Abstract
Although extensive phosphoproteomic information is available for renal epithelial cells, previous emphasis has been on phosphorylation of serines and threonines with little focus on tyrosine phosphorylation. Here we have carried out large-scale identification of phosphotyrosine sites in pervanadate-treated native inner medullary collecting ducts of rat, with a view towards identification of physiological processes in epithelial cells that are potentially regulated by tyrosine phosphorylation. The method combined antibody-based affinity purification of tyrosine phosphorylated peptides coupled with immobilized metal ion chromatography to enrich tyrosine phosphopeptides, which were identified by LC-MS/MS. A total of 418 unique tyrosine phosphorylation sites in 273 proteins were identified. A large fraction of these sites have not been previously reported on standard phosphoproteomic databases. All results are accessible via an online database: http://helixweb.nih.gov/ESBL/Database/iPY/. Analysis of surrounding sequences revealed four overrepresented motifs: [D/E]xxY*, Y*xxP, DY*, and Y*E, where the asterisk symbol indicates the site of phosphorylation. These motifs plus contextual information, integrated using the NetworKIN tool, suggest that the protein tyrosine kinases involved include members of the insulin- and ephrin-receptor kinase families. Analysis of the gene ontology (GO) terms and KEGG pathways whose protein elements are overrepresented in our data set point to structures involved in epithelial cell-cell and cell-matrix interactions (“adherens junction,” “tight junction,” and “focal adhesion”) and to components of the actin cytoskeleton as major sites of tyrosine phosphorylation in these cells. In general, these findings mesh well with evidence that tyrosine phosphorylation plays a key role in epithelial polarity determination.
Keywords: mass spectrometry, kidney, adherens junction, tight junction, cytoskeleton
global phosphoproteomic profiling is providing important new information regarding the signaling pathways involved in functional regulation in renal epithelia (3, 11, 20). The general strategy is to isolate the cells or tissue of interest, extract proteins, proteolyze the proteins with trypsin, and select phosphopeptides using chromatographic techniques, followed by large-scale peptide identification and sequencing using protein mass spectrometry (LC-MS/MS) (25, 41). In general, three amino acid moieties are phosphorylated in eukaryotic tissues, viz. serine, threonine, and tyrosine. The techniques employed in renal epithelial phosphoproteomic analysis, however, have been tilted toward serine and threonine, and not tyrosine phosphorylation. For example, in the current inner medullary collecting duct (IMCD) phosphoproteome database (3) (http://dir.nhlbi.nih.gov/papers/lkem/mpkccdprot/), <2% of the identified phosphorylation sites are on tyrosines. Nevertheless, tyrosine phosphorylation is physiologically important in epithelia in the action of growth factor receptors and in cell adhesion-associated signaling, which are instrumental in the determination of the state of cell differentiation and polarity. Consequently, it was of interest to adapt phosphoproteomics techniques developed for the study of tyrosine phosphorylation in cancer cells (51) to investigation of signaling in the renal IMCD. Such techniques, based on the use of phosphotyrosine-specific antibodies for peptide immunoprecipitation, have allowed the identification and quantification of hundreds of tyrosine phosphopeptides (33, 69). The goal of the present studies was to utilize such approaches to expand the number of tyrosine phosphorylation sites known to be present in native IMCD cells, thereby enhancing the size and value of our IMCD phosphoproteome database. To do this, we treated IMCD cells with two general protein tyrosine phosphatase (PTP) inhibitors, vanadate and pervanadate, to maximize the number of phosphorylation site identifications. In the “systems biology paradigm,” proteomic databases, such as the one reported here, can provide a basis for hypothesis generation and meta-analysis by researchers in the renal physiology community. The results yielded 503 unique phosphotyrosine-containing peptides, corresponding to 418 tyrosine phosphorylation sites. These sites are shown by computational analyses to be associated with pathways involved in regulation of adherens junctions, tight junctions, and the actin cytoskeleton.1
METHODS
Animals and IMCD preparation.
Animal procedures were approved by the National Heart, Lung, and Blood Institute Animal Care and Use Committee (no. H-0110). Male Sprague-Dawley rats (200–300 g) were injected intraperitoneally with furosemide (5 mg/rat) 20 min before decapitation and removal of kidneys. Furosemide dissipates the hypertonic medullary osmolality toward plasma osmolality level (∼290 mosmol/kgH2O), thereby preventing osmotic shock to the cells upon isolation of IMCDs (54). IMCDs were isolated from renal medullas as described (8) after digestion (3 mg/ml collagenase B, 2,000 U/ml hyaluronidase, 250 mM sucrose, 10 mM triethanolamine, pH 7.6) at 37°C with continuous stirring for 75–90 min. IMCDs were sedimented by low-speed centrifugation (70 g for 20 s), separating them from the lighter non-IMCD cells. IMCD pellets were washed and sedimented twice in sucrose buffer (250 mM sucrose, 10 mM triethanolamine, pH 7.6), followed by buffer exchange into 290 mosmol/kgH2O bicarbonate buffer (9). In previous studies, an IMCD purity of >80% was achieved by this isolation technique (64).
Pervanadate preparation and treatment.
Preparation of pervanadate has been previously described (30, 38). Briefly, a 30 mM stock solution of pervanadate was prepared using 100 mM sodium orthovanadate (New England BioLabs, Ipswich, MA) and 3% (wt/wt) H2O2 (Fisher Scientific, Hampton, NH) mixed in 1× PBS at 2:1 molar ratio of H2O2:orthovanadate. The mixture was incubated in the dark at room temperature for 15 min. Five minutes prior to treatment, pervanadate was diluted in bicarbonate buffer (118 mM NaCl, 25 mM NaHCO3, 5 mM KCl, 4 mM Na2HPO4, 2 mM CaCl2, 1.2 mM MgSO4, 5.5 mM glucose, 5 mM acetate, gassed with 95% air-5% CO2 for 20 min before use). IMCD suspensions were treated immediately with the diluted pervanadate (final pervanadate concentration: 100 μM) to minimize decomposition of the H2O2-vanadate complex. For the comparison of effects of different treatments, the IMCD suspension was treated with 100 μM pervanadate, 1 mM vanadate, 180 μM H2O2, or 100 μM pervanadate with 100 μg/ml catalase for 10 min. After treatment, the IMCD suspensions were solubilized and denatured with lysis buffer [final concentrations: 8 M urea, 50 mM Tris·HCl, 75 mM NaCl, 1× HALT protease/phosphatase inhibitor cocktail (Thermo Scientific, Rockford, IL), 1 mM sodium orthovanadate]. Samples were sonicated on ice for 30 s. Lysates for immunoblot analysis were resuspended in Laemmli buffer while lysates for proteomic analysis were resuspended in 8 M urea, 75 mM NaCl, and 50 mM Tris·HCl. The protein concentration of the lysate was determined with the BCA assay (Pierce, Rockford, IL).
Antibodies.
Antiphosphotyrosine monoclonal mouse PY100 (Cell Signaling Technology, Danvers, MA) and PY66 (Sigma-Aldrich, St. Louis, MO) antibodies were used for immunoblotting and immunoprecipitation. The species-specific secondary antibodies conjugated with fluorophores were obtained from Rockland Immunochemicals (Gilbertsville, PA).
Immunoblot analysis.
Immunoblotting of IMCD proteins followed procedures described by Pisitkun et al. (48). Sixteen micrograms of protein in Laemmli buffer were loaded onto a 4–20% gradient SDS-PAGE gel, and electrophoresis was performed at 200 V. Proteins were then transferred onto a nitrocellulose membrane (0.2 μm pore size) under 80 V for 45 min. After incubating in Odyssey Blocking Buffer (LI-COR, Lincoln, NE) for 1 h, primary antibody was added to the membrane and the membrane was incubated overnight. The membrane was washed three times using 1× PBS with 0.1% Tween-20 followed by the application of secondary antibody for 1 h. The membrane was washed three times with 1× PBS with 0.1% Tween-20 followed by a final rinse with 1× PBS. The protein bands on the membrane were scanned using the LI-COR Odyssey Scanner and further analyzed with Odyssey software v2.1.
In-solution trypsin digestion.
Reduction, alkylation, and trypsinization were performed as previously described (25) with modifications. Samples were reduced with 10 mM DTT for 1 h at 55°C and alkylated with 40 mM iodoacetamide for 1 h in the dark at room temperature. Unreacted iodoacetamide was quenched with 40 mM DTT, and the solution was incubated for at least 15 min. Samples were diluted to <1 M urea with 25 mM ammonium bicarbonate and digested overnight at 37°C with trypsin [1:40 (wt/wt)]. Samples were acidified with 0.5% formic acid and spun at 1,000 g for 10 min at 4°C. Finally, the samples were desalted using Oasis 1cc HLB columns (Waters, Milford, MA).
Peptide immunoprecipitation and immobilized metal affinity chromatography.
Phosphotyrosine peptide enrichment was performed with antiphosphotyrosine peptide immunoprecipitation as described by White and colleagues (69) with modifications. A protein G agarose bead slurry (20 μl, IP04, Calbiochem, EMD Chemicals, Darmstadt, Germany) was mixed with 200 μl IP buffer (100 mM Tris, 0.3% Nonidet P-40, pH 7.4) and 12 μg of each antiphosphotyrosine antibody (PY100 and PY66) and incubated for 8 h at 4°C. The beads were washed with IP buffer. Samples, resuspended in 400 μl IP buffer, were added to the beads and incubated overnight at 4°C. The beads were washed three times with rinse buffer (100 mM Tris, pH 7.4), and the phosphotyrosine peptides were eluted from the antibody with 200 μl of elution buffer (100 mM glycine, pH 2.5) for 30 min at room temperature.
Phosphopeptide enrichment using Ga3+ immobilized metal affinity chromatography (IMAC) (Phosphopeptide Isolation Kit, Pierce) was performed as previously described (25) either before or after the immunoprecipitation procedure. For IMAC followed by immunoprecipitation, an extra step of desalting with a Graphite Spin Column (Pierce, Thermo Scientific) to remove excess ammonium bicarbonate between the two enrichment procedures was performed. Enriched peptides were desalted with PepClean C-18 Spin Columns (Pierce, Thermo Scientific) and resuspended in 0.1% formic acid.
LC-MS/MS analysis.
All samples were analyzed on an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific) interfaced with a NanoLC-1D Plus system (Eksigent Technologies). Fragmentation of peptide ions was achieved via collision-induced dissociation. Samples were loaded onto an Agilent Zorbax 300SB-C18 trap column [0.3 mm inner diameter (ID) × 5 mm length, 5-μm particle size] at a flow rate of 10 μl/min for 15 min. Reversed-phase C18 chromatographic separation of trapped peptides was carried out on a prepacked BetaBasic C18 PicoFrit column (75 μm ID × 10 cm length; New Objective) at 300 nl/min using the following gradient: 2–5% solvent B (balance, solvent A) for 2 min; 5–45% solvent B for 45 min; 45–50% solvent B for 5 min; 50–95% solvent B for 5 min (solvent A: 0.1% formic acid in 98% water, 2% acetonitrile; solvent B: 0.1% formic acid in 100% acetonitrile). A data-dependent mode was employed such that a single survey MS1 scan for precursor ions was followed by six data-dependent MS2 scans. Survey MS scans were acquired in the Orbitrap component with a resolution of 30,000, and MS2 scanning was performed in the linear ion trap.
Data searching and scoring.
Searches were performed using the latest version of the rat RefSeq database [National Center for Biotechnology Information (NCBI)] with concatenated forward and reversed sequences to allow for target-decoy analysis. The database also contained sequences for common MS contaminants, such as human keratin and porcine trypsin. Spectra were searched with SEQUEST (18), InsPecT (63), and OMSSA (22). MS searches were aided by the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (http://biowulf.nih.gov). Spectra were filtered to obtain a false discovery rate (FDR) of 1% as described previously (26). Phosphorylation site localization was performed using PhosphoScore (52) for the SEQUEST data and Phosphate Localization Score for the InsPecT data (2). Venn diagrams for visualizing the results of the three search algorithms were generated using Venn Diagram Plotter v1.4.3740. The online database PhosphoSitePlus (28) (http://www.phosphosite.org/) was used to determine whether phosphorylation sites identified in this study had been previously archived.
Phosphorylation site functional analyses.
The conservation of the phosphorylated amino acids among multiple mammalian species was determined using the NCBI HomoloGene database v64 for obtaining the orthologous proteins and ClustalW2 v2.0.12 for aligning the sequences. Phosphorylation motifs were extracted using Motif-X v1.2 (56) (http://motif-x.med.harvard.edu/motif-x.html). Only peptides containing phosphorylated tyrosine were analyzed, and the sequences were prealigned with in-house software before submission to Motif-X. Sequence logos were generated with Weblogo (10) (http://weblogo.berkeley.edu/). Kinase predictions were performed using NetworKIN v2.0 (31, 32) (http://networkin.info/) with phosphoprotein sequence and corresponding tyrosine phosphorylation sites as inputs. The human database was used because this was the database available with the greatest homology to Rattus norvegicus. Prediction is applicable to non-human species because the NetworKIN algorithm considers both consensus motifs and context data that are expected to be conserved through evolution (62). A cutoff NetworKIN score of 0.7 was used. The predicted kinases were cross-referenced with the IMCD transcriptome (http://dir.nhlbi.nih.gov/papers/lkem/imcdtr/).
Gene ontology and network analyses.
Gene ontology (GO) enrichment was assessed and visualized with the Biological Network Gene Ontology (BiNGO) plugin v2.44 (34) for Cytoscape v2.8.1 (58). The level of enrichment was assessed by hypergeometric distribution tests. The P value was corrected for multiple hypothesis testing with the Benjamini-Hochberg FDR algorithm (4), implemented in BiNGO. Enriched “Biological Process,” “Molecular Function,” and “Cellular Component” terms with a significance level of < 0.01 after correction were used in the visualization. Functional clustering was performed with Functional Annotation Clustering in DAVID v6.7 (12, 29) (http://david.abcc.ncifcrf.gov/) using the following databases: GO FAT, KEGG pathway, SMART, InterPro, Swiss-Pro/Protein Information Resource Keywords, and UniProt Sequence Features. Enrichment score was reported as the minus log transformation of the geometric mean of P values (modified Fisher's exact test). The list of proteins for enriched GO terms and list of enriched KEGG pathways were also generated with DAVID. The level of enrichment was assessed by modified Fischer's exact test (EASE). The P value was corrected for multiple hypothesis testing with the Benjamini-Hochberg FDR algorithm. Terms with a corrected P < 0.01 were considered significant. For the control, the proteins corresponding to the nonphosphorylated peptides identified in our mass spectrometry analysis were used. For both BiNGO and DAVID analyses, the reference set used was the IMCD transcriptome (http://dir.nhlbi.nih.gov/papers/lkem/imcdtr/). The phosphotyrosine data set was also imported into Ingenuity Pathway Analysis (IPA) v9.0 (Ingenuity Systems, Redwood City, CA) to identify top canonical pathways containing enriched components.
RESULTS
Tyrosine phosphatase inhibition.
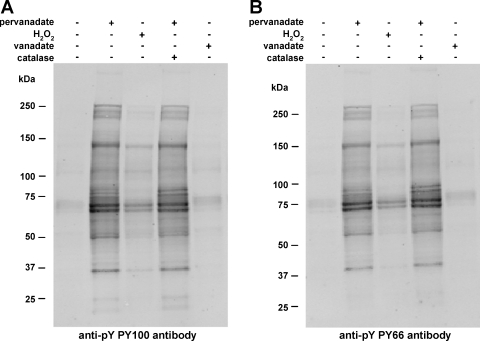
The objective of this paper was to identify protein substrates of tyrosine kinases in IMCD cells and to localize the sites of phosphorylation. To maximize the identifications, we used tyrosine phosphatase inhibitors. Vanadate and pervanadate are two widely used general PTP inhibitors. In previous studies, pervanadate, generated as a complex of vanadate and H2O2, was found to be an irreversible inhibitor of PTP and to be a much more potent inhibitor than vanadate in raising the cellular tyrosine phosphorylation level (30, 49, 57). Our findings in IMCD cells agree with these observations (Fig. 1). Protein tyrosine phosphorylation was not augmented with the no-treatment control and treatment with H2O2 or vanadate alone but was markedly increased upon pervanadate treatment in the absence or presence of catalase. Thus, the IMCD suspensions were treated with pervanadate for the rest of our study.
Fig. 1.
Immunoblot analysis illustrating effective tyrosine phosphorylation enrichment with pervanadate treatment on rat inner medullary collecting duct (IMCD) samples. IMCD suspension was incubated with 100 μM pervanadate, 1 mM vanadate, 180 μM H2O2, or 100 μM pervanadate + 100 μg/ml catalase for 10 min. A no-treatment control was also used. Equal amounts of lysate (16 μg) were loaded into each well for immunoblot analysis using two antiphosphotyrosine antibodies, PY100 (A) and PY66 (B).
Phosphotyrosine enrichment.
Phosphotyrosine-containing peptides were enriched using antiphosphotyrosine immunoprecipitation. This technique has been successfully used for studying tyrosine phosphorylation dynamics in epidermal growth factor receptor signaling networks (69). The combination of immunoprecipitation and IMAC on peptides from pervanadate-treated IMCD proteins was found to be effective in enriching phosphotyrosine with high specificity. In addition, the ordering of immunoprecipitation and IMAC was assessed. Although IMAC followed by immunoprecipitation yielded higher numbers of peptides, the results from both approaches were combined to maximize the yield. Altogether, these results demonstrate that the enrichment techniques described succeeded at enriching tyrosine phosphorylation in IMCD.
Phosphoproteomic profiling of inner medullary collecting duct.
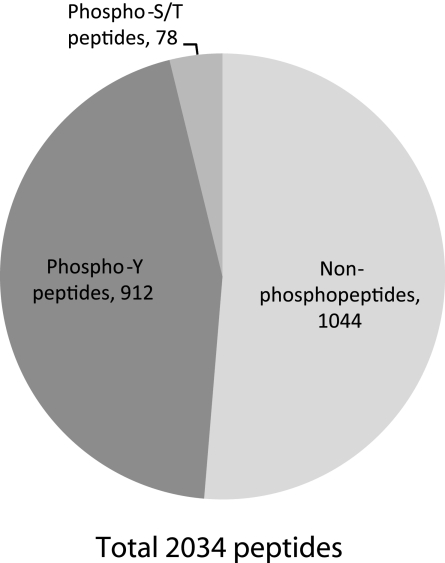
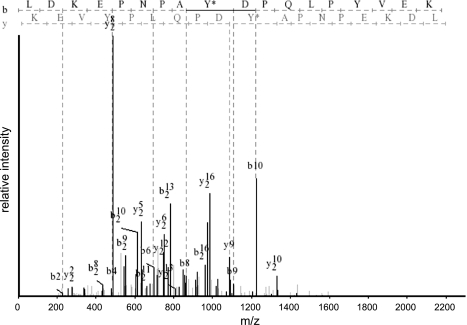
Inner medullary collecting ducts were subjected to phosphoproteomic analysis as described in methods. Peptide samples were analyzed on an LTQ Orbitrap XL mass spectrometer, and the resulting spectra were searched using three different search algorithms—SEQUEST, InsPecT, and OMSSA—to maximize the number of phosphopeptide identifications from high-quality spectra (3, 20, 23, 50). The data set was filtered using a target-decoy approach (16, 17) to limit false-positive identifications to <1%. A total of 2,034 peptides were identified, of which 1,044 were nonphosphopeptides and 990 were phosphopeptides (Fig. 2). A total of 912 phosphotyrosine-containing peptides, corresponding to 503 unique peptides and 273 proteins, were identified by the combination of the three search algorithms. These peptides contain 453 unique phosphorylation sites, of which 418 were unique tyrosine phosphorylation sites (see Supplemental Table S1; Supplemental Material for this article is available online at the Journal website). Approximately 19% of the tyrosine phosphorylation sites identified were not found in the PhosphoSitePlus database and are thus considered novel. Annotated phosphopeptide data from all searches are accessible online via http://helixweb.nih.gov/ESBL/Database/iPY/. A representative mass spectrum (showing Y1027 phosphorylation in the tight junction protein ZO-1) is shown in Fig. 3.
Fig. 2.
Number of nonphosphopeptides, phosphoserine/threonine peptides, and phosphotyrosine peptides identified in rat IMCD samples.
Fig. 3.
Typical mass spectrum showing identification of phosphorylation of Y1027 in the tight junction protein ZO-1. m/z, mass-to-charge ratio.
Computational analyses of phosphotyrosine peptides.
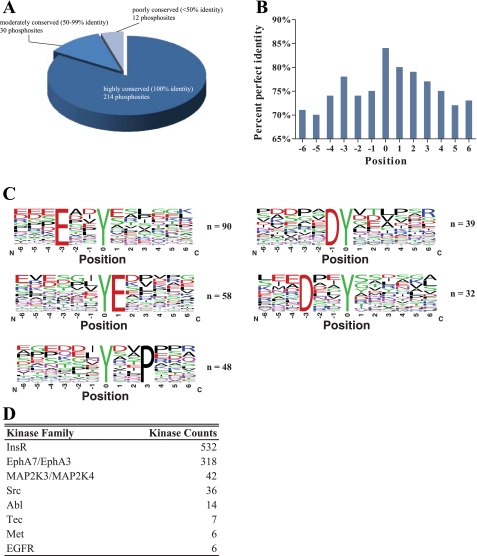
We analyzed the level of conservation for each tyrosine phosphorylation site among mammalian species other than rat (Supplemental Table S2). A total of 256 phosphorylation sites for which the corresponding proteins have orthologs (based on HomoloGene database) were included and analyzed. Approximately 84% of sites analyzed were 100% conserved among species examined, while 12% (50–99% of species) were moderately conserved and 5% (<50% of species) were poorly conserved (Fig. 4A). The degree of conservation for the tyrosine phosphorylation site was also higher than those in the neighboring amino acids, for which the percentage of sites that are 100% conserved ranged from 70% to 80% (Fig. 4B). Overrepresented motifs were also identified in sequences flanking the phosphorylation sites using Motif-X (Fig. 4C). Four motifs were identified: [D/E]xxY*, Y*xxP, DY*, and Y*E, where the asterisk symbol indicates the site of phosphorylation. This suggests that multiple tyrosine kinases are responsible for the phosphorylation at the identified sites.
Fig. 4.
Conservation of tyrosine phosphorylation sites detected by MS in rat IMCD samples. A: number of sites that are highly conserved, moderately conserved, or poorly conserved among mammalian species. B: percent perfect identity (100% conservation) of neighboring amino acids of phosphorylated tyrosine residue (position 0). C: sequence logos showing overrepresented phosphorylation motifs extracted using Motif-X. D: a list of potential protein kinase families that phosphorylate these tyrosine sites as analyzed by NetworKIN.
We next used NetworKIN (32) to identify potential protein kinases that phosphorylate these tyrosine sites. The algorithm combines kinase consensus motifs, extracted from the NetPhorest atlas (39), with contextual information of the kinase and substrate in protein association networks, extracted from the STRING database (61). After cross-referencing the results with the IMCD transcriptome, this analysis reveals a large number of kinases belonging to the insulin and ephrin receptor kinase families (Fig. 4D). Other kinase groups included MAP2K, Src, Tec, Abl, EGFR, and Met. A list of all predicted kinases with the corresponding phosphoprotein substrates and phosphorylation sites is provided in Supplemental Table S3.
Functional classification enrichment.
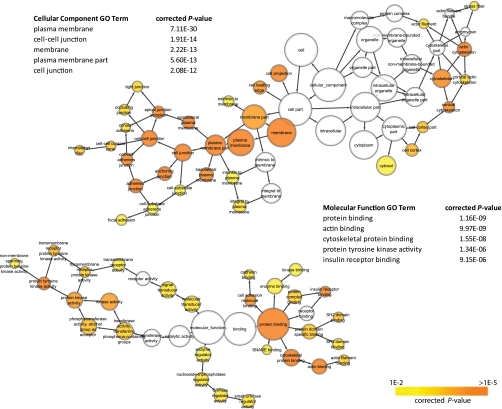
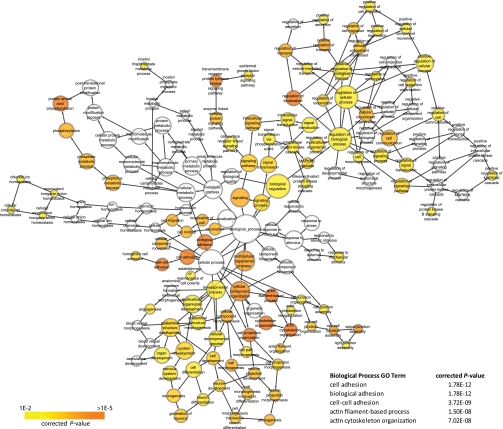
To assess the functional roles of tyrosine phosphorylated IMCD proteins, enrichment of GO terms relative to the IMCD transcriptome (64) (http://dir.nhlbi.nih.gov/papers/lkem/imcdtr/) were analyzed. Since GO terms are categorized into different hierarchies with interdependent relationships, visualization of GO as a network helps in understanding the relationship of the lower level terms with the broader terms (Figs. 5 and 6). The network also enables the identification of functional modules/clusters. The nodes of a network represent GO terms while the directed edges describe the relationship between terms. Distinct clusters of Biological Process, Molecular Function, and Cellular Component GO terms were immediately apparent. For Cellular Component terms, the clusters relating to “cell junction” and “cytoskeleton” were enriched. Significantly enriched Molecular Function GO terms contained two clusters of terms involved with “protein tyrosine kinase activity” and “protein binding” (Fig. 5). For Biological Process, the highly enriched GO terms fall under groups that describe “cytoskeleton organization,” “cell adhesion,” “protein amino acid phosphorylation,” “transmembrane receptor protein tyrosine signaling pathway,” and “regulation of localization” (Fig. 6).
Fig. 5.
Gene Ontology (GO) Cellular Component and Molecular Function terms enrichment of IMCD phosphotyrosine proteins relative to IMCD transcriptome using BINGO. Enrichment was assessed with hypergeometric distribution tests, corrected for multiple hypothesis testing with Benjamini-Hochberg false discovery rate (FDR) algorithm. Only GO terms with P < 0.01 are displayed. Nodes represent GO terms while directed edges represent the relationship between terms. Node size is proportional to the number of IMCD proteins categorized to the GO term. Node color corresponds to P value, with the highly enriched terms shown in orange.
Fig. 6.
GO Biological Process terms enrichment of IMCD phosphotyrosine proteins relative to IMCD transcriptome using BINGO. Enrichment was assessed with hypergeometric distribution tests, corrected for multiple hypothesis testing with Benjamini-Hochberg FDR. Only GO terms with P < 0.01 are displayed. Nodes represent GO terms while directed edges represent the relationship between terms. Node size is proportional to the number of IMCD proteins categorized to the GO term. Node color corresponds to P value, with the highly enriched terms shown in orange.
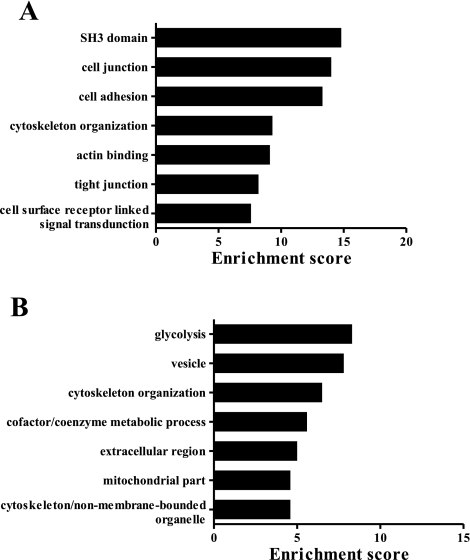
We complemented our GO singular enrichment analysis above with a modular enrichment analysis approach using DAVID functional annotation clustering. The GO singular enrichment approach extracts biological meaning by determining whether each individual GO term is overrepresented in the data set. This approach may miss functional modules that are only apparent when classes of genes or terms are considered. The DAVID functional annotation clustering utilizes a set of fuzzy clustering techniques to classify the input data into functionally related groups with integration across multiple databases (see methods for details). The top seven functional clusters were “SH3 domain,” “cell junction,” “cell adhesion,” “cytoskeleton organization,” “actin binding,” “tight junction,” and “cell surface receptor linked signal transduction” (Fig. 7A). To demonstrate that the enrichment is specific to the phosphotyrosine data set, we performed the same analysis on proteins containing nonphosphorylated peptides identified in our IMCD sample. The highly enriched terms in this control data set (“glycolysis,” “vesicle,” “cytoskeleton organization,” “cofactor/coenzyme metabolic process,” etc.) (Fig. 7B) were largely different than those for the phosphotyrosine data set. The term “cytoskeleton organization” is common to both data sets because of the existence of shared proteins (23% of the proteins in the nonphosphorylated control for this term were also found in the phosphotyrosine data set). All functional clusters with enrichment score > 1.3 (corresponding to minus log of P = 0.05) were included in Supplemental Table S4.
Fig. 7.
Top seven functional annotation clusters of rat IMCD phosphotyrosine proteins (A) and nonphosphorylated proteins (B) relative to IMCD transcriptome using DAVID analysis. See methods for details on the approach and the list of databases used. Enrichment score is reported as the minus log transformation of the geometric mean of P values (modified Fisher's exact test).
On the basis of these two analyses, detailed descriptions of proteins in selected GO categories were generated using DAVID. Tables 1–4 include proteins related to “cell junction,” “cytoskeleton organization,” “cell surface receptor linked signal transduction,” and “protein tyrosine kinase activity,” respectively. Table 5 contains proteins with either the SH2 or SH3 domain, or both.
Table 1.
Gene Ontology Cellular Component: “cell junction”
| Protein Name | Gene Symbol | RefSeq Number | Phosphopeptide Sequence | Phosphorylation Sites |
|---|---|---|---|---|
| Abl interactor 1 | Abi1 | NP_077373 | NT*PYKTLEPVKPPTVPNDY*M@TSPAR | Y213 |
| NTPY*KTLEPVKPPTVPNDY*M@TSPAR | Y198, Y213 | |||
| NTPY*KTLEPVKPPTVPNDY*MTSPAR | Y198, Y213 | |||
| NTPY*KTLEPVKPPTVPNDYM@T*SPAR | Y198 | |||
| TLEPVKPPTVPNDY*MTS*PAR | Y213 | |||
| TLEPVKPPTVPNDY*MTSPAR | Y213 | |||
| Breast cancer anti-estrogen resistance protein 1 | Bcar1 | NP_037063 | DPLLDVY*DVPPSVEK | Y385 |
| GLLPNQYGQEVY*DTPPMAVK | Y365 | |||
| GPNGRDPLLDVY*DVPPSVEK | Y385 | |||
| HLLAPGSQDIY*DVPPVR | Y347 | |||
| RPGPGTLY*DVPR | Y485 | |||
| TQQGLY*QAPGPNPQFQSPPAK | Y226 | |||
| VGQGYVYEASQAEQDEY*DTPR | Y332 | |||
| VLPPEVADGSVIDDGVY*AVPPPAER | Y508 | |||
| Cell adhesion molecule 1 | Cadm1 | NP_001012201 | GADDAADADTAIINAEGGQNNSEEKKEY*FI | Y474 |
| Serine/threonine-protein kinase MRCK-β | Cdc42bpb | NP_446072 | LPDFQDPIFEY*FNTAPLAHDLTFR | Y954 |
| Cingulin | Cgn | XP_001059265 | GSPGALSSDSELPENPY*SQVK | Y238 |
| Claudin-3 | Cldn3 | NP_113888 | ILY*SAPR | Y197 |
| Tyrosine-protein kinase CSK | Csk | NP_001025210 | VM@EGTVAAQDEFY*R | Y184 |
| VMEGTVAAQDEFY*R | Y184 | |||
| Catenin α-1 | Ctnna1 | NP_001007146 | NAGNEQDLGIQY*R | Y177 |
| Catenin (cadherin-associated protein), δ2 (neural plakophilin-related arm-repeat protein) | Ctnnd2 | XP_001065606 | ALQSPEHHIDPIY*EDR | Y262† |
| ASYAAGPASNY*ADPYR | Y337 | |||
| Junctional adhesion molecule A precursor | F11r | NP_446248 | KVIY*SQPSAR | Y281 |
| VIY*SQPS*AR | Y281 | |||
| VIY*SQPSAR | Y281 | |||
| ARF GTPase-activating protein GIT1 | Git1 | NP_114002 | HSTELEDDAIY*SVHVPAGLYR | Y554 |
| LQPFHSTELEDDAIY*SVHVPAGLYR | Y554 | |||
| Integrin β-1 | Itgb1 | NP_058718 | WDTGENPIY*K | Y783 |
| Junction plakoglobin | Jup | NP_112309 | ISEDKNPDY*R | Y660 |
| TTTYTQGVPQSQGDLEY*QMSTTAR | Y74 | |||
| LIM and SH3 domain protein 1 | Lasp1 | NP_116002 | HIPTSAPVY*QQPQQQQVTPSYGGYK | Y173 |
| HIPTSAPVY*QQPQQQQVTPSYGGYKEPAAPVSIQR | Y173 | |||
| LIM domain only protein 7 | Lmo7 | NP_001001515 | AGDEDRGQLEDDAVY*EDQSQK | Y1358† |
| GQLEDDAVY*EDQSQK | Y1358† | |||
| QSSWHSQPAGVY*ASSSVQDFSR | Y1588† | |||
| WIDTTSGIY*STEK | Y1169 | |||
| Tyrosine-protein kinase Lyn isoform A | Lyn | NP_110484 | VIEDNEY*TAR | Y397 |
| Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 3 | Magi3 | NP_620784 | IEDPQY*GTYYVDHLNQK | Y356† |
| Afadin | MIIt4 | NP_037349 | DLQY*ITISK | Y1502 |
| EY*FTFPASK | Y1237 | |||
| LAAEVY*KDMPETSFTR | Y203 | |||
| SPGTWTIYFEGADY*ESHLMR | Y993 | |||
| SQEELREEKVY*QLER | Y1292 | |||
| Myosin-9 | Myh9 | NP_037326 | YLY*VDK | Y11 |
| Occludin | Ocln | NP_112619 | SYPDSLY*K | Y342 |
| Oligophrenin-1 | Ophn1 | NP_001101318 | LWMEAMDGKEPIY*HSPITK | Y370 |
| Partitioning defective 3 homolog | Pard3 | NP_112514 | ISHSLY*SGIEGLDESPTR | Y719 |
| Partitioning defective 6 homolog β | Pard6b | NP_001102079 | EEADY*SAFGTDTLIR | Y101 |
| KEEADY*SAFGTDTLIR | Y101 | |||
| Plakophilin-3 | Pkp3 | NP_001099785 | ADY*DTLSLR | Y201 |
| Focal adhesion kinase 1 | Ptk2 | NP_037213 | THAVSVSETDDY*AEIIDEEDTYTMPSTR | Y397 |
| YM@EDSTY*YK | Y576 | |||
| YMEDSTY*Y*KASK | Y576, Y577 | |||
| YMEDSTY*YK | Y576 | |||
| Poliovirus receptor-related protein 2 | Pvrl2 | NP_001012064 | Y*HELPTLEER | Y445 |
| Paxillin | Pxn | NP_001012147 | AGEEEHVY*SFPNK | Y118 |
| YAHQQPPS*PSPIY*SSSTK | Y88 | |||
| Protein Shroom2 | Shroom2 | NP_001041358 | RDS*AY*GSFSTCSSTPDHTLPK | Y215† |
| Sorbin and SH3 domain containing 1 | Sorbs1 | XP_002725845 | TPVDY*IDLPYSSSPSR | Y948† |
| Spectrin α chain, brain | Spna2 | NP_741984 | VAEDLESEGLM@AEEVQAVQQQEVY*GM@M@PR | Y1176 |
| VAEDLESEGLMAEEVQAVQQQEVY*GMM@PR | Y1176 | |||
| VAEDLESEGLMAEEVQAVQQQEVY*GMMPR | Y1176 | |||
| Tight junction protein ZO-1 | Tjp1 | NP_001099736 | AEQLASVQY*TLPK | Y576 |
| FEEPAPLPY*DSRPR | Y1127 | |||
| HEEQPTSGY*EVHNR | Y1152 | |||
| LDKEPNPAY*DPQLPYVEK | Y1027† | |||
| LDKEPNPAYDPQLPY*VEK | Y1033† | |||
| SNHY*DPEEDEEYYRK | Y1333 | |||
| SNHYDPEEDEEY*YRK | Y1341 | |||
| SSEPKQY*FDQYPR | Y1178 | |||
| TLY*RLPEPQKPQAKPPEDIVR | Y1311 | |||
| YESSSY*TDQFSR | Y1054 | |||
| YRPEAQPY*APAGPK | Y1165 | |||
| Tight junction protein ZO-2 | Tjp2 | NP_446225 | HPDIY*AVPIK | Y1093 |
| IEIAQKHPDIY*AVPIK | Y1093 | |||
| M@SYLTAM@GADY*LSCDSR | Y891 | |||
| MSYLTAMGADY*LSCDSR | Y891 | |||
| SYHQAYEPDY*EGR | Y234 | |||
| SYHQAYEPDYEGRY*S*PSYDRR | Y238 | |||
| VFLRPSPEDEAIY*GPNTK | Y486 | |||
| Vinculin | Vcl | NP_001100718 | SFLDSGY*R | Y822 |
Phosphorylated amino acid.
Novel phosphorylation site (not present on Phosphosite Plus database, for all available species).
Table 4.
Gene Ontology Molecular Function: “protein tyrosine kinase activity”
| Protein Name | Gene Symbol | RefSeq Number | Phosphopeptide Sequence | Phosphorylation Sites |
|---|---|---|---|---|
| Tyrosine-protein kinase | CSK | NP_001025210 | VM@EGTVAAQDEFY*R | Y184 |
| VMEGTVAAQDEFY*R | Y184 | |||
| Epithelial discoidin domain-containing receptor 1 | Ddr1 | NP_037269 | GTPTHSAPCVPNGSALLLSNPAY*R | Y510 |
| Dual specificity tyrosine-phosphorylation-regulated kinase 1A | Dyrk1a | NP_036923 | IYQY*IQSR | Y321 |
| Epidermal growth factor receptor | Egfr | NP_113695 | GPTAENAEY*LR | Y1196 |
| GSHQM@SLDNPDY*QQDFFPK | Y1171 | |||
| GSHQMSLDNPDY*QQDFFPK | Y1171 | |||
| LLGAEEKEY*HAEGGK | Y870 | |||
| RPAGSVQNPVY*HNQPLHPAPGR | Y1109 | |||
| Receptor tyrosine-protein kinase erbB-2 | Erbb2 | NP_058699 | LLDIDETEY*HADGGKVPIK | Y881 |
| Tyrosine-protein kinase FRK | Frk | NP_077344 | SVVIENPGAY*CPPEANR | Y35† |
| VDNEDIY*ESKHEIK | Y388 | |||
| Insulin-like growth factor 1 receptor precursor | Igf1r | NP_434694 | DIYETDY*YR | Y1166 |
| Tyrosine protein kinase Lyn isoform A | Lyn | NP_110484 | VIEDNEY*TAR | Y397 |
| Hepatocyte growth factor receptor precursor | Met | NP_113705 | DMYDKEY*YSVHNK | Y1235 |
| Neuropilin-1 precursor | Nrp1 | NP_659566 | DKLNPQSNY*SEA | Y919 |
| Focal adhesion kinase 1 | Ptk2 | NP_037213 | THAVSVSETDDY*AEIIDEEDTYTMPSTR | Y397 |
| YM@EDSTY*YK | Y576 | |||
| YMEDSTY*Y*KASK | Y576, Y577 | |||
| YMEDSTY*YK | Y576 | |||
| Proto-oncogene tyrosine-protein kinase Src | Src | NP_114183 | YFTSTEPQY*QPGENL | Y536 |
| Angiopoietin-1 receptor | Tek | NP_001099207 | KTY*VNTTLY*EK | Y1098, Y1104 |
Phosphorylated amino acid.
Novel phosphorylation site (not present on Phosphosite Plus database, for all available species).
Table 5.
Tyrosine phosphorylated proteins that contain SH2 or SH3 domains
| Protein Name | Gene Symbol | RefSeq Number | Phosphopeptide Sequence | Phosphorylation Sites |
|---|---|---|---|---|
| Src Homology 2 domain | ||||
| B-cell linker protein | Blnk | NP_001020938 | DLLEDEADY*VVPVEDNDENYIHPR | Y178 |
| DLLEDEADYVVPVEDNDENY*IHPR | Y189 | |||
| Tyrosine-protein kinase CSK | Csk | NP_001025210 | VM@EGTVAAQDEFY*R | Y184 |
| VMEGTVAAQDEFY*R | Y184 | |||
| Tyrosine-protein kinase FRK | Frk | NP_077344 | SVVIENPGAY*CPPEANR | Y35† |
| VDNEDIY*ESKHEIK | Y388 | |||
| Phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 2 | Inppl1 | NP_075233 | ERLY*EWISIDKDDTGAK | Y887 |
| LY*EWISIDKDDTGAK | Y887 | |||
| NSFNNPAY*YVLEGVPHQLLPLEPTSFAR | Y987 | |||
| TLSEVDY*SPGPGR | Y1136 | |||
| Tyrosine-protein kinase Lyn isoform A | Lyn | NP_110484 | VIEDNEY*TAR | Y397 |
| Cytoplasmic protein NCK1 | Nck1 | NP_001100321 | LY*DLNM@PAFVK | Y105 |
| LY*DLNMPAFVK | Y105 | |||
| 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase γ-1 | Plcg1 | NP_037319 | IGTAEPDY*GALYEGR | Y771 |
| Tyrosine-protein phosphatase nonreceptor type 11 isoform 1 | Ptpn11 | NP_001171064 | IQNTGDY*YDLYGGEK | Y62 |
| VVLHDGDPNEPVSDY*INANIIMPEFETK | Y304 | |||
| VY*ENVGLMQQQR | Y584 | |||
| SHC-transforming protein 1 isoform a | Shc1 | NP_001157532 | ELFDDPSY*VNIQNLDK | Y423 |
| Proto-oncogene tyrosine-protein kinase Src | Src | NP_114183 | YFTSTEPQY*QPGENL | Y536 |
| Signal transducer and activator of transcription 3 | Stat3 | NP_036879 | YCRPESQEHPEADPGSAAPY*LK | Y705 |
| Src Homology 3 domain | ||||
| Abl interactor 1 | Abi1 | NP_077373 | NT*PYKTLEPVKPPTVPNDY*M@TSPAR | Y213 |
| NTPY*KTLEPVKPPTVPNDY*M@TSPAR | Y198, Y213 | |||
| NTPY*KTLEPVKPPTVPNDY*MTSPAR | Y198, Y213 | |||
| NTPY*KTLEPVKPPTVPNDYM@T*SPAR | Y198 | |||
| TLEPVKPPTVPNDY*MTS*PAR | Y213 | |||
| TLEPVKPPTVPNDY*MTSPAR | Y213 | |||
| Rho GTPase-activating protein 27 | Arhgap27 | NP_942054 | QVDEPPEPVY*ANVER | Y227 |
| SDSENVY*EAIPDLR | Y201† | |||
| Rho guanine nucleotide exchange factor (GEF) 5 | Arhgef5 | XP_001073085 | TAIY*GSLIPR | Y636† |
| Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1 | Baiap2l1 | NP_001029312 | EIEY*VETVTSR | Y163 |
| M@IGKDY*DTLSK | Y274 | |||
| MIGKDY*DTLSKYS*PK | Y274 | |||
| SSVVIPPPDY*LECLSMGATSDK | Y440† | |||
| Breast cancer anti-estrogen resistance protein 1 | Bcar1 | NP_037063 | DPLLDVY*DVPPSVEK | Y385 |
| GLLPNQYGQEVY*DTPPMAVK | Y365 | |||
| GPNGRDPLLDVY*DVPPSVEK | Y385 | |||
| HLLAPGSQDIY*DVPPVR | Y347 | |||
| RPGPGTLY*DVPR | Y485 | |||
| TQQGLY*QAPGPNPQFQSPPAK | Y226 | |||
| VGQGYVYEASQAEQDEY*DTPR | Y332 | |||
| VLPPEVADGSVIDDGVY*AVPPPAER | Y508 | |||
| CD2-associated protein | Cd2ap | NP_852140 | DAGY*SSKPSLSAPSSASK | Y541 |
| ISTY*GLPAGGIQPHPQTK | Y88 | |||
| Tyrosine-protein kinase CSK | Csk | NP_001025210 | VM@EGTVAAQDEFY*R | Y184 |
| VMEGTVAAQDEFY*R | Y184 | |||
| Cortactin isoform B | Cttn | NP_068640 | KQT*PPASPS*PQPAEDRPPSS*PIY*EDAAPLK | Y384 |
| NASTFEEVVQVPSAY*QK | Y297 | |||
| SAVGFDY*QGK | Y178 | |||
| SAVGFEY*QGKTEK | Y141 | |||
| VDQSAVGFEY*QGK | Y141 | |||
| Disks large homolog 3 | Dlg3 | NP_113827 | RDNEVDGQDY*HFVVSR | Y705 |
| Epidermal growth factor receptor kinase substrate 8-like protein 2 | Eps8l2 | NP_001101978 | AEDSY*SNVHTSPEAEGAPHL | Y669 |
| RAEDSY*SNVHTSPEAEGAPHL | Y669 | |||
| VGLPLPASFSEPGY*R | Y192† | |||
| VY*SQLTVQK | Y636 | |||
| Tyrosine-protein kinase FRK | Frk | NP_077344 | SVVIENPGAY*CPPEANR | Y35† |
| VDNEDIY*ESKHEIK | Y388 | |||
| LIM and SH3 domain protein 1 | Lasp1 | NP_116002 | HIPTSAPVY*QQPQQQQVTPSYGGYK | Y173 |
| HIPTSAPVY*QQPQQQQVTPSYGGYKEPAAPVSIQR | Y173 | |||
| P55 protein | LOC652956 | NP_001032748 | KGEADGAEY*HFVSAEEMAR | Y332† |
| Tyrosine-protein kinase Lyn isoform A | Lyn | NP_110484 | VIEDNEY*TAR | Y397 |
| Membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) | Mpp5 | NP_001101504 | VY*ESIGHY*GGETVK | Y243, Y249 |
| VY*ESIGHYGGETVK | Y243 | |||
| Myosin-Ie | Myo1e | NP_775124 | SLY*TSMARPPLPR | Y988 |
| Cytoplasmic protein NCK1 | Nck1 | NP_001100321 | LY*DLNM@PAFVK | Y105 |
| LY*DLNMPAFVK | Y105 | |||
| 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-1 | Plcg1 | NP_037319 | IGTAEPDY*GALYEGR | Y771 |
| Endophilin-B1 | Sh3 glb1 | NP_001011929 | IEEFVY*EKLDR | Y80 |
| SH3 domain-containing YSC84-like protein 1 | Sh3yl1 | NP_001102175 | NEY*KLYPELSSYHEK | Y264† |
| Sorting nexin 9 | Snx9 | NP_001121109 | SSS*PY*FKDSEPAEAGGIQR | Y177 |
| SSSPY*FKDSEPAEAGGIQR | Y177 | |||
| Spectrin α chain, brain | Spna2 | NP_741984 | VAEDLESEGLM@AEEVQAVQQQEVY*GM@M@PR | Y1176 |
| VAEDLESEGLMAEEVQAVQQQEVY*GMM@PR | Y1176 | |||
| VAEDLESEGLMAEEVQAVQQQEVY*GMMPR | Y1176 | |||
| Proto-oncogene tyrosine-protein kinase Src | Src | NP_114183 | YFTSTEPQY*QPGENL | Y536 |
| Tight junction protein ZO-2 | Tjp2 | NP_446225 | HPDIY*AVPIK | Y1093 |
| IEIAQKHPDIY*AVPIK | Y1093 | |||
| M@SYLTAM@GADY*LSCDSR | Y891 | |||
| MSYLTAMGADY*LSCDSR | Y891 | |||
| SYHQAYEPDY*EGR | Y234 | |||
| SYHQAYEPDYEGRY*S*PSYDRR | Y238 | |||
| VFLRPSPEDEAIY*GPNTK | Y486 | |||
Phosphorylated amino acid.
Novel phosphorylation site (not present on Phosphosite Plus database, for all available species).
Signaling network analyses.
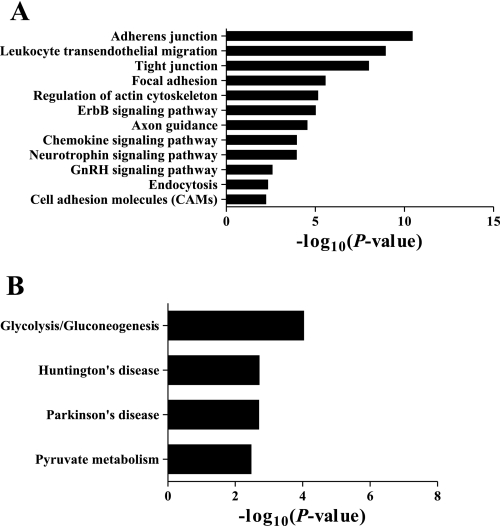
We next wanted to see whether specific pathways are enriched in the set of proteins present in our phosphotyrosine database. DAVID analysis with P value determined using a modified Fisher's exact test with Benjamini-Hochberg FDR correction and a cutoff of P < 0.01 was used. The top KEGG pathways included “adherens junction,” “tight junction,” “focal adhesion,” and “regulation of actin cytoskeleton” (Fig. 8A). As expected, our control data set with only proteins containing the nonphosphorylated peptides did not show any enriched pathways associated with cell junction. Instead, the top pathways were found to be involved in glycolysis and pyruvate metabolism (Fig. 8B).
Fig. 8.
Enriched KEGG pathways based on rat IMCD phosphotyrosine proteins (A) and nonphosphorylated proteins (B) relative to rat IMCD transcriptome using DAVID analysis. P values were determined with modified Fisher's exact test and corrected for multiple hypothesis testing with Benjamini-Hochberg FDR. Pathways with corrected P < 0.01 are presented.
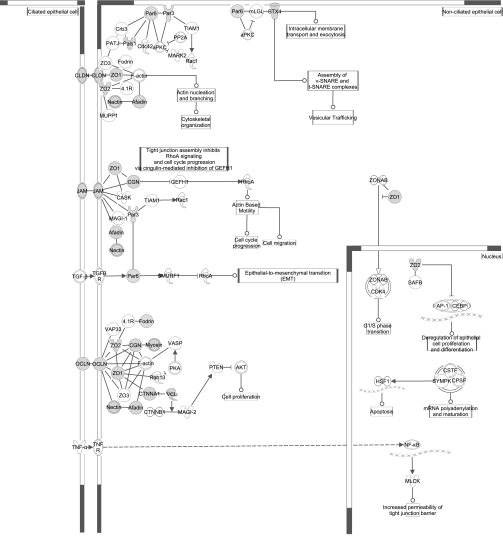
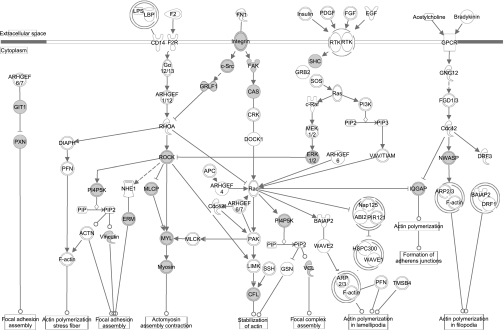
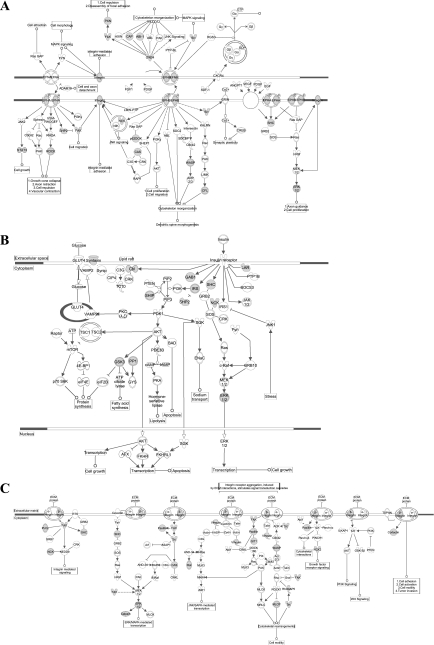
To examine the signaling and protein-protein interactions involved in the enriched canonical pathways, the same phosphotyrosine data set was imported into Ingenuity Pathway Analysis. “Tight junction” and “actin cytoskeleton signaling” pathways were also found to be highly enriched in this analysis. Proteins with tyrosine phosphorylation found in our study are highlighted in these two pathways in Figs. 9 and 10. Additional enriched signaling pathways, viz. “ephrin receptor,” “insulin receptor,” and “integrin signaling,” are included in Fig. 11.
Fig. 9.
Diagram of canonical tight junction signaling pathway. Enriched phosphotyrosine proteins are shown in gray. Analysis and visualization using IPA (see methods).
Fig. 10.
Diagram of canonical actin cytoskeleton signaling pathway. Enriched phosphotyrosine proteins are shown in gray. Analysis and visualization using IPA (see methods).
Fig. 11.
Diagrams of canonical ephrin (A), insulin receptor (B), and integrin (C) signaling pathways. Enriched phosphotyrosine proteins are shown in gray. Analysis and visualization using IPA (see methods).
DISCUSSION
Systems biology approach to the study of cell physiology.
An understanding of the complex regulatory machinery of a cell is increasingly reliant on the use of a systematic and integrative approach. Cell physiology in particular involves a complicated network of interactions of proteins and other molecules. As such, a comprehensive understanding involves first, the identification and quantification of all components of this network; and second, the analysis of the functional roles and interactions of these components to elucidate how the cell functions as a whole.
The enumeration of the list of components has been greatly benefited from the recent advancements in high-throughput techniques such as DNA and oligonucleotide microarrays, deep sequencing, protein mass spectrometry, multiplex affinity-based assays, and cell sorting-based assays. We have previously used mass spectrometry to perform phosphoproteomic profiling in several cell types in the kidney (3, 20, 23, 50). Although tyrosine phosphorylation plays a critical role in cellular signaling and has been the target of dysregulation in a number of diseases, its extremely low abundance [estimated to constitute only 1.8% of the phosphoproteome (43)] makes its detection in phosphoproteomic analyses challenging. In fact, before this study, our IMCD phosphoproteome database contained only 55 unique phosphotyrosine sites (3). Therefore, in this study, we applied phosphotyrosine enrichment techniques in combination with protein mass spectrometry for large-scale profiling of tyrosine phosphorylation sites of proteins from the native epithelium of the renal IMCD. Overall, we identified 418 unique tyrosine phosphorylation sites. Over 19% of these sites were not found in the PhosphoSitePlus database and are therefore labeled “novel.” The entire database can be found in Supplemental Table S1 and is also available online at http://helixweb.nih.gov/ESBL/Database/iPY/.
Tyrosine signaling networks in IMCD epithelia.
An understanding on the functional role of phosphotyrosine-mediated signaling in IMCD epithelial cells can potentially be achieved by integrating our database with functional annotation data sets. The renal IMCD is the terminal portion of the renal tubule and the final site for adjusting urinary composition and volume. Therefore, the IMCD is critical for solute transport and in maintenance of homeostasis. Especially important are anchoring junctions such as adherens and focal adhesions that provide cell-cell and cell-extracellular matrix (ECM) interaction and attachment, as well as tight junctions that (together with anchoring junctions) establish and maintain cell polarity (7). Interestingly, the proteins found in our phosphotyrosine database are highly enriched in GO Cellular Component annotations “plasma membrane,” “cell junction,” and “cytoskeleton” (Figs. 5 and 7 and Table 1). Functional classification analyses also suggest tyrosine phosphorylation to be involved in “cytoskeletal protein binding,” “cytoskeleton organization,” “cell adhesion,” and “cell surface receptor linked signal transduction” (Figs. 5–7 and Tables 2 and 3). This is in agreement with the mounting evidence demonstrating the coordination of junctional proteins with signaling molecules (particularly receptor tyrosine kinases) to regulate cellular processes (37, 44). A large number of non-receptor tyrosine kinases, receptor tyrosine kinases, non-receptor tyrosine phosphatases, and receptor tyrosine phosphatases have also been identified at epithelial adherens junctions (37, 40). Not surprisingly, phosphotyrosine staining in epithelial cells are most prominent at junctional regions (35, 60).
Table 2.
Gene Ontology Biological Process: “cytoskeleton organization”
| Protein Name | Gene Symbol | RefSeq Number | Phosphopeptide Sequence | Phosphorylation Sites |
|---|---|---|---|---|
| Breast cancer anti-estrogen resistance protein 1 | Bcar1 | NP_037063 | DPLLDVY*DVPPSVEK | Y385 |
| GLLPNQYGQEVY*DTPPMAVK | Y365 | |||
| GPNGRDPLLDVY*DVPPSVEK | Y385 | |||
| HLLAPGSQDIY*DVPPVR | Y347 | |||
| RPGPGTLY*DVPR | Y485 | |||
| TQQGLY*QAPGPNPQFQSPPAK | Y226 | |||
| VGQGYVYEASQAEQDEY*DTPR | Y332 | |||
| VLPPEVADGSVIDDGVY*AVPPPAER | Y508 | |||
| Adenylyl cyclase-associated protein 1 | Cap1 | NP_071778 | LEAVSHTSDMHCGY*GDSPSK | Y31 |
| Serine/threonine-protein kinase MRCK β | Cdc42bpb | NP_446072 | LPDFQDPIFEY*FNTAPLAHDLTFR | Y954 |
| Cofilin-1 | Cfl1 | NP_058843 | HELQANCY*EEVKDR | Y140 |
| Calponin-3 | Cnn3 | NP_062232 | GMSVY*GLGR | Y261 |
| EH domain-containing protein 2 | Ehd2 | NP_001020068 | SKYDEIFY*NLAPADGK | Y458 |
| Band4.1-like protein 1 isoform L | Epb4.lll | NP_067713 | AVVY*RETDPSPEER | Y1534 |
| HLTQQDTRPAEQSLDMEEKDY*CEADGLSER | Y68 | |||
| IRPGEY*EQFESTIGFK | Y343 | |||
| PAEQSLDMEEKDY*CEADGLSER | Y68 | |||
| Ezrin | Ezr | NP_062230 | SQEQLAAELAEY*TAK | Y424 |
| Integrin β-1 | Itgb1 | NP_058718 | WDTGENPIY*K | Y783 |
| Keratin, type I cytoskeletal 19 | Krt19 | NP_955792 | SLLEGQEAHY*NSLSIAK | Y394 |
| Microtubule-associated protein 1B | Map1b | NP_062090 | EEDHEPDKTEAEDY*VMAVVDK | Y1038† |
| Myosin-10 | Myh10 | NP_113708 | AVIY*NPATQADWTAK | Y22 |
| TGLEDPERY*LFVDR | Y13 | |||
| Myosin-9 | Myh9 | NP_037326 | YLY*VDK | Y11 |
| Cytoplasmic protein NCK1 | Nck1 | NP_001100321 | LY*DLNM@PAFVK | Y105 |
| LY*DLNMPAFVK | Y105 | |||
| Cytoplasmic protein NCK2 | Nck2 | NP_001101686 | IY*DLNIPAFVK | Y110 |
| Oligophrenin-1 | Ophn1 | NP_001101318 | LWMEAMDGKEPIY*HSPITK | Y370 |
| Focal adhesion kinase 1 | Ptk2 | NP_037213 | THAVSVSETDDY*AEIIDEEDTYTMPSTR | Y397 |
| YM@EDSTY*YK | Y576 | |||
| YMEDSTY*Y*KASK | Y576, Y577 | |||
| YMEDSTY*YK | Y576 | |||
| Rho-guanine nucleotide exchange factor | Rgnef | NP_001102012 | IQEEEWDDY*IIPAK | Y605† |
| Rho-associated protein kinase 2 | Rock2 | NP_037154 | IY*ESIEEAK | Y713 |
| NKIY*ESIEEAK | Y713 | |||
| SHC-transforming protein 1 isoform a | Shc1 | NP_001157532 | ELFDDPSY*VNIQNLDK | Y423 |
| Tyrosine-protein phosphatase nonreceptor type substrate 1 | Sirpa | NP_037148 | LPRPEDTLTY*ADLDMVHLNR | Y477 |
| Sorbin and SH3 domain containing 1 | Sorbs1 | XP_002725845 | TPVDY*IDLPYSSSPSR | Y948† |
| Spectrin α chain, brain | Spna2 | NP_741984 | VAEDLESEGLM@AEEVQAVQQQEVY*GM@M@PR | Y1176 |
| VAEDLESEGLMAEEVQAVQQQEVY*GMM@PR | Y1176 | |||
| VAEDLESEGLMAEEVQAVQQQEVY*GMMPR | Y1176 | |||
| Neural Wiskott-Aldrich syndrome protein | Wasl | NP_001103835 | VIY*DFIEK | Y253 |
Phosphorylated amino acid.
Novel phosphorylation site (not present on Phosphosite Plus database, for all available species).
Table 3.
Gene Ontology Biological Process: “cell surface receptor linked signal transduction”
| Protein Name | Gene Symbol | RefSeq Number | Phosphopeptide Sequence | Phosphorylation Sites |
|---|---|---|---|---|
| Breast cancer anti-estrogen resistance protein 1 | Bcar1 | NP_037063 | DPLLDVY*DVPPSVEK | Y385 |
| GLLPNQYGQEVY*DTPPMAVK | Y365 | |||
| GPNGRDPLLDVY*DVPPSVEK | Y385 | |||
| HLLAPGSQDIY*DVPPVR | Y347 | |||
| RPGPGTLY*DVPR | Y485 | |||
| TQQGLY*QAPGPNPQFQSPPAK | Y226 | |||
| VGQGYVYEASQAEQDEY*DTPR | Y332 | |||
| VLPPEVADGSVIDDGVY*AVPPPAER | Y508 | |||
| E3 ubiquitin-protein ligase CBL-like isoform 2 | Cbl | XP_002727095 | IKPSSSANAIY*SLAARPLPVPK | Y656 |
| LPPGEQGDSEEDTEY*MTPTSRPVGVQKPEPK | Y682 | |||
| C-X-C chemokine receptor type 7 | Cxcr7 | NP_445804 | VSETEY*SALEQNTK | Y354 |
| Epithelial discoidin domain-containing receptor 1 | Ddr1 | NP_037269 | GTPTHSAPCVPNGSALLLSNPAY*R | Y510 |
| Endothelin B receptor precursor | Ednrb | NP_059029 | ANDHGY*DNFR | Y430 |
| Ephrin-B1 precursor | Efnb1 | NP_058785 | TTENNYCPHY*EK | Y316 |
| Ephrin B2 | Efnb2 | NP_001100798 | TADSVFCPHY*EK | Y307 |
| Epidermal growth factor receptor | Egfr | NP_113695 | GPTAENAEY*LR | Y1196 |
| GSHQM@SLDNPDY*QQDFFPK | Y1171 | |||
| GSHQMSLDNPDY*QQDFFPK | Y1171 | |||
| LLGAEEKEY*HAEGGK | Y870 | |||
| RPAGSVQNPVY*HNQPLHPAPGR | Y1109 | |||
| Ephrin type-A receptor 2 | Epha2 | NP_001102447 | IAY*SLLGLK | Y961 |
| QSSEDVY*FSK | Y576 | |||
| T*YVDPHTY*EDPNQAVLK | Y595 | |||
| TY*VDPHT*YEDPNQAVLK | Y589 | |||
| TYVDPHTY*EDPNQAVLK | Y595 | |||
| VLEDDPEATY*TTSGGK | Y773 | |||
| Receptor tyrosine-protein kinase erbB-2 | Erbb2 | NP_058699 | LLDIDETEY*HADGGKVPIK | Y881 |
| GRB2-associated-binding protein 1 | Gab1 | NP_001101914 | DAGSQDCY*DIPR | Y406 |
| EDPQDY*LLLINCQSK | Y183† | |||
| GDKQVEY*LDLDLESGK | Y627† | |||
| NSGSGSSM@SDERVDY*VVVDQQK | Y659 | |||
| NSGSGSSMSDERVDY*VVVDQQK | Y659 | |||
| QVEY*LDLDLESGK | Y627† | |||
| VDY*VVVDQQK | Y659 | |||
| VTSVSGESGLY*NLPR | Y259 | |||
| Retinoic acid-induced protein 3 | Gprc5a | NP_001073359 | AHTPASPY*NDYEGR | Y348 |
| AHTPASPYNDY*EGR | Y351 | |||
| AYSQEEITQGLEEMGDTLYAPY*STHFQMQNR | Y323 | |||
| G protein-coupled receptor, family C, group 5, member C isoform 1 | Gprc5c | XP_001081657 | AEDM@Y*M@VQSHQVATPTK | Y413 |
| AEDMY*M@VQSHQVATPTK | Y413 | |||
| AEDMY*MVQSHQVAT*PTKDGK | Y413 | |||
| AEDMY*MVQSHQVATPTK | Y413 | |||
| GPSEGAY*DVILPR | Y386 | |||
| GVGY*ETILK | Y323 | |||
| RPVSPYSGYNGQLLTSVY*QPTEMALMHK | Y369 | |||
| Glycogen synthase kinase-3 α | Gsk3a | NP_059040 | GEPNVSY*ICSR | Y279 |
| Homeodomain-interacting protein kinase 1 | Hipk1 | NP_001094456 | AVCSTY*LQSR | Y352 |
| Insulin-like growth factor 1 receptor precursor | Igflr | NP_434694 | DIYETDY*YR | Y1166 |
| Insulin receptor substrate 2 | Irs2 | NP_001162104 | VAYNPYPEDY*GDIEIGSHK | Y629 |
| Integrin β-1 | Itgb1 | NP_058718 | WDTGENPIY*K | Y783 |
| Tyrosine-protein kinase Lyn isoform A | Lyn | NP_110484 | VIEDNEY*TAR | Y397 |
| Mitogen-activated protein kinase 1 | Mapk1 | NP_446294 | VADPDHDHTGFLT*EY*VATR | Y185 |
| VADPDHDHTGFLTEY*VATR | Y185 | |||
| Mitogen-activated protein kinase 14 | Mapk14 | NP_112282 | HTDDEM@TGY*VATR | Y182 |
| HTDDEMTGY*VATR | Y182 | |||
| Mitogen-activated protein kinase 3 | Mapk3 | NP_059043 | IADPEHDHTGFLT*EY*VATR | Y205 |
| IADPEHDHTGFLTEY*VATR | Y205 | |||
| Hepatocyte growth factor receptor precursor | Met | NP_113705 | DMYDKEY*YSVHNK | Y1235 |
| Myosin-Ie | Myole | NP_775124 | SLY*TSMARPPLPR | Y988 |
| Neuropilin-1 precursor | Nrp1 | NP_659566 | DKLNPQSNY*SEA | Y919 |
| 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase γ-1 | Plcg1 | NP_037319 | IGTAEPDY*GALYEGR | Y771 |
| Pleckstrin homology domain-containing family A member 1 | Plekha1 | NP_001073363 | SQSHLPY*FAPKPPADSAVIK | Y181 |
| Focal adhesion kinase 1 | Ptk2 | NP_037213 | THAVSVSETDDY*AEIIDEEDTYTMPSTR | Y397 |
| YM@EDSTY*YK | Y576 | |||
| YMEDSTY*Y*KASK | Y576, Y577 | |||
| YMEDSTY*YK | Y576 | |||
| Tyrosine-protein phosphatase nonreceptor type 11 isoform 1 | Ptpn11 | NP_001171064 | IQNTGDY*YDLYGGEK | Y62 |
| VVLHDGDPNEPVSDY*INANIIMPEFETK | Y304 | |||
| VY*ENVGLMQQQR | Y584 | |||
| Receptor-type tyrosine-protein phosphatase α precursor | Ptpra | NP_036895 | VVQEYIDAFSDY*ANFK | Y792 |
| SHC-transforming protein 1 isoform a | Shc1 | NP_001157532 | ELFDDPSY*VNIQNLDK | Y423 |
| Sorbin and SH3 domain containing 1 | Sorbs1 | XP_002725845 | TPVDY*IDLPYSSSPSR | Y948† |
| Proto-oncogene tyrosine-protein kinase Src | Src | NP_114183 | YFTSTEPQY*QPGENL | Y536 |
| Signal transducer and activator of transcription 3 | Stat3 | NP_036879 | YCRPESQEHPEADPGSAAPY*LK | Y705 |
| Angiopoietin-1 receptor | Tek | NP_001099207 | KTY*VNTTLY*EK | Y1098, Y1104 |
| Transient receptor potential cation channel subfamily V member 4 | Trpv4 | NP_076460 | GVPNPIDLLESTLY*ESSVVPGPK | Y91 |
| KAPMDSLFDY*GTYR | Y110 | |||
| KGVPNPIDLLESTLY*ESSVVPGPK | Y91 |
Phosphorylated amino acid.
Novel phosphorylation site (not present on Phosphosite Plus database, for all available species).
These enriched functions agree with the model for which cell-cell adhesion (mediated principally by E-cadherin) and cell-ECM adhesion (mediated by integrins) initiate spatial cues for the establishment of physical and molecular asymmetry in epithelial cells—a process that is largely dependent on cytoskeletal reorganization (68). This asymmetry/polarization occurs both along the apico-basal axis, which defines the apical and basolateral membrane domains, and a second axis commonly referred to as the planar cell polarity axis that is perpendicular to the apico-basal axis.
Involvement of tight junction and adherens junction in cell polarity.
An important process in establishing apico-basal polarity is in defining the apico-basal axis and forming the tight junction (65). Therefore, we expect signaling networks involved in tight junction formation to be potentially overrepresented. This was indeed observed with our phosphotyrosine data set (Figs. 7 and 8). A critical component of this network is a group of widely studied transmembrane proteins, viz. occludin, claudin, and junctional adhesion molecule (JAM). Phosphorylation of these proteins is a major mode of regulation that affects the binding affinity to adaptors such as the zonula occludens proteins ZO-1, ZO-2, and ZO-3 and subsequently tight junction assembly (19, 21, 46). Although the phosphorylation of occludin on Y342 has been previously reported according to the PhosphoSitePlus database, this site has not been well characterized. Our analyses indicate that this site is highly conserved and is predicted to be modulated by a number of different kinases, including ephrin and insulin receptors and MAP2K family of kinases (Supplemental Table S3). Similar to sites Y398 and Y402 of occludin that are known to destabilize binding to ZO-1 (15), Y342 may potentially serve a regulatory role in its interaction with tight junction proteins. We have also identified a number of phosphotyrosine sites for ZO-1 (Y576, Y1027, Y1033, Y1054, Y1127, Y1152, Y1165, Y1178, Y1311, Y1333, Y1341) and ZO-2 (Y234, Y238, Y486, Y891, Y1093). Interestingly, sites Y1027 and Y1033 of ZO-1 have not been previously reported and are highly conserved. Site Y1027 is predicted to be modulated by insulin and/or insulin-like growth factor 1 receptor while Y1033 can be potentially modulated by ephrin receptor A7 and/or A3.
We have also identified a number of other phosphotyrosine proteins involved in the tight junction signaling network, including afadin (Y203, Y993, Y1237, Y1292, Y1502), vinculin (Y822), cingulin (Y238), MAGI-1 (Y373), PALS1 (also known as MPP5) (Y243, Y249), Par-3 (Y719), and Par-6 beta (Y101) (Fig. 9). Together with aPKC, Par-3 and Par-6 constitute the Par complex. PALS1, Crumbs, and PATJ constitute the Crumbs complex. Both of these complexes along with the Scribble complex are the three highly conserved core polarity complexes known to play a critical role in mediating apico-basal polarity (47). In addition, the proteins of the Crumbs and Par complex are colocalized in the apical domain at epithelial tight junctions and are thought to be involved in the assembly of tight junction (46). Members of the MAGI family of proteins have also been found at the tight junction (46). In particular, MAGI-3 has recently been shown to be localized with ZO-1 and cingulin in epithelial cells and serves as a scaffolding protein by recruiting phosphatases PTEN and RPTPβ to their substrates (1, 67). The novel site Y356 of MAGI-3 found in our study may serve a regulatory role in activating/deactivating the protein's recruitment activities. Moreover, this site is within the [D/E]xxY motif, which is among several motifs (e.g., YxxP and Y[A/G/S/T/E/D]) recognized by the kinase c-Src (56). However, NetworKIN analysis, which combines motif with contextual information, predicts the kinase to be insulin and/or insulin-like growth factor 1 receptor. This may suggest the importance of subcellular localization, in this case to the tight junction, in controlling the specificity of signaling and information flow.
Several components present in the tight junction signaling network are actually part of the adherens junction. Thus, it was expected to see enrichment in both “adherens junction” and “tight junction” in the KEGG pathway enrichment analysis (Fig. 8). In particular, α-catenin is an adherens junction protein that, aside from its role in mediating the association of E-cadherin/β-catenin to actin, also serves as an adaptor in mediating the interaction of ZO-1 and ZO-2 to actin (46). We have identified Y177 of α-catenin to be phosphorylated, and this agrees with the phosphorylation previously reported for other species according to PhosphoSitePlus. The functional significance of this site remains to be determined, although a recent study suggests the tyrosine phosphorylation of α-catenin to be involved in its translocation to the plasma membrane and subsequent association with β-catenin (5). Hence, one speculation is the involvement of α-catenin Y177 in the positive regulation of adherens and tight junction assembly and the establishment of apico-basal polarity. Another binding partner of α-catenin is the F-actin-binding protein afadin, which is known to associate with a number of other cell adhesion molecules, including nectins and Lim domain only 7 (LMO7) (14). We found two phosphorylation sites, Y1358 and Y1588, on LMO7 that have not been archived on PhosphoSitePlus. Of these two sites, Y1588 is highly conserved and is a potential target for phosphorylation by Tec and/or MAP2K3/MAP2K4. LMO7 associates not only with afadin involved in the nectin/afadin complex, but also with α-actinin, which is one of several actin-associated proteins that interact with α-catenin in the E-cadherin/catenin complex (14). Therefore, the novel sites identified on LMO7 may serve regulatory roles in modulating the interaction and signaling between the nectin/afadin and E-cadherin/catenin adhesion complexes at adherens junctions.
Remarkably, all core planar cell polarity proteins have been found to colocalize with junctional apical complexes (66). Recent studies also support the existence of interactions between components involved in apico-basal polarity and planar cell polarity (13). Thus, this suggests potential cross talk among the signaling networks for these two types of cell polarity, and the discussion above on apico-basal polarity may be also relevant for the regulation of planar cell polarity.
Regulation of cytoskeletal organization.
Much of the regulatory mechanism in initiating and establishing cell polarity involves the reorganization of cytoskeleton. In particular, cell adhesion induces localized assembly of cytoskeletal networks that promote the recruitment of signaling proteins and trafficking of domain-specific proteins [e.g., aquaporin (AQP) 2 and AQP-4] to the apical or basolateral membrane (36, 65, 68). Many proteins found in our profiling are highly enriched in the actin cytoskeleton signaling pathway (Figs. 8 and 10). One in particular, focal adhesion kinase (FAK), is a cytoplasmic protein tyrosine kinase and plays a major role in the regulation of the actin cytoskeleton at both cell-cell junctions and cell-ECM adhesions and affects cellular processes such as proliferation, motility, cell polarization, and adhesion (24, 55). The site Y397 of FAK, also identified in our profiling, is autophosphorylated in response to integrin clustering, and its activation facilitates the association of c-Src and phosphatidylinositol 3-kinase via their SH2 domains (6). It is of note that autophosphorylation and interactions via the SH2 domain are common in phosphotyrosine-mediated signaling networks. Table 4 lists the tyrosine kinases found in our study. All phosphotyrosine sites identified are potential autophosphorylation sites that may play important signaling roles in the IMCD. Table 5 lists the IMCD phosphotyrosine proteins with SH2 domains, which recognizes phosphorylated tyrosine within certain motifs (e.g., Y*xxP) (27, 59), and SH3 domains, which recognizes certain proline-rich ligands.
Recently, integrin αvβ3 has been found to colocalize with nectin at adherens junction and is involved in the reorganization of cytoskeleton and subsequent adherens and tight junction formation and maintenance via PKC/FAK/c-Src activation (42, 45). Among the effectors of c-Src are Rho guanine-nucleotide exchange factors (GEFs) that regulate the Rho family of GTPases (Cdc42 and Rac) that are critical in regulating actin dynamics (53). We have identified novel phosphorylation sites on a number of Rho GEFs, including Y1128 of Arhgef11, Y636 of Arhgef5, and Y605 of Rgnef. GTPase activating protein (GAP) and guanine nucleotide dissociation inhibitors (GDI) are also upstream regulators of Rho GTPases. We found a novel phosphorylation site Y201 on Arhgap27 that can be potentially regulated by insulin and/or ephrin receptors. Because many of the novel phosphorylation sites discussed are likely substrates of the insulin and ephrin receptors, this may suggest a critical role for these families of kinases in modulating cytoskeleton organization and cell junction interactions. The insulin, insulin-like growth factor, and ephrin receptors have been previously identified in the IMCD transcriptome (64). Our database provides a foundation for further study of the role of phosphotyrosine-mediated signaling in the renal inner medullary collecting duct and other epithelia.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: B.Z., M.A.K., C.-L.C., and T.P. conception and design of research; B.Z., C.-L.C., and T.P. performed experiments; B.Z., M.A.K., and T.P. analyzed data; B.Z., M.A.K., and T.P. interpreted results of experiments; B.Z. and T.P. prepared figures; B.Z. and M.A.K. drafted manuscript; B.Z., M.A.K., C.-L.C., and T.P. edited and revised manuscript; B.Z., M.A.K., C.-L.C., and T.P. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Jennifer Huling from National Heart, Lung, and Blood Institute (NHLBI) for help in setting up the Rat Inner Medullary Collecting Duct Phosphotyrosine Database and Aleksandra Nita-Lazar from National Institute of Allergy and Infectious Diseases for information on the antiphosphotyrosine peptide immunoprecipitation protocol. This study was supported by the Intramural Budget of the NHLBI (NHLBI Project Z01-HL-001285). Mass spectrometry was done in the NHLBI Proteomics Core Facility (Marjan Gucek, Director). Boyang Zhao was a student intern from the University of Michigan in the National Institutes of Health Biomedical Engineering Summer Internship Program, funded by the National Institute of Biomedical Imaging and Bioengineering (present e-mail: bozhao@mit.edu).
Footnotes
This article is the topic of an Editorial Focus by Carolyn M. Ecelbarger (14a).
REFERENCES
- 1. Adamsky K, Arnold K, Sabanay H, Peles E. Junctional protein MAGI-3 interacts with receptor tyrosine phosphatase beta (RPTP beta) and tyrosine-phosphorylated proteins. J Cell Sci 116: 1279–1289, 2003 [DOI] [PubMed] [Google Scholar]
- 2. Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics 7: 1389–1396, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bansal AD, Hoffert JD, Pisitkun T, Hwang S, Chou CL, Boja ES, Wang G, Knepper MA. Phosphoproteomic profiling reveals vasopressin-regulated phosphorylation sites in collecting duct. J Am Soc Nephrol 21: 303–315, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300, 1995 [Google Scholar]
- 5. Burks J, Agazie YM. Modulation of alpha-catenin Tyr phosphorylation by SHP2 positively effects cell transformation induced by the constitutively active FGFR3. Oncogene 25: 7166–7179, 2006 [DOI] [PubMed] [Google Scholar]
- 6. Carragher NO, Frame MC. Focal adhesion and actin dynamics: a place where kinases and proteases meet to promote invasion. Trends Cell Biol 14: 241–249, 2004 [DOI] [PubMed] [Google Scholar]
- 7. Cereijido M, Valdes J, Shoshani L, Contreras RG. Role of tight junctions in establishing and maintaining cell polarity. Annu Rev Physiol 60: 161–177, 1998 [DOI] [PubMed] [Google Scholar]
- 8. Chou CL, DiGiovanni SR, Luther A, Lolait SJ, Knepper MA. Oxytocin as an antidiuretic hormone. II. Role of V2 vasopressin receptor. Am J Physiol Renal Fluid Electrolyte Physiol 269: F78–F85, 1995 [DOI] [PubMed] [Google Scholar]
- 9. Chou CL, Rapko SI, Knepper MA. Phosphoinositide signaling in rat inner medullary collecting duct. Am J Physiol Renal Physiol 274: F564–F572, 1998 [DOI] [PubMed] [Google Scholar]
- 10. Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Del Rosario AM, White FM. Quantifying oncogenic phosphotyrosine signaling networks through systems biology. Curr Opin Genet Dev 20: 23–30, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: 3, 2003 [PubMed] [Google Scholar]
- 13. Djiane A, Yogev S, Mlodzik M. The apical determinants aPKC and dPatj regulate Frizzled-dependent planar cell polarity in the Drosophila eye. Cell 121: 621–631, 2005 [DOI] [PubMed] [Google Scholar]
- 14. Ebnet K. Organization of multiprotein complexes at cell-cell junctions. Histochem Cell Biol 130: 1–20, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a. Ecelbarger CM. The elusive phosphotyrosine: pinning down a rare species. Focus on “Large-scale phosphotyrosine proteomic profiling of rat renal collecting duct epithelium reveals predominance of proteins involved in cell polarity determination.” Am J Physiol Cell Physiol (October 5, 2011). doi:10.1152/ajpcell.00363.2011 [DOI] [PubMed] [Google Scholar]
- 15. Elias BC, Suzuki T, Seth A, Giorgianni F, Kale G, Shen L, Turner JR, Naren A, Desiderio DM, Rao R. Phosphorylation of Tyr-398 and Tyr-402 in occludin prevents its interaction with ZO-1 and destabilizes its assembly at the tight junctions. J Biol Chem 284: 1559–1569, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elias JE, Gibbons FD, King OD, Roth FP, Gygi SP. Intensity-based protein identification by machine learning from a library of tandem mass spectra. Nat Biotechnol 22: 214–219, 2004 [DOI] [PubMed] [Google Scholar]
- 17. Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry 2. Nat Methods 4: 207–214, 2007 [DOI] [PubMed] [Google Scholar]
- 18. Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989, 1994 [DOI] [PubMed] [Google Scholar]
- 19. Feldman GJ, Mullin JM, Ryan MP. Occludin: structure, function and regulation. Adv Drug Deliv Rev 57: 883–917, 2005 [DOI] [PubMed] [Google Scholar]
- 20. Feric M, Zhao B, Hoffert JD, Pisitkun T, Knepper MA. Large-scale phosphoproteomic analysis of membrane proteins in renal proximal and distal tubule. Am J Physiol Cell Physiol 300: C755–C770, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Findley MK, Koval M. Regulation and roles for claudin-family tight junction proteins. IUBMB Life 61: 431–437, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J Proteome Res 3: 958–964, 2004 [DOI] [PubMed] [Google Scholar]
- 23. Gunaratne R, Braucht DW, Rinschen MM, Chou CL, Hoffert JD, Pisitkun T, Knepper MA. Quantitative phosphoproteomic analysis reveals cAMP/vasopressin-dependent signaling pathways in native renal thick ascending limb cells. Proc Natl Acad Sci USA 107: 15653–15658, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hall JE, Fu W, Schaller MD. Focal adhesion kinase: exploring Fak structure to gain insight into function. Int Rev Cell Mol Biol 288: 185–225, 2011 [DOI] [PubMed] [Google Scholar]
- 25. Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc Natl Acad Sci USA 103: 7159–7164, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoffert JD, Wang G, Pisitkun T, Shen RF, Knepper MA. An automated platform for analysis of phosphoproteomic datasets: application to kidney collecting duct phosphoproteins. J Proteome Res 6: 3501–3508, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holland SJ, Gale NW, Gish GD, Roth RA, Songyang Z, Cantley LC, Henkemeyer M, Yancopoulos GD, Pawson T. Juxtamembrane tyrosine residues couple the Eph family receptor EphB2/Nuk to specific SH2 domain proteins in neuronal cells. EMBO J 16: 3877–3888, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: a bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 4: 1551–1561, 2004 [DOI] [PubMed] [Google Scholar]
- 29. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57, 2009 [DOI] [PubMed] [Google Scholar]
- 30. Huyer G, Liu S, Kelly J, Moffat J, Payette P, Kennedy B, Tsaprailis G, Gresser MJ, Ramachandran C. Mechanism of inhibition of protein-tyrosine phosphatases by vanadate and pervanadate. J Biol Chem 272: 843–851, 1997 [DOI] [PubMed] [Google Scholar]
- 31. Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, Metalnikov P, Nguyen V, Pasculescu A, Jin J, Park JG, Samson LD, Woodgett JR, Russell RB, Bork P, Yaffe MB, Pawson T. Systematic discovery of in vivo phosphorylation networks. Cell 129: 1415–1426, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Linding R, Jensen LJ, Pasculescu A, Olhovsky M, Colwill K, Bork P, Yaffe MB, Pawson T. NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res 36: D695–D699, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luo W, Slebos RJ, Hill S, Li M, Brabek J, Amanchy R, Chaerkady R, Pandey A, Ham AJ, Hanks SK. Global impact of oncogenic Src on a phosphotyrosine proteome. J Proteome Res 7: 3447–3460, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449, 2005 [DOI] [PubMed] [Google Scholar]
- 35. Maher PA, Pasquale EB, Wang JY, Singer SJ. Phosphotyrosine-containing proteins are concentrated in focal adhesions and intercellular junctions in normal cells. Proc Natl Acad Sci USA 82: 6576–6580, 1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mazzochi C, Benos DJ, Smith PR. Interaction of epithelial ion channels with the actin-based cytoskeleton. Am J Physiol Renal Physiol 291: F1113–F1122, 2006 [DOI] [PubMed] [Google Scholar]
- 37. McLachlan RW, Yap AS. Not so simple: the complexity of phosphotyrosine signaling at cadherin adhesive contacts. J Mol Med (Berl) 85: 545–554, 2007 [DOI] [PubMed] [Google Scholar]
- 38. Mikalsen SO, Kaalhus O. A characterization of pervanadate, an inducer of cellular tyrosine phosphorylation and inhibitor of gap junctional intercellular communication. Biochim Biophys Acta 1290: 308–318, 1996 [DOI] [PubMed] [Google Scholar]
- 39. Miller ML, Jensen LJ, Diella F, Jorgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, Olhovsky M, Pasculescu A, Alexander J, Knapp S, Blom N, Bork P, Li S, Cesareni G, Pawson T, Turk BE, Yaffe MB, Brunak S, Linding R. Linear motif atlas for phosphorylation-dependent signaling. Sci Signal 1: ra2, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Niessen CM, Leckband D, Yap AS. Tissue organization by cadherin adhesion molecules: dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol Rev 91: 691–731, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nita-Lazar A, Saito-Benz H, White FM. Quantitative phosphoproteomics by mass spectrometry: past, present, and future. Proteomics 8: 4433–4443, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ogita H, Rikitake Y, Miyoshi J, Takai Y. Cell adhesion molecules nectins and associating proteins: Implications for physiology and pathology. Proc Jpn Acad Ser B Phys Biol Sci 86: 621–629, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648, 2006 [DOI] [PubMed] [Google Scholar]
- 44. Orian-Rousseau V, Ponta H. Adhesion proteins meet receptors: a common theme? Adv Cancer Res 101: 63–92, 2008 [DOI] [PubMed] [Google Scholar]
- 45. Ozaki M, Ogita H, Takai Y. Involvement of integrin-induced activation of protein kinase C in the formation of adherens junctions. Genes Cells 12: 651–662, 2007 [DOI] [PubMed] [Google Scholar]
- 46. Paris L, Tonutti L, Vannini C, Bazzoni G. Structural organization of the tight junctions. Biochim Biophys Acta 1778: 646–659, 2008 [DOI] [PubMed] [Google Scholar]
- 47. Pieczynski J, Margolis B. Protein complexes that control renal epithelial polarity. Am J Physiol Renal Physiol 300: F589–F601, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pisitkun T, Jacob V, Schleicher SM, Chou CL, Yu MJ, Knepper MA. Akt and ERK1/2 pathways are components of the vasopressin signaling network in rat native IMCD. Am J Physiol Renal Physiol 295: F1030–F1043, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reiland J, Ott VL, Lebakken CS, Yeaman C, McCarthy J, Rapraeger AC. Pervanadate activation of intracellular kinases leads to tyrosine phosphorylation and shedding of syndecan-1. Biochem J 319: 39–47, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rinschen MM, Yu MJ, Wang G, Boja ES, Hoffert JD, Pisitkun T, Knepper MA. Quantitative phosphoproteomic analysis reveals vasopressin V2-receptor-dependent signaling pathways in renal collecting duct cells. Proc Natl Acad Sci USA 107: 3882–3887, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol 23: 94–101, 2005 [DOI] [PubMed] [Google Scholar]
- 52. Ruttenberg BE, Pisitkun T, Knepper MA, Hoffert JD. PhosphoScore: an open-source phosphorylation site assignment tool for MSn data. J Proteome Res 7: 3054–3059, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Samarin S, Nusrat A. Regulation of epithelial apical junctional complex by Rho family GTPases. Front Biosci 14: 1129–1142, 2009 [DOI] [PubMed] [Google Scholar]
- 54. Sands JM, Knepper MA. Urea permeability of mammalian inner medullary collecting duct system and papillary surface epithelium. J Clin Invest 79: 138–147, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schaller MD. Cellular functions of FAK kinases: insight into molecular mechanisms and novel functions. J Cell Sci 123: 1007–1013, 2010 [DOI] [PubMed] [Google Scholar]
- 56. Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol 23: 1391–1398, 2005 [DOI] [PubMed] [Google Scholar]
- 57. Secrist JP, Burns LA, Karnitz L, Koretzky GA, Abraham RT. Stimulatory effects of the protein tyrosine phosphatase inhibitor, pervanadate, on T-cell activation events. J Biol Chem 268: 5886–5893, 1993 [PubMed] [Google Scholar]
- 58. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ. SH2 domains recognize specific phosphopeptide sequences. Cell 72: 767–778, 1993 [DOI] [PubMed] [Google Scholar]
- 60. Staddon JM, Herrenknecht K, Smales C, Rubin LL. Evidence that tyrosine phosphorylation may increase tight junction permeability. J Cell Sci 108: 609–619, 1995 [DOI] [PubMed] [Google Scholar]
- 61. Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39: D561–D568, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tan CS, Bodenmiller B, Pasculescu A, Jovanovic M, Hengartner MO, Jorgensen C, Bader GD, Aebersold R, Pawson T, Linding R. Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci Signal 2: ra39, 2009 [DOI] [PubMed] [Google Scholar]
- 63. Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem 77: 4626–4639, 2005 [DOI] [PubMed] [Google Scholar]
- 64. Uawithya P, Pisitkun T, Ruttenberg BE, Knepper MA. Transcriptional profiling of native inner medullary collecting duct cells from rat kidney. Physiol Genomics 32: 229–253, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Valenti G, Procino G, Tamma G, Carmosino M, Svelto M. Minireview: aquaporin 2 trafficking. Endocrinology 146: 5063–5070, 2005 [DOI] [PubMed] [Google Scholar]
- 66. Wu J, Mlodzik M. A quest for the mechanism regulating global planar cell polarity of tissues. Trends Cell Biol 19: 295–305, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wu Y, Dowbenko D, Spencer S, Laura R, Lee J, Gu Q, Lasky LA. Interaction of the tumor suppressor PTEN/MMAC with a PDZ domain of MAGI3, a novel membrane-associated guanylate kinase. J Biol Chem 275: 21477–21485, 2000 [DOI] [PubMed] [Google Scholar]
- 68. Yeaman C, Grindstaff KK, Nelson WJ. New perspectives on mechanisms involved in generating epithelial cell polarity. Physiol Rev 79: 73–98, 1999 [DOI] [PubMed] [Google Scholar]
- 69. Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics 4: 1240–1250, 2005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.