Abstract
In the last years the use of a multistarter fermentation process has been proposed to improve the organoleptic characteristics of wines. In the present study the fermentation performances and the interactions of mixed and sequential cultures of Hanseniaspora uvarum, Candida zemplinina, and a strain of Saccharomyces cerevisiae isolated from organic musts were investigated. To evaluate the oenological performances of the tested strains microvinifications in pasteurized red grape juice from Montepulciano d’Abruzzo cultivar were compared. The course of fermentation has been controlled through classical determinations (CO2 evolution, ethanol, glycerol, pH, total titratable acidity, sugar content, free sulfur dioxide (SO2), dry extract, sugars, organic acids, and volatile compounds). Moreover, the yeast population was determined by both culture-dependent and independent approaches. In particular, the pure culture of H. uvarum and C. zemplinina did not end the fermentation. On the contrary, when S. cerevisiae was added, fermentations were faster confirming that yeast interactions influence the fermentation kinetics. Moreover, C. zemplinina showed a good interaction with S. cerevisiae by increasing the fermentation kinetic in high gravity Montepulciano must, with low ethyl acetate and acetic acid production. This study confirmed that non-Saccharomyces yeasts play a crucial role also in organic wines and their activity could be modulated through the selection of appropriate strains that correctly interact with S. cerevisiae.
Keywords: non-Saccharomyces wine yeast, mixed fermentation, metabolic profile, multistarter
Introduction
Over the last decade, the phenomenon of organic products has taken hold of large segments of consumers. At its most basic level, organic wine is made from grapes that have been grown with as little human impact as possible. Organic wine is a wine obtained from organically growing grapes without the help of or need for synthetic fertilizers, synthetic plant treatments, or herbicides (Trioli and Hofmann, 2009). Accurate studies have been carried out on soil and vineyard management in organic wine making whereas, as far as we know, no or few data are available on microbial populations of grape berries from organic vineyard as well as of those from organic wines. In a previous paper (Tofalo et al., 2011) the yeast populations present on grape berries and must from organic vineyards of red Montepulciano d’Abruzzo and white Trebbiano cultivars were studied. In particular non-Saccharomyces (NS) wine yeasts were identified at species level. Moreover the strains were typed and characterized for some oenological parameters. In recent years, a lot of studies evaluated the NS species present in wine ecosystem, and demonstrated the impact of grape conditions on NS populations (Fernández et al., 2000; Raspor et al., 2006; González et al., 2007). The role of NS yeasts in wine production has been debated extensively and several researchers have shown that NS yeasts survive during fermentation and could reach cell concentrations similar to those reached by Saccharomyces cerevisiae 106–108 cells/ml (Fleet et al., 1984; Gafner and Schultz, 1996). In fact, as suggested by several authors (Zironi et al., 1993; Gil et al., 1996; Lema et al., 1996; Toro and Vazquez, 2002; Ciani et al., 2006; Viana et al., 2008), there is growing evidence that NS yeasts play an important role in wine quality. Fleet (2008) discussed the possibilities of using yeasts other than those from the genus Saccharomyces for future wine fermentations and the commercial viability of mixed cultures, because NS species have great potential to introduce appealing characteristics to wine that may improve its organoleptic quality. Consequently, the impact of NS yeasts on wine fermentation cannot be ignored. The major NS yeasts present during organic must fermentation of Trebbiano and Montepulciano cultivars were Hanseniaspora uvarum, Metschnikowia fructicola and Candida zemplinina, representing 43, 31 and 11%, respectively, of the total NS population isolated. Although the population size of these species was reduced throughout the wine fermentations, their growth was not completely suppressed and NS yeasts were still present at the end of the fermentation process (Tofalo et al., 2011). These yeasts from organic wine shared many characteristics which suggest that the strong selection pressure exerted by farming system and vine variety could have generated variability at different levels. Knowledge about the biodiversity of native yeasts is essential for the preservation and exploitation of the oenological potential of wine grape growing regions. The use of a selected multistarter (controlled mixed cultures) was proposed several years ago. In the middle of the last century, to reduce the acetic acid content of wine, Cantarelli (1955), Castelli (1969) encouraged the sequential use of Torulaspora delbrueckii (formerly known as Saccharomyces rosei) and S. cerevisiae. Later on, mixed cultures were proposed also for other objectives such as the biological deacidification of must or increase the glycerol content but one of the most investigated uses of mixed cultures relates to confer greater complexity to a wine, enhancing its organoleptic profile (Ciani et al., 2010). Several studies on mixed fermentations containing S. cerevisiae and NS wine yeasts have been carried out to evaluate the possibility of using controlled multistarter cultures to improve wine quality (Mora et al., 1990; Zironi et al., 1993; Toro and Vazquez, 2002; Ciani et al., 2006; Andorrà et al., 2010). For this purpose different NS species were used to study mixed fermentation such as Hanseniaspora guilliermondii, H. uvarum, Candida pulcherrima (Metschnikowia), Pichia kluyveri, Pichia fermentans, Candida cantarellii, T. delbrueckii, Kluyveromyces thermotolerans, Candida stellata (recently reclassified as Starmerella bombicola). The yeasts present on grape berries and must from organic vineyards could have a unique composition and these indigenous yeasts impart distinct regional and desired characteristics to wines. In this context, autochthonous NS strains could be selected to conferment organic musts alongside S. cerevisiae.
The aim of this research is to evaluate the fermentation performance and the interactions of mixed and sequential cultures of H. uvarum and C. zemplinina and a strain of S. cerevisiae isolated from organic must. The results of fermentation kinetics, secondary compound formation and sensorial analysis could be useful to formulate mixed starter cultures.
Materials and Methods
Yeast strains
Non-Saccharomyces strains (C. zemplinina STS12 and H. uvarum STS45) have been isolated in a previous study from spontaneous fermentation of organic Montepulciano d’Abruzzo and Trebbiano grapes (Tofalo et al., 2011). All of the strains have been previously analyzed by sequencing the approximately 600 bp D1/D2 region of the large (26S) ribosomal subunit using primers NL1 and NL4. These natural wine strains belong to the culture collection of the Food Science Department (University of Teramo, Italy). An autochthonous starter culture S. cerevisiae (STS1) was also used as starter culture (Tofalo et al., 2011). The strains were maintained at −80°C in glycerol 20% (v/v) and, in parallel, on agar slants under paraffin oil at 4°C.
Microvinifications
To evaluate oenological performances, the strains were tested in microvinification trials using pasteurized must without grape skin from Montepulciano d’Abruzzo cultivar (280 g/l fermentable sugars, 7.4 g/l titratable acidity (TTA) and a pH 3.2). Fermentations were conducted using several combinations of the strain S. cerevisiae STS1 with strains C. zemplinina STS12 and H. uvarum STS45. The musts were inoculated with 106 cells/ml as follows: S. cerevisiae (S), C. zemplinina (C), H. uvarum (H), C. zemplinina/S. cerevisiae (CS), H. uvarum/S. cerevisiae (HS), C. zemplinina/H. uvarum/S. cerevisiae (CHS), moreover the must was inoculated with all the three strains as follows C. zemplinina/H. uvarum/S. cerevisiae (RCHS) ratios of 25:25:50. A sequential fermentation (QS) was inoculated with 106 cells/ml C. zemplinina/H. uvarum and after 48 h S. cerevisiae was added for each strain.
The must samples (95 ml) were inoculated with 5 ml of a pre-culture grown for 48 h in the same must, as described by Tofalo et al. (2011). Fermentations were carried out in duplicate for each strain at a controlled temperature of 25°C. The kinetic fermentations were monitored daily by gravimetric determinations, evaluating the loss of weight due to the production of CO2. When the CO2 evolution stopped (i.e., at constant weight), the samples were refrigerated for 2 days at 4°C, racked, and stored at −20°C until analysis. Non-inoculated must was used as negative control.
Determination of microbial growth and differential enumeration
From each flask, samples were taken along the fermentation process to evaluate viable cell counts of the inoculated species. One hundred microliters aliquots of serial dilutions of each sample were plated on Lysine Agar (LA medium; Oxoid Unipath, Hampshire, UK) and Wallerstein Laboratory nutrient agar medium (WLN medium; Oxoid; Pallmann et al., 2001) to estimate the NS yeast and the total yeast population, respectively. Moreover, a culture-independent approach was used. S. cerevisiae, H. uvarum, C. zemplinina specific quantitative PCR (qPCR) tests were carried out according Hierro et al. (2007) and Zott et al. (2010), respectively. DNA was extracted using the DNA PowerSoil® Isolation Kit (Mobio Laboratories, Inc.) according to the manufacturer’s instructions.
Real-time amplifications were carried out in a 25 μl reaction volume, using 1× 2XIQ SYBR Green PCR Supermix (Bio-Rad, Hercules, CA, USA), 0.2 μM of each primer and 5 μl of DNA suspension. All amplifications were carried out in optical-grade 96-well plates on a Cycle IQ system (Bio-Rad). The qPCR threshold cycle (Ct) was determined automatically by the instrument. Samples with known quantities of yeast cells were prepared to generate standard curves. Sterilized grape juice was inoculated with the yeast strains, plated on WLN media for viable counts. The counted samples were immediately extracted (triplicate), as described above. The DNA obtained was used to prepare serial dilutions, from 108 to 10 cell/ml. The correlation coefficient between Ct and count values was analyzed and interpreted using the appropriate Microsoft Excel function. Each Ct was the average of four measures obtained by amplifying four DNA extracts from the same artificially inoculated sample. In all PCR runs, negative controls (sterilized water), positive controls, and samples were run in triplicate. Sensitivity of qPCR assays was evaluated with reference to other reports (Hierro et al., 2006).
Physico-chemical determinations
The main products (ethanol, glycerol, pH, TTA, sugar content, free sulfur dioxide (SO2) and dry extract of wine), and must under fermentation were determined on samples taken at the end of fermentation following the official International Organization of Vine and Wine (2011) methods of analysis. Organic acid, glucose, and fructose concentrations were determined according to Tofalo et al. (2011) and Lopez-Tamames et al. (1996), respectively. Biogenic amines production was determined according to Tofalo et al. (2007). All analyses were performed in triplicate.
Solid phase microextraction–gas chromatography analysis of volatile compounds
Five milliliters of wine samples were placed in 10 ml glass vials with 1 g NaCl and 10 μl of 4-methyl-2-pentanol (final concentration 4 mg/l) were added as internal standard. Both equilibration and adsorption phases were carried out by stirring for 30 min at 40°C. A carboxen–polydimethylsiloxane-coated fiber (85 μm) was used (Sigma-Aldrich, St. Louis, MO, USA). Under the extraction conditions described above, the recovery of the volatile compounds was between 88.9 and 103.5%. For quantitative determination, a CP 380 capillary gas chromatograph equipped with a 8200 autosampler SPME III (Varian, Italy) was used. The fused silica capillary column was a CP-Wax 52 CB (50 m × 0.32 mm) by Crompack (Netherlands), coated with polyethylene glycol (film thickness 1.2 μm), as stationary phase. The injector and FID temperature was 250°C. After extraction, the fiber was placed in the injector of the GC for 15 min. The temperature program was the following: initial temperature (50°C) held for 2 min; first ramp, 1°C min to 65°C (0 min hold); second ramp, 10°C min to 150°C (10 min hold); third ramp 10°C min to 200°C (1 min hold). The carrier gas (N2) flow rate was 2.5 ml/min. The aroma compounds were identified by comparing the retention time of standards and their identification was confirmed by using GC–MS. GC–MS analysis was performed using a GC–mass spectrometer Finnigan Trace DSQ Quadrupole (Thermo Finnigan, San Jose, CA, USA). Mass spectrometer conditions were: Ion Source: electron ionization (EI), Ion Polarity: POS, Ion Source Temperature: 250°C, MS transfer line: 220°C, Turbomolecular Pump: 70 l/s, Acquisition: full Scan, Mass range: 30–400 m/z, Carrier gas: He. The data were processed using Xcalibur Data System Software 1.4.1 SP3 (Thermo Finnigan, San Jose, CA, USA). The quantitative analysis of wine aroma compounds was carried out on the basis of the relative peak area (Qi) calculated from head space SPME (HS/SPME) gas chromatograms after addition of know amounts of analyte standards, as well as the internal standard according to De la Calle-Garcia et al. (1998). The chemical analyses were carried out in the same period of the sensory analysis. Each determination was carried out in duplicate. The data presented are the means of three determinations. All reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA), with a purity greater than 99%.
Statistical analysis
The mean and SD were calculated for each experimental parameter. Principal component analysis (PCA) was performed using statistical software STATISTICA for Windows (STAT. version 8.0, StatSoft Inc., Tulsa, OK, USA).
Results
Course of fermentation and development of yeasts during fermentations
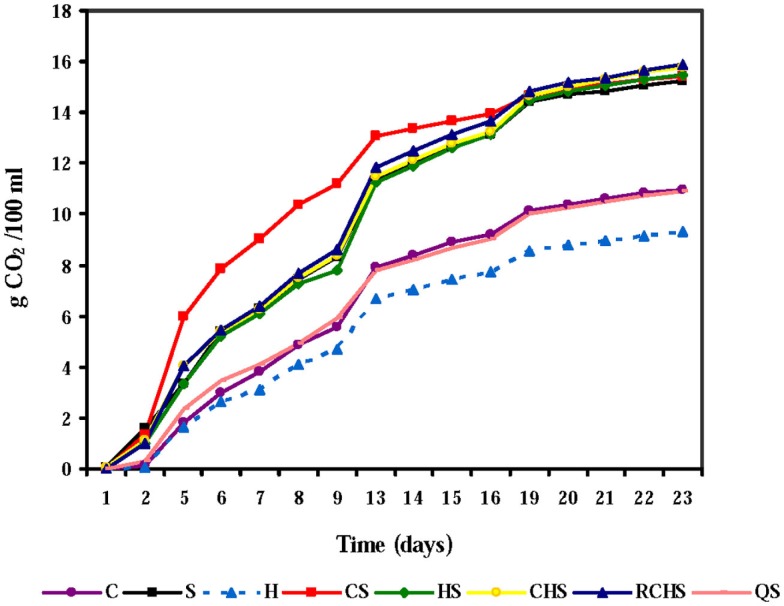
The course of fermentation rates with pure, mixed, and sequential cultures is given in Figure 1. The pure culture of H. uvarum was unable to finish fermentation according to the poor fermentative capacity of this species, but also the pure culture of C. zemplinina did not end the fermentation. The yeast interactions had a clear impact on the fermentation kinetics and the presence of S. cerevisiae gave faster fermentations.
Figure 1.
Fermentation kinetics of pure, mixed, and sequential starter cultures in Montepulciano d’Abruzzo musts. C, C. zemplinina; S, S. cerevisiae; H, H. uvarum; CS, C. zemplinina/S. cerevisiae; HS, H. uvarum/S. cerevisiae; CHS, C. zemplinina/H. uvarum/S. cerevisiae; RCHS, C. zemplinina/H. uvarum/S. cerevisiae; QS, sequential fermentation.
In fact, fermentation kinetics of mixed cultures, HS, CHS, RCHS were comparable to those of S. cerevisiae pure culture. Only the trial CS showed a great improvement of fermentation kinetic during the first 15 days of fermentation highlighting the good association between C. zemplinina and S. cerevisiae. The RCHS trial, characterized by the inoculum of C. zemplinina and H. uvarum together with S. cerevisiae, showed a worst fermentation kinetic than CS trial probably for the lower proportion of cells of the two NS with respect to S. cerevisiae. The sequential trial QS presented a dramatic decrease of fermentation rate probably because S. cerevisiae was inoculated 48 h after the inoculum of C. zemplinina and H. uvarum.
The viable counts of yeast populations of the eight trials are reported in Table 1. The yeast populations of pure cultures of S. cerevisiae and C. zemplinina were similar, reaching maximum population around 107 CFU/ml after 48 h. In the case of H. uvarum pure culture the maximum population (around 106 CFU/ml) was reached after 48 h, but starting from the 14th day it started to decrease at 105 CFU/ml and was not more countable on the 24th day. Regarding the mixed fermentations HS and CS, NS yeasts were not found at the 14th day, whereas S. cerevisiae delayed the reaching of maximum population. As regards the viable cells of the three species during mixed fermentation CHS and RCHS, differences were observed only in the latest one. In fact H. uvarum disappeared after 48 h, whereas C. zemplinina increased up to 105 CFU/ml and S. cerevisiae reached maximum population of about 107 CFU/ml on the 14th day. In general, S. cerevisiae quickly reached its maximum population and kept stable during the vinifications, with the exception of sequential fermentation QS during which S. cerevisiae was not able to grow after the 48 h growth of NS wine yeast. In fact, the number of viable cells of S. cerevisiae decreased up to 105 CFU/ml at 24th day. H. uvarum and C. zemplinina reached the maximum population of 107 CFU/ml instead after 48 h, but during the progress of fermentation the first one was not more countable whereas C. zemplinina decreased up to 106 CFU/ml.
Table 1.
Yeast counts (log CFU/ml) and quantification by qPCR (in brackets) in pure, mixed, and sequential fermentation of organic Montepulciano d’Abruzzo musts.
| Trial | Time (days) | Strains |
||
|---|---|---|---|---|
| S. cerevisiae | C. zemplinina | H. uvarum | ||
| C | 0 | 6.79 ± 0.05* | ||
| 2 | 7.09 ± 0.04 | |||
| 14 | 7.36 ± 0.04 | |||
| 24 | 5.64 ± 0.06 | |||
| S | 0 | 6.88 ± 0.03 | ||
| 2 | 7.21 ± 0.03 | |||
| 14 | 7.27 ± 0.01 | |||
| 24 | 7.15 ± 0.04 | |||
| H | 0 | 6.68 ± 0.02 | ||
| 2 | 6.87 ± 0.03 | |||
| 14 | 5.32 ± 0.03 | |||
| 24 | nd | |||
| CS | 0 | 6.82 ± 0.04 | 6.85 ± 0.02 | |
| 2 | 6.50 ± 0.02 | 6.08 ± 0.11 | ||
| 14 | 6.74 ± 0.07 | nd | ||
| 24 | 7.11 ± 0.04 | nd | ||
| HS | 0 | 6.89 ± 0.04 | 6.68 ± 0.02 | |
| 2 | 6.67 ± 0.02 | 5.32 ± 0.03 | ||
| 14 | 7.08 ± 0.02 | nd | ||
| 24 | 6.78 ± 0.014 | nd | ||
| CHS | 0 | 6.88 ± 0.03 (6.98 ± 0.03) | 6.81 ± 0.02 (6.34 ± 0.03) | 6.67 ± 0.01 (6.45 ± 0.0.4) |
| 2 | 6.87 ± 0.03 (7.24 ± 0.01) | 6.69 ± 0.03 (6.45 ± 0.09) | 6.29 ± 0.02 (6.39 ± 0.01) | |
| 14 | 7.09 ± 0.04 (7.48 ± 0.04) | nd (4.03 ± 0.03) | nd (5.28 ± 0.01) | |
| 24 | 6.72 ± 0.03 (7.48 ± 0.01) | nd (3.76 ± 0.01) | nd (4.83 ± 0.02) | |
| RCHS | 0 | 6.88 ± 0.03 (6.98 ± 0.04) | 3.84 ± 0.01 (3.36 ± 0.05) | 3.68 ± 0.01 (3.50 ± 0.11) |
| 2 | 7.08 ± 0.02 (7.24 ± 0.01) | 5.06 ± 0.08 (5.01 ± 0.01) | nd (3.58 ± 0.03) | |
| 14 | 7.44 ± 0.02 (7.48 ± 0.04) | 5.04 ± 0.04 (5.09 ± 0.04) | nd (5.60 ± 0.07) | |
| 24 | 7.46 ± 0.04 (7.48 ± 0.01) | nd (4.91 ± 0.02) | nd (5.40 ± 0.09) | |
| QS | 0 | – | 6.85 ± 0.02 (6.45 ± 0.02) | 6.68 ± 0.02 (6.34 ± 0.03) |
| 2 | 6.9 ± 0.01 (6.98 ± 0.01) | 7.16 ± 0.03 (7.24 ± 0.01) | 7.19 ± 0.01 (7.35 ± 0.02) | |
| 14 | 6.14 ± 0.04 (6.15 ± 0.02) | 6.89 ± 0.01 (6.90 ± 0.04) | nd (4.43 ± 0.01) | |
| 24 | 5.92 ± 0.04 (7.16 ± 0.01) | 6.08 ± 0.02 (6.29 ± 0.01) | nd (4.05 ± 0.1) | |
C, C. zemplinina; S, S. cerevisiae; H, H. uvarum; CS, C. zemplinina/S. cerevisiae; HS, H. uvarum/S. cerevisiae; CHS, C. zemplinina/H. uvarum/S. cerevisiae; RCHS, C. zemplinina/H. uvarum/S. cerevisiae; QS, Sequential Fermentation.
*Data are expressed as average ± SD; nd, not inoculated.
Quantification of multistarter population during fermentation by qPCR
As culture-dependent techniques can underestimate the size and the diversity of a given population because they do not account for non-cultivable populations, the data on S. cerevisiae and NS populations during mixed fermentation obtained by plating were compared with those obtained by qPCR, as reported in Section “Materials and Methods.” In particular the trials CHS, RCHS, and QS were analyzed by real-time PCR and the obtained results are reported in Table 1. S. cerevisiae resulted to be always present at high level (106–107 cells/ml), but also H. uvarum and C. zemplinina were found at the end of fermentation, generally at about 104 cells/ml. In particular H. uvarum at the end of fermentation of trail CHS showed a Ct value of 25, corresponding to 104 cells/ml (data not shown).
Main fermentation products and volatile compounds
Table 2 reports some oenological parameters of the pure culture, mixed, and sequential fermentations. The pure cultures of H. uvarum and C. zemplinina did not finish the fermentations, leaving in the medium glucose and fructose, and only glucose, respectively. Also trial QS had 50 g/l of residual glucose, as expected by the analysis of fermentation kinetics. The highest ethanol concentration was determined in pure culture of S. cerevisiae. All the multistarter fermentations reached a lower ethanol concentration ranging from 8.34 to 9.38%. On the contrary the production of glycerol was greater by NS yeasts and in mixed cultures. Despite the high production of acetic acid in pure culture, H. uvarum did not increase volatile acidity in multistarter fermentations. No significant differences were found in the other compounds. Some discrepancies were found among ethanol, residual sugars, and other secondary metabolic compounds. What is stricking in our experiments was that all the strains showed an extremely poor ethanol yield from sugar consumed, which cannot be explained by the overproduction of any other metabolic products investigated in this study. Similar results have been previously obtained by Magyar and Toth (2011) and Tofalo et al. (2012).
Table 2.
Chemical profile of wines obtained with pure, mixed, and sequential starter cultures.
| Parameters | Wines |
|||||||
|---|---|---|---|---|---|---|---|---|
| C | S | H | CS | HS | CHS | RCHS | QS | |
| pH | 3.14 ± 0.02a | 3.06 ± 0.01bc | 3.26 ± 0.01d | 3.19 ± 0.01a | 3.05 ± 0.06bc | 3.03 ± 0.01c | 3.08 ± 0.01b | 3.08 ± 0.01b |
| Ethanol (% v/v) | 8.96 ± 0.02b | 9.43 ± 0.03e | 5.64 ± 0.02c | 9.38 ± 0.01f | 9.14 ± 0.03g | 8.93 ± 0.02b | 8.78 ± 0.01d | 8.35 ± 0.01a |
| Residual sugars (g/l) | 41.83 ± 0.04d | 21.67 ± 0.03b | 76.82 ± 0.02h | 24.57 ± 0.03a | 23.15 ± 0.01c | 43.95 ± 0.04f | 42.55 ± 0.01g | 48.25 ± 0.05e |
| Total titratable acidity (g/l tartaric acid) | 5.55 ± 0.04d | 5.33 ± 0.02a | 5.47 ± 0.02b | 5.82 ± 0.01f | 5.15 ± 0.04c | 5.65 ± 0.04g | 5.44 ± 0.01b | 6.05 ± 0.05e |
| Volatile acidity (g/l acetic acid) | 0.78 ± 0.04b | 0.67 ± 0.01a | 1.23 ± 0.03g | 0.84 ± 0.01bc | 0.86 ± 0.05c | 0.84 ± 0.01bc | 0.98 ± 0.01d | 0.72 ± 0.01a |
| Glycerol (g/l) | 10.20 ± 0.01e | 5.24 ± 0.05b | 10.33 ± 0.03h | 9.26 ± 0.04f | 8.14 ± 0.05a | 7.88 ± 0.03c | 8.25 ± 0.05d | 8.92 ± 0.01g |
| Dry extract (g/l) | 89.34 ± 0.04b | 87.76 ± 0.03f | 83.37 ± 0.02d | 89.29 ± 0.00b | 91.97 ± 0.01e | 78.72 ± 0.01c | 83.28 ± 0.01a | 84.64 ± 0.04g |
| Free SO2 (mg/l) | 6.10 ± 0.03d | 4.09 ± 0.01b | 3.23 ± 0.04c | 4.05 ± 0.06b | 4.05 ± 0.05b | 3.87 ± 0.04a | 6.44 ± 0.06g | 3.23 ± 0.04c |
| Malic acid (g/l) | 0.42 ± 0.01bc | 0.47 ± 0.01a | 0.44 ± 0.01cd | 0.40 ± 0.02b | 0.48 ± 0.02a | 0.45 ± 0.01ad | 0.39 ± 0.01b | 0.41 ± 0.01bc |
| Tartaric acid (g/l) | 5.85 ± 0.03g | 5.53 ± 0.04a | 5.26 ± 0.03b | 5.71 ± 0.03d | 6.55 ± 0.02f | 5.34 ± 0.05bc | 5.34 ± 0.01c | 5.48 ± 0.04a |
| Citric acid (g/l) | 0.15 ± 0.01b | 0.16 ± 0.01b | 0.15 ± 0.01b | 0.15 ± 0.01b | 0.16 ± 0.01b | 0.15 ± 0.02b | 0.16 ± 0.01b | 0.14 ± 0.01b |
| Acetic acid (g/l) | 0.62 ± 0.01b | 0.66 ± 0.03b | 0.66 ± 0.03b | 0.73 ± 0.01c | 0.54 ± 0.01d | 0.84 ± 0.02g | 0.74 ± 0.02ba | 0.78 ± 0.01a |
C, C. zemplinina; S, S. cerevisiae; H, H. uvarum; CS, C. zemplinina/S. cerevisiae; HS, H. uvarum/S. cerevisiae; CHS, C. zemplinina/H. uvarum/S. cerevisiae; RCHS, C. zemplinina/H. uvarum/S. cerevisiae; QS, Sequential Fermentation.
*Data are expressed as average ± SD. Different letters (a–h) in the same row correspond to statistically significant differences (P < 0.05).
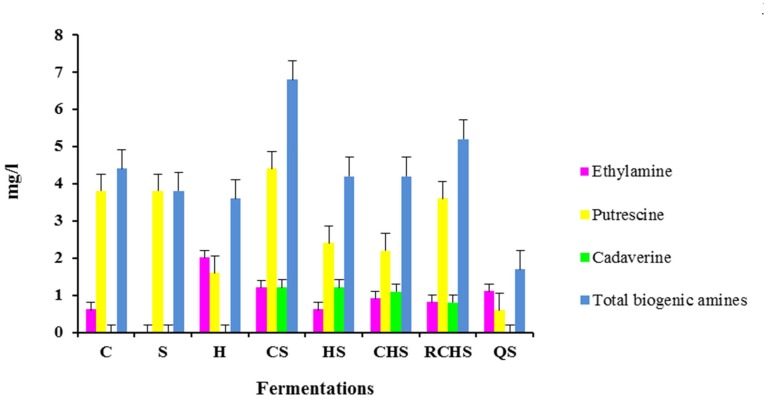
Biogenic amines were detected at low levels in all eight microvinifications (Figure 2). In particular, the trial CS has the highest value of biogenic amines and QS has the lowest value. The pure cultures of H. uvarum, C. zemplinina, and S. cerevisiae did not produce cadaverine that was formed in all multistarter fermentations with the exception of QS. Tyramine, histamine, 2-phenylethylamine, spermidine, and methylamine were not detectable in any the samples analyzed.
Figure 2.
Biogenic amines content in wines obtained with pure, mixed, and sequential starter cultures. C, C. zemplinina; S, S. cerevisiae; H, H. uvarum; CS, C. zemplinina/S. cerevisiae; HS, H. uvarum/S. cerevisiae; CHS, C. zemplinina/H. uvarum/S. cerevisiae; RCHS, C. zemplinina/H. uvarum/S. cerevisiae; QS, sequential fermentation.
Table 3 shows some volatile compounds that well discriminated the aromatic profiles of the three wine yeast species. Pure cultures of C. zemplinina and S. cerevisiae produced low quantities of ethyl acetate and acetoin and high amounts of isoamyl alcohols and β-phenylethanol, having C. zemplinina the highest productions with respect to S. cerevisiae. In this study H. uvarum pure culture produced high amounts of ethyl acetate and acetoin and very low amounts of isoamyl alcohols and β-phenylethanol. The fermentations of mixed cultures CS, HS, and RCHS presented low levels of ethyl acetate and decreased the quantity of isoamylic alcohols and 2-phenylethanol with respect to S. cerevisiae. The fermentation of mixed culture CHS presented the highest formation of isoamyl alcohols and ethyl acetate. The sequential fermentation QS showed a completely different situation among the multistarter fermentations, with the lowest content of ethyl acetate and isoamyl alcohols and the highest one of acetoin, 2-phenylethanol, and isobutyl alcohol.
Table 3.
Aromatic compounds (mg/l) of the wines obtained with pure, mixed, and sequential starter cultures.
| Aromatic compounds (%) | Wines |
|||||||
|---|---|---|---|---|---|---|---|---|
| C | S | H | CS | HS | CHS | RCHS | QS | |
| Ethyl acetate | 0.644 ± 0.001a | 1.850 ± 0.003b | 5.384 ± 0.002c | 0.441 ± 0.003d | 0.674 ± 0.001e | 1.959 ± 0.001f | 0.511 ± 0.003g | 0.369 ± 0.004h |
| Acetoin | 0.020 ± 0.001dg | 0.027 ± 0.002h | 0.533 ± 0.002b | 0.028 ± 0.004h | 0.075 ± 0.001a | 0.023 ± 0.003h | 0.014 ± 0.004d | 0.479 ± 0.003e |
| Isoamyl alcohols | 8.673 ± 0.003f | 5.104 ± 0.001e | 1.237 ± 0.004h | 4.590 ± 0.001a | 3.750 ± 0.001g | 9.283 ± 0.004c | 5.174 ± 0.004b | 2.528 ± 0.001d |
| n-Heptane | 0.264 ± 0.069d | 0.839 ± 0.001e | 1.283 ± 0.002f | 0.761 ± 0.001a | 0.913 ± 0.003b | 1.663 ± 0.002c | 0.106 ± 0.004h | 0.692 ± 0.002g |
| n-Butanol | 0.107 ± 0.001b | 0.058 ± 0.001d | 0.025 ± 0.004h | 0.082 ± 0.003a | 0.018 ± 0.001g | 0.093 ± 0.003e | 0.062 ± 0.001d | 0.028 ± 0.001h |
| 1-Hexanol | 0.031 ± 0.0dg | 0.026 ± 0.003hd | 0.027 ± 0.003hd | 0.018 ± 0.002e | 0.00 ± 0.0a | 0.025 ± 0.001h | 0.00 ± 0.0a | 0.034 ± 0.005g |
| 1-Octanol | 16.933 ± 0.004f | 0.170 ± 0.003a | 0.049 ± 0.003h | 0.085 ± 0.002g | 0.218 ± 0.001e | 0.043 ± 0.003h | 0.059 ± 0.003d | 0.407 ± 0.003b |
| Phenylethyl alcohol | 2.928 ± 0.004c | 2.268 ± 0.002b | 0.00 ± 0.00h | 1.383 ± 0.003d | 1.859 ± 0.002e | 1.469 ± 0.003g | 1.684 ± 0.001a | 2.394 ± 0.002f |
| Isobutyl alcohol | 0.277 ± 0.003e | 0.285 ± 0.002b | 0.219 ± 0.001g | 0.190 ± 0.001d | 0.663 ± 0.002f | 0.267 ± 0.003a | 0.156 ± 0.001h | 1.020 ± 0.001c |
C, C. zemplinina; S, S. cerevisiae; H, H. uvarum; CS, C. zemplinina/S. cerevisiae; HS, H. uvarum/S. cerevisiae; CHS, C. zemplinina/H. uvarum/S. cerevisiae; RCHS, C. zemplinina/H. uvarum/S. cerevisiae; QS, sequential fermentation.
Data are expressed as average ± SD. Different letters (a–h) in the same row correspond to statistically significant differences (P < 0.05).
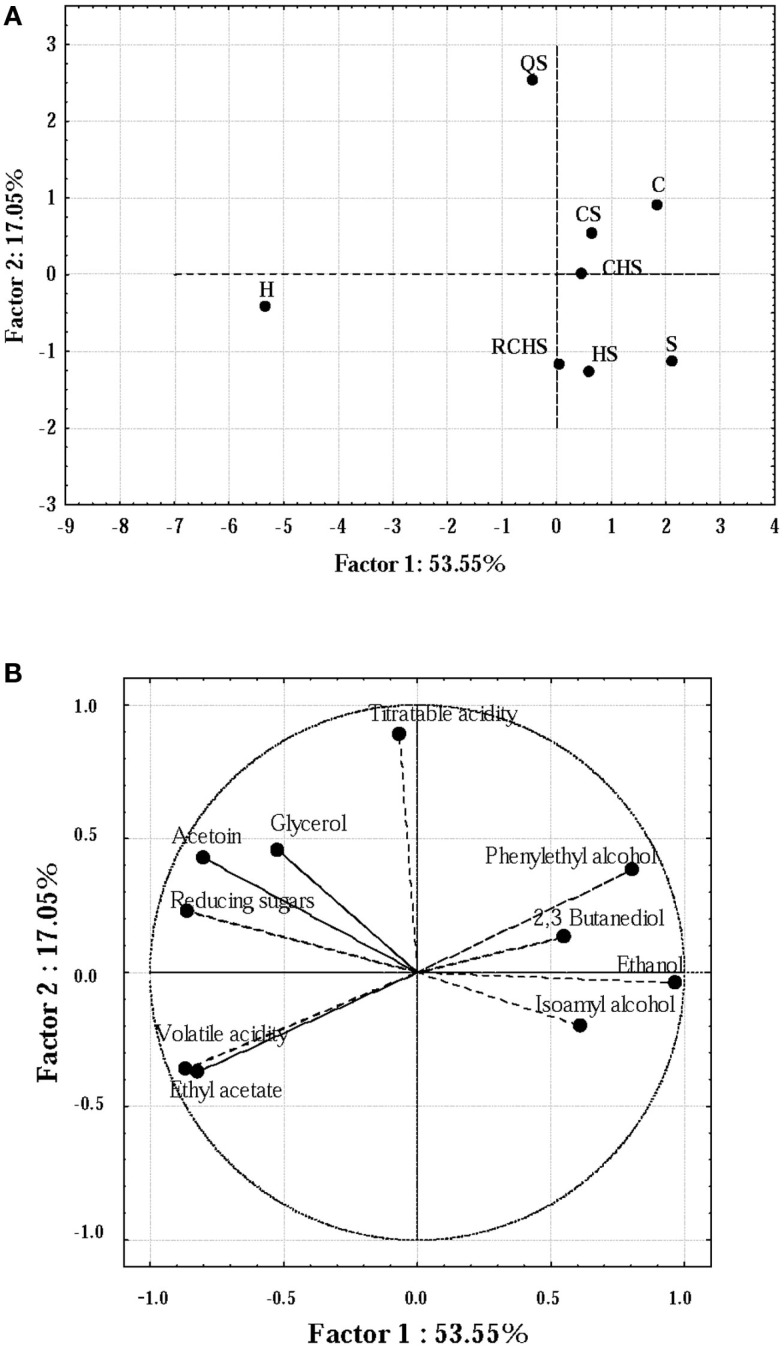
Principal component analysis has been used to obtain biochemical fingerprints of wines and to elucidate differences in the different components of wine fermented by different strains of Saccharomyces or by multistarter cultures (Nurgel et al., 2002; Howell et al., 2006).
Some compounds produced by the yeast pure and multistarter cultures were analyzed using PCA (Figures 3A,B). Firstly, the correlation matrix was computed in order to discriminate the variables, thus selecting 10 parameters (acetoin, 2,3 butanediol, ethanol, ethyl acetate, titratable and volatile acidity, glycerol, reducing sugars, phenylethyl alcohol and isoamyl alcohols). The PCA explained 70.6% of the total variance. PC 1 accounted for 53.55% of the variance and the negative segment of loading plot for this dimension (Figure 3B) was closely related to the levels of volatile acidity, whereas its positive counterpart was mainly related to ethanol. PC2 explained 17.05% of the variance; this dimension was mainly related positively with titratable acidity. Then, in score plot (Figure 3A) it is possible to distinguish three different groups of wines produced by strains. The fermentation carried out by H. uvarum was well differentiated from the others for the highest concentration of ethyl acetate, glycerol, acetoin, and volatile acidity; whereas C, CS, and CHS were very similar except for the concentration of isoamyl alcohols, 2,3 butanediol and reducing sugars (in C and CS, the values were higher than in CHS).
Figure 3.
Score plot (A) and loading plot (B) of the first and second principal components (PC) after PC analysis by the yeast pure and multistarter cultures. C, C. zemplinina; S, S. cerevisiae; H, H. uvarum; CS, C. zemplinina/S. cerevisiae; HS, H. uvarum/S. cerevisiae; CHS, C. zemplinina/H. uvarum/S. cerevisiae; RCHS, C. zemplinina/H. uvarum/S. cerevisiae; QS, sequential fermentation.
Discussion
One of the most studied technologically advances in wine making is the inoculation of grape juice with mixed cultures of S. cerevisiae and NS yeasts. In this study the fermentation kinetics and metabolic compounds produced by multistarters during fermentation of organic musts were compared. Generally the fermentation kinetics of pure cultures of S. cerevisiae, H. uvarum, and C. zemplinina were in agreement with those expected and reported in other studies (Egli et al., 1998; Toro and Vazquez, 2002; Zohre and Erten, 2002; Mendoza et al., 2007; Fleet, 2008; Ciani et al., 2010). The pure culture of H. uvarum was unable to finish fermentation according to the poor fermentative capacity of this species, but also the pure culture of C. zemplinina did not end the fermentation. These two strains were selected on the basis of their fermentative capacity in a must containing 180 g/l sugars (Tofalo et al., 2011) and probably the metabolic fermentative products from the high sugar Montepulciano must (280 g/l) affected their performances. In particular, C. zemplinina has been reported to complete fermentation of Macabeo must containing 180 g/l sugars (Andorrà et al., 2010) even if with a slight delay compared to the S. cerevisiae fermentation (Sipiczki et al., 2005; Tofalo et al., 2009, 2012). The yeast interactions had a clear impact on the fermentation kinetics and the presence of S. cerevisiae gave faster fermentations. However C. zemplinina and S. cerevisiae association (trial CS) showed a great improvement of kinetic during the first 15 days of fermentation. C. zemplinina is an osmotolerant and fructophilic yeast, generally producing low amounts of acetic acid and relevant quantities of glycerol from sugar fermentation (Sipiczki et al., 2005; Magyar and Toth, 2011; Tofalo et al., 2011), it could be suggested that it will be able to consume sugars at the very beginning of the fermentation, alleviating the S. cerevisiae from the osmotic stress, thereby improving also the fermentation kinetics (Rantsiou et al., 2012). As previously demonstrated for S. cerevisiae species, different strains of C. zemplinina can have a specific effect, as reported by Tofalo et al. (2012). Moreover this positive interaction on fermentation kinetics between S. cerevisiae and C. zemplinina can have an important application on must with high sugar contents or special wines such as icewine, “passito,” botrytized wines (Rantsiou et al., 2012). On the contrary the inoculum of C. zemplinina and H. uvarum together with S. cerevisiae (RCHS) did not produce the same results of CS trial, probably for the lower proportion of cells of the two NS with respect to S. cerevisiae.
The inoculum of S. cerevisiae after 48 h since that of C. zemplinina and H. uvarum (trial QS) produced a stuck fermentation Similar results have been reported in other studies, even if the mechanisms of this performance reduction of S. cerevisiae have not yet been explained. When S. cerevisiae and C. cantarelli interact during fermentation, the maximum S. cerevisiae population decreases (Toro and Vazquez, 2002). This fact may be due to amino acid and vitamin consumption during the first days of fermentation that can disable the subsequent growth and fermentative capacity of S. cerevisiae (Fleet, 2003). On the other hand these data were confirmed by the plating counts. S. cerevisiae dominated all the multistarter fermentations a part trial QS in which C. zemplinina had its best performance. The higher ability of S. cerevisiae to withstand the stress conditions such as increasing ethanol, decreasing pH, nutrition depletion is generally considered to drive the wine yeast population dynamics (Pretorius, 2000). These selective pressures are currently questioned, whereas cell–cell interactions are being put forward as significant in affecting yeast succession (Ciani and Pepe, 2002; Fleet, 2003; Nissen et al., 2003). The early death of C. zemplinina and H. uvarum during mixed fermentation, even if with quantitative differences, appeared to be due to the antagonistic effect of S. cerevisiae as reported also by Andorrà et al. (2010) in the same species. The NS, in mixed fermentations, decreased as fermentation proceeded. On the contrary other authors (Ciani et al., 2006; Mendoza et al., 2007) reported that the presence of both Saccharomyces and NS yeasts promotes an increase in the persistence of NS yeasts during fermentation process. However it is clear that S. cerevisiae has a relevant antagonist effect upon C. zemplinina and H. uvarum, but also that the same NS can affect S. cerevisiae, depending on the sequence of growth, the number of viable cells and strain specificity. Few studies have been carried out to elucidate the mechanisms of these antagonistic phenomena (Nissen et al., 2003; Arneborg et al., 2005; Pérez-Nevado et al., 2006). During alcoholic fermentation yeasts can produce compounds that can have inhibitory effects against other yeast species or strains, such as short to medium-chain fatty acids (Ludovico et al., 2001; Fleet, 2003), killer toxins (Schmitt and Breinig, 2002), growth and nutritional conditions (Fleet, 2003). However in the above-mentioned studies, cell–cell contact-mediated mechanism appear to effect antagonism among yeasts. To confirm the data obtained by plating counts qPCR was used as culture-independent method. In fact sub lethally injured and/or viable but non-culturable (VBNC) cells, may fail to grow on plates and are common in wine (Millet and Lonvaud-Funel, 2000; Andorrà et al., 2011). The qPCR method used was previously developed to monitor the yeast evolution during alcoholic fermentations (Hierro et al., 2007; Andorrà et al., 2011). However, qPCR targeted at DNA quantifies also dead yeasts because of the DNA’s stability (Hierro et al., 2006). Our data confirmed those of Andorrà et al. (2011) who found high populations of H. uvarum (up to 108 cells/ml) throughout the mixed fermentations by qPCR methods. Of course these findings do not resolve the question if these populations correspond to VBNC, injured and/or dead cells or how many of these cells are metabolically active and how these in turn can influence the final wines (Andorrà et al., 2011). This aspect needs further researches and advances. When some yeasts develop together under fermentation conditions, they do not passively coexist, but rather they interact and can produce different levels of fermentation products, which can affect the chemical and aromatic composition of wines (Howell et al., 2006; Anfang et al., 2009). Grape must fermentations performed by pure, mixed or sequential cultures of NS yeasts with S. cerevisiae can produce wines with significant differences in the chemical composition (Herraiz et al., 1990; Ciani and Picciotti, 1995; Gil et al., 1996; Lambrechts and Pretorius, 2000; Rojas et al., 2003; Romano et al., 2003; Moreira et al., 2008). A relevant consequence of the NS yeasts was the increased presence of glycerol in all the multistarter fermentations, due the intrinsic characteristic of C. zemplinina and H. uvarum. In similar way acetic acid production is considered as a common pattern in apiculate yeasts that for this reason have been considered for long time as spoilage yeasts (Romano et al., 2003). Despite the high production of acetic acid in pure culture, H. uvarum did not increase volatile acidity in multistarter fermentations. These results are in complete agreement with those reported by other authors (Ciani et al., 2006; Mendoza et al., 2007; Andorrà et al., 2010). As regards biogenic amines, few studies have been conducted on their formation by yeasts, comparing different yeast species and quantifying only histamine (Torrea and Ancín, 2002). Their presence in wine ranged from a few milligrams per liter to about 50 mg/l depending on the wine. This study confirmed the low amino-decarboxylase of wine yeasts, even if some differences were determined in the eight trials. Even if a number of authors suggest that yeasts do not appear to be responsible for the production of most amines found in industrial commercial red wines (Marcobal et al., 2006; Smit et al., 2008), the yeast contribution to biogenic amine production could therefore be indirect: yeasts can alter the composition of grape musts by using some amino acids and secreting others during alcoholic fermentation and autolysis, thereby changing the concentration of precursor amino acids in the wine that can be used by other microorganisms in subsequent fermentation steps (Soufleros et al., 1998). According to the intrinsic characteristic of the single species, the production of main aromatic compounds was well differentiated in the trials. Low quantities of ethyl acetate and acetoin and high amounts of isoamyl alcohols and β-phenylethanol were produced by pure cultures of C. zemplinina and S. cerevisiae, as similarly reported by Andorrà et al. (2010). In this study H. uvarum pure culture produced high amounts of ethyl acetate and acetoin and very low amounts of isoamyl alcohols and β-phenylethanol. The production of large quantities of ethyl acetate and acetic acid by H. uvarum has always been considered a negative characteristic (Ciani and Picciotti, 1995), whereas there is controversy concerning the production of higher alcohols. H. uvarum and K. apiculata were found to be the main producers of higher alcohols by Gil et al. (1996). However, some authors (Herraiz et al., 1990; Rojas et al., 2003; Romano et al., 2003) reported that apiculate yeasts were low producers of higher alcohols and can promote the esterification of various alcohols such as ethanol, geraniol, isoamyl alcohols, and 2-phenylethanol. Probably high production of higher alcohols could be a strain character dependent also in some NS wine yeasts (Capece et al., 2005). The secondary volatile compounds produced by mixed cultures were a combination of the different strains. PCA well highlighted these differences, indicating that the final wine characteristics can be modulated by different combinations or species and sequence of inoculum (Nurgel et al., 2002; Howell et al., 2006).
Candida zemplinina showed interesting features for mixed fermentation with S. cerevisiae, in particularly increasing the fermentation kinetic in high gravity Montepulciano must, with low ethyl acetate and acetic acid production. The combined use of three starter cultures (CSH and RCSH) could allow the improvement of the organoleptic characteristics of a wine. Further studies are needed to clarify the interaction among the different starters and to optimize the fermentation and the modalities of inoculation. Effectively the use of NS wine yeasts together with Saccharomyces strains in mixed fermentations might be recommended as a tool to obtain the advantages of spontaneous fermentation of organic wines such as those obtained with Montepulciano, while avoiding the risks of stuck fermentation (Rojas et al., 2003; Romano et al., 2003; Jolly et al., 2006; Ciani et al., 2010). Because of the NS yeast strains biodiversity about their production level of enzymatic activities (Manzanares et al., 1999, 2000; Mendes-Ferreira et al., 2001; Strauss et al., 2001) and fermentation metabolites (Romano et al., 1992, 2003; Capece et al., 2005) of enological importance, suitable strains should be selected in order to be able to design mixed starter able to provide beneficial contributions also for wine obtained by organic viticulture. In particular C. zemplinina showed a good interaction with S. cerevisiae by increasing the fermentation kinetic in high gravity Montepulciano must, with low ethyl acetate and acetic acid production.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is part of the project “Oenological microbiota: selection to identify the wine character and to improve the competitiveness of Montepulciano d’Abruzzo wineries” supported by grant from Cassa di Risparmio di Teramo.
References
- Andorrà I., Berradre M., Rozès N., Mas A., Guillamón J. M., Esteve-Zarzoso B. (2010). Effect of pure and mixed cultures of the main wine yeast species on grape must fermentations. Eur. Food Res. Technol. 231, 215–224 10.1007/s00217-010-1272-0 [DOI] [Google Scholar]
- Andorrà I., Monteiro M., Esteve-Zarzoso B., Albergaria H., Mas A. (2011). Analysis and direct quantification of Saccharomyces cerevisiae and Hanseniaspora guilliermondii populations during alcoholic fermentation by fluorescence in situ hybridization, flow cytometry and quantitative PCR. Food Microbiol. 28, 1483–1491 10.1016/j.fm.2011.08.009 [DOI] [PubMed] [Google Scholar]
- Anfang N., Brajkovich M., Goddard M. R. (2009). Co-fermentation with Pichia kluyveri increases varietal thiol concentrations in Sauvignon Blanc. Aust. J. Grape Wine Res. 15, 1–8 10.1111/j.1755-0238.2008.00031.x [DOI] [Google Scholar]
- Arneborg N., Siegumfeldt H., Andersen G., Nissen P., Daria V., Rodrigo P., Gluckstad J. (2005). Interactive optical trapping shows that confinement is a determinant of growth in a mixed yeast. FEMS Microbiol. Lett. 245, 155–159 10.1016/j.femsle.2005.03.008 [DOI] [PubMed] [Google Scholar]
- Cantarelli C. (1955). Studio comparativo dei lieviti apiculati dei generi Kloeckera (Janke) ed Hanseniaspora (Zikes). Ann. Microbiol. 6, 85 [Google Scholar]
- Capece A., Fiore C., Maraz A., Romano P. (2005). Molecular and technological approaches to evaluate strain biodiversity in Hanseniaspora uvarum of wine origin. J. Appl. Microbiol. 98, 136–144 10.1111/j.1365-2672.2004.02434.x [DOI] [PubMed] [Google Scholar]
- Castelli T. (1969). Vino al microscopio. Roma: Scialpi Editore [Google Scholar]
- Ciani M., Beco L., Comitini F. (2006). Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int. J. Food Microbiol. 108, 239–245 10.1016/j.ijfoodmicro.2005.11.012 [DOI] [PubMed] [Google Scholar]
- Ciani M., Comitini F., Mannazzu I., Domizio P. (2010). Controlled mixed culture fermentation: a new perspective on the use of non-Saccharomyces yeasts in winemaking. FEMS Yeast Res. 10, 123–133 10.1111/j.1567-1364.2009.00579.x [DOI] [PubMed] [Google Scholar]
- Ciani M., Pepe V. (2002). The influence of pre-fermentative practices on the dominance of inoculated yeast starter under industrial conditions. J. Sci. Food Agric. 82, 573–578 10.1002/jsfa.1082 [DOI] [Google Scholar]
- Ciani M., Picciotti G. (1995). The growth kinetics and fermentation behaviour of some non-Saccharomyces yeasts associated with wine-making. Biotechnol. Lett. 17, 1247–1250 10.1007/BF00128395 [DOI] [Google Scholar]
- De la Calle-Garcia D., Reichenbacher M., Dancer K., Hurlbeck C., Bartzsch C., Feller K. H. (1998). Analysis of wine bouquet components using headspace space solid-phase microextraction-capillary gas chromatography. J. High Resolut. Chromatogr. 21, 373–377 [DOI] [Google Scholar]
- Egli C. M., Edinger W. D., Mitrakul C. M., Henick-Kling T. (1998). Dynamics of indigenous and inoculated yeast populations and their effect on the sensory character of riesling and Chardonnay wines. J. Appl. Microbiol. 85, 779–789 10.1046/j.1365-2672.1998.00521.x [DOI] [PubMed] [Google Scholar]
- Fernández M. T., Ubeda J. F., Briones A. I. (2000). Typing of non-Saccharomyces yeasts with enzymatic activities of interest in winemaking. Int. J. Food Microbiol. 59, 29–36 10.1016/S0168-1605(00)00283-X [DOI] [PubMed] [Google Scholar]
- Fleet G. H. (2003). Yeast interactions and wine flavour. Int. J. Food Microbiol. 86, 11–22 10.1016/S0168-1605(03)00245-9 [DOI] [PubMed] [Google Scholar]
- Fleet G. H. (2008). Wine yeast for the future. FEMS Yeast Res. 8, 979–995 10.1111/j.1567-1364.2008.00427.x [DOI] [PubMed] [Google Scholar]
- Fleet G. H., Lafon-Lafourcade S., Ribéreau-Gayon P. (1984). Evolution of yeasts and lactic acid bacteria during fermentation and storage of Bordeaux Wines. Appl. Environ. Microbiol. 48, 1034–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafner J., Schultz M. (1996). Impact of glucose-fructose ratio on stuck fermentations: practical experiences to restart stuck fermentation. Vitic. Enol. Sci. 51, 214–218 [Google Scholar]
- Gil J., Mateo J., Jiménez M., Pastor A., Huerta T. (1996). Aroma compounds in wine as influenced by apiculate yeasts. J. Food Sci. 61, 1247–1266 10.1111/j.1365-2621.1996.tb14749.x [DOI] [Google Scholar]
- González S. S., Barrio E., Querol A. (2007). Molecular identification and characterization of wine yeast isolated from Tenerife (Canary Island, Spain). J. Appl. Microbiol. 102, 1018–1025 [DOI] [PubMed] [Google Scholar]
- Herraiz T., Reglero G., Herraiz M., Martín-Alvarez P., Cabezudo M. D. (1990). The influence of the yeast and type of culture on the volatile composition of wines fermented without sulfur dioxide. Am. J. Enol. Vitic. 41, 313–318 [Google Scholar]
- Hierro N., Esteve-Zarzoso B., González A., Mas A., Guillamón J. M. (2006). Real-time quantitative PCR (QPCR) and reverse transcription-QPCR for detection and enumeration of total yeasts in wine. Appl. Environ. Microbiol. 72, 7148–7155 10.1128/AEM.00388-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierro N., Esteve-Zarzoso B., Mas A., Guillamón J. M. (2007). Monitoring of Saccharomyces and Hanseniaspora populations during alcoholic fermentation by real-time quantitative PCR. FEMS Yeast Res. 7, 1340–1349 10.1111/j.1567-1364.2007.00304.x [DOI] [PubMed] [Google Scholar]
- Howell K. S., Cozzolino D., Bartowsky E. J., Fleet G. H., Henschke P. A. (2006). Metabolic profiling as a tool for revealing Saccharomyces interactions during wine fermentation. FEMS Yeast Res. 6, 91–101 10.1111/j.1567-1364.2005.00010.x [DOI] [PubMed] [Google Scholar]
- International Organization of Vine and Wine (2011). Compendium of International Methods of Wine and must Analysis. [Google Scholar]
- Jolly N. P., Augustyn O. P. H., Pretorius I. S. (2006). The role and use of non-Saccharomyces yeasts in wine production. S. Afr. J. Enol. Vitic. 27, 15–39 [Google Scholar]
- Lambrechts M. G., Pretorius I. S. (2000). Yeast and its importance to wine aroma. A review. South Afr. J. Enol. Vitic. 21, 97–129 [Google Scholar]
- Lema C., Garcia-Jares C., Orriols I., Angulo L. (1996). Contribution of Saccharomyces and non-Saccharomyces populations to the production of some components of Albarino wine aroma. Am. J. Enol. Vitic. 47, 206–216 [Google Scholar]
- Lopez-Tamames E., Puig-Deu M., Teixeira E., Buxaderas S. (1996). Organic acids, sugars, and glycerol content in white winemaking products determined by HPLC: relationship to climate and varietal factors. Am. J. Enol. Vitic. 47, 193–198 [Google Scholar]
- Ludovico P., Sousa M. J., Silva M. T., Leao C., Corte-Real M. (2001). Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology 147, 2409–2415 [DOI] [PubMed] [Google Scholar]
- Magyar I., Toth T. (2011). Comparative evaluation of some oenological properties in wine strains of Candida stellata, Candida zemplinina, Saccharomyces uvarum and Saccharomyces cerevisiae. Food Microbiol. 28, 94–100 10.1016/j.fm.2010.08.011 [DOI] [PubMed] [Google Scholar]
- Manzanares P., Ramon D., Querol A. (1999). Screening of non-Saccharomyces wine yeasts for the production of β-D-xylosidase activity. Int. J. Food Microbiol. 46, 105–112 10.1016/S0168-1605(98)00186-X [DOI] [PubMed] [Google Scholar]
- Manzanares P., Rojas V., Genovés S., Vallés S. (2000). A preliminary search for anthocyanin-β-D-glucosidase activity in non-Saccharomyces wine yeasts. Int. J. Food Sci. Technol. 35, 95–103 10.1046/j.1365-2621.2000.00364.x [DOI] [Google Scholar]
- Marcobal Á., Martín-Álvarez P. J., Polo C., Muñoz R., Moreno-Arribas M. V. (2006). Formation of biogenic amines throughout the industrial manufacture of red wine. J. Food Prot. 69, 397–404 [DOI] [PubMed] [Google Scholar]
- Mendes-Ferreira A., Climaco M. C., Mendes-Faia A. (2001). The role of non-Saccharomyces species in releasing glycosidic bound fraction of grape aroma components – a preliminary study. J. Appl. Microbiol. 91, 67–71 10.1046/j.1365-2672.2001.01348.x [DOI] [PubMed] [Google Scholar]
- Mendoza L. M., Manca de Nadra M. C., Farías M. E. (2007). Kinetics and metabolic behavior of a composite culture of Kloeckera apiculata and Saccharomyces cerevisiae wine related strains. Biotechnol. Lett. 29, 1057–1063 10.1007/s10529-007-9355-0 [DOI] [PubMed] [Google Scholar]
- Millet V., Lonvaud-Funel A. (2000). The viable but non-cultivable state of wine microorganisms during storage. Lett. Appl. Microbiol. 30, 136–141 10.1046/j.1472-765x.2000.00684.x [DOI] [PubMed] [Google Scholar]
- Mora J., Barbas J. I., Mulet A. (1990). Growth of yeast species during the fermentation of musts inoculated with Kluyveromyces thermotolerans and Saccharomyces cerevisiae. Am. J. Enol. Vitic. 41, 156–159 [Google Scholar]
- Moreira N., Mendes F., Guedes de Pinho P., Hogg T., Vasconcelos I. (2008). Heavy sulphur compounds, higher alcohols and esters production profile of Hanseniaspora uvarum and Hanseniaspora guilliermondii grown as pure and mixed cultures in grape must. Int. J. Food Microbiol. 124, 231–238 10.1016/j.ijfoodmicro.2008.03.025 [DOI] [PubMed] [Google Scholar]
- Nissen P., Nielsen D., Arneborg N. (2003). Saccharomyces cerevisiae cells at high concentrations cause early growth arrest of non-Saccharomyces yeasts in mixed cultures by a cell-cell contact-mediated mechanism. Yeast 20, 331–341 10.1002/yea.965 [DOI] [PubMed] [Google Scholar]
- Nurgel C., Erten H., Canbas A., Cabaroglu T., Selli S. (2002). Influence of Saccharomyces cerevisiae strains on fermentation and flavor compounds of white wines made from cv. Emir grown in Central Anatolia, Turkey. J. Ind. Microbiol. Biotechnol. 29, 28–33 10.1038/sj.jim.7000258 [DOI] [PubMed] [Google Scholar]
- Pallmann C. L., Brown J. A., Olineka T. L., Cocolin L., Mills D. A., Bisson L. F. (2001). Use of WL medium to profile native flora fermentations. Am. J. Enol. Vitic. 52, 198–203 [Google Scholar]
- Pérez-Nevado F., Albergaria H., Hogg T., Girio F. (2006). Cellular death of two non-Saccharomyces wine-related yeasts during mixed fermentation with Saccharomyces cerevisiae. Int. J. Food Microbiol. 108, 336–345 [DOI] [PubMed] [Google Scholar]
- Pretorius I. S. (2000). Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16, 675–729 [DOI] [PubMed] [Google Scholar]
- Rantsiou K., Dolci P., Giacosa S., Torchio F., Tofalo R., Torriani S., Suzzi G., Rolle L., Cocolin L. (2012) Candida zemplinina can reduce acetic acid production by Saccharomyces cerevisiae in sweet wine fermentations. Appl. Environ. Microbiol. 78, 1987–1994 10.1128/AEM.06768-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspor P., Milek D. M., Polanc J., Mozina S. S., Cadez N. (2006). Yeasts isolated from three varieties of grapes cultivated in different locations of the Dolenjska vine-growing region, Slovenia. Int. J. Food Microbiol. 109, 97–102 10.1016/j.ijfoodmicro.2006.01.017 [DOI] [PubMed] [Google Scholar]
- Rojas V., Gil J., Piñaga F., Manzanares P. (2003). Acetate ester formation in wine by mixed cultures in laboratory fermentations. Int. J. Food Microbiol. 86, 181–188 10.1016/S0168-1605(03)00255-1 [DOI] [PubMed] [Google Scholar]
- Romano P., Fiore C., Paraggio M., Caruso M., Capece A. (2003). Function of yeasts species and strains in wine flavour. Int. J. Food Microbiol. 86, 169–180 10.1016/S0168-1605(03)00290-3 [DOI] [PubMed] [Google Scholar]
- Romano P., Suzzi G., Comi G., Zironi R. (1992). Higher alcohol and acetic acid production by apiculate wine yeasts. J. Appl. Bacteriol. 73, 126–130 10.1111/j.1365-2672.1992.tb01698.x [DOI] [Google Scholar]
- Schmitt M. J., Breinig F. (2002). The viral killer system in yeast: from molecular biology to application. FEMS Microbiol. Rev. 26, 257–276 10.1111/j.1574-6976.2002.tb00614.x [DOI] [PubMed] [Google Scholar]
- Sipiczki M., Ciani M., Csoma H. (2005). Taxonomic reclassification of Candida stellata DBVPG 3827. Folia Microbiol. (Praha) 50, 494–498 10.1007/BF02931436 [DOI] [PubMed] [Google Scholar]
- Smit A. Y., du Toit W. J., du Toit M. (2008). Biogenic amines in wine: understanding the headache. South Afr. J. Enol. Vitic. 29, 109–127 [Google Scholar]
- Soufleros E., Barrios M. L., Bertrand A. (1998). Correlation between the content of biogenic amines and other wine compounds. Am. J. Enol. Vitic. 49, 266–269 [Google Scholar]
- Strauss M. L. A., Jolly N. P., Lambrechts M. G., van Rensburg P. (2001). Screening for the production of extracellular hydrolytic enzymes by non-Saccharomyces wine yeasts. J. Appl. Microbiol. 91, 182–190 10.1046/j.1365-2672.2001.01379.x [DOI] [PubMed] [Google Scholar]
- Tofalo R., Chaves-López C., Di Fabio F., Schirone M., Felis G. E., Torriani S., Paparella A., Suzzi G. (2009). Molecular identification and osmotolerant profile of wine yeasts that ferment a high sugar grape must. Int. J. Food Microbiol. 130, 179–187 10.1016/j.ijfoodmicro.2009.01.024 [DOI] [PubMed] [Google Scholar]
- Tofalo R., Schirone M., Telera G. C., Manetta A. C., Corsetti A., Suzzi G. (2011). Influence of organic viticulture on non-Saccharomyces wine yeast populations. Ann. Microbiol. 61, 57–66 10.1007/s13213-010-0102-8 [DOI] [Google Scholar]
- Tofalo R., Schirone M., Torriani S., Rantsiou K., Cocolin L., Perpetuini G., Suzzi G. (2012). Diversity of Candida zemplinina strains from grapes and Italian wines. Food Microbiol. 29, 18–26 10.1016/j.fm.2011.08.014 [DOI] [PubMed] [Google Scholar]
- Tofalo R., Torriani S., Chaves-López C., Martuscelli M., Paparella A., Suzzi G. (2007). A survey of Saccharomyces populations associated with wine fermentations from the Apulia region (South Italy). Ann. Microbiol. 57, 545–552 10.1007/BF03175353 [DOI] [Google Scholar]
- Toro M. E., Vazquez F. (2002). Fermentation behavior of controlled mixed and sequential cultures of Candida cantarellii and Saccharomyces cerevisiae wine yeasts. World J. Microbiol. Biotechnol. 18, 347–354 10.1023/A:1015242818473 [DOI] [Google Scholar]
- Torrea D., Ancín C. (2002). Content of biogenic amines in a Chardonnay wine obtained through spontaneous and inoculated fermentation. J. Agric. Food Chem. 50, 4895–4899 10.1021/jf011653h [DOI] [PubMed] [Google Scholar]
- Trioli G., Hofmann U. (2009). ORWINE: Code of Good Organic Viticulture and Wine-Making. Oppenheim: ECOVIN-Federal Association of Organic Wine-Producer [Google Scholar]
- Viana F., Gil J. V., Genovés S., Vallés S., Manzanares P. (2008). Rational selection of non-Saccharomyces wine yeasts for mixed starters based on ester formation and enological traits. Food Microbiol. 25, 778–785 10.1016/j.fm.2008.04.015 [DOI] [PubMed] [Google Scholar]
- Zironi R., Romano P., Suzzi G., Battistuta F., Comi G. (1993). Volatile metabolites produced in wine by mixed and sequential cultures of Hanseniaspora guilliermondii or Kloeckera apiculata and Saccharomyces cerevisiae. Biotechnol. Lett. 15, 235–238 10.1007/BF00128311 [DOI] [Google Scholar]
- Zohre D. E., Erten H. (2002). The influence of Kloeckera apiculata and Candida pulcherrima yeasts on wine fermentation. Process. Biochem. 38, 319–324 10.1016/S0032-9592(02)00086-9 [DOI] [Google Scholar]
- Zott K., Claisse O., Lucas P., Coulon J., Lonvaud-Funel A., Masneuf-Pomarede I. (2010). Characterization of the yeast ecosystem in grape must and wine using real-timePCR. Food Microbiol. 27, 559–567 10.1016/j.fm.2010.01.006 [DOI] [PubMed] [Google Scholar]