Abstract
Investigation of physiological mechanisms at a cellular level often requires production of high-quality antibodies, frequently using synthetic peptides as immunogens. Here we describe a new, web-based software tool called NHLBI-AbDesigner that allows the user to visualize the information needed to choose optimal peptide sequences for peptide-directed antibody production (http://helixweb.nih.gov/AbDesigner/). The choice of an immunizing peptide is generally based on a need to optimize immunogenicity, antibody specificity, multispecies conservation, and robustness in the face of posttranslational modifications (PTMs). AbDesigner displays information relevant to these criteria as follows: 1) “Immunogenicity Score,” based on hydropathy and secondary structure prediction; 2) “Uniqueness Score,” a predictor of specificity of an antibody against all proteins expressed in the same species; 3) “Conservation Score,” a predictor of ability of the antibody to recognize orthologs in other animal species; and 4) “Protein Features” that show structural domains, variable regions, and annotated PTMs that may affect antibody performance. AbDesigner displays the information online in an interactive graphical user interface, which allows the user to recognize the trade-offs that exist for alternative synthetic peptide choices and to choose the one that is best for a proposed application. Several examples of the use of AbDesigner for the display of such trade-offs are presented, including production of a new antibody to Slc9a3. We also used the program in large-scale mode to create a database listing the 15-amino acid peptides with the highest Immunogenicity Scores for all known proteins in five animal species, one plant species (Arabidopsis thaliana), and Saccharomyces cerevisiae.
Keywords: immunogenicity, posttranslational modification, specificity, epitope, conservation
the advent of genome sequencing projects for multiple animal and plant species at the beginning of this century has led to a broadened view of physiological and biochemical mechanisms at a cellular level, owing to the recognition of many poorly studied proteins that may play key roles in cellular regulation. The data from these sequencing projects also provide the information needed for facile generation of reagents, including antibodies, necessary for investigation of these proteins and the cellular pathways that they are involved with. Such antibodies are typically used for identification of protein localization in cells (e.g., by immunocytochemistry), purification of protein complexes (e.g., by immunoprecipitation), and routine quantification (e.g., by immunoblotting). However, the acquisition or production of antibodies for such investigations remains a trial-and-error undertaking in many cases.
The antibody-design task involves the choice of an immunogen that is predicted to evoke a vigorous immunogenic response in the host species. Frequently, the immunogen consists of a short synthetic peptide that is conjugated to a carrier protein. In this setting, the initial task involves the choice of a potentially immunogenic peptide sequence that corresponds to a portion of the target protein. In many cases, suitable antibodies have been obtained using the relative hydropathy of candidate peptides as the sole predictor of immunogenicity [e.g., via Kyte-Doolittle hydropathy index (22)]. Jameson and Wolf (14) have presented an objective function, the so-called “antigenic index,” that has also been widely used as a predictor of immunogenicity. The Jameson-Wolf antigenic index is a weighted sum of several determinants including hydropathy, secondary structure predictors [e.g., via the Chou-Fasman method (4)], and surface probability [via Janin method (15)]. However, the decision of what immunizing peptides to use often depends on factors other than imputed antigenicity (or more properly “immunogenicity”). For example, an immunizing peptide that is identical to or similar to sequences in other proteins is likely to produce an antibody that is not specific, recognizing not only the target protein but also these other proteins. Thus, there may be a trade-off between immunogenicity and “uniqueness” of a given synthetic peptide that may complicate the choice. Other trade-offs can also be recognized. For example, an investigator may wish to produce an antibody that recognizes a given protein in more than one species. Under this circumstance, he or she may wish to choose an immunizing peptide that is common to all species of interest. Another potential problem with selecting the immunizing peptide comes when posttranslational modifications occur within the corresponding region of the target protein. Under this circumstance, the antibody may recognize the protein in the absence of the posttranslational modification but not in the presence of the modification if a key epitope is obliterated. When multiple trade-offs must be considered in the production of the synthetic peptide to be used for immunization, it can be difficult to take all of the relevant information into consideration, since such information must be culled from multiple sources using multiple software applications.
To address this task, we present a fully integrated online software tool, NHLBI-AbDesigner, for the design of peptide-directed antibodies. This program is implemented as a web application (http://helixweb.nih.gov/AbDesigner/) that displays information relevant to the choice of the optimal immunizing peptide for a given biological application, including predicted immunogenicity, uniqueness (predictor of specificity), conservation (predictor of multispecies cross-reactivity), and relevant protein features such as posttranslational modifications, domain architecture, sites of sequence variation due to alternative splicing, and other regions or sites of interest culled from the corresponding Swiss-Prot record. AbDesigner was also employed in batch mode to generate a genome-wide list of top-scoring immunizing peptides for selected animal and plant species (viz. Homo sapiens, Rattus norvegicus, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Saccharomyces cerevisiae, and Arabidopsis thaliana) as a resource for large-scale antibody design.
EXPERIMENTAL PROCEDURES
Peptide synthesis and antibody production.
Based on analysis of rat sodium/hydrogen exchanger 3 (NHE3; gene symbol: Slc9a3) using AbDesigner, a 20-aa synthetic peptide corresponding to amino acids 621–640 of the COOH-terminal tail of NHE3, was produced by standard solid-phase peptide synthesis techniques followed by HPLC purification to >95% (sequence: YLYKPRQEYKHLYSRHELTP with an added amino-terminal cysteine for conjugation, Lofstrand, Gaithersburg, MD). The peptide was purified by HPLC and was conjugated to maleimide-activated keyhole limpet hemocyanin via covalent linkage to the NH2-terminal cysteine. Two rabbits were immunized (Antibodies, Davis, CA) with this conjugate using a combination of Freund's complete and incomplete adjuvants. One of these antisera (7644) was used for the present studies after affinity purification on a column made with the same synthetic peptide used for immunizations (SulfoLink Immobilization Kit, Pierce, Rockford, IL).
Immunoblotting.
Immunoblotting was carried out as described (31) to assess the ability of antibodies to recognize the target proteins using whole homogenates (or membrane fractions) from the kidney cortex and outer medulla of rats or humans. Rat kidneys were resected from pathogen-free male Sprague-Dawley rats (Taconic Farms) weighing 180–220 g after euthanization [Animal protocol no. H-0110, approved by the Institutional Animal Care and Use Committee, National Heart, Lung, and Blood Institute (NHLBI)]. Human kidney tissue was obtained from a nephrectomy specimen (approved as exempt from review by the National Institutes of Health Office of Human Subjects Research) (32). In brief, kidney tissue was homogenized using a tissue homogenizer (Omni 1000 fitted with a micro-sawtooth generator) in ice-cold isolation solution (pH 7.6) containing 250 mM sucrose, 10 mM triethanolamine, 1 μg/ml leupeptin, and 0.1 mg/ml phenylmethylsulfonyl fluoride, and total protein concentration (BCA Protein Assay Kit, Pierce) was adjusted to 1–2 μg/μl with isolation solution. The samples were solubilized in 5× Laemmli sample buffer (1 vol per 4 vol of sample) followed by heating to 60°C for 15 min. After solubilization in Laemmli buffer, protein samples (15–20 μg) were resolved by SDS-PAGE gel electrophoresis on 10 or 4–15% polyacrylamide gels (Bio-Rad, Hercules, CA) and transferred electrophoretically onto nitrocellulose membranes. The membranes were then blocked with Odyssey blocking buffer (Li-Cor, Lincoln, NE), rinsed, and probed with primary antibody overnight at room temperature. After a washing, blots were incubated with species-specific fluorescently labeled secondary antibodies Alexa Fluor 680 (Molecular Probes, Eugene, OR) used at 1:5,000 for detection of all primary antibodies. Fluorescence was imaged using an Odyssey Imaging System (Li-Cor).
Software implementation.
The main workflow of AbDesigner is illustrated in Fig. 1. AbDesigner was written in Java (Java Development Kit 6 Update 23, Oracle) using NetBeans IDE 7.0 as an integrated development environment. The web application of AbDesigner was developed using Java Servlet as a controller and using Java Applet and JavaServer Pages for presentation. The web application was implemented using Apache Tomcat 7.0.12 hosted by the National Institutes of Health (NIH) Biowulf cluster (http://biowulf.nih.gov/) and the NHLBI Center for Biomedical Informatics (http://www.nhlbi.nih.gov/about/cbi/index.htm). A mirror site has been established at https://javaapps.nhlbi.nih.gov/AbDesigner/.
Fig. 1.
The main workflow of AbDesigner.
To run the web version of AbDesigner, a user needs the Java SE Runtime Environment version 6 (http://www.java.com/en/download/). Earlier versions may work with reduced functionality. The web version of AbDesigner has been tested on multiple operating systems and web browsers. The operating systems tested are Microsoft Windows XP (version 5.1.2600), Microsoft Windows 7 (version 6.1.7600), and Mac OS X 10.6.4 (10F569). The web browsers tested are Firefox 4 (Windows), Internet Explorer 9 (Windows), and Safari 5 (Macintosh and Windows).
The calculation of “Immunogenicity Score” (Ig Score), “Uniqueness Score,” and “Conservation Score” is described in the appendix. To display “Protein Features” (including structural domains, variable regions, and annotated PTMs), AbDesigner extracts the relevant information from the Swiss-Prot Protein Database downloaded from the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/protein).
RESULTS
Software description: submission page.
The object of AbDesigner is to display the features of a protein relevant to the choice of a synthetic peptide sequence to be used as an immunogen in antibody production. It does so in a manner that allows the user to judge trade-offs for candidate peptide sequences with respect to multiple factors including hydropathy, secondary structure, uniqueness, conservation among species, and the presence or absence of posttranslational modifications. The online submission page can be found at http://helixweb.nih.gov/AbDesigner/ and is illustrated in Fig. 2. To specify a protein for analysis, the program accepts the following types of input: Gene Symbol, Swiss-Prot Accession Number, or Swiss-Prot Entry Name from any of the following seven species: Homo sapiens, Rattus norvegicus, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Saccharomyces cerevisiae, or Arabidopsis thaliana (see Fig. 2, A and B). Proteins from other species can be analyzed by entering the sequence of that protein FASTA format (this should be avoided when analyzing proteins from the above seven species because of the limitations associated with submitting a FASTA amino acid sequence as described in the appendix). The amino acid sequence and protein annotations for the input protein are extracted from the Swiss-Prot Protein Database downloaded from NCBI. A user-defined sequence length for production of synthetic peptides for immunization can be set from 5 to 50 amino acids (“Peptide Length”; see part C of Fig. 2). (A peptide of 10–25 amino acids is typically used. A longer sequence can provide a greater likelihood of producing a potent antibody by virtue of the fact that it contains more potential epitopes, but by the same token there is a greater chance that it will produce IgG clones with lower specificity. Furthermore, the cost of peptide synthesis often increases substantially with larger peptides.) Finally, a user-defined linear “Epitope Length” can be set from five amino acids to the full length of the peptide (to be used for determination of uniqueness and conservation of a peptide; see part D of Fig. 2). In the example in Fig. 2, Gene Symbol (Rat) is selected as an input type, AQP2 (the gene symbol for water channel aquaporin-2) is the input, a peptide length of 15 amino acids is entered, and an epitope length of 7 amino acids is selected.
Fig. 2.
An online submission page of AbDesigner. The page contains four elements that need to be entered. A: input type (i.e., gene symbol, Swiss-Prot accession number, Swiss-Prot entry name, or FASTA amino acid sequence). B: input. C: peptide length (i.e., user-defined sequence length to be used for production of synthetic peptides for immunization). D: epitope length (i.e., theoretical length of linear epitopes to be used for determination of uniqueness and conservation of a peptide, default = 7 amino acids).
Software description: output page.
After the input values are entered, AbDesigner calculates and displays (Fig. 3) the Immunogenicity Score (Ig Score) [modified from the principle of the Jameson-Wolf antigenic index (14)], which is aligned with the calculated Uniqueness Score, the calculated Conservation Score, and Protein Features extracted from the corresponding Swiss-Prot record. Uniqueness Score allows the user to predict the specificity of an antibody produced by a peptide. Conservation Score allows the user to predict the likelihood that the ortholog of the target protein (i.e., from alternative species) will be recognized by the antibody. How the scores are calculated from the sequence data is described in the appendix. The Protein Features reported are position-dependent annotations of regions or sites of interest such as posttranslational modifications, binding sites, enzyme active sites, local secondary structure, sequence conflicts, and other characteristics culled from the appropriate Swiss-Prot protein record. The parallel linear display shown in Fig. 3 allows the user to evaluate the various trade-offs among factors that may impinge on the choice of an immunizing peptide sequence, according to the intended user-specific purpose. In the following, we describe the features of the output page in greater detail.
Fig. 3.
An online graphical output of AbDesigner. The following information is displayed in the interactive Java Applet component of the web page (main panel). A: protein sequence. B: Chou-Fasman secondary structure prediction. C: Kyte-Doolittle (K-D) hydropathy index. D: Immunogenicity Score (Ig Score). E: Uniqueness Score. F: Conservation Score. G: Protein Features. The three bottom panels display lists of peptides sorted by Immunogenicity Score rank (H), Uniqueness-optimized rank (I), and Conservation-optimized rank (J).
The online graphical output of AbDesigner consists of an interactive Java Applet embedded in an html page (Fig. 3). The main panel (the Applet) displays a variety of information and allows users to “mouse-over” each element to obtain further data. Annotations in Fig. 3 are designated by letters A–J (not part of the actual output). In the main panel, the upper line (A) shows the input amino acid sequence. When users mouse-over the main panel, the peptide of interest of the appropriate length is highlighted in yellow with the central amino acid underlined. The low-complexity regions of a protein identified by the segmasker program [based on the SEG filtering algorithm (36), obtained from the NCBI C++ Toolkit, ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/] are displayed in red font and on mouse-over along the protein sequence row.
The next line (B) of the main panel displays Chou-Fasman secondary structure prediction. The Chou-Fasman prediction is performed as described (4) except for slight modifications (see appendix). The secondary structure predicted, i.e., alpha helix, beta sheet, strong beta turn, or weak beta turn, is displayed by different colors and on mouse-over. High immunogenicity correlates with a lack of alpha helices or beta sheets, and presence of beta turns. The chief practical value of this analysis is that it identifies locations of prolines (the chief determinant of a prediction of “beta-turn”), which aid immunogenicity by interfering with α-helix formation.
The next line (C) of the main panel displays the Kyte-Doolittle hydropathy index (KDHI) of each peptide along a protein sequence. The KDHI is displayed in 8-bit scale (0–255) RGB color heat map and on mouse-over. Given that lower KDHI values correlate with greater immunogenicity, the 8-bit transformation of the KDHI is performed in the reverse direction so that the lowest value (−4.5, most hydrophilic) is transformed to 255 (displayed in the brightest cyan, by default) while the highest value (4.5, most hydrophobic) is transformed to 0.
Parts D–F of the main panel display Immunogenicity (Ig) Score, Uniqueness Score, and Conservation Score, respectively, using the desired peptide length as the window for these calculations. These scores are displayed in 8-bit RGB color heat maps. By default, the highest Immunogenicity Score, Uniqueness Score, and Conservation Score are displayed in the brightest green, yellow, and red, respectively. However, user-defined colors can be selected from the menu bar. In addition, for each peptide, the Immunogenicity Score value and rank are displayed on mouse-over in the Immunogenicity Score heat map, whereas multiple sequence alignments are displayed on mouse-over in the Uniqueness and Conservation heat maps.
Part G of the main panel displays various useful Protein Features. The position-dependent annotations of regions or sites of interest in the sequence (including known sites of posttranslational modifications), extracted from the Swiss-Prot database, are displayed by various distinct colors and on mouse-over along a protein sequence. Figure 4 illustrates the mouse-over feature of the output page.
Fig. 4.
The main panel of the graphical output with interactive details. In the main panel, users can mouse-over the display to obtain further details for each element of the output.
At the bottom of the results page (H–J) are three separate lists of peptides sorted by Immunogenicity Score rank, Uniqueness-optimized rank, and Conservation-optimized rank (Fig. 3). “Ig Score rank” shows peptides from most immunogenic to least. “Uniqueness-optimized rank” shows primary ranking of peptides from most “unique” (i.e., specific for target protein versus all other proteins in that species) to least unique, with secondary ranking based on Immunogenicity Score. “Conservation-optimized rank” shows primary ranking of peptides from most “conserved” (i.e., sequence is conserved among multiple orthologous species) to least conserved, with secondary ranking based on Immunogenicity Score. These lists provide an alternative to the heat maps for identification of candidate peptides for immunization. However, use of these lists alone has the pitfall that they do not take into account the protein annotations provided in the main display.
Analysis of previously produced antibodies.
Our laboratory has made extensive use of the principles incorporated into AbDesigner to produce a substantial number of antibodies that have been successfully used for research applications over the past 15 years (Table 1). These peptide sequences were chosen based on calculation of the various protein characteristics by a series of analyses using the GCG suite of protein analysis programs (3), BLAST (1), and manual inspection of protein records available on GenBank as described by Knepper and Masilamani (21). The main advantage of AbDesigner is in the visualization of this information in a parallel arrangement, allowing facile recognition of the trade-offs involved in the consideration of alternative peptide sequences for immunization. In most cases, viewed retrospectively, peptides chosen for immunization were within the top 10th percentile of AbDesigner Immunogenicity Scores (Table 1). However, as illustrated in the examples described in Figs. 5–8, predicted immunogenicity cannot be practically employed alone in the choice of immunizing peptides.
Table 1.
A list of the peptide-directed antibodies and the synthetic peptides used for the generation of these antibodies by our laboratory
| Target Protein (Ref. no.) | Swiss-Prot No. | Peptide Sequence | Position | Length | Ig Score | Ig Score Percentile Rank |
|---|---|---|---|---|---|---|
| NHE3 (16) | P26433 | DSFLQADGPEEQLQPASPESTHM | 809–831 | 23 | 8.69 | 9.52 |
| NHE3 (this paper) | P26433 | YLYKPRQEYKHLYSRHELTP | 621–640 | 20 | 9.18 | 6.40 |
| NaPi-2a (17) | Q06496 | LEELPPATPSPRLALPAHHNATRL | 614–637 | 24 | 7.17 | 10.75 |
| NKCC2 (8) | P55016 | EYYRNTGSVSGPKVNRPSLQE | 109–129 | 21 | 9.9 | 1.49 |
| NKCC2 (28) | P55016 | SDSTDPPHYEETSFGDEAQNRLK | 33–55 | 23 | 10.5 | 0.09 |
| NCC (18) | P55018 | DGRPGHELTDGLVEDETGANSEK | 104–126 | 23 | 9.67 | 0.41 |
| NCC (2) | P55018 | PGEPRKVRPTLADLHSFLKQEG | 74–95 | 22 | 8.32 | 5.50 |
| aENaC (26) | P37089 | LGKGDKREEQGLGPEPSAPRQPT | 45–67 | 23 | 10.47 | 3.55 |
| bENaC (26) | P37090 | NYDSLRLQPLDTMESDSEVEAI | 617–638 | 22 | 7.76 | 12.48 |
| gENaC (26) | P37091 | NTLRLDRAFSSQLTDTQLTNEL | 629–650 | 22 | 7.57 | 14.94 |
| Na-K-ATPase, a1-subunit (30) | P06685 | DEVRKLIIRRRPGGWVEKETYY | 1002–1023 | 22 | 7.43 | 5.99 |
| Aquaporin-1 (35) | P29975 | GQVEEYDLDADDINSRVEMKPK | 248–269 | 22 | 8.84 | 0.81 |
| Aquaporin-2 (5) | P34080 | EVRRRQSVELHSPQSLPRGSKA | 250–271 | 22 | 8.81 | 5.20 |
| Aquaporin-2 (12) | P34080 | LKGLEPDTDWEEREVRRRQ | 237–255 | 19 | 9.43 | 1.19 |
| Aquaporin-3 (7) | P47862 | HLEQPPPSTEAENVKLAHMKHKEQI | 268–292 | 25 | 8.01 | 1.12 |
| Aquaporin-4 (34) | P47863 | IDIDRGDEKKGKDSSGE | 302–318 | 17 | 11.03 | 0.65 |
| Aquaporin-4 (34) | P47863 | TKGSYMEVEDNRSQVETED | 273–291 | 19 | 9.4 | 2.62 |
| P2Y2 purinoceptor (19) | P41232 | SISSDDSRRTESTPAGSETKDIRL | 351–374 | 24 | 9.46 | 9.12 |
| NSF (9) | Q9QUL6 | DPEYRVRKFLALMREEGASPLDFD | 721–744 | 24 | 5.01 | 19.28 |
| VAMP-2 (24) | P63045 | SATAATVPPAAPAGEGG | 2–18 | 17 | 6.77 | 61.00 |
| Syntaxin-3 (25) | Q08849 | KDRLEQLKAKQLTQDDDTDEVE | 2–23 | 22 | 9.09 | 3.73 |
| Syntaxin-4 (24) | Q08850 | RDRTHELRQGDNISDDEDEVRV | 2–23 | 22 | 9.93 | 1.81 |
| Synaptotagmin VIII (20) | Q9R0N6 | PREVDRVLALQPRLPLLRPRS | 375–395 | 21 | 7.19 | 46.13 |
| ROMK1 (6) | P35560 | KRGYDNPNFVLSEVDETDDTQM | 370–391 | 22 | 9.39 | 1.62 |
| UT-A1 (29) | Q62668 | QEKNRRASMITKYQAYDVS | 911–929 | 19 | 8.56 | 7.94 |
Ig Score, Immunogenicity Score; NHE3, sodium/hydrogen exchanger 3; NKCC, Na-K-2Cl cotransporter; NCC, Na-Cl cotransporter; ENaC, epithelial sodium channel; NSF, N-ethyl maleimide-sensitive fusion protein; VAMP-2, vesicle-associated membrane protein 2; ROMK1, regulation of potassium channel Kir 1.1.
Fig. 5.
AbDesigner output for rat thiazide-sensitive Na-Cl cotransporter (NCC). A: a low-magnification view of the output. B and C: magnified views of selected portions of the amino-terminal region. D: immunoblots of rat and human kidney cortex probed with the antibodies against the relatively poorly conserved peptide (peptide 104–126, top) and the highly conserved peptide (peptide 74–95, bottom).
Fig. 6.
AbDesigner output for rat syntaxin-4. Top: a low-magnification view of the output. Bottom: a magnified view of the amino-terminal region. Region A contains peptides with the best Immunogenicity Scores (brightest green region) but it is also within a coiled-coil region (purple bar). Region B has the second-best Immunogenicity Score and is outside of the coiled-coil region.
Fig. 7.
AbDesigner output for rat sodium/hydrogen exchanger 3 (NHE3). A: a low-magnification view of the output. B: a magnified view of the carboxyl-terminal region with high Immunogenicity Score, high Uniqueness Score, and high Conservation Score (bright green, yellow, and red, respectively). C: an immunoblot of rat kidney cortical and outer medullary tissues probed with the newly produced antibody against a peptide YLYKPRQEYKHLYSRHELTP of NHE3.
Fig. 8.
AbDesigner output for rat aquaporin-2. Top: a low-magnification view of the output. Bottom: a magnified view of the carboxyl-terminal region. Peptide 237–255 and peptide 250–271 are highlighted.
Maximizing multispecies coverage.
The first example comes from the production of antibodies to the thiazide-sensitive Na-Cl cotransporter (NCC; gene symbol: Slc12a3), which is expressed in the renal distal convoluted tubule and is important for regulation of blood pressure and extracellular fluid volume. An initial antibody made to a 23 amino acid peptide corresponding to rat NCC produced an effective antibody that has been utilized for studies in rat (18). However, this region is poorly conserved among mammalian species and the rat antibody only poorly recognized human NCC. Figure 5 shows the AbDesigner output for rat NCC at low magnification at the top (A), and magnified views of the amino-terminal region at the bottom (B and C). The original antibody was made from amino acids 104–126 (Fig. 5C), which can be seen to have a relatively low Conservation Score (dark region on heat map). A region of high conservation and with reasonable predicted immunogenicity was identified in the amino acid range 74–95 of rat NCC (Fig. 5B). To obtain an antibody that would efficiently recognize human NCC, a peptide from this region was utilized to make a new antibody to NCC (2). Figure 5D shows an immunoblot demonstrating that the antibody against the highly conserved peptide (bottom) recognizes an ∼160 kDa band in both rat and human kidney cortex (human > rat). In contrast, as seen previously, the original antibody (Fig. 5D, top) recognized rat NCC much more strongly than human NCC.
Avoiding nonunique regions and special protein domains.
The next example comes from the antibodies targeting SNARE proteins, i.e., syntaxins and VAMPs (synaptobrevins) (Table 1). These are type II integral membrane proteins with a single membrane span near the COOH terminus and an extracellular COOH-terminal tail. The AbDesigner output for syntaxin-4 is illustrated in Fig. 6 with a low-magnification view at the top and a magnified view of the amino-terminal region at the bottom. The best Immunogenicity Scores were found amid amino acids 105–128 (region A). However, this region was within a coiled-coil region (purple bar) that forms very stable SNARE complexes with other SNARE proteins. This factor may limit the use of the antibody to situations that would require exhaustive denaturation of protein samples prior to the analysis to effectively break up the coiled-coil interactions with other SNARE proteins. As seen in Fig. 6, the second best Immunogenicity Score region where these circumstances could be reasonably avoided is the NH2-terminal 23 amino acids (region B). This region also exhibited a high degree of specificity (high Uniqueness Score as displayed in brightest yellow). Using a syntaxin-4 peptide from this region, an antibody was successfully produced and utilized for immunoblotting and immunocytochemistry (24).
Another example comes from the production of antibodies to NHE3 (gene symbol: Slc9a3). To produce an effective antibody for NHE3, a region of high Immunogenicity Score, high Uniqueness Score (specificity), and high Conservation Score was identified (Fig. 7A, low-magnification view; Fig. 7B, magnified view) in the carboxyl-terminal region. We have now utilized a peptide from this region (sequence: YLYKPRQEYKHLYSRHELTP, amino acids 621–640 with an added NH2-terminal cysteine) to make a new antibody to NHE3 (previously unpublished). Figure 7C shows an immunoblot demonstrating that the antibody successfully recognizes an 84-kDa band in rat kidney cortical and outer medullary tissues, matching the mass seen previously with other NHE3 antibodies. As anticipated from the high Uniqueness Score, the new antibody appears to be highly specific, i.e., it labeled no major bands other than that expected for NHE3.
Avoiding posttranslational modifications.
Another example comes from two antibodies targeting aquaporin-2 (gene symbol: Aqp2) (Table 1), a water channel expressed in the kidney and responsible for regulation of water excretion. Figure 8 shows the AbDesigner output at low magnification at the top and a magnified view of the carboxyl-terminal region of aquaporin-2 at the bottom. The original antibodies to aquaporin-2 arbitrarily targeted the carboxyl-terminal 22 amino acids of the protein (amino acids 250–271), producing antibodies that have been extensively used in the localization of the protein (5, 27). Other laboratories made antibodies to the same region (10, 33). Subsequent studies, however, revealed that this region encompasses several serines that are variably phosphorylated (12, 13). Phosphorylation at these sites is likely to alter the ability of the antibody to recognize the aquaporin-2 protein if one or more of the phosphorylated serines lies within the major epitopes recognized by the antibody. This led us to produce a new antibody (12) upstream from this poly-phosphorylated region (amino acids 237–255, in an area that is also highly conserved among rat, mouse, and human), allowing the use of the antibody for precise quantification of aquaporin-2 abundance (37).
Large-scale implementation.
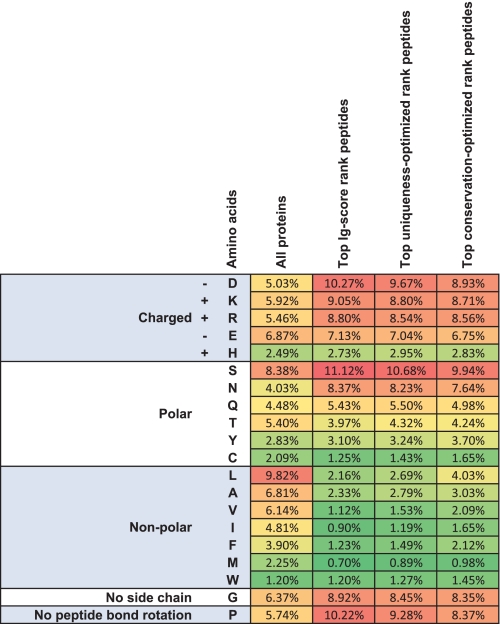
We used a command line version of AbDesigner running on the NIH Biowulf cluster (http://biowulf.nih.gov/) to identify the top scoring 15 amino acid peptides against all known members of the proteomes of Homo sapiens, Rattus norvegicus, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Saccharomyces cerevisiae, and Arabidopsis thaliana. A database of the top Immunogenicity Score peptide, the top Uniqueness-optimized peptide (top Immunogenicity Score among top-ranked Uniqueness Scores), and the top Conservation-optimized peptide (top Immunogenicity Score among top-ranked Conservation Scores) for each protein is included as Dataset 1 (available for download at http://helixweb.nih.gov/ESBL/Database/AbDesignerPeptides/). An analysis of the amino acid composition of these top candidate peptides in comparison to the background composition of all 67,660 proteins is shown in Fig. 9. As expected (21), the top-ranking peptides have higher percentages of charged amino acids (D, E, R, K, and H), polar amino acids (S, N, Q, and Y), glycines, and prolines compared with the background (all amino acids in all input proteins).
Fig. 9.
A heat map summarizing the amino acid frequencies of the top scoring peptides from a full proteome analysis of multiple organisms. The amino acid composition of top Immunogenicity Score rank peptides, top Uniqueness-optimized rank peptides, and top Conservation-optimized rank peptides were compared with the background composition of all 67,660 proteins from the seven species.
DISCUSSION
In this paper, we describe a web-based software tool, AbDesigner, created by biologists to help biologists design peptide-directed antibodies. Its design is based on our own experience in peptide-directed antibody design (Table 1) using principles discussed by Knepper and Masilamani (21) to obtain peptide sequences that are most likely to be highly immunogenic, while producing antibodies that are specific for the target protein (based on the calculated Uniqueness Score). Beyond this, AbDesigner reports Conservation Scores for all candidate peptides, which reflect the likelihood that an antibody made in a given mammalian species will recognize the ortholog in other species. Additional Protein Features are displayed, which allow the user to avoid regions in the target protein that undergo posttranslational modifications (possibly obliterating the epitope) or splicing variations that may make some isoforms of the target protein invisible to the antibody. The web interface is designed to provide the user with a “minimalist,” intuitive tool that will allow successful use without having to read complicated instructions or to install files on the user's computer (other than perhaps Java, which is usually preinstalled on most computers). The multiple “mouse-over” features that were included in the AbDesigner display provide a great deal of ancillary information without a cluttered presentation. Overall, the software allows the user to display the information needed to recognize the trade-offs that may exist for alternative choices of synthetic peptide sequences to use in antibody production. Software with a similar goal called Bishop has been previously reported (23); it shares some features of AbDesigner but is not available as an online tool. Although AbDesigner is designed for peptide-directed antibody production in which the immunogen is a synthetic peptide conjugated to a carrier protein, it can also be used to visualize the same type of information for candidate recombinant fusion proteins, an approach that is employed by many investigators for creation of immunogens for antibody production. AbDesigner is also well-suited for researchers who wish to evaluate existing custom made or purchased antibodies. Before purchase, the user can predict whether or not a given commercial antibody will work for his or her purpose.
We emphasize that the online version of AbDesigner is not designed for unsupervised choices of immunizing peptides. The final sequence chosen in a particular context depends on subjective factors that must be furnished by the user, reflecting the nature of the intended experimental use of the antibody. It is clear from the analysis of the successfully produced antibodies described in Table 1 that excellent antibodies can be produced from sequences that do not necessarily rank highest in terms of Immunogenicity Score. Thus, the purpose of AbDesigner is to organize and display information, not to make predictions. In that regard, AbDesigner may find broader use as a means of displaying an overview of the properties of a given protein, for example, that users may encounter in their reading or may newly identify in discovery studies. Future development should include addition of other types of information including three-dimensional protein structure and location of epitopes for existing antibodies.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: T.P. and M.A.K. conception and design of research; T.P. performed experiments; T.P. and M.A.K. analyzed data; T.P. and M.A.K. interpreted results of experiments; T.P. prepared figures; T.P. and M.A.K. drafted manuscript; T.P., J.D.H., F.S., and M.A.K. edited and revised manuscript; T.P., J.D.H., F.S., and M.A.K. approved final version of manuscript.
ACKNOWLEDGMENTS
This work was supported by the Intramural Budget of the National Heart, Lung, and Blood Institute (Project Z01-HL-001285).
APPENDIX
Immunogenicity Score.
Immunogenicity Score is a predictor of immunogenicity. The higher the score, the greater the predicted immunogenicity. The Immunogenicity Score of a peptide is calculated from the following formula:
where KDHI is Kyte-Doolittle hydropathy index, Pt(average) is the average Chou-Fasman conformational parameters of beta turn, and Tail bonus is a value ranging from 1.0 to 1.5.
KDHI is an average value of the hydropathy indices of consecutive amino acids in a peptide. It is calculated using the hydropathy scale (a range of −4.5 to 4.5) (22), with a negative KDHI predicted to be more immunogenic than a positive KDHI. Thus, the negative value of the KDHI is used in the formula. A value of 4.5 is added to keep the Immunogenicity Score in a positive range. Pt(average) is an average value of the Chou-Fasman conformational parameters of a beta turn (Pt) of amino acids in a peptide. It is calculated using Pt values from a reference database of 29 proteins as described (4). The “topological domain” information of a protein extracted from the Swiss-Prot database is used for determining the Tail bonus. Tail bonus is only given to a peptide that resides in NH2- or COOH-terminal tail of an integral membrane protein. Tail bonus values can range from 1.0 to 1.5. A Tail bonus value of 1.5 corresponds to the full length of a peptide being present in a tail region, while a peptide whose full length is present in a non-tail region is given a tail bonus value of 1.0. Values are linearly correlated with the number of amino acids contained in the tail compared with the entire length of the peptide.
Uniqueness Score.
A typical immunogenic peptide is assumed to contain multiple linear, overlapping potential epitopes [∼5 amino acids (11)], each of which can theoretically invoke an immune response. The uniqueness of these linear epitope sequences compared with other proteins of the same species helps determine the specificity of an antibody produced against that peptide. In AbDesigner, the sequence of each successive linear epitope (shifting by one amino acid) along a peptide sequence is compared against the entire protein sequence database of that species. The length of a linear epitope can be set from five amino acids to the full length of the peptide. The total number of linear epitopes in other proteins that have sequences identical to the linear epitopes in a given peptide is calculated as follows:
where M is the total number of linear epitope matches, n is the number of successive linear epitopes along a peptide sequence, and mi is the number of the linear epitopes in the other proteins that have sequences identical to the linear epitope i. Redundant linear epitopes in a protein are counted only once. A higher value of M corresponds to a less unique peptide and vice versa. The Uniqueness Score of a peptide is then calculated on the basis of the following formula:
where Mc is the cutoff value for M. The highest Uniqueness Score is equal to Mc (when there is no match to any linear epitopes in a peptide, M = 0), and the lowest Uniqueness Score is equal to 0 (when the total number of linear epitope matches is equal to or above the cutoff value, M ≥ Mc). Mc is arbitrarily set to three times of n.
Conservation Score.
Conservation Score predicts the likelihood that the target protein will be detectable by the antibody in multiple orthologous species. The higher the score, the greater the predicted conservation. The Conservation Score of a peptide is determined in a comparable manner to the Uniqueness Score. To begin with, the sequence of each successive linear epitope (shifting by one amino acid) along a peptide in a protein is compared against the sequences of the orthologous proteins, based on a mnemonic protein identification code in the Swiss-Prot entry name (http://www.uniprot.org/manual/entry_name), among three commonly studied species (i.e., human, rat, and mouse). The Conservation Score of a peptide is then calculated from the total number of linear epitopes in orthologous proteins that have sequences identical to the linear epitopes in a given peptide as follows:
where n is the number of successive linear epitopes along a peptide sequence and ci is the number of the linear epitopes in the orthologous proteins that have sequences identical to the linear epitope i. Redundant linear epitopes in a protein are counted only once. The highest Conservation Score is reached when a peptide is completely conserved across all three species and is equal to the total number of the orthologous species evaluated multiplied by n. The lowest Conservation Score is equal to 0 (when there is no conservation).
Heat maps.
The Immunogenicity Score, Uniqueness Score, and Conservation Score of each peptide are displayed in 8-bit RGB color heat maps. The transformation of those values into a density representation (D) on an 8-bit scale (0–255) is performed using linear scaling: D = 255 * (X − Xmin)/(Xmax − Xmin), where X is the value, Xmin is the minimum value, and Xmax is the maximum value as defined in Table A1.
Table A1.
Variables used for 8-bit transformation of Ig Score, Uniqueness Score, and Conservation Score
| X | Xmin | Xmax |
|---|---|---|
| Ig Score | The lowest Ig Score in that protein | The highest Ig Score in that protein |
| Uniqueness Score | 0 | 3 times of n |
| Conservation Score | 0 | The total number of the orthologous species evaluated multiplied by n |
n, no. of successive linear epitopes along a peptide sequence.
Limitations associated with submitting a FASTA amino acid sequence.
By submitting just a FASTA amino acid sequence, the program assumes that the input protein is not present in the local Swiss-Prot protein database (from the following seven species: Homo sapiens, Rattus norvegicus, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Saccharomyces cerevisiae, or Arabidopsis thaliana). Thus, the following processes that make use of the database cannot be executed: 1) extraction of Protein Features; 2) calculation of Tail bonus; 3) calculation of Uniqueness Score; and 4) calculation of Conservation Score. This leads to the following results: 1) Protein Features will not be displayed in the graphical output; 2) Immunogenicity Score will be calculated without factoring in Tail bonus; 3) Uniqueness Score will not be calculated, the graphical output of Uniqueness Score will be the default brightest yellow, and Uniqueness-optimized rank will be the same as Immunogenicity Score rank; and 4) Conservation Score will not be calculated, the graphical output of Conservation Score will be the default black, and Conservation-optimized rank will be the same as Immunogenicity Score rank.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 215: 403–410, 1990 [DOI] [PubMed] [Google Scholar]
- 2. Biner HL, Arpin-Bott MP, Loffing J, Wang X, Knepper M, Hebert SC, Kaissling B. Human cortical distal nephron: distribution of electrolyte and water transport pathways. J Am Soc Nephrol 13: 836–847, 2002 [DOI] [PubMed] [Google Scholar]
- 3. Butler BA. Sequence analysis using GCG. Methods Biochem Anal 39: 74–97, 1998 [DOI] [PubMed] [Google Scholar]
- 4. Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol 47: 45–148, 1978 [DOI] [PubMed] [Google Scholar]
- 5. DiGiovanni SR, Nielsen S, Christensen EI, Knepper MA. Regulation of collecting duct water channel expression by vasopressin in Brattleboro rat. Proc Natl Acad Sci USA 91: 8984–8988, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ecelbarger CA, Kim GH, Knepper MA, Liu J, Tate M, Welling PA, Wade JB. Regulation of potassium channel Kir 1.1 (ROMK) abundance in the thick ascending limb of Henle's loop. J Am Soc Nephrol 12: 10–18, 2001 [DOI] [PubMed] [Google Scholar]
- 7. Ecelbarger CA, Terris J, Frindt G, Echevarria M, Marples D, Nielsen S, Knepper MA. Aquaporin-3 water channel localization and regulation in rat kidney. Am J Physiol Renal Fluid Electrolyte Physiol 269: F663–F672, 1995 [DOI] [PubMed] [Google Scholar]
- 8. Ecelbarger CA, Terris J, Hoyer JR, Nielsen S, Wade JB, Knepper MA. Localization and regulation of the rat renal Na+-K+-2Cl− cotransporter, BSC-1. Am J Physiol Renal Fluid Electrolyte Physiol 271: F619–F628, 1996 [DOI] [PubMed] [Google Scholar]
- 9. Frank AE, Wingo CS, Andrews PM, Ageloff S, Knepper MA, Weiner ID. Mechanisms through which ammonia regulates cortical collecting duct net proton secretion. Am J Physiol Renal Physiol 282: F1120–F1128, 2002 [DOI] [PubMed] [Google Scholar]
- 10. Fushimi K, Uchida S, Hara Y, Hirata Y, Marumo F, Sasaki S. Cloning and expression of apical membrane water channel of rat kidney collecting tubule. Nature 361: 549–552, 1993 [DOI] [PubMed] [Google Scholar]
- 11. Geysen HM, Mason TJ, Rodda SJ. Cognitive features of continuous antigenic determinants. J Mol Recognit 1: 32–41, 1988 [DOI] [PubMed] [Google Scholar]
- 12. Hoffert JD, Fenton RA, Moeller HB, Simons B, Tchapyjnikov D, McDill BW, Yu MJ, Pistikun T, Chen F, Knepper MA. Vasopressin-stimulated increase in phosphorylation at ser-269 potentiates plasma membrane retention of aquaporin-2. J Biol Chem 283: 24617–24627, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc Natl Acad Sci USA 103: 7159–7164, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jameson BA, Wolf H. The antigenic index: a novel algorithm for predicting antigenic determinants. Comput Appl Biosci 4: 181–186, 1988 [DOI] [PubMed] [Google Scholar]
- 15. Janin J, Wodak S. Conformation of amino acid side-chains in proteins. J Mol Biol 125: 357–386, 1978 [DOI] [PubMed] [Google Scholar]
- 16. Kim GH, Ecelbarger C, Knepper MA, Packer RK. Regulation of thick ascending limb ion transporter abundance in response to altered acid/base intake. J Am Soc Nephrol 10: 935–942, 1999 [DOI] [PubMed] [Google Scholar]
- 17. Kim GH, Martin SW, Fernandez-Llama P, Masilamani S, Packer RK, Knepper MA. Long-term regulation of renal Na-dependent cotransporters and ENaC: response to altered acid-base intake. Am J Physiol Renal Physiol 279: F459–F467, 2000 [DOI] [PubMed] [Google Scholar]
- 18. Kim GH, Masilamani S, Turner R, Mitchell C, Wade JB, Knepper MA. The thiazide-sensitive Na-Cl cotransporter is an aldosterone-induced protein. Proc Natl Acad Sci USA 95: 14552–14557, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kishore BK, Ginns SM, Krane CM, Nielsen S, Knepper MA. Cellular localization of P2Y2 purinoceptor in rat renal inner medulla and lung. Am J Physiol Renal Physiol 278: F43–F51, 2000 [DOI] [PubMed] [Google Scholar]
- 20. Kishore BK, Wade JB, Schorr K, Inoue T, Mandon B, Knepper MA. Expression of synaptotagmin VIII in rat kidney. Am J Physiol Renal Physiol 275: F131–F142, 1998 [DOI] [PubMed] [Google Scholar]
- 21. Knepper MA, Masilamani S. Targeted proteomics in the kidney using ensembles of antibodies. Acta Physiol Scand 173: 11–21, 2001 [DOI] [PubMed] [Google Scholar]
- 22. Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132, 1982 [DOI] [PubMed] [Google Scholar]
- 23. Lindskog M, Rockberg J, Uhlen M, Sterky F. Selection of protein epitopes for antibody production. Biotechniques 38: 723–727, 2005 [DOI] [PubMed] [Google Scholar]
- 24. Mandon B, Chou CL, Nielsen S, Knepper MA. Syntaxin-4 is localized to the apical plasma membrane of rat renal collecting duct cells: possible role in aquaporin-2 trafficking. J Clin Invest 98: 906–913, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mandon B, Nielsen S, Kishore BK, Knepper MA. Expression of syntaxins in rat kidney. Am J Physiol Renal Physiol 273: F718–F730, 1997 [DOI] [PubMed] [Google Scholar]
- 26. Masilamani S, Kim GH, Mitchell C, Wade JB, Knepper MA. Aldosterone-mediated regulation of ENaC α, β, and γ subunit proteins in rat kidney. J Clin Invest 104: R19–R23, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nielsen S, Chou CL, Marples D, Christensen EI, Kishore BK, Knepper MA. Vasopressin increases water permeability of kidney collecting duct by inducing translocation of aquaporin-CD water channels to plasma membrane. Proc Natl Acad Sci USA 92: 1013–1017, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nielsen S, Maunsbach AB, Ecelbarger CA, Knepper MA. Ultrastructural localization of Na-K-2Cl cotransporter in thick ascending limb and macula densa of rat kidney. Am J Physiol Renal Physiol 275: F885–F893, 1998 [DOI] [PubMed] [Google Scholar]
- 29. Nielsen S, Terris J, Smith CP, Hediger MA, Ecelbarger CA, Knepper MA. Cellular and subcellular localization of the vasopressin-regulated urea transporter in rat kidney. Proc Natl Acad Sci USA 93: 5495–5500, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pisitkun T, Bieniek J, Tchapyjnikov D, Wang G, Wu WW, Shen RF, Knepper MA. High-throughput identification of IMCD proteins using LC-MS/MS. Physiol Genomics 25: 263–276, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pisitkun T, Jacob V, Schleicher SM, Chou CL, Yu MJ, Knepper MA. Akt and ERK1/2 pathways are components of the vasopressin signaling network in rat native IMCD. Am J Physiol Renal Physiol 295: F1030–F1043, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA 101: 13368–13373, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sabolic I, Katsura T, Verbavatz JM, Brown D. The AQP2 water channel: effect of vasopressin treatment, microtubule disruption, and distribution in neonatal rats. J Membr Biol 143: 165–175, 1995 [DOI] [PubMed] [Google Scholar]
- 34. Terris J, Ecelbarger CA, Marples D, Knepper MA, Nielsen S. Distribution of aquaporin-4 water channel expression within rat kidney. Am J Physiol Renal Fluid Electrolyte Physiol 269: F775–F785, 1995 [DOI] [PubMed] [Google Scholar]
- 35. Terris J, Ecelbarger CA, Nielsen S, Knepper MA. Long-term regulation of four renal aquaporins in rats. Am J Physiol Renal Fluid Electrolyte Physiol 271: F414–F422, 1996 [DOI] [PubMed] [Google Scholar]
- 36. Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol 266: 554–571, 1996 [DOI] [PubMed] [Google Scholar]
- 37. Xie L, Hoffert JD, Chou CL, Yu MJ, Pisitkun T, Knepper MA, Fenton RA. Quantitative analysis of aquaporin-2 phosphorylation. Am J Physiol Renal Physiol 298: F1018–F1023, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]