Abstract
The recurrent t(1;22)(p13;q13) translocation is exclusively associated with infant acute megakaryoblastic leukemia. We have identified the two genes involved in this translocation. Both genes possess related sequences in the Drosophila genome. The chromosome 22 gene (megakaryocytic acute leukemia, MAL) product is predicted to be involved in chromatin organization, and the chromosome 1 gene (one twenty-two, OTT) product is related to the Drosophila split-end (spen) family of proteins. Drosophila genetic experiments identified spen as involved in connecting the Raf and Hox pathways. Because almost all of the sequences and all of the identified domains of both OTT and MAL proteins are included in the predicted fusion protein, the OTT-MAL fusion could aberrantly modulate chromatin organization, Hox differentiation pathways, or extracellular signaling.
Molecular characterization of chromosomal translocations present in human malignancies showed that many of them result in the creation of chimeric genes and the expression of fusion products, mRNA, and protein from one of the rearranged chromosomes. Those studies have led to the discovery of a large number of genes, the products of which are involved in controlling both malignant and normal cellular processes (1, 2). Several of these genes encode transcription factors whose counterparts are involved in embryogenesis and establishment of the body plan in Drosophila or mouse model organisms (3). In addition to the direct involvement in leukemogenesis of HOX genes themselves, disregulation of homeotic genes expression or function is an expected result in two examples of chromosomal translocations. One is the t(1;19)(q23;p13), which is highly specific of a subset of human childhood B cell acute lymphoblastic leukemia (ALL) and fuses the E2A gene on chromosome 19 to the PBX1 gene on chromosome 1. The PBX1 protein, the mammalian counterpart of the Drosophila extradenticle (exd), interacts with and modulates the activity of specific subsets of HOX proteins (4). The second well established example is the MLL/HRX gene, located on human chromosome band 11q23, implicated in fusion with more than 40 other genes (2, 5, 6). HRX is the mammalian counterpart of the Drosophila trithorax (trx) gene, which, like other trx-G members, has been shown to exert a positive role in the maintenance of cell-specific patterns of hom-c gene expression in the fly (7). An analogous role for HRX is supported by phenotypic analyses of HRX knockout mice (8). Strikingly, HRX is rearranged in various hematological malignancies, acute myeloblastic leukemia (AML), ALL, therapy-related leukemia, and frequently in infant leukemia, which exhibit an altered HRX locus in more than half of the samples (9–11).
We and others identified the t(1;22)(p13;q13) translocation as a recurrent translocation exclusively associated with infant AML of the megakaryocytic lineage (M7) (12–14). We now report the characterization of the two genes involved in this translocation.
Materials and Methods
Materials.
Patient 1 is a 9-mo-old infant with a t(1;22;4) (p13;q13;q35) translocation, and patient 2 is a 12-mo-old infant with a common t(1;22)(p13;q13), both suffering from AML-M7. Informed consent of patients' parents was obtained. Fluorescence in situ hybridization (FISH) analysis was performed as described (15). The chromosome 22 P1 artificial chromosome (PAC) clone, which generates a split signal in patient metaphase chromosomes, is RP5–1042k10 (AC AL022238). It was selected from the chromosome 22 map (16) after a series of FISH experiments on the patient's metaphase chromosomes. Bacterial artificial chromosome (BAC) B640H2 was isolated from the CEPH library and mapped, by end nucleotide analyses, to extend telomeric to nucleotide 156743 of AL022238. Because it gave split signals on metaphase chromosomes, it allowed mapping of the breakpoint between nucleotide 156743 and the telomeric end of 1042K10 (nucleotide 214870). BAC clones from chromosome 1p13 were selected from sequences databases (260A24; AC025987) or after PCR screening of the CEPH library (H629B6).
Nucleic Acid Methods.
DNA techniques were performed according to standard protocols. The cDNA library (Stratagene) is a kind gift from S. Gisselbrecht and G. Courtois, Hôpital Cochin, Paris, and the Multiple Tissues Northern Blot was purchased from CLONTECH. Anchored PCR was performed by using the 5′/3′ rapid amplification of cDNA ends kit (Roche Biochemicals) with the following primers: [retrotranscription (22–3:cggctagtctggctctcttca), first PCR (22–4:ctcagccgaggtctcttccaa); second PCR (22–108: gcggatccgtttgagatagtcctctgtcctgg. Underlined nucleotides were added for cloning purposes.)]. Bispecific reverse transcription–PCR (RT-PCR) experiments were performed starting from random-primed cDNA by using 1–4 (tcctggattcccctgccaag) and 22–6 (caagctccttctctgctcatg). The specificity of PCR products was checked by nucleotide sequencing.
Computer analyses were performed locally or at the National Center for Biotechnology Information site (http://www.ncbi.nlm.nih.gov/). Sequence data were derived from the following accession nos.: megakaryocytic acute leukemia (MAL) 16 (AC 012626, AC040173, HSU91322), MAL 17 (AC005358), Dmal (AE003475), one twenty-two (OTT) 3 (AC012263, AC067763, NM013286), Msx2-interacting nuclear target (MINT) (AL096858), spen (AF221715), and D ott (AI114303).
Results
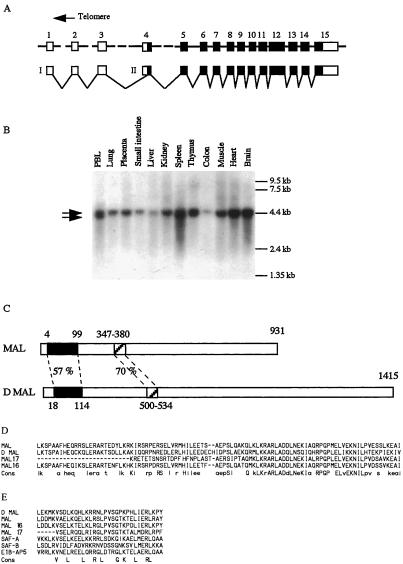
We recently observed a variant t(1;22;4) translocation in a young child suffering from AML-M7 (patient 1). We took advantage of the data generated by the human chromosome 22 mapping and sequencing project (16) to identify a chromosome 22 P1 artificial chromosome (PAC) clone (1042K10), which was shown to encompass the chromosome 22 breakpoint in this patient, by using FISH techniques (data not shown). This clone was then shown to span the chromosome 22 breakpoint in a common t(1;22) translocation (patient 2). Additional BAC clones covering this region were isolated from the CEPH library and mapped with respect to the 1042K10 sequences by end sequencing. Selected clones were used in FISH experiments on the patient's metaphase chromosomes, which allowed us to map the breakpoint within a 58-kb region of the 1042k10 PAC. We used PCR-amplified probes derived from sequences of the telomeric half of the BAC to isolate several cDNAs from a human blood cell library. Nucleotide analyses of these cDNAs showed that the telomeric half of the PAC was covered by a single gene, the structure of which is represented in Fig. 1A. Because it is involved in a chromosomal translocation specific for megakaryocytic acute leukemia, we named it MAL. By Northern blot analysis, MAL was found to be transcribed as one or two RNA species in all tissues tested (Fig. 1B).
Figure 1.
The MAL gene on chromosome 22. (A) The MAL gene comprises 15 exons spanning 226 kb. Predicted coding sequences appear as black boxes and noncoding sequences as empty boxes. (B) Two putative promoters would direct transcription of 4.5 (I) and 4 kb (II) RNA species (shown by arrows) with a variable ratio, depending on the tissues. Translational initiation would occur at the most 5′ in frame ATG located within exon 4. See also Fig. 3C for nucleotide sequences. (C) Representation of the predicted MAL protein along with its closest related Drosophila protein (D mal). The percentage of amino acid identity is shown. The two highest regions of similarity between those two proteins are aligned together with related human predicted proteins. (D) The 95-aa region of unknown function of MAL is aligned with D mal and with human MAL 16 and MAL 17. Amino acids present in at least three of the sequences appear in lowercase letters in the consensus, and those present in the four sequences appear in uppercase letters. The MAL 16 sequences are derived from genomic, EST, and specific RT-PCR products, and MAL 17 sequences are derived from human genomic sequences. (E) The predicted SAF box of MAL is compared with those of D mal, MAL 16, MAL 17. SAF A, SAF B, E1B-AP5, and the SAF consensus sequences are from ref. 17.
When compared with the translated human sequences present in the databases, the MAL-predicted 931-aa protein demonstrated close similarity with the predicted product of two human genes, here named MAL 16 on chromosome 16 and MAL 17 on chromosome 17. Interestingly, comparison of the MAL protein sequences with translation of Drosophila genomic sequences identified its putative Drosophila ortholog. Comparison between the two proteins is represented Fig. 1C, and amino acid alignments between two regions of high similarity are shown in Fig. 1 D and E. The first region, which spans amino acids 4–99 of MAL and 18–113 of Drosophila mal (D mal), is 57% identical between both proteins. It also exhibits high similarity to MAL 16- and MAL 17-predicted products but not to other known human proteins.
Examination of the second region of similarity between human and Drosophila proteins revealed that it corresponds to the recently described scaffold attachment factor (SAF) box (17). This 31-aa motif is endowed with minor grove DNA-binding properties that have been shown to be specific for AT-rich scaffold attachment region sequences but with low sequence specificity. SAF boxes are present in many different proteins, ranging from yeast to human origins, and appear to be associated with other protein motifs, such as RNA-binding RGG motifs in SAF-A. These proteins are supposed to attach DNA to nuclear scaffold or matrix and to be involved in chromatin organization. The MAL subgroup defines a SAF box subfamily that is associated with a motif of unknown function. Indirect support for the importance of this protein family in normal biological processes is provided by gene-trapping experiments that affected Gt4–1, the putative MAL 16 murine gene, and led to perinatal lethality of the homozygous mice (18).
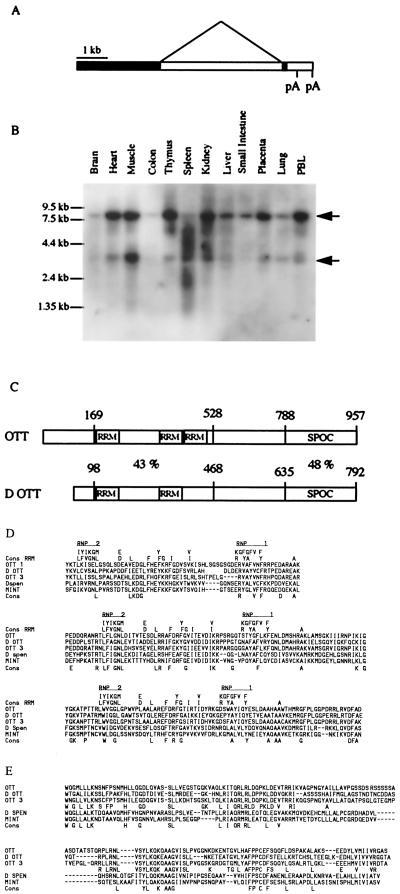
Because the chromosome 22 breakpoint of the t(1;22;4) is located within intron 3 or 4 of the MAL gene, we performed anchored RT-PCR to isolate putative fusion transcripts between MAL and a chromosome 1 partner gene. A cDNA fragment, spliced upstream of MAL exon 4, was identified that did not originate from human chromosome 22 sequences but was similar to a human chromosome 1 BAC clone sequences present in the databases (260A24). This clone, together with another BAC clone isolated from the CEPH library and corresponding to the same region, was confirmed to correspond to chromosome 1 and span the translocation breakpoint in patient 1 (data not shown). The fusion between chromosome 1 and 22 sequences was confirmed by using RT-PCR (Fig. 3A). The isolated chromosome 1 sequences were then used as a probe to isolate several cDNAs from a human library. Comparison of genomic and cDNA sequences allowed us to determine the structure of this gene (Fig. 2A), which was named one twenty-two (OTT). By Northern blot analysis, OTT was found to be transcribed as one or two RNA species in all tissues tested (Fig. 2B). An 8.2-kb single exon transcript and a second smaller (3.9-kb) species seem to be caused by the splicing of 4,039 bp. When compared with the human sequences present in the databases, the predicted OTT protein was found to be closely related to a chromosome 3 gene product and somewhat more distantly to the human homolog of the MINT gene located on chromosomal band 1p36 (19). Comparison of the OTT predicted protein with translation of Drosophila sequences identified two related proteins, the Drosophila spen protein and a putative OTT Drosophila ortholog. Two regions of similarity could be identified from comparisons between human and Drosophila OTTs. The first region spans amino acids 169–528 of human OTT and exhibits 43% identity between the two proteins. Careful examination of the sequences revealed three RNA recognition motifs (20) within the human sequences, whereas only two could be identified in the Drosophila ott. The second region of similarity spans the 741–957 C-terminal amino acids of OTT and 596–792 of D ott, and 45% of amino acid identity is observed within the two sequences. This region corresponds to the previously described Spen Paralog and Ortholog C-terminal (SPOC) domain (21–23).
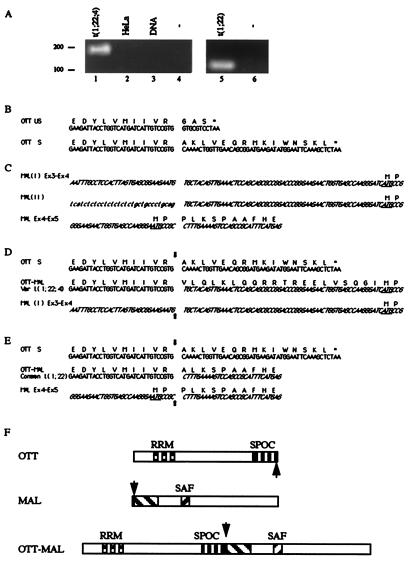
Figure 3.
OTT-MAL fusion transcripts in infant AML-M7. (A) RT-PCR amplification of OTT-MAL fusion transcripts. A specific OTT-MAL fusion cDNA could be amplified from variant translocation (patient 1, lane 1, 179 bp) or common translocation (patient 2, lane 5, 113 bp) t(1;22), but not from negative control cDNA (HeLa; lane 2), genomic DNA (lane 3), or in the absence of template (lanes 4 and 6). (B) Nucleotide and deduced amino acid sequences of unspliced (US) and spliced (S) normal OTT transcripts. Both species are identifiable in EST and RT-PCR experiments. (C) Nucleotide and deduced amino acid sequences of types I and II normal MAL transcripts. Here and below, chromosome 22 sequences appear in italics, and 5′ untranslated (UT) sequences of type II MAL transcripts absent from type I appear in lowercase letters. (D) Comparison of normal and fused OTT and MAL transcripts, as observed in variant t(1;22;4). The fusion sites are indicated by arrows. The ATG codon underlined in MAL exon 4 sequences is the predicted translational start site of MAL proteins. Stop codons are shown by asterisks. Because the OTT-MAL fusion is in frame and devoid of stop codon, the 5′ UT sequence of MAL exon 4 should be translated in this case. (E) Comparison of normal and fused OTT and MAL transcripts, as observed in common t(1;22). (F) Representation of predicted normal and fusion proteins, as deduced from the common t(1;22) structural analysis. Interspecies conserved regions are indicated, and fusion points are shown by arrows.
Figure 2.
The OTT gene on chromosome 1. (A) Genomic organization of OTT. (B) Northern blot analysis of OTT expression in human tissues by using a probe corresponding to coding sequences. Note that a probe corresponding to the noncoding sequences reacts only with the larger OTT RNA species (data not shown). See also Fig. 3B for nucleotide sequences. (C) Schematic comparison of OTT with its putative Drosophila homolog (D ott). The percentage of amino acid identity is shown. (D) Amino acid alignment of OTT with RNA recognition motif (RRM) consensus (20) and with the similar region of human (OTT3 and MINT) and Drosophila (spen and D ott) proteins. Note that the spacer between RNP2 and RNP1 in the last predicted RRM motif is shorter than the consensus. (E) Amino acid alignment of the same proteins as in D but within the SPOC region. An OTT family consensus is shown in addition to a global one.
The nucleotide sequence of the OTT-MAL fusion transcript present in patient 1 (Fig. 3A, line 1) is aligned with normal OTT and MAL transcripts in Fig. 3D. The fusion occurs in frame with MAL exon 4 sequences, adding 20 amino acids, coded by MAL sequences but predicted not to be normally translated, at the junction of OTT and MAL sequences.
Because FISH analysis suggested the involvement of OTT and MAL genes in the common t(1;22) observed in patient 2 (data not shown), we used RT-PCR to isolate a similar OTT-MAL fusion transcript. A specific fragment could be amplified from patient 2 material (Fig. 3A, line 5) but not from control (line 6) and could be checked by nucleotide sequencing. Alignment of normal and fusion sequences in the common example (Fig. 3E), showing that, in the common case, the OTT-MAL fusion occurs in frame upstream of exon 5.
Discussion
We have identified the two genes involved in the recurrent and specific t(1;22)(p13;q13) translocation. Both the MAL gene on chromosome 22 and the OTT gene on chromosome 1 were previously undescribed, but several clues regarding the OTT function could be deduced from its similarity to the Drosophila spen protein. Genetic studies in the fly model implicated spen in indirectly modulating both hox function (22) and ras signaling (23, 24). At least in the Drosophila R7 eye cells, spen could repress raf signaling and assist p21 in blocking S-phase entry (25). This could be achieved through the modulation of some nuclear targets of intracellular signaling pathways, such as suppressor of hairless [Su(H)], a known transcriptional effector of NOTCH signaling, or yan, a Drosophila ETS family member closely related to the human translocated, ETS, leukemia (TEL) gene (26, 27), and a target of the epidermal growth factor receptor (EGFR)/Ras/Raf/mitogen-activated protein kinase pathway. Spen mutants indeed exhibit altered level of yan and Su(H) in neuronal precursors (21). Whether spen acts as a transcriptional factor or through RNA metabolism, regulation is as yet unclear, because this protein possesses RNA-binding domains, but its putative mammalian homolog protein, MINT, exhibits DNA-binding activity and transcriptional regulation properties (19).
A fusion OTT-MAL transcript could be detected in two infants suffering from AML-M7 and bearing variant and common t(1;22) translocation, demonstrating the recurrence of this fusion. Our structural analyses show that the OTT promoter initiates transcription of a fusion RNA that would encode a fusion protein comprising almost all OTT and MAL products, whereas the reciprocal fusion transcript would code for a 17-aa peptide in the common translocation. This is in keeping with cytogenetic data from the first patient analyzed here, which exhibit a variant t(1;22;4) translocation that created only the OTT-MAL fused gene on der 22 but not the MAL-OTT. The OTT-MAL product is represented Fig. 3F. It presents several common features with the HRX fusions, the other fusion frequently observed in infant acute leukemias. Both fusion proteins are expected to participate in chromatin organization through the binding of AT-rich DNA sequences, recognized by the AT Hook motif in HRX fusions (28) and by the SAF box in the OTT-MAL fusion. The HRX gene product has been shown to regulate HOX gene expression (29) and, on the basis of strong structural similarities between spen and OTT proteins, this is also likely to be the case for the latter protein. Establishment of mouse models will shed some light on chromatin structure regulation and control of cellular differentiation during normal and pathological processes and on the molecular bases of the unique association between t(1;22) and AML-M7.
Acknowledgments
We thank V. Lacronique and S. Gisselbrecht for critical reading of the manuscript, J. Zucman-Rossi, and V. Della Valle for helpful discussions, and the CEPH facilities for sequencing. This work was partially supported by Institut National de la Santé et de la Recherche Médicale, Fondation Saint-Louis, and the Ligue Nationale Contre le Cancer. We stress that the maps and sequences of the human genome made available by the public Human Genome Project facilitated this work.
Abbreviations
- AML
acute myeloblastic leukemia
- FISH
fluorescence in situ hybridization
- MAL
megakaryocytic acute leukemia
- OTT
one twenty-two
- RT-PCR
reverse transcription–PCR
- EST
expressed sequence tag
Footnotes
References
- 1.Rabbitts T H. Nature (London) 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 2.Rowley J D. Annu Rev Genet. 1998;32:495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- 3.Look A T. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 4.Mann R S, Chan S K. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 5.Rowley J D. Semin Cancer Biol. 1993;4:377–385. [PubMed] [Google Scholar]
- 6.Dimartino J F, Cleary M L. Br J Haematol. 1999;106:614–626. doi: 10.1046/j.1365-2141.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- 7.Gould A. Curr Opin Genet Dev. 1997;7:488–494. doi: 10.1016/s0959-437x(97)80075-5. [DOI] [PubMed] [Google Scholar]
- 8.Yu B D, Hess J L, Horning S E, Brown G A, Korsmeyer S J. Nature (London) 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 9.Rubnitz J E, Link M P, Shuster J J, Carroll A J, Hakami N, Frankel L S, Pullen D J, Cleary M L. Blood. 1994;84:570–573. [PubMed] [Google Scholar]
- 10.Hilden J M, Frestedt J L, Kersey J H. Leuk Lymphoma. 1997;25:191–199. doi: 10.3109/10428199709114159. [DOI] [PubMed] [Google Scholar]
- 11.Cimino G, Rapanotti M C, Elia L, Biondi A, Fizzotti M, Testi A M, Tosti S, Croce C M, Canaani E, Mandelli F, et al. Cancer Res. 1995;55:1625–1628. [PubMed] [Google Scholar]
- 12.Baruchel A, Daniel M T, Schaison G, Berger R. Cancer Genet Cytogenet. 1991;54:239–243. doi: 10.1016/0165-4608(91)90213-e. [DOI] [PubMed] [Google Scholar]
- 13.Bernstein J, Dastugue N, Haas O A, Harbott J, Heerema N A, Huret J L, Landman-Parker J, LeBeau M M, Leonard C, Mann G, et al. Leukemia. 2000;14:216–218. doi: 10.1038/sj.leu.2401639. [DOI] [PubMed] [Google Scholar]
- 14.Carroll A, Civin C, Schneider N, Dahl G, Pappo A, Bowman P, Emami A, Gross S, Alvarado C, Phillips C, et al. Blood. 1991;78:748–752. [PubMed] [Google Scholar]
- 15.Romana S P, Cherif D, Le Coniat M, Derre J, Flexor M A, Berger R. Genes Chromosomes Cancer. 1993;8:98–103. doi: 10.1002/gcc.2870080206. [DOI] [PubMed] [Google Scholar]
- 16.Dunham I, Shimizu N, Roe B A, Chissoe S, Hunt A R, Collins J E, Bruskiewich R, Beare D M, Clamp M, Smink L J, et al. Nature (London) 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- 17.Kipp M, Gohring F, Ostendorp T, van Drunen C M, van Driel R, Przybylski M, Fackelmayer F O. Mol Cell Biol. 2000;20:7480–7489. doi: 10.1128/mcb.20.20.7480-7489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skarnes W C, Auerbach B A, Joyner A L. Genes Dev. 1992;6:903–918. doi: 10.1101/gad.6.6.903. [DOI] [PubMed] [Google Scholar]
- 19.Newberry E P, Latifi T, Towler D A. Biochemistry. 1999;38:10678–10690. doi: 10.1021/bi990967j. [DOI] [PubMed] [Google Scholar]
- 20.Burd C G, Dreyfuss G. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 21.Kuang B, Wu S C, Shin Y, Luo L, Kolodziej P. Development (Cambridge, UK) 2000;127:1517–1529. doi: 10.1242/dev.127.7.1517. [DOI] [PubMed] [Google Scholar]
- 22.Wiellette E L, Harding K W, Mace K A, Ronshaugen M R, Wang F Y, McGinnis W. Development (Cambridge, UK) 1999;126:5373–5385. doi: 10.1242/dev.126.23.5373. [DOI] [PubMed] [Google Scholar]
- 23.Rebay I, Chen F, Hsiao F, Kolodziej P A, Kuang B H, Laverty T, Suh C, Voas M, Williams A, Rubin G M. Genetics. 2000;154:695–712. doi: 10.1093/genetics/154.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen F, Rebay I. Curr Biol. 2000;10:943–946. doi: 10.1016/s0960-9822(00)00625-4. [DOI] [PubMed] [Google Scholar]
- 25.Staehling-Hampton K, Ciampa P J, Brook A, Dyson N. Genetics. 1999;153:275–287. doi: 10.1093/genetics/153.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golub T R, Barker G F, Stegmaier K, Gilliland D G. Curr Top Microbiol Immunol. 1997;220:67–79. doi: 10.1007/978-3-642-60479-9_5. [DOI] [PubMed] [Google Scholar]
- 27.Salomon-Nguyen F, Della-Valle V, Mauchauffe M, Busson-Le Coniat M, Ghysdael J, Berger R, Bernard O A. Proc Natl Acad Sci USA. 2000;97:6757–6762. doi: 10.1073/pnas.120162297. . (First Published May 30, 2000; 10.1073/pnas.120162297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeleznik-Le N J, Harden A M, Rowley J D. Proc Natl Acad Sci USA. 1994;91:10610–10614. doi: 10.1073/pnas.91.22.10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu B D, Hanson R D, Hess J L, Horning S E, Korsmeyer S J. Proc Natl Acad Sci USA. 1998;95:10632–10636. doi: 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]