Abstract
Background:
The dopamine D2 receptor gene (DRD2) plays a role in many diseases such as schizophrenia, Parkinson’s disease, and addictive behaviour. Methods currently available for the detection of DRD2 polymorphisms are costly and cannot detect all 8 polymorphisms of our research interest simultaneously (Val96Ala, Leu141Leu, Val154Ile, Pro310Ser, Ser311Cys, TaqI A, A-241G, and −141C Ins/Del). Therefore, we developed a nested multiplex polymerase chain reaction (PCR) for simultaneous detection of these polymorphisms.
Methods:
Genomic DNA was extracted from blood using standardised methods. Primers specific at the 3′-end for the polymorphic sites were designed. A two-step PCR method was developed. In the first PCR, a region from exon 3 to 4, exon 7, the promoter region, and the 3′-region of DRD2 were specifically amplified. The products were subsequently used as templates in the second PCR. Sequencing was performed to validate the test results.
Results:
Specific bands corresponding to the amplified product of interest were obtained. The method was reproducible and specific when used to genotype patients with schizophrenia. The amplified sequences showed 100% homology to the DRD2 sequence.
Conclusion:
The method was found to be simple, rapid, specific, and reproducible for the simultaneous detection of the DRD2 polymorphisms.
Keywords: dopamine D2 receptor, genetics, genetic polymorphism, methods, nested PCR, reproducibility of results, specificity
Introduction
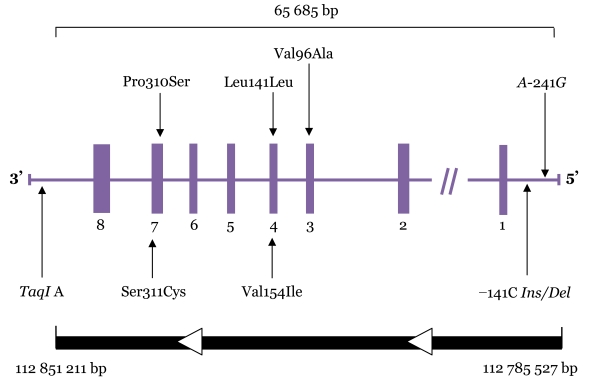
The dopamine D2 receptor (DRD2) belongs to the G protein–coupled receptor superfamily located on postsynaptic dopaminergic neurons (1). It is mainly expressed in the striatum, cortex, and limbic system (2). The human DRD2 gene is located at 11 q22-q23 (3), as depicted in Figure 1. This gene was previously found to contain 8 exons that span over 270 kb (4), but it has recently been found to span over only 65.69 kb (GenBank accession number NG_00884.1). Of the more than 200 polymorphisms at the DRD2 locus that may have important clinical and physiological implications, the most important polymorphisms are Val96Ala, Leu141Leu, Val154Ile, Pro310Ser, Ser311Cys, DRD2/ANKK1 TaqI A (referred to here as TaqI A), A-241G, and −141C Ins/Del (3,5–10), as detailed in Table 1.
Figure 1:
Dopamine D2 receptor gene (DRD2) structure and polymorphisms studied. Boxes represent exons; horizontal lines connecting boxes represent introns, promoter, and untranslated regions. Arrows indicate relative locations of the polymorphisms.
Table 1:
Characteristics and positions of 8 dopamine D2 receptor polymorphisms
| Polymorphisms | Characteristic mutation | Position | Accession no. |
|---|---|---|---|
| Val96Ala | Substitution of alanine for valine at codon 96 | 14858 | AF050737 |
| Leu141Leu | Silent substitution at codon 141 (C T) | 16019 | AF050737 |
| Val154Ile | Substitution of isoleucine for valine at codon 154 | 16058 | AF050737 |
| Pro310Ser | Substitution of serine for proline at codon 310 | 20225 | AF050737 |
| Ser311Cys | Substitution of cysteine for serine at codon 311 | 20229 | AF050737 |
| TaqI A | Alteration of a TaqI restriction site at 10 541 kb downstream of DRD2 stop codon (exon 8) | 32806 | AF050737 |
| A-241G | Substitution of guanine for adenosine at −241 | 6091 | AF148806 |
| −141C Ins/Del | Single base pair cytosine insertion/deletion at position −141 | 6191 | AF148806 |
The TaqI A1 allele of the TaqI A polymorphism is associated with reduced DRD2 density (9,10). The TaqI A1 allele has been shown to be associated with addictive behaviour, including alcoholism (11) and smoking (12). This allele has also been implicated in the development of motor fluctuations in patients with Parkinson’s disease in response to levodopa (13). Previous studies have shown that the −141C Ins/Del polymorphism leads to reduced promoter activity in vitro (8). The −141C Del allele was found to be associated with a high striatal dopamine receptor density in healthy volunteers (10) as well as with schizophrenia (14) and the clinical response to antipsychotics in the first episode of schizophrenia (15).
Previously described methods for the detection of DRD2 polymorphisms include polymerase chain reaction (PCR)–restriction fragment length polymorphism (RFLP) (5,6,8,16–19), denaturing gradient gel electrophoresis (6), Southern blot analysis (3,16,20), and direct sequencing (3,16,20). These methods are generally costly and tedious, and some require specialised equipment and manpower that may not be available or appropriate for all research groups. The objective of this paper is to describe a novel method for the detection of these 8 DRD2 polymorphisms that is simple and relatively rapid while maintaining sensitivity. This assay was developed for a bigger study to investigate the influence of DRD2 polymorphisms on treatment outcomes in patients with schizophrenia. Application of this method will enable smaller research groups to increase the scale of their DRD2 genotyping throughput by less specialised personnel in less specialised settings.
Materials and Methods
Genomic DNA
Genomic DNA was obtained from peripheral leucocytes extracted from 10 mL of blood taken from patients with schizophrenia attending the psychiatry clinic at Hospital Universiti Sains Malaysia, using previously described methods (21). The protocol for this study was approved by the Research and Ethics Committee, Universiti Sains Malaysia, Kelantan, Malaysia.
Primer design
To improve sensitivity, the PCR was designed as a two-step nested PCR where, in the first PCR, primers were designed to amplify specific regions of the DRD2 gene containing the mutations of interest. The products were then used as templates for the second allele-specific reactions. For the second PCR, primers were designed to have specific 3′-ends, manipulated to differentiate single nucleotide changes at the specific locus during PCR amplifications. We identified the location of the flanking regions of the primers against the DNA sequences published by Hauge et al. (16) and Murakami et al. (22) that are available at http://www.ncbi.nlm.nih.gov/ (accession number: AF050737 and AF148806).
We also used the Basic Local Alignment Search Tool (BLAST) programme at http://www.ncbi.nlm.nih.gov/blast to ascertain the specificity of the primers. The primers were also designed to have similar annealing temperatures and appropriate length as well as GC contents for multiplexing reactions to avoid incompatibility of the primer sets. Table 2 lists the sequences of the primers used for both the first and the second PCRs.
Table 2:
List of primer sequences, fragment sizes, calculated melting temperature (Tm), and primer concentration of the first and second PCRs used for the detection of 8 dopamine D2 receptor polymorphisms
| PCR | Primer | Sequence (5′ – 3′) | Fragment size (bp) | Calculated Tm (°C) | Primer concentration (μM) |
|---|---|---|---|---|---|
| 1st PCR | |||||
| Set A | DRD2EX3&4FW | cag ctg cct cct gag tct gt | 1497 | 64 | 0.30 |
| DRD2EX3&4RV | cca tat ctg tgc cag gga ct | 62 | 0.30 | ||
| Set B | DRD2EX7FW | ctg atg cct ggg aac ttg tc | 566 | 62 | 0.15 |
| DRD2EX7RV | gcc cat ctg taa agt gag ca | 60 | 0.15 | ||
| D2TAQ1AFW | acg gct ggc caa gtt gtc t | 305 | 60 | 0.25 | |
| D2TAQ1ARV | acc ttc ctg agt gtc atc aac | 62 | 0.25 | ||
| Set C | D2PRFW | act ggc gag cag acg gtg a | 276 | 62 | 0.40 |
| D2PRRV | tga agc tgg aca gct ctg c | 60 | 0.40 | ||
| 2nd PCR | |||||
| Set D | DRD2EX7RV | cca tat ctg tgc cag gga ct | 0.25 | ||
| D2TAQ1AFW | acg gct ggc caa gtt gtc t | 0.25 | |||
| D2WT311 | tga ctc tcc ccg acc cgt c | 409 | 60 | 0.25 | |
| D2MT311 | tga ctc tcc ccg acc cgt g | 60 | 0.25 | ||
| D2TAQ1AWT | atc ctc aaa gtg ctg gtc g | 197 | 58 | 0.25 | |
| D2TAQ1AMUT | atc ctc aaa gtg ctg gtc a | 56 | 0.25 | ||
| Set E | DRD2EX3&4RV | cca tat ctg tgc cag gga ct | 0.25 | ||
| DRD2EX7RV | gcc cat ctg taa agt gag ca | 0.25 | |||
| D2WT310 | gct gac tct ccc cga cc | 411 | 58 | 0.25 | |
| D2MT310 | gct gac tct ccc cga ct | 56 | 0.25 | ||
| D2WT141 | ctg tgg cca tgc cca tgc | 225 | 60 | 0.25 | |
| D2MT141 | ctg tgg cca tgc cca tgt | 58 | 0.25 | ||
| Set F | DRD2EX3&4RV | cca tat ctg tgc cag gga ct | 0.25 | ||
| D2WT96 | tgt tgc ttt gtc ccc agg t | 1387 | 58 | 0.25 | |
| D2MT96 | tgt tgc ttt gtc ccc agg c | 60 | 0.25 | ||
| D2WT154 | caa gcg ccg ggt cac cg | 185 | 60 | 0.25 | |
| D2MT154 | caa gcg ccg ggt cac ca | 58 | 0.25 | ||
| Set G | D2PRRV | tga agc tgg aca gct ctg c | 0.25 | ||
| D2-141WT | aac ccc tcc tac ccg ttc c | 151 | 62 | 0.25 | |
| D2-141MUT | aac ccc tcc tac ccg ttc a | 60 | 0.25 | ||
| Set H | D2PRRV | tga agc tgg aca gct ctg c | 0.25 | ||
| D2-241WT | cag cct gca atc aca gct ta | 252 | 60 | 0.25 | |
| D2-241MUT | cag cct gca atc aca gct tg | 62 | 0.25 | ||
Method development for nested allele-specific multiplex PCR
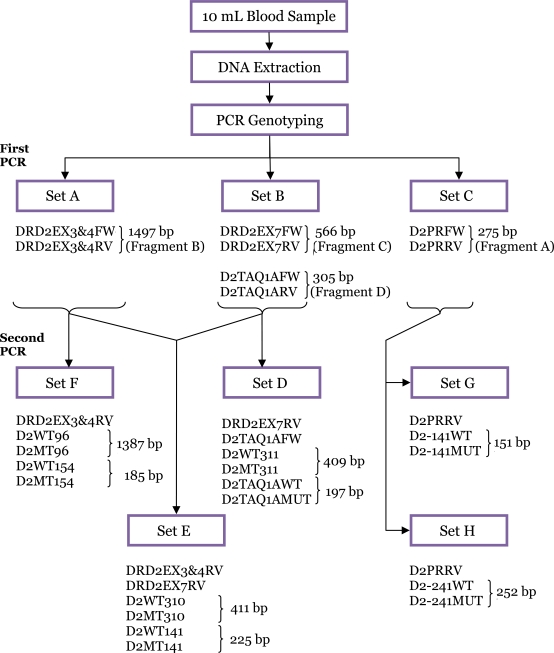
The schematic chart for the detection of DRD2 polymorphisms using a nested allele-specific multiplex PCR is shown in Figure 2. A total of 276 bp (fragment A) of the 5′-untranslated region (i.e., promoter region) of DRD2 was amplified with the primers D2PRFW and D2PRRV. It was then used as template for the second PCR of the A-241G and −141C Ins/Del polymorphisms. For identification of Val96Ala, Leu141Leu, and Val154Ile polymorphisms, the larger first PCR fragment was amplified by using primers DRD2EX3&4FW and DRD2EX3&4RV spanning exons 3 to 4. Fragment B, as large as 1497 bp, was then used as a template for the second PCR. Primers DRD2EX7FW and DRD2EX7RV were used to amplify exon 7 for fragment C. It was then used as template for the second PCR of Pro310Ser and Ser311Cys. Fragment D spanning the 3′-untranslated region (i.e., TaqI A region) of DRD2 was amplified using primers D2TAQ1AFW and D2TAQ1ARV. It was used as a template for the second PCR of the TaqI A polymorphism.
Figure 2:
Schematic chart of SNP genotyping of DRD2 polymorphisms using a nested allele-specific multiplex PCR.
Initially, 4 PCR procedures were performed using singlet pairs of primers to determine a PCR programme that would allow optimal amplifications of all the loci when performed individually. The initial reaction condition was determined empirically. When these initial experiments produced a successful uniplex PCR, a multiplex PCR was attempted first by combining the 4 primer sets in a single reaction. The PCR protocols initially employed were exactly the same as they were with the reactions with individual primer pairs.
To improve the amplification of certain loci while eliminating the amplifications of non-specific products, further adjustments to the PCR protocols were made. The concentrations of MgCl2 and Taq polymerase were varied. Different combinations of primer pairs and template dilutions were also tried to improve the reproducibility, specificity, and sensitivity. The same strategies were used for the optimisation of the second PCR.
Optimised PCR method for genotyping DRD2
After numerous experiments, the most robust protocol that also gave equal amplification of all alleles was achieved in a total volume of 25.0 μL, containing 200 ng DNA template, 1.0 mM MgCl2, 0.2 mM dNTPs (Promega, Madison, Wisconsin, USA), 0.5 U Biotool® DNA Taq Polymerase (B&M Labs, Madrid, Spain), and 1× Biotool® PCR buffer (B&M Labs, Madrid, Spain). The optimal primer (Invitrogen, California, USA) concentrations were found to be 0.15–0.40 μM (Table 1). All the PCRs were done in standard 0.2 mL Eppendorf PCR tubes and ran in an Eppendorf Mastercycler Gradient® Cycler (Eppendorf, Hamburg, Germany).
The first PCR amplified a region from exon 3 to 4 of DRD2, exon 7, the promoter region, and the downstream region of the DRD2 stop codon (exon 8) using specifically designed primers (Table 2). This step was performed to isolate regions of interest containing the relevant DRD2 polymorphisms that were later used for the second allele-specific PCR to avoid amplification of similar sequences in the human genome that may be located outside the gene. The protocols involved 2 uniplex and 1 duplex PCRs for improved specificity and sensitivity. The uniplex reactions amplified the region from exon 3 to exon 4 (Set A) and the promoter region (Set C) of DRD2, and the duplex reaction amplified exon 7 and the 3′-region (Set B) of the gene. Four primer sets were used in the first PCR, yielding fragments of sizes 1497, 566, 276, and 305 bp. For all the reactions, DNA was denaturated initially at 94 °C for 2 minutes before the cycling programme, followed by 35 cycles of DNA denaturing step at 94 °C for 1 minute, annealing at 65 °C for 1 minute, extension at 72 °C for 2 minutes, and a final extension period at 72 °C for 5 minutes. The PCR products were analysed on a 2.0% agarose gel (LE, analytical grade; Promega, Madison, Wisconsin, USA) stained in ethidium bromide in 1× Tris-borate-EDTA (TBE) buffer at 130 V for 90 minutes.
After a successful first PCR, 2.0 μL of diluted PCR product were used as a template for the detection of wild-type or mutant-type alleles in the second PCR. The second PCR was carried out in a reaction mixture that was identical to that described for the first PCR, with the exception of the primer concentrations shown in Table 2. The second PCR was comprised of 15 cycles of DNA denaturing at 94 °C for 1 minutes, annealing at 63 °C for 1 minutes, and extension at 72 °C for 2 minutes. Next, 10 mL of the second PCR product was analysed on a 2.0% agarose gel and 1× TBE at 130 V for 90 minutes. The expected fragment size for each of the products is listed in Table 2.
The optimised method was validated for reproducibility and specificity. This method was tested against DNA samples obtained from patients with schizophrenia. Samples detected with DRD2 polymorphisms were identified, re-amplified, and sent for direct sequencing. These were later used as positive controls for the alleles.
Direct DNA sequencing
Specificity of the primer sets used in this study was confirmed using a panel of single PCR products from the first PCR as positive controls that contained either heterozygous or homozygous wild-type/mutant-type, wild-type/wild-type, or mutant-type/mutant-type alleles. Direct sequencing was used to validate each positive control sample. The DNA samples were purified using QIAquick® PCR Purification Kit (Qiagen, Hilden, Germany) and sequenced on an ABI 3700 using Big Dye® Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, California, USA).
Results
Our final method was the result of the optimisation of many factors such as primer pair selection, magnesium amount, Taq polymerase amount, and annealing temperatures. In general, the PCR conditions were optimised separately for each polymorphism and then combined using the following steps: (i) design of specific primers, (ii) selection of annealing temperature at which the primers were specific, (iii) determination of magnesium amount and Taq polymerase amount, and (iv) different combinations of primer pairs and template dilution.
As shown in Figure 3, the primers designed for the first PCR successfully produced the desired products from the genomic DNA extracted. These were intended as templates for our second allele-specific PCR. The primers designed for the second PCR were also later found to produce the desired second PCR products (Figure 4). Furthermore, the primers designed were found to be compatible with each other with no evidence of misannealing, and the primers produced the specific products for both PCRs (Figures 3 and 4).
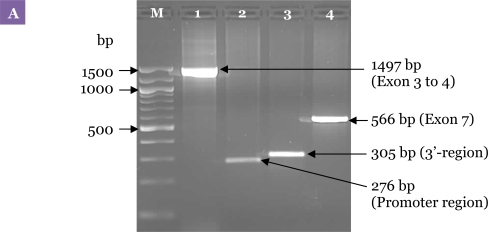
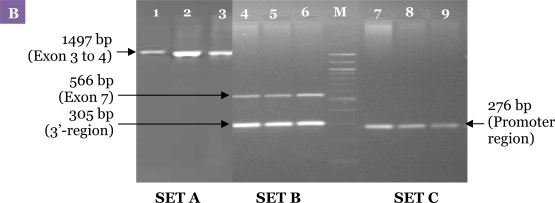
Figure 3:
Electrophoresis pattern for the first PCR carried out with the specific primers. (A) Uniplex amplification of the 4 regions of DRD2. Lane 1: exon 3 to 4 with primer sets DRD2EX3&4FW and DRD2EX3&4RV; Lane 2: the promoter region (i.e., 5′-untranslated region) with D2PRFW and D2PRRV; Lane 3: the 3′-untranslated region (i.e., TaqI A) with D2TAQ1AFW and D2TAQ1ARV; Lane 4: exon 7 with DRD2EX7FW and DRD2EX7RV. (B) Result of the first PCR of 3 DNA samples that were obtained using the optimised PCR method. Lanes 1–3: PCR amplification of Set A, exon 3 to 4; Lanes 4–6: Set B, exon 7 and the 3′-untranslated region simultaneously; Lanes 7–9: Set C, the promoter region. M: 100-bp DNA ladder.
Figure 4:
Uniplex second PCR products carried out with the specific primers for Val96Ala, Leu141Leu, Val154Ile, Pro310Ser, Ser311Cys, TaqI A, −141C Ins/Del, and A-241G polymorphisms. The genotypes of the samples are homozygous wild-type for Val96Ala, Leu141Leu, Val154Ile, Pro310Ser, Ser311Cys, and −141C Ins/Del polymorphisms, and heterozygotes for TaqI A and A-241G polymorphisms. M: 100-bp DNA ladder; Wt: wild-type; Mt: mutant-type.
A uniplex first PCR for each single locus was performed to amplify the region from exon 3 to 4, the promoter region, the 3′-region and exon 7 of DRD2 with primer sets DRD2EX3&4FW and DRD2EX3&4RV, D2PRFW and D2PRRV, D2TAQ1AFW and D2TAQ1ARV, and DRD2EX7FW and DRD2EX7RV, respectively, as shown in Figure 3A. Subsequently, a multiplex first PCR was performed using exactly the same PCR programme as with individual primer pairs except that the primer sets for all 4 regions of DRD2 were combined in a single reaction. Optimisation of the multiplex PCR method was unsuccessful due to problems such as non-reproducible amplification of the promoter region and exon 3 to 4. Finally, the problematic primer sets were separated resulting in 2 uniplex and 1 duplex first PCR sets. The methods were further optimised by reducing the amount of magnesium and Taq polymerase. The MgCl2 concentration was varied between 2.0 and 1.0 mM. The Taq polymerase concentrations used for the assay were varied from 1.0 U to 0.5 U. Figure 3B shows the agarose gel electrophoresis of the first PCR products using the final optimised method.
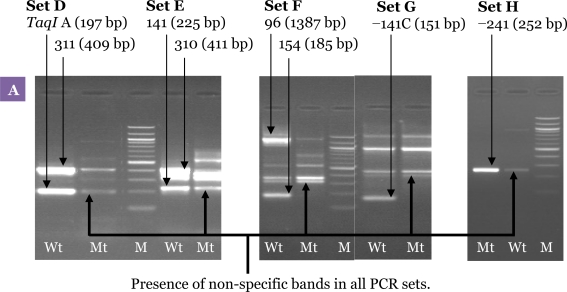
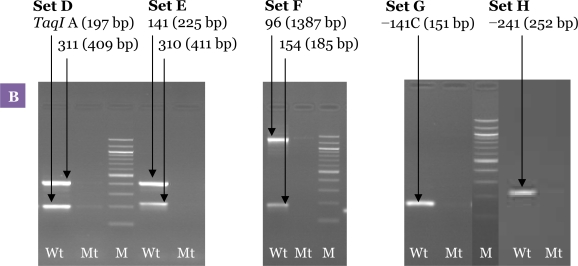
The second PCR condition was optimised separately for each polymorphism before being combined. Figure 4 shows the result of the uniplex second PCR amplification performed using the same cycling conditions. A multiplex second PCR was developed by combining the desired primer pairs at equimolar concentration in a single reaction. The first parameter that was tested and found to be important was annealing temperature (Tm). The annealing temperature was increased step-wise from 50 °C to 65 °C to eliminate the non-specific products and to avoid false negative results. We found that increasing the annealing temperature decreased the intensity of the amplified non-specific bands.
The optimum amount of MgCl2 was 1.0 mM (Figure 5). Although higher concentrations of MgCl2 increased the intensity of the desired bands, it also produced more non-specific backgrounds. A range of Taq polymerase concentrations were also tested during the experiments. With 1.0 U of Taq polymerase, although all the desired bands were amplified, non-specific bands were also observed. The non-specific bands were successfully eliminated by reducing the amount of Taq polymerase to 0.5 U (Figure 5).
Figure 5:
Optimisation of the allele-specific multiplex PCR. Agarose gel electrophoresis of the products from the second PCR showing the effect of different MgCl2 and Taq polymerase concentrations. (A) Results of the second PCR products amplified at concentration of MgCl2 2.0 mM and Taq polymerase 1.0 U showing the presence of non-specific bands in all PCR sets. (B) Second PCR products amplified at a concentration of MgCl2 1.0 mM and Taq polymerase 0.5 U. Results showed that MgCl2 and Taq polymerase concentrations have a significant effect on elimination of the non-specific bands, and all the expected bands were visualised clearly. M: 100-bp DNA ladder; Wt: wild-type; Mt: mutant-type.
Thus, the condition for optimum sensitivity, specificity, and robustness for all the desired polymorphisms for the second PCR involved the use of 1.0 mM MgCl2, 0.2 mM dNTPs, 0.5 U Taq polymerase, 2.0 μL (1:100) template, and 0.25 μM of each primer pair (Set D, E, F, G, and H) in a final volume of 25.0 μL at an annealing temperature of 63 °C (Figure 6).
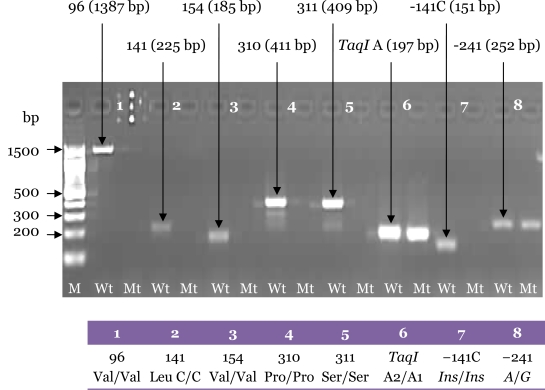
Figure 6:
Electrophoresis pattern for the second PCR amplification from 3 genomic DNA samples carried out with the specific primers using the final optimised PCR for Set D (Ser311Cys and TaqI A), Set E (Pro310Ser, Leu141Leu), Set F (Val96Ala and Val154Ile), Set G (−141C Ins/Del) and Set H (A-241G). Each pair of lanes represents 1 sample. The genotypes for each sample are shown in the tables. M: 100-bp DNA ladder; Wt: wild-type; Mt: mutant-type.
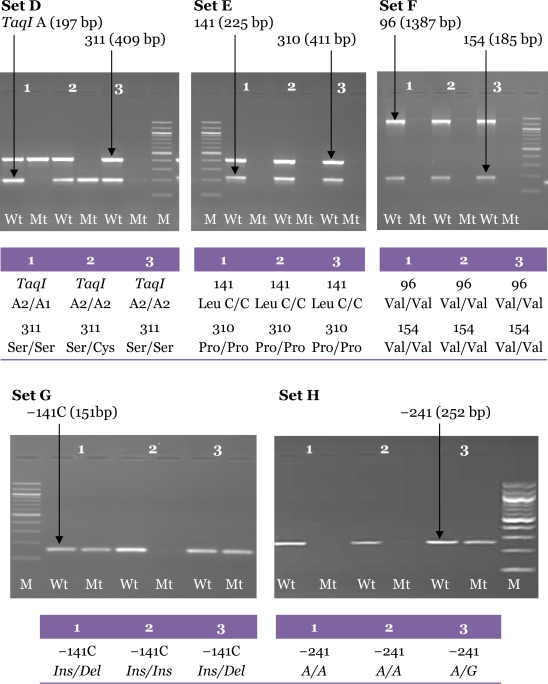
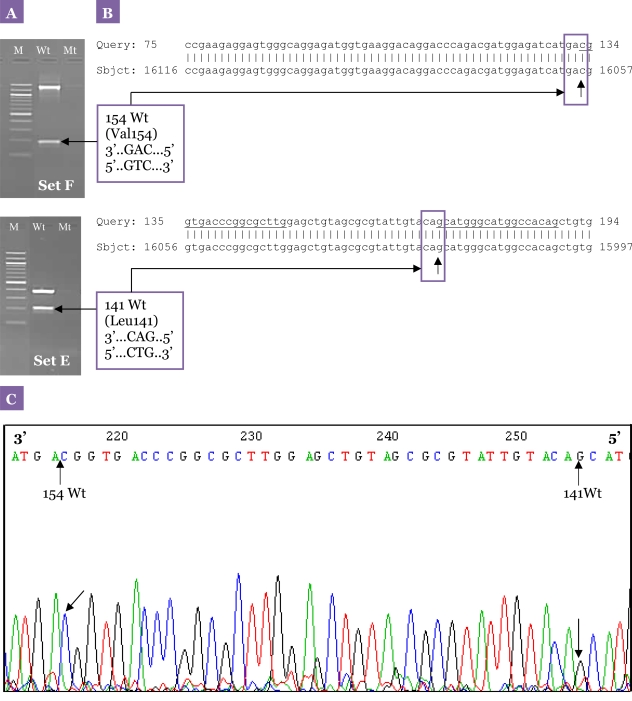
The optimised method was shown to be specific, sensitive, and reproducible when validated against the 156 DNA samples obtained from patients with schizophrenia (23). The mutation sites detected by this method were further confirmed with sequencing results. The sequencing analysis showed 100% homology to DRD2, and the results from the second PCR obtained from the samples also corresponded with the sequencing analysis. An example of our sequencing results is illustrated in Figure 7.
Figure 7:
Specificity of the primer sets for the optimised nested allele-specific PCR for DRD2 polymorphisms. (A) A representative agarose gel electrophoresis of the multiplex allele-specific PCR analysis of Set F and Set E showing no mutant-type alleles detected. M: 100-bp DNA ladder; Wt: wild-type; Mt: mutant-type. (B) Nucleotide sequence of the PCR product obtained from sequencing was aligned using BLAST. The result was then compared with gel electrophoresis results of the selected sample, which confirmed the variation sites at codon 141 and 154 of the human DRD2 for Leu141Leu and Val154Ile polymorphisms, respectively. The locations of the allele-specific second PCR primers are underlined. The nucleotide sequence is numbered from the 3′-end of DRD2 as indicated at both sides of each line. (C) Chromatogram of nucleotide sequences from the sample showing homozygous wild-type alleles for Leu141Leu and Val154Ile polymorphisms. The position of the Leu141Leu and Val154Ile polymorphisms is indicated by the mark symbols and the vertical arrows.
Discussion
Currently, the time-consuming and labour-intensive PCR–RFLP is one of the most commonly used methods for the determination of genetic polymorphisms of the DRD2 gene. Apart from requiring highly skilled personnel, if the polymorphism does not involve a restriction site change, PCR–RFLP cannot be used. In contrast, allele-specific multiplex PCRs do not require restriction enzyme cleavage and are less time-consuming as well as more user-friendly.
Apart from its advantages over PCR–RFLP, multiplex PCR, as developed in this study, is a promising technique to overcome other shortcomings of single PCR reactions and thus can increase the diagnostic capacity of PCR. In a multiplex PCR, more than 1 target sequence is amplified by including more primer pairs in a PCR reaction (24). Multiplex PCR thus has the potential to save time and reduce tedious laboratory work without compromising test accuracy. To our knowledge, this is the first report of a method for the simultaneous detection of multiple DRD2 polymorphisms using nested allele-specific multiplex PCR with the same conditions of thermal cycling.
Three main components that are important for the success of multiplex PCR are the cycling conditions, the PCR mixture, and the characteristics of the designed primers (25,26). Primers are the primary components of a PCR. The choice of specific primers will ensure the specificity of the PCR method because a non-exact match with the sample DNA may cause false amplification. We opted to use a nested multiplex PCR approach to amplify the DRD2 regions of interest in the first PCR for use as a template in the second allele-specific PCR. Primers were designed to yield PCR products of different sizes so that identification was easier by gel electrophoresis. As a rule-of-thumb, it is advisable to design primers with a similar Tm of 55 to 58 °C or higher, of 18 to 30 bp or higher in length, and of GC content of 35% to 60% for easier optimisation (24). These designed primers were aligned using BLAST programme and were shown to be 100% specific for DRD2. The second PCR gel pictures (Figure 4) showed no significant primer dimers or large allelic noise bands, which indicated that the designed primer sets were PCR compatible. The specificity of the primers was further confirmed with sequencing analysis.
Alterations in the PCR mixture and the cycling conditions resulted in improvement in the sensitivity and specificity of the method developed. The recommended PCR cycles were between 25 and 35 (27). Thirty-five cycles of PCR were used for the first PCR to ensure sensitivity. However, increasing the number of cycles will also increase amplification of non-specific products and analysis time. The cycles chosen for the first PCR were sufficient to allow for the second PCR. Subsequently, 15 cycles of PCR were performed for detection of variants.
Annealing temperature is an important parameter for PCR optimisation. If the annealing temperature is too high, no annealing can occur, and an annealing temperature that is too low will result in an increase in non-specific annealing (27). Taq polymerase is another important factor to be optimised along with magnesium and dNTPs. One of the reasons for its importance is the expense, and another reason is the fact that fidelity (nucleotide mis-incorporation frequency) of Taq polymerase depends upon the concentration of free Mg2+ and dNTP. For most PCRs, the optimum amount of Taq polymerase will be between 0.5 to 2.5 U in a 50.0 μL reaction volume (27).
In this study, 3 separate sets of the first PCR mixture were designed to amplify a region from exon 3 to 4 (Set A), exon 7 and TaqI A (Set B), and the promoter region (Set C) instead of just 1 PCR mixture. The initial 4 primer pairs in 1 set resulted in problems with the second PCR. The problems included amplification of non-specific products, multiple product yield (first PCR product carried over), high molecular weight smears, primer dimmers, and failure to amplify.
Similarly, combining the designed primer pairs to yield just 3 reaction mixtures for use in a multiplex second PCR was not successful. The combinations of primer pairs attempted were Val96Ala, Ser311Cys, and TaqI A (Set 1); Pro310Ser, Leu141Leu, and Val154Ile (Set 2); and the promoter region polymorphisms, A-241G, and −141C Ins/Del (Set 3). The resulting multiplexes were not reproducible especially for Val96Ala, Val154Ile, and −141C Ins/Del. Val96Ala and Val154Ile were taken out from the sets and combined in a separate reaction mixture. Similarly, −141C Ins/Del was separated from A-241G, which resulted in 2 uniplex mixtures for the promoter region. Other primer pair combinations were tried but were also not reproducible. Splitting the multiplex reactions into 5 final sets made optimisation of the PCR easier, and the results obtained were more specific and reproducible.
The optimised methods were found to be robust when repeated by several individuals at the laboratory at the Faculty of Pharmacy, Universiti Teknologi MARA, to test for inter-individual robustness. The gel images were analysed by 2 independent, blinded reviewers to ensure unequivocal results. Samples that did not have clear bands or equivocal results were repeated. The PCR results were subsequently confirmed by direct sequencing.
Overall, the final allele-specific multiplex PCR method was the result of condition optimisation of factors such as primer selection, quality of the DNA, magnesium amount, Taq polymerase amount, and annealing temperature. Our method was found to be specific in detecting 8 DRD2 polymorphisms (Val96Ala, Leu141Leu, Val154Ile, Ser311Cys, Pro310Ser, TaqI A, A-241G, and −141C Ins/Del). The specificity of the method was further confirmed with sequencing analysis (Figure 7).
Recently, a new genotyping method has been described that uses microchip electrophoresis for the analysis of PCR products for sizing, mutations, or polymorphisms. For example, this method can be used in detecting the methylated p16 gene in cancer patients (28), the analysis of the dopamine D4 receptor gene polymorphism (29) and the detection and identification of yeast strains (30). The microchip electrophoretic system is faster and more specific when compared with agarose gel electrophoresis (28). Barta et al. (29) confirmed the reliability of the new separation method by comparing it to the conventional slab gel and the ultra-thin-layer techniques in genotyping the 48 bp repeat polymorphism in the dopamine D4 receptor gene (DRD4). On the other hand, no differences in discriminating power or sensitivity were observed between the PCR-agarose gel electrophoresis method and the PCR-microchip electrophoresis method in identification of common and uncommon yeast strains (30). To the best of our knowledge, there has been no study in which the microchip electrophoresis analysis system was used in a clinical or research laboratory for determining the lengths of PCR products for simultaneous detection of DRD2 polymorphisms. This new approach requires sophisticated techniques and expensive equipment that are not usually available in most research and diagnostic settings, hence the limited implementation of this method. The nested multiplex allele-specific PCR in this study offers a simple, inexpensive, and specific method that does not require additional equipment or additional complexity. The only step involved after PCR is the conventional agarose gel electrophoresis for interpretation of the PCR products. Furthermore, agarose gel electrophoresis is the technique of choice for many laboratories without the need for costly reagents and equipment.
Conclusion
We have developed a nested multiplex allele-specific PCR for the simultaneous detection of DRD2 polymorphisms and sufficient information about the assay has been provided. Our ability to simultaneously amplify more than 1 locus in the same reaction in a multiplex PCR is desirable for population studies.
Acknowledgements
This study was supported by the IRPA PR (RM8) Grant No. 06-02-05-1015PR002, Ministry of Science, Technology, and Environment Malaysia under the project “Value Added Therapeutic Drug Monitoring: Application of Pharmacogenomics in the Management of Mental Illness”. We thank all the members in Pharmacogenetics Research Group, Institute for Research in Molecular Medicine, Universiti Sains Malaysia, and in Faculty of Pharmacy, Universiti Teknologi MARA, for their invaluable contributions to the project.
Footnotes
Authors’ Contributions
Conception and design, provision of study materials, analysis and interpretation of the data, critical revision and final approval of the article: ZZ, MRS, MKZJ, NM, RI
Obtaining of funding: MRS, RI
Collection and assembly of data, drafting of the article: ZZ
Administrative, technical, or logistic support: RI
References
- 1.Neville MJ, Johnstone EC, Walton RT. Identification and characterization of ANKK1: A novel kinase gene closely linked to DRD2 on chromosome band 11q23.1. Hum Mutat. 2004;23(6):540–545. doi: 10.1002/humu.20039. [DOI] [PubMed] [Google Scholar]
- 2.Jackson DM, Westlind-Danielsson A. Dopamine receptors: Molecular biology, biochemistry and behavioural aspects. Pharmacol Ther. 1994;64(2):291–370. doi: 10.1016/0163-7258(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 3.Grandy DK, Marchionni MA, Makam H, Stofko RE, Alfano M, Frothingham L, et al. Cloning of the cDNA and gene for a human D2 dopamine receptor. Proc Natl Acad Sci U S A. 1989;86(24):9762–9766. doi: 10.1073/pnas.86.24.9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eubanks JH, Djabali M, Selleri L, Grandy DK, Civelli O, McElligott DL, et al. Structure and linkage of the D2 dopamine receptor and neural cell adhesion molecule genes on human chromosome 11q23. Genomics. 1992;14(4):1010–1018. doi: 10.1016/s0888-7543(05)80124-7. [DOI] [PubMed] [Google Scholar]
- 5.Itokawa M, Arinami T, Futamura N, Hamaguchi H, Toru M. A structural polymorphism of human dopamine D2 receptor, D2 (Ser311→Cys) Biochem Biophys Res Commun. 1993;196(3):1369–1375. doi: 10.1006/bbrc.1993.2404. [DOI] [PubMed] [Google Scholar]
- 6.Gejman PV, Ram A, Gelernter J, Friedman E, Cao Q, Pickar D, et al. No structural mutation in the dopamine D2 receptor gene in alcoholism or schizophrenia. Analysis using denaturing gradient gel electrophoresis. JAMA. 1994;271(3):204–208. [PubMed] [Google Scholar]
- 7.Cravchik A, Sibley DR, Gejman PV. Functional analysis of the human D2 dopamine receptor missense variants. J Biol Chem. 1996;271(42):26013–26017. doi: 10.1074/jbc.271.42.26013. [DOI] [PubMed] [Google Scholar]
- 8.Arinami T, Gao M, Hamaguchi H, Toru M. A functional polymorphism in the promoter region of the dopamine D2 receptor gene is associated with schizophrenia. Hum Mol Genet. 1997;6(4):577–582. doi: 10.1093/hmg/6.4.577. [DOI] [PubMed] [Google Scholar]
- 9.Thompson J, Thomas N, Singleton A, Piggott M, Lloyd S, Perry EK, et al. D2 dopamine receptor gene (DRD2) Taq1 A polymorphism: Reduced dopamine D2 receptor binding in the human striatum associated with the A1 allele. Pharmacogenetics. 1997;7(6):479–484. doi: 10.1097/00008571-199712000-00006. [DOI] [PubMed] [Google Scholar]
- 10.Jonsson EG, Nothen MM, Grunhage F, Farde L, Nakashima Y, Propping P, et al. Polymorphisms in the dopamine D2 receptor gene and their relationships to striatal dopamine receptor density of healthy volunteers. Mol Psychiatry. 1999;4(3):290–296. doi: 10.1038/sj.mp.4000532. [DOI] [PubMed] [Google Scholar]
- 11.Blum K, Noble EP, Sheridan PJ, Montgomery A, Ritchie T, Jagadeeswaran P, et al. Allelic association of human dopamine D2 receptor gene in alcoholism. JAMA. 1990;263(15):2055–2060. [PubMed] [Google Scholar]
- 12.Li MD, Ma JZ, Beuten J. Progress in searching for susceptibility loci and genes for smoking-related behaviour. Clin Genet. 2004;66(5):382–392. doi: 10.1111/j.1399-0004.2004.00302.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Liu ZL, Chen B. Association study of dopamine D2, D3 receptor gene polymorphisms with motor fluctuations in PD. Neurology. 2001;56(12):1757–1759. doi: 10.1212/wnl.56.12.1757. [DOI] [PubMed] [Google Scholar]
- 14.Schindler KM, Pato MT, Dourado A, Macedo A, Azevedo MH, Kennedy JL, et al. Association and linkage disequilibrium between a functional polymorphism of the dopamine-2 receptor gene and schizophrenia in a genetically homogeneous Portuguese population. Mol Psychiatry. 2002;7(9):1002–1005. doi: 10.1038/sj.mp.4001126. [DOI] [PubMed] [Google Scholar]
- 15.Lencz T, Robinson DG, Xu K, Ekholm J, Sevy S, Gunduz-Bruce H, et al. DRD2 promoter region variation as a predictor of sustained response to antipsychotic medication in first-episode schizophrenia patients. Am J Psychiatry. 2006;163(3):529–531. doi: 10.1176/appi.ajp.163.3.529. [DOI] [PubMed] [Google Scholar]
- 16.Hauge XY, Grandy DK, Eubanks JH, Evans GA, Civelli O, Litt M. Detection and characterization of additional DNA polymorphisms in the dopamine D2 receptor gene. Genomics. 1991;10(3):527–530. doi: 10.1016/0888-7543(91)90431-d. [DOI] [PubMed] [Google Scholar]
- 17.Grandy DK, Zhang Y, Civelli O. PCR detection of the TaqA RFLP at the DRD2 locus. Hum Mol Genet. 1993;2(12):2197. doi: 10.1093/hmg/2.12.2197-a. [DOI] [PubMed] [Google Scholar]
- 18.Arinami T, Itokawa M, Enguchi H, Tagaya H, Yano S, Shimizu H, et al. Association of dopamine D2 receptor molecular variant with schizophrenia. Lancet. 1994;343(8899):703–704. doi: 10.1016/s0140-6736(94)91581-4. [DOI] [PubMed] [Google Scholar]
- 19.Hori H, Ohmori O, Shinkai T, Kojima H, Nakamura J. Association analysis between two functional dopamine D2 receptor gene polymorphisms and schizophrenia. Am J Med Genet. 2001;105(2):176–178. doi: 10.1002/ajmg.1196. [DOI] [PubMed] [Google Scholar]
- 20.Grandy DK, Litt M, Allen L, Bunzow JR, Marchionni M, Makam H, et al. The human dopamine D2 receptor gene is located on chromosome 11 at q22-q23 and identifies a TaqI RFLP. Am J Hum Genet. 1989;45(5):778–785. [PMC free article] [PubMed] [Google Scholar]
- 21.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami N, Sirugo G, Seaman MI, Bray-Ward P, Pakstis AJ, Kidd KK. Characterization of the 5′-flanking region of the DRD2 gene including a (GAAA)n STRP. Am J Hum Genet. 1998;63(Suppl):A302(1748). [Google Scholar]
- 23.Zahari Z, Teh LK, Ismail R, Razali SM. Influence of DRD2 polymorphisms on the clinical outcomes of patients with schizophrenia. Psychiatr Genet. 2011;21(4):183–189. doi: 10.1097/YPG.0b013e3283437250. [DOI] [PubMed] [Google Scholar]
- 24.Henegariu O, Heerema NA, Dlouhy SR, Vance GH, Vogt PH. Multiplex PCR: Critical parameters and step-by-step protocol. Biotechniques. 1997;23(3):504–511. doi: 10.2144/97233rr01. [DOI] [PubMed] [Google Scholar]
- 25.Zainuddin Z, Teh LK, Suhaimi AW, Salleh MZ, Ismail R. A simple method for the detection of CYP2C9 polymorphisms: Nested allele-specific multiplex polymerase chain reaction. Clin Chim Acta. 2003;336(1–2):97–102. doi: 10.1016/s0009-8981(03)00319-x. [DOI] [PubMed] [Google Scholar]
- 26.Richards CS, Bradley LA, Amos J, Allitto B, Grody WW, Maddalena A, et al. Standards and guidelines for CFTR mutation testing. Genet Med. 2002;4(5):379–391. doi: 10.1097/00125817-200209000-00010. [DOI] [PubMed] [Google Scholar]
- 27.Rolf A, Schuller I, Finckh U, Weber-Rolfs I. PCR: Diagnostics and research. Berlin (DE): Springer-Verlag Telos; 1992. [Google Scholar]
- 28.Zhou XM, Shao SJ, Xu GD, Zhong RT, Liu DY, Tang JW, et al. Highly sensitive determination of the methylated p16 gene in cancer patients by microchip electrophoresis. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;816(1–2):145–151. doi: 10.1016/j.jchromb.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 29.Barta C, Ronai Z, Nemoda Z, Szekely A, Kovacs E, Sasvari-Szekely M, et al. Analysis of dopamine D4 receptor gene polymorphism using microchip electrophoresis. J Chromatogr A. 2001;924(1–2):285–290. doi: 10.1016/s0021-9673(01)00915-3. [DOI] [PubMed] [Google Scholar]
- 30.Fujita SI, Senda Y, Nakaguchi S, Hashimoto T. Multiplex PCR using internal transcribed spacer 1 and 2 regions for rapid detection and identification of yeast strains. J Clin Microbiol. 2001;39(10):3617–3622. doi: 10.1128/JCM.39.10.3617-3622.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]