriptase-HAI-1 and 140-kDa complex, the LNCaP cell lysates also contains the 100-kDa matriptase complex (Fig. 6). The 120-kDa matriptase-HAI-1 complex was removed by HAI-1 mAb immunoaffinity chromatography, and then both 140- and 100-kDa complexes were purified using M69 mAb Sepharose. Immunoblot analysis using the total matriptase mAb M24 revealed that the eluates from M69-Sepharose contain the 100-kDa complexes and 70-kDa matriptase dissociated from the 140-kDa complex (Fig. 8, lane 1, total matriptase), consistent with the differential stability and reversibility of both matriptase complexes as shown in Fig. 6. The 100-kDa complex was dissociated by incubation in SDS sample buffer containing no reducing agents at 95°C for 5 min (Fig. 8, lane 2, total matriptase), suggesting the noncovalent nature of the subunit interaction in the 100-kDa complex. Although immunoblot analysis clearly showed the presence of the 100-kDa complex in the eluates, we only saw a major band at 70-kDa and a 60-kDa minor band but no 100-kDa band by Commassie blue staining (Fig. 8, lane 1, ProtoBlue). Nevertheless, proteomic protein identification was carried out using the three gel strips sliced out at the sizes equivalent to 100-, 70-, and 60-kDa. As expected, the 70-kDa slice contained matriptase and the 60-kDa slice contained prosapsin (sequences not shown), similar to the results obtained with lysates from the 184 A1N4 cells (Fig. 7). These data further support the hypothesis that the 140-kDa complex is indeed a matriptase homodimer. Taken together, the purification and proteomic protein identification data provide strong evidence that the 140-kDa complex is a homodimer of activated matriptase.
Fig. 8.
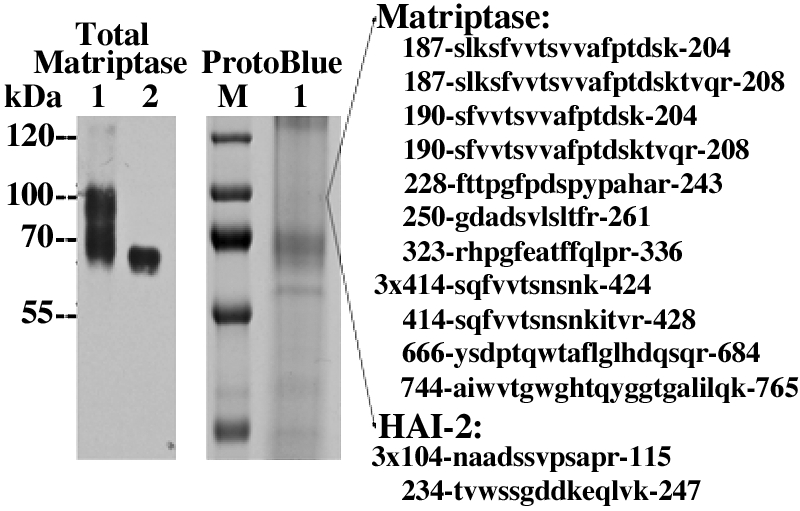
The 100-kDa matriptase complex contains HAI-2. Both 140-kDa and 100-kDa activated matriptase complexes were purified from LNCaP prostate cancer cells (see Fig. 6A, lane 1, LNCaP) by a 3-step purification scheme, including WGA lectin chromatography, immunodepletion of 120-kDa matriptase-HAI-1 complex, and immunoaffinity purification using the mAb M69. Eluted proteins were analyzed by immunoblot using matriptase mAb M24 under nonboiled condition (left, lane 1) or boiled conditions (left, lane 2). The 100-kDa matriptase complexes were eluted as major proteins (lane 1, total matriptase) and were completely dissociated after boiling and detected as 70-kDa bands by matriptase mAb M24 mAb (lane 2, total matriptase). Eluted proteins were also examined by SDS-PAGE under nonboiled conditions and stained by ProtoBlue (right). Protein bands at 60-, 70-, and 100-kDa were excised, trypsinized, and then subjected to LC-MS/MS for protein identification. Database search revealed that the 100-kDa spots yielded 17 tryptic fragments, which matched to 2 different proteins. Among these tryptic fragments, 13 fragments matched to 10 stretches of amino acid sequences of matriptase and 4 fragments matched to 2 stretches of amino acid sequences of HAI-2, as indicated. The 70-kDa proteins contain matriptase, and the 60-kDa band was identified as prosapsin, similar to that seen in Fig. 7.
Analysis of proteins in the 100-kDa gel slice yielded 13 peptides matching to matriptase and 4 peptides matching to HAI-2 (Fig. 8). An attempt to further determine whether the 100-kDa complex contains HAI-2 has proven to be much more difficult than expected. Three different HAI-2 antibodies, including two HAI-2 mAbs named 2N9 and 2N12 (8), were used to demonstrate high expression of the Kunitz serine protease inhibitor in OVCAR-3 ovarian cancer cells and LNCaP prostate cancer cells in which the 100-kDa complexes were clearly seen. Since the epitope recognized by the HAI-2 mAbs can, however, only be significantly exposed after heat treatment, under which the 100-kDa complexes is dissociated, we cannot examine whether the 100-kDa complexes contain HAI-2 by direct immunoblot analysis. With the use of purified 100-kDa matriptase complexes, these HAI-2 antibodies failed to detect HAI-2 in the heat-treated 100-kDa complexes. Although the discrepancy could result from different detection sensitivity between immunoblotting and proteomic protein identification and possible heterogeneity of the 100-kDa complexes, these data suggest that the vast majority of the 100-kDa complex might be an activated matriptase complex with an unidentified serine protease inhibitor(s). Further study remained needed for identification of what protease inhibitors are in the 100-kDa complexes.
DISCUSSION
Through an investigation of the fate of activated matriptase resulting from the massive zymogen activation of the enzyme in the cells with a low HAI-1-to-matriptase protein ratio, we were able to identify three complexes containing activated matriptase. Two of these complexes are comprised of two-chain matriptase associated with the protease inhibitor HAI-1 or a yet to be identified serine protease inhibitor. The putative serine protease inhibitor is biochemically similar to Kunitz-type serine protease inhibitors, such as HAI-1 and HAI-2. The third appears to be comprised of a homodimer of two-chain matriptase. None of these matriptase complexes exhibit gelatinolytic activity in spite of the fact that all three contain matriptase molecules that have been activated by cleavage at the canonical activation motif. Furthermore, the matriptase molecule in these complexes also exposes the epitope associated with zymogen activation and recognized by the activated matriptase mAb M69. The observation that massive and robust zymogen activation is immediately followed by rapid and effective sequestering of the two-chain matriptase into the three inactive complexes indicates that matriptase-expressing cells are equipped with several independent strategies for keeping matriptase activity under tight control. Although the need for such tightly regulated mechanisms governing matriptase activity remains unclear, the maintenance of matriptase activity in an appropriate balance with its cognate inhibitors is critical for the physiological function of the epithelium and the deregulation of this regulation causes disease (4, 18, 20, 21, 31, 32, 34).
The partnership between matriptase and HAI-1 as a cognate pair of a protease and protease inhibitor is very unusual compared with that of other proteases and their cognate inhibitors. Proteases and their cognate inhibitors are commonly expressed by different cell types, likely so that active proteases have time to act on their substrates before inactivation by interaction with their inhibitors. For example, in human colon adenocarcinomas, urokinase plasminogen activator is expressed by fibroblast-like cells (7) and plasminogen activator inhibitor-1 is expressed by tumor endothelial cells (26). In contrast, matriptase and HAI-1 are almost ubiquitous coexpressed in many cells and codistributed along the secretory pathway (30, 40). Although the maintenance of higher levels of protease inhibitors than proteases is common and seems necessary, the overwhelming molar excess of HAI-1 relative to matriptase in most cells may be due to the unique features of matriptase autoactivation, which involves interactions between two matriptase molecules (11, 25). Given that the cells can activate matriptase in response to a variety of stimuli and that spontaneous matriptase activation can occur under cell-free conditions, matriptase autoactivation appears, therefore, to be an autonomous process. Furthermore, the activation is susceptible to environmental changes and cellular conditions. As a transmembrane protein, matriptase is present throughout the secretory pathway and is detected in secretory vesicles/granules and on the cell surface, at both of which locations, matriptase activation can take place (40). The autonomous nature of matriptase activation and its susceptibility to induction by diverse stimuli, in conjunction with the confinement of this potent trypsin-like serine protease within secretory pathway, present a tremendous potential for harm to the cells that express it. Indeed, exogenous expression of matriptase in cells that express no endogenous HAI-1 results in accumulation of low levels of matriptase in the Golgi apparatus (23). This inability of matriptase to traffic out of the Golgi apparatus can be rescued by the coexpression of HAI-1. Thus the interaction between HAI-1 and matriptase is required for matriptase expression and intracellular trafficking. Since matriptase mutants with a nonfunctional active site triad have no problem trafficking out of Golgi apparatus (23), it seems likely that the HAI-1-mediated rescue of matriptase intracellular trafficking results from the suppression of matriptase proteolytic activity. The predominant role of HAI-1 in matriptase inhibition is consistent with the fact that the vast majority of activated matriptase is present in HAI-1 complexes as we have shown in human body fluids, including milk, urine, and semen and in the conditioned media from a variety of cells (40). The presence of another HAI-1-like serine protease inhibitor in those cells with low level of HAI-1 further supports the hypothesis that matriptase activity must be under tight control.
In addition to the inhibition of matriptase activity by membrane-associated serine protease inhibitors, the two-chain matriptase can apparently be “locked” in an enzymatically inactive 140-kDa homodimer. The interaction between the two matriptase molecules is noncovalent and susceptible to dissociation in pH 2.4 buffer, similar to the interaction between activated matriptase and HAI-1. The interaction is, however, not reversible once dissociated and returned to a neutral pH. In contrast, the binding between active matriptase and HAI-1 is reversible (1). The irreversibility suggests that the structural basis for the binding between the two cleaved matriptase molecules might be altered during dissociation. Thus it seems that a transient, less stable conformation of matriptase might be generated during the process of matriptase zymogen activation. Through this transient conformation, the matriptase molecules could bind to one another to form the complex. While the structural basis for the interaction remained to be further defined, the two CUB (Complement Clr/Cls, Uegf, Bmp1) domains of matriptase might be involved in the formation of the 140-kDa complex. This hypothesis is based on the fact that the seminal plasma spermadhesins PSP-I and PSP-II are built by a single CUB domain architecture and can form heterodimer (38). Furthermore, matriptase contains four LDL receptor class A domains, many of which have been identified in membrane receptors belonged to the LDL receptor family. These domains in the membrane receptors provide the structural basis for interactions between the receptors and their ligands. More importantly, the conformations of the LDL receptor class A domains and the binding between the receptor and their ligands are pH dependent (27, 28). It is tempting to propose that an activation-permissive mildly acidic pH environment might confer conformational changes to the matriptase LDL receptor domains and that these conformational changes might in turn be critical for matriptase activation. This hypothesis is supported by the fact that both the CUB and the LDL receptor class A domains are required for matriptase activation (25).
While the mechanism by which the proteolytic activity of the activated matriptase is suppressed in the homodimer remains unclear, the formation of such homodimers may represent an additional effective mechanism to reduce the potentially hazardous effects of matriptase activity, particularly in those cells that express lower levels of the protease inhibitors. Consistent with this hypothesis, matriptase homodimers are only seen to accumulate during acid-induced activation using intact cells in the three cells with very low levels of HAI-1 (Fig. 2). Furthermore, matriptase homodimer may also serve as an important check-point in response to suboptimal conditions in which the subsequent inhibition of active matriptase by HAI-1 may not be warranted, such as in the cell-free activation system. In the cell-free activation system, the 140-kDa matriptase homodimer was detected in all of these cells tested. In common with the intact-cell system, the levels of matriptase homodimer that accumulated in the cell-free activation system are inversely correlated with the ratio of HAI-1 to matriptase (Fig. 4). The ubiquitous appearance of the matriptase homodimer in the cell-free activation system may have some mechanistic and regulatory implications. The most interesting speculation is that the matriptase homodimer is an intermediate of matriptase autoactivation. Although autoactivation dictates that interaction must occur between two matriptase zymogen molecules resulting in proteolytic cleavages at the activation motif, the matriptase homodimer identified in the current study provides the first critical evidence for the interaction between two matriptase molecules during autoactivation.
In Fig. 4, the levels of matriptase complexes and the matriptase zymogen were comparable as determined by the total matriptase mAb M24 (Fig. 4A, lane 1, OVCAR-3, total matriptase). In contrast, the levels of the 25-kDa matriptase fragment released from these matriptase complexes are very low compared with the levels of matriptase zymogen in the analysis of diagonal gel electrophoresis using the ST-14 antibody (Fig. 5A). This discrepancy could result from the epitope recognized by the ST-14 antibody being differentially exposed in the 25-kDa fragment and 70-kDa matriptase zymogen. Alternatively, the 25-kDa fragment might be transferred to and/or retained on nitrocellulose membrane less efficiently compared with 70-kDa matriptase zymogen due to the size and mobility of this fragment.
In summary, by examining those matriptase-expressing cells that express low levels of HAI-1, we have identified two novel mechanisms for control of matriptase activity. These matriptase-expressing cells can lock activated matriptase in an inactive homodimer and rapidly inactivate active matriptase by interaction with HAI-1 and other unidentified serine protease inhibitors. With these novel mechanisms, matriptase proteolytic activity can be tightly regulated.
GRANTS
This study was supported by National Cancer Institute Grants R01-CA-104944 (to C.-Y. Lin) and R01-CA-123223 (to M. D. Johnson and C.-L. Lin) and by the Maryland Cigarette Restitution Fund Program (to C.-Y. Lin).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: H.X., Z.X., I.-C.T., F.-P.C., and Y.-W.C. performed experiments; H.X., Z.X., I.-C.T., Y.-W.C., J.-K.W., and C.-Y.L. analyzed data; H.X., H.K., and C.-Y.L. interpreted results of experiments; H.X., Z.X., F.-P.C., and Y.-W.C. prepared figures; H.X., J.-K.W., M.D.J., H.K., and C.-Y.L. edited and revised manuscript; H.X., Z.X., I.-C.T., F.-P.C., Y.-W.C., J.-K.W., M.D.J., H.K., and C.-Y.L. approved final version of manuscript; C.-Y.L. conception and design of research; C.-Y.L. drafted manuscript.
REFERENCES
- 1. Benaud C, Dickson RB, Lin CY. Regulation of the activity of matriptase on epithelial cell surfaces by a blood-derived factor. Eur J Biochem 268: 1439–1447, 2001 [DOI] [PubMed] [Google Scholar]
- 2. Benaud C, Oberst M, Hobson JP, Spiegel S, Dickson RB, Lin CY. Sphingosine 1-phosphate, present in serum-derived lipoproteins, activates matriptase. J Biol Chem 277: 10539–10546, 2002 [DOI] [PubMed] [Google Scholar]
- 3. Bhatt AS, Welm A, Farady CJ, Vasquez M, Wilson K, Craik CS. Coordinate expression and functional profiling identify an extracellular proteolytic signaling pathway. Proc Natl Acad Sci USA 104: 5771–5776, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carney TJ, von der HS, Sonntag C, Amsterdam A, Topczewski J, Hopkins N, Hammerschmidt M. Inactivation of serine protease Matriptase1a by its inhibitor Hai1 is required for epithelial integrity of the zebrafish epidermis. Development 134: 3461–3471, 2007 [DOI] [PubMed] [Google Scholar]
- 5. Chen YW, Wang JK, Chou FP, Chen CY, Rorke EA, Chen LM, Chai KX, Eckert RL, Johnson MD, Lin CY. Regulation of the matriptase-prostasin cell surface proteolytic cascade by hepatocyte growth factor activator inhibitor-1 (HAI-1) during epidermal differentiation. J Biol Chem 285: 31755–31762, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chou FP, Xu H, Lee MS, Chen YW, Richards OX, Swanson R, Olson ST, Johnson MD, Lin CY. Matriptase is inhibited by extravascular antithrombin in epithelial cells but not in most carcinoma cells. Am J Physiol Cell Physiol 301: C1093–C1103, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grondahl-Hansen J, Ralfkiaer E, Kirkeby LT, Kristensen P, Lund LR, Dano K. Localization of urokinase-type plasminogen activator in stromal cells in adenocarcinomas of the colon in humans. Am J Pathol 138: 111–117, 1991 [PMC free article] [PubMed] [Google Scholar]
- 8. Kataoka H, Shimomura T, Kawaguchi T, Hamasuna R, Itoh H, Kitamura N, Miyazawa K, Koono M. Hepatocyte growth factor activator inhibitor type 1 is a specific cell surface binding protein of hepatocyte growth factor activator (HGFA) and regulates HGFA activity in the pericellular microenvironment. J Biol Chem 275: 40453–40462, 2000 [DOI] [PubMed] [Google Scholar]
- 9. Kilpatrick LM, Harris RL, Owen KA, Bass R, Ghorayeb C, Bar-Or A, Ellis V. Initiation of plasminogen activation on the surface of monocytes expressing the type II transmembrane serine protease matriptase. Blood 108: 2616–2623, 2006 [DOI] [PubMed] [Google Scholar]
- 10. Kiyomiya KI, Lee MS, Tseng IC, Zuo H, Barndt RJ, Johnson MD, Dickson RB, Lin CY. Matriptase activation and subsequent shedding with HAI-1 is induced by steroid sex hormones in human prostate cancer cells, but not in breast cancer cells. Am J Physiol Cell Physiol 291: C40–C49, 2006 [DOI] [PubMed] [Google Scholar]
- 11. Lee MS, Tseng IC, Wang Y, Kiyomiya K, Johnson MD, Dickson RB, Lin CY. Autoactivation of matriptase in vitro: requirement for biomembrane and LDL receptor domain. Am J Physiol Cell Physiol 293: C95–C105, 2007 [DOI] [PubMed] [Google Scholar]
- 12. Lee MS, Kiyomiya K, Benaud C, Dickson RB, Lin CY. Simultaneous activation and HAI-1-mediated inhibition of matriptase induced at activation foci in immortal human mammary epithelial cells. Am J Physiol Cell Physiol 288: C932–C941, 2005 [DOI] [PubMed] [Google Scholar]
- 13. Lin CY, Anders J, Johnson M, Dickson RB. Purification and characterization of a complex containing matriptase and a Kunitz-type serine protease inhibitor from human milk. J Biol Chem 274: 18237–18242, 1999 [DOI] [PubMed] [Google Scholar]
- 14. Lin CY, Tseng IC, Chou FP, Su SF, Chen YW, Johnson MD, Dickson RB. Zymogen activation, inhibition, and ectodomain shedding of matriptase. Front Biosci 13: 621–635, 2008 [DOI] [PubMed] [Google Scholar]
- 15. Lin CY, Wang JK, Torri J, Dou L, Sang QA, Dickson RB. Characterization of a novel, membrane-bound, 80-kDa matrix-degrading protease from human breast cancer cells. Monoclonal antibody production, isolation, and localization. J Biol Chem 272: 9147–9152, 1997 [PubMed] [Google Scholar]
- 16. List K, Bugge TH, Szabo R. Matriptase: potent proteolysis on the cell surface. Mol Med 12: 1–7, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. List K, Kosa P, Szabo R, Bey AL, Wang CB, Molinolo A, Bugge TH. Epithelial integrity is maintained by a matriptase-dependent proteolytic pathway. Am J Pathol 175: 1453–1463, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. List K, Szabo R, Molinolo A, Sriuranpong V, Redeye V, Murdock T, Burke B, Nielsen BS, Gutkind JS, Bugge TH. Deregulated matriptase causes ras-independent multistage carcinogenesis and promotes ras-mediated malignant transformation. Genes Dev 19: 1934–1950, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luthy JA, Praissman M, Finkenstadt WR, Laskowski M., Jr Detailed mechanism of interaction of bovine -trypsin with soybean trypsin inhibitor (Kunitz). I. Stopped flow measurements. J Biol Chem 248: 1760–1771, 1973 [PubMed] [Google Scholar]
- 20. Mathias JR, Dodd ME, Walters KB, Rhodes J, Kanki JP, Look AT, Huttenlocher A. Live imaging of chronic inflammation caused by mutation of zebrafish Hai1. J Cell Sci 120: 3372–3383, 2007 [DOI] [PubMed] [Google Scholar]
- 21. Nagaike K, Kawaguchi M, Takeda N, Fukushima T, Sawaguchi A, Kohama K, Setoyama M, Kataoka H. Defect of hepatocyte growth factor activator inhibitor type 1/serine protease inhibitor, kunitz type 1 (Hai-1/Spint1) leads to ichthyosis-like condition and abnormal hair development in mice. Am J Pathol 173: 1464–1475, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oberst M, Anders J, Xie B, Singh B, Ossandon M, Johnson M, Dickson RB, Lin CY. Matriptase and HAI-1 are expressed by normal and malignant epithelial cells in vitro and in vivo. Am J Pathol 158: 1301–1311, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oberst MD, Chen LY, Kiyomiya KI, Williams CA, Lee MS, Johnson MD, Dickson RB, Lin CY. Hepatocyte growth factor activator inhibitor 1 (HAI-1) regulates activation and expression of matriptase, a membrane-bound serine protease. Am J Physiol Cell Physiol 289: C462–C470, 2005 [DOI] [PubMed] [Google Scholar]
- 24. Oberst MD, Johnson MD, Dickson RB, Lin CY, Singh B, Stewart M, Williams A, al Nafussi A, Smyth JF, Gabra H, Sellar GC. Expression of the serine protease matriptase and its inhibitor HAI-1 in epithelial ovarian cancer: correlation with clinical outcome and tumor clinicopathological parameters. Clin Cancer Res 8: 1101–1107, 2002 [PubMed] [Google Scholar]
- 25. Oberst MD, Williams CA, Dickson RB, Johnson MD, Lin CY. The activation of matriptase requires its noncatalytic domains, serine protease domain, and its cognate inhibitor. J Biol Chem 278: 26773–26779, 2003 [DOI] [PubMed] [Google Scholar]
- 26. Pyke C, Kristensen P, Ralfkiaer E, Grondahl-Hansen J, Eriksen J, Blasi F, Dano K. Urokinase-type plasminogen activator is expressed in stromal cells and its receptor in cancer cells at invasive foci in human colon adenocarcinomas. Am J Pathol 138: 1059–1067, 1991 [PMC free article] [PubMed] [Google Scholar]
- 27. Rias-Moreno X, Velazquez-Campoy A, Rodriguez JC, Pocovi M, Sancho J. Mechanism of low density lipoprotein (LDL) release in the endosome: implications of the stability and Ca2+ affinity of the fifth binding module of the LDL receptor. J Biol Chem 283: 22670–22679, 2008 [DOI] [PubMed] [Google Scholar]
- 28. Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, Deisenhofer J. Structure of the LDL receptor extracellular domain at endosomal pH. Science 298: 2353–2358, 2002 [DOI] [PubMed] [Google Scholar]
- 29. Saleem M, Adhami VM, Zhong W, Longley BJ, Lin CY, Dickson RB, Reagan-Shaw S, Jarrard DF, Mukhtar H. A novel biomarker for staging human prostate adenocarcinoma: overexpression of matriptase with concomitant loss of its inhibitor, hepatocyte growth factor activator inhibitor-1. Cancer Epidemiol Biomarkers Prev 15: 217–227, 2006 [DOI] [PubMed] [Google Scholar]
- 30. Szabo R, Hobson JP, List K, Molinolo A, Lin CY, Bugge TH. Potent inhibition and global co-localization implicate the transmembrane kunitz-type serine protease inhibitor hai-2 in the regulation of epithelial matriptase activity. J Biol Chem 283: 29495–504, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Szabo R, Kosa P, List K, Bugge TH. Loss of matriptase suppression underlies spint1 mutation-associated ichthyosis and postnatal lethality. Am J Pathol 174: 2015–2022, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Szabo R, Molinolo A, List K, Bugge TH. Matriptase inhibition by hepatocyte growth factor activator inhibitor-1 is essential for placental development. Oncogene 26: 1546–1556, 2007 [DOI] [PubMed] [Google Scholar]
- 33. Szabo R, Wu Q, Dickson RB, Netzel-Arnett S, Antalis TM, Bugge TH. Type II transmembrane serine proteases. Thromb Haemost 90: 185–193, 2003 [DOI] [PubMed] [Google Scholar]
- 34. Tanaka H, Nagaike K, Takeda N, Itoh H, Kohama K, Fukushima T, Miyata S, Uchiyama S, Uchinokura S, Shimomura T, Miyazawa K, Kitamura N, Yamada G, Kataoka H. Hepatocyte growth factor activator inhibitor type 1 (HAI-1) is required for branching morphogenesis in the chorioallantoic placenta. Mol Cell Biol 25: 5687–5698, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tseng IC, Chou FP, Su SF, Oberst M, Madayiputhiya N, Lee MS, Wang JK, Sloane DE, Johnson M, Lin CY. Purification from human milk of matriptase complexes with secreted serpins: mechanism for inhibition of matriptase other than HAI-1. Am J Physiol Cell Physiol 295: C423–C431, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tseng IC, Xu H, Chou FP, Li G, Vazzano AP, Kao JP, Johnson MD, Lin CY. Matriptase activation, an early cellular response to acidosis. J Biol Chem 285: 3261–3270, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uhland K. Matriptase and its putative role in cancer. Cell Mol Life Sci 63: 2968–2978, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Varela PF, Romero A, Sanz L, Romao MJ, Topfer-Petersen E, Calvete JJ. The 2.4 A resolution crystal structure of boar seminal plasma PSP-I/PSP-II: a zona pellucida-binding glycoprotein heterodimer of the spermadhesin family built by a CUB domain architecture. J Mol Biol 274: 635–649, 1997 [DOI] [PubMed] [Google Scholar]
- 39. Vogel LK, Saebo M, Skjelbred CF, Abell K, Pedersen ED, Vogel U, Kure EH. The ratio of Matriptase/HAI-1 mRNA is higher in colorectal cancer adenomas and carcinomas than corresponding tissue from control individuals. BMC Cancer 6: 176–183, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang JK, Lee MS, Tseng IC, Chou FP, Chen YW, Fulton A, Lee HS, Chen CJ, Johnson MD, Lin CY. Polarized epithelial cells secrete matriptase as a consequence of zymogen activation and HAI-1-mediated inhibition. Am J Physiol Cell Physiol 297: C459–C470, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Warren M, Twohig M, Pier T, Eickhoff J, Lin CY, Jarrard D, Huang W. Protein expression of matriptase and its cognate inhibitor HAI-1 in human prostate cancer: a tissue microarray and automated quantitative analysis. Appl Immunohistochem Mol Morphol 17: 23–30, 2008 [DOI] [PubMed] [Google Scholar]