Abstract
Increasing evidence demonstrated the involvement of ammonia-oxidizing archaea (AOA) in the global nitrogen cycle, but the relative contributions of AOA and ammonia-oxidizing bacteria (AOB) to ammonia oxidation are still in debate. Previous studies suggest that AOA would be more adapted to ammonia-limited oligotrophic conditions, which seems to be favored by protonation of ammonia, turning into ammonium in low-pH environments. Here, we investigated the autotrophic nitrification activity of AOA and AOB in five strongly acidic soils (pH<4.50) during microcosm incubation for 30 days. Significantly positive correlations between nitrate concentration and amoA gene abundance of AOA, but not of AOB, were observed during the active nitrification. 13CO2-DNA-stable isotope probing results showed significant assimilation of 13C-labeled carbon source into the amoA gene of AOA, but not of AOB, in one of the selected soil samples. High levels of thaumarchaeal amoA gene abundance were observed during the active nitrification, coupled with increasing intensity of two denaturing gradient gel electrophoresis bands for specific thaumarchaeal community. Addition of the nitrification inhibitor dicyandiamide (DCD) completely inhibited the nitrification activity and CO2 fixation by AOA, accompanied by decreasing thaumarchaeal amoA gene abundance. Bacterial amoA gene abundance decreased in all microcosms irrespective of DCD addition, and mostly showed no correlation with nitrate concentrations. Phylogenetic analysis of thaumarchaeal amoA gene and 16S rRNA gene revealed active 13CO2-labeled AOA belonged to groups 1.1a-associated and 1.1b. Taken together, these results provided strong evidence that AOA have a more important role than AOB in autotrophic ammonia oxidation in strongly acidic soils.
Keywords: acidic soil, ammonia-oxidizing archaea, ammonia-oxidizing bacteria, dicyandiamide, nitrification, stable isotope probing
Introduction
Nitrification in acidic soils has been substantially investigated since the early twentieth century, but few studies have provided direct evidence on the specific group of microorganisms serving as the dominating ammonia oxidizers in low-pH environments. Previously, the widespread presence of ammonia-oxidizing bacteria (AOB) and the observation of nitrification activity at low pH values have led to speculations that AOB was primarily responsible for the autotrophic nitrifying activity under acidic conditions (De Boer and Kowalchuk, 2001), but questions were raised regarding the tolerance of AOB to low-pH stresses, because most cultured isolates of AOB could not nitrify in batch culture below pH 5.5 (Hankinson and Schmidt, 1988; Jiang and Bakken, 1999). The sensitivity of AOB to acidic conditions was generally explained by the exponential ionization of ammonia to ammonium with decreasing pH (Suzuki et al., 1974), because it is assumed that ammonia rather than ammonium could directly fuel AOB. Further studies suggested that autotrophic AOB had a minor role in acidic soils subjected to high nitrogen deposition (Schmidt et al., 2007), and showed that the community structure of bacterial amoA genes remained relatively stable with changing nitrogen mineralization in acidic forest soils (Mintie et al., 2003). An acetylene inhibition study revealed that heterotrophic nitrifiers other than AOB may have contributed to the nitrification in an acidic forest soil (Jordan et al., 2005), and laboratory incubation using 15N-labeling techniques found heterotrophic nitrification accounted for <20% of the total nitrification in two acidic pasture soils (Islam et al., 2007). Thus, the direct links between nitrification activity and specific ammonia oxidizers in acidic soils are still missing. It remains to be known which nitrifier groups are mainly responsible for the nitrate production in low-pH environments.
Recent progress in ammonia-oxidizing archaea (AOA) studies (Venter et al., 2004; Könneke et al., 2005; Hallam et al., 2006) greatly suggested their putative contributions to the global nitrogen cycle by possessing the potential capacity to oxidize ammonia (Nicol and Schleper, 2006), and phylogenetic analysis categorized AOA into the Thaumarchaeota lineage, a novel archaeal phylum (Brochier-Armanet et al., 2008; Spang et al., 2010). A previous study across a soil pH gradient from 4.9 to 7.5 indicated thaumarchaeal amoA gene abundance and transcript activity, but not bacterial, increased with decreasing pH (Nicol et al., 2008). A majority of acidic soils selected higher AOA abundance over AOB (Leininger et al., 2006; He et al., 2007) implying a strong ability of AOA ecotypes adapting to low-pH soils. Moreover, high nitrification rates were presumably associated with growth of AOA in an acidic forest soil in the absence of AOB (Stopnisek et al., 2010), and a significant correlation between AOA community abundance and nitrification potential was observed in Chinese acidic tea orchard soils (Yao et al., 2011). In addition, physiological and genomic studies of the first isolate AOA, Nitrosopumilus maritimus, demonstrated a low-level ammonia substrate threshold that was significantly lower than the requirement by cultivated AOB (Martens-Habbena et al., 2009), and significantly divergent pathways of ammonia oxidation from AOB offering ecological advantage in ammonia-limited environments (Walker et al., 2010). Although the soil-isolated AOA strain, Nitrososphaera viennensis, revealed a relatively higher tolerance of ammonia substrates than marine isolate, it is still considerably lower than the reported maximum ammonia tolerance of AOB strains (Tourna et al., 2011). Very recently, an obligate acidophilic thaumarchaeal ammonia oxidizer was enriched from an acidic agricultural soil, demonstrating the autotrophic growth of ammonia-oxidizing thaumarchaea at extremely low ammonia concentration (Lehtovirta et al., 2011). These findings were supported by soil microcosm experiments where autotrophic ammonia oxidation was driven mainly by AOA rather than AOB under low ammonia conditions without external nitrogen supply (Offre et al., 2009; Zhang et al., 2010). A recent study found available ammonia concentrations determined the niche separation of AOA and AOB in soil microcosms with AOA preferring low levels of ammonia supply (Verhamme et al., 2011), and growth of AOA has been presumably correlated with high rates of nitrification in an acetylene inhibition study of two agricultural acidic soils (Gubry-Rangin et al., 2010). The lower pH values could drive the conversion from ammonia to ammonium, decreasing substrate availability and increasing energy stress (Valentine, 2007) that might be able to selectively choose AOA over AOB as the dominating ammonia oxidizer. Thus, we hypothesize that AOA outcompete AOB in acidic soils, which is tested in the present study.
Acidic soils are extensively distributed around the world, and global expanse of acidic soils (pH<5.5 in the surface layers) is estimated to be 3950 million Ha, occupying ∼30% of the world's total ice-free lands, including forests, pastures, woodlands, wetlands and croplands (Vonuexkull and Mutert, 1995). In the past several decades, high levels of nitrogen-based fertilization have aggravated large-scale soil acidification worldwide (Wallace, 1994; Vitousek et al., 1997) as well as in major Chinese croplands (Guo et al., 2010), which could significantly alter the biogeochemical cycles. Therefore, it is of great significance to understand nitrogen cycling in acidic soils, thus providing possible strategies for managing the global nitrification process and mitigating nitrous oxide emissions and nitrate leaching. However, considerably less efforts have been placed on the exploration of the mechanisms of ammonia oxidation in acidic soils. Because of the development of new technologies like DNA-stable isotope probing (SIP), we could now be able to gain a deep insight into the autotrophic nitrification activity in complex soil environments. 13CO2-DNA-SIP has shown its powerful potential to directly link CO2-incorporating ammonia oxidizers with nitrification activity in several neutral or alkaline soils (Jia and Conrad, 2009; Zhang et al., 2010; Pratscher et al., 2011; Xia et al., 2011) and provided evidence for autotrophic growth of N. devanaterra in acidic soil, whereas to the best of our knowledge, no study has been able to clearly identify the dominating ammonia oxidizers in the autotrophic nitrification of acidic soils.
This study was designed to investigate the relative contributions of AOA and AOB to autotrophic ammonia oxidation in five strongly acidic agricultural soils (pH<4.50). A combined approach of 13CO2-DNA-SIP and application of the nitrification inhibitor dicyandiamide (DCD, C2H4N4) was employed to identify the ammonia-oxidizing community responsible for the nitrification in the acidic soils. Previous studies found only AOB to be significantly affected by addition of DCD in grazed dairy pastures (Di et al., 2009, 2010; O'Callaghan et al., 2010) and, as far as we know, currently there is no published study demonstrating inhibition of AOA by DCD. Therefore, it remains to be known whether DCD could inhibit the nitrification activity and growth of AOA in acidic soils.
Materials and methods
Soil sampling
Soil samples for microcosm incubation were collected from five distinct sites across South China, that is, Hangzhou (HZ), Yuhang (YH), Qujing (QJ), Qiyang (QY) and Taoyuan (TY). HZ soil was collected from a 50-year-old tea orchard characterized by a subtropical wet monsoon climate with mean annual rainfall of 1500 mm, which received a high-rate nitrogen fertilizer input, averaging about 450 kg N Ha−1 per year (Xue et al., 2006). YH, QJ, QY and TY soils are also located in a subtropical monsoon climate, with a mean annual temperature of 16.2 °C, 14.5 °C, 18.1 °C, and 16.5 °C, respectively, and mean annual rainfall of 1550 mm, 1030 mm, 1288 mm, and 1437 mm, respectively. Soil samples were collected from 0 to 20 cm surface soils by mixing nine random soil cores. All samples were passed through a 2.0-mm sieve and stored at 4 °C before construction of microcosms. Soil pH (H2O and 1 KCl) was determined using a Delta 320 pH-meter (Mettler-Toledo Instruments Co., Shanghai, China). Soil moisture content was determined after oven drying at 105 °C for 24 h. Total N and total C were determined using the Dumas method by an Element Analyser (Vario EL III, Elementar, Hanau, Germany). Soil organic carbon was determined using the K2Cr2O7 oxidation–reduction titration method. Particle size was measured by the rapid sieving procedure (Kettler et al., 2001). Cation exchange capacity was determined by BaCl2 replacement method. The detailed properties of soils were listed in Table 1.
Table 1. Basic properties of soils used in this study.
| HZ | YH | QJ | QY | TY | |
|---|---|---|---|---|---|
| Location | N 30°10′ | N 30°25′ | N 25°24′ | N 26°45′ | N 28°25′ |
| E 120°05′ | E 119°44′ | E 103°48 | E 111°52′ | E 111°26′ | |
| pH (H2O)a | 4.20 | 4.21 | 4.36 | 4.43 | 4.47 |
| pH (KCl)b | 3.29 | 3.34 | 3.66 | 3.87 | 3.89 |
| Total N (g kg−1) | 5.02 | 2.07 | 2.46 | 1.27 | 1.79 |
| Total C (g kg−1) | 54.9 | 22.7 | 22.0 | 12.7 | 17.9 |
| SOC (g kg−1) | 31.9 | 12.1 | 12.8 | 7.60 | 9.54 |
| NH4+-N (mg kg−1) | 39.2 | 9.45 | 22.1 | 11.5 | 12.9 |
| NO3−-N (mg kg−1) | 31.3 | 13.2 | 203 | 2.77 | 2.87 |
| CEC (cmol kg−1) | 24.1 | 18.0 | 10.7 | 22.1 | 19.0 |
| H2O % | 28.8 | 29.5 | 24.0 | 22.7 | 27.4 |
| Particle size (%) | |||||
| Sand | 43.4 | 14.1 | 14.2 | 8.2 | 11.5 |
| Silt | 41.2 | 45.0 | 32.7 | 63.9 | 68.3 |
| Clay | 15.4 | 40.9 | 53.1 | 28.9 | 20.2 |
Abbreviations: CEC, cation exchange capacity; HZ, Hangzhou; SOC, soil organic carbon; TY, Taoyuan; QJ, Qujing; QY, Qiyang; YH, Yuhang.
pH (H2O) was determined with a soil to water ratio of 1:2.5.
pH (KCl) was determined with a soil to 1 KCl ratio of 1:2.5.
Soil microcosm incubation
The laboratory microcosms for the five acidic soils were performed in 120-ml serum bottles containing 10 g of the sieved fresh soil samples and sealed with rubber stoppers and aluminum caps. Each soil had two treatments in the presence or absence of 50 mg DCD kg−1 soil. This rate of DCD applied is typical for the amount applied to grazed pasture (Zaman and Blennerhassett, 2010). Microcosms were incubated at 28 °C in the dark, and the serum bottles were opened every 2 days for air exchange to maintain aerobic conditions. The water content of soils was maintained throughout the incubation by resupplying the lost water every 2 days. Three replicates of each treatment were destructively sampled after 0, 7, 15 and 30 days and then frozen immediately at −80 °C before further analysis. Ammonium and nitrate were extracted from all microcosms with 1 KCl and determined by a Continuous Flow Analyser (SAN++, Skalar, Breda, Holland).
DNA-SIP microcosm studies
HZ soil sample with the lowest pH value and a high-rate nitrification activity was then chosen for DNA-SIP microcosm studies. Twenty-four triplicate SIP microcosms were performed in 120-ml serum bottles containing 10 g of the sieved fresh soil sample and sealed with rubber stoppers and aluminum caps. Four sets of treatments employed in the incubation were 5% (v/v) 12C-CO2, 5% 12C-CO2 + 50 mg DCD kg−1 soil, 5% 13C-CO2 (99 atoms%, Sigma-Aldrich Co., St Louis, MO, USA), 5% 13C-CO2 + 50 mg DCD kg−1 soil, respectively. Aerobic conditions were maintained by opening microcosms for air refreshing every 2 days, then reestablishing the CO2 concentration in the headspace. Microcosms were incubated at 28 °C in the dark, and three replicates of each treatment were destructively sampled at 0, 15 and 30 days, and then frozen immediately at −80 °C.
DNA extraction and SIP fractionation
Nucleic acids were extracted from 0.5 g of samples using the FASTDNA SPIN Kit for soil (Q BIOgene Inc., Carlsbad, CA, USA) according to the manufacturer's instructions. The concentration of extracted nucleic acids was determined photometrically using Nanodrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Density gradient centrifugation was performed in 4.9-ml OptiSeal polyallomer tubes (Beckman Coulter, Palo Alto, CA, USA) in a VTi 90 vertical rotor (Beckman Coulter), subject to centrifugation at 228 166 gav (56 200 r.p.m.) for 24 h at 20 °C (Freitag et al., 2006; Zhang et al., 2010). Extracted DNA (∼2 μg) was added into CsCl gradients with an initial density of 1.696 g ml−1, prepared by adjusting the refractive index to 1.399 with an ATAGO-R-5000 hand-held refractometer (UNI-IT, Tokyo, Japan). Centrifuged gradients were fractionated into 25 equal volumes (∼200 μl) by displacing CsCl solution with sterile water at the top of the tube, using a fraction recovery system (Beckman Coulter) and a MPP-100 Mini Peristaltic Pump (CBS Scientific Co., Del Mar, CA, USA). Buoyant density of each collected fraction was measured by determining the refractive index of 25 μl aliquots. Nucleic acids were precipitated from CsCl overnight by adding two volumes of PEG 6000 in 1.6 NaCl and 70% ethanol washing (Griffiths et al., 2000), then dissolved in 30 μl of sterile water.
Real-time PCR assay of amoA genes
Abundance of thaumarchaeal amoA genes was determined on an iCycler iQ 5 thermocycler (Bio-Rad Laboratories, Hercules, CA, USA) using primer pairs Arch-amoAF (5′-STAATGGTCTGGCTTAGACG-3′) and Arch-amoAR (5′-GCGGCCATCCATCTGTATGT-3′) (Francis et al., 2005). Each reaction was performed in a 25 μl volume containing 12.5 μl SYBR Premix Ex Taq (TaKaRa Biotechnology, Otsu, Shiga, Japan), 1 μ of each primer and 2 μl of 10-fold dilution DNA template (1–10 ng). Amplification conditions were as follows: 95 °C for 1 min, 40 cycles of 10 s at 95 °C, 30 s at 53 °C, 1 min at 72 °C and followed by plate reads at 83 °C. Bacterial amoA genes were quantified using the primers amoA1F (5′-GGGGTTTCTACTGGTGGT-3′) and amoA2R (5′-CCCCTCKGSAAAGCCTTCTTC-3′) (Rotthauwe et al., 1997) with the same reaction conditions except 0.25 mg ml−1 bovine serum albumin was added to each reaction and the annealing temperature was 55 °C. Melting curve analysis was performed at the end of each real-time PCR run to check the specificity of amplification products, before confirmation by standard agarose gel eletrophoresis.
Denaturing gradient gel electrophoresis analysis, cloning, sequencing and phylogenetic analysis
Denaturing gradient gel electrophoresis (DGGE) analysis of thaumarchaeal and bacterial ammonia oxidizers was performed using a DCode Universal Mutation Detection System (Bio-Rad Laboratories). Thaumarchaeal amoA genes were amplified with the primers CrenamoA23f (5′-ATGGTCTGGCTWAGACG-3′) and CrenamoA616r (5′-GCCATACABCKRTANGTCCA-3′) as described by Tourna et al. (2008) and used for DGGE. A touchdown PCR procedure was used for the amplification of bacterial amoA genes with the primer pair amoA1F-GC/amoA2R. PCR products were loaded onto 6% polyacrylamide gel with a linear gradient of 20–50% and 40–60% denaturant for thaumarchaeal and bacterial amoA genes, respectively. Gels were electrophoresed at 90 V for 12 h with a constant temperature of 60 °C, and stained by SYBR Gold Nucleic Acid Gel Stain (Invitrogen-Molecular Probes, Eugene, OR, USA) for 30 min before scanning using a GBOX/HR-E-M (Syngene, Cambridge, UK).
Eight bands representing active populations of AOA after 30 days of incubation in the heavy fractions around a buoyant density of 1.73 g ml−1 were excised and re-amplified with the primers CrenamoA23f/CrenamoA616r for cloning and sequencing. Clone libraries of archaeal 16S rRNA genes were also constructed from the light fractions and the heavy fractions (each containing 60 clones) in the 13CO2 microcosms at day 30. Archaeal 16S rRNA genes were amplified using the primers Arch21f (5′-TTCCGGTTGATCCYGCCGGA-3′) and Arch958R (5′-YCCGGCGTTGAMTCCAATT-3′) (Delong, 1992). PCR products were gel-purified and cloned into pGEM-T Easy vector (Promega, Madison, WI, USA), and the resulting ligation products were transformed into Escherichia coli JM109 competent cells according to the manufacturer's instructions. Clones were checked following re-amplification with the primers T7 and SP6, and positive ones were selected for sequencing. Phylogenetic analysis was conducted with MEGA version 4.0 through neighbor-joining tree using Kimura 2-parameter distance with 1000 replicates to produce Bootstrap values (Tamura et al., 2007). Nucleotide sequences retrieved in this study have been deposited at GenBank under accession numbers JF681783 to JF681790 for thaumarchaeal amoA genes and JF917243 to JF917267 for archaeal 16S rRNA genes, respectively.
Statistical analysis
Nitrification rates and log-transformed amoA gene copy numbers were compared by one-way analysis of variance followed by Student–Newman–Keulstest to check for quantitative variance between different treatments. All analyses were conducted using SPSS version 13.0 (IBM Co., Armonk, NY, USA), and P<0.05 was considered to be statistically significant.
Results
Microcosm incubation with five acidic soils
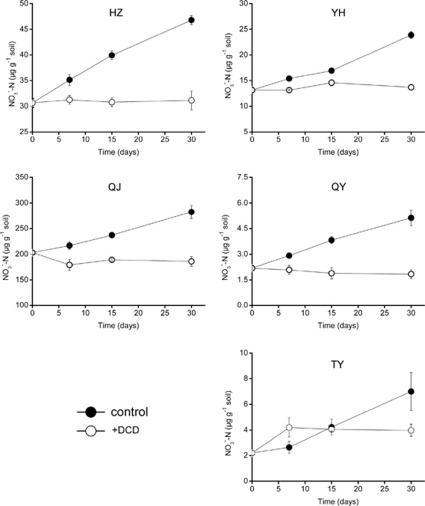
In control microcosms without amendment of DCD, nitrate concentration increased continuously over time indicating a sustaining nitrification activity in all five soils (Figure 1). QJ soil had the highest net nitrification rate of ∼2.64 μg NO3−-N g−1 soil per day, whereas QY soil had the lowest net nitrification rate of ∼0.10 μg NO3−-N g−1 soil per day. However, although different net nitrification rates were observed during incubation, addition of DCD could effectively inhibit the production of nitrate and only partially reduced the nitrification rate of TY soil.
Figure 1.
Changes in nitrate concentrations during incubation of five acidic soils in the presence or absence of DCD. Error bars represent standard errors of triplicate samples.
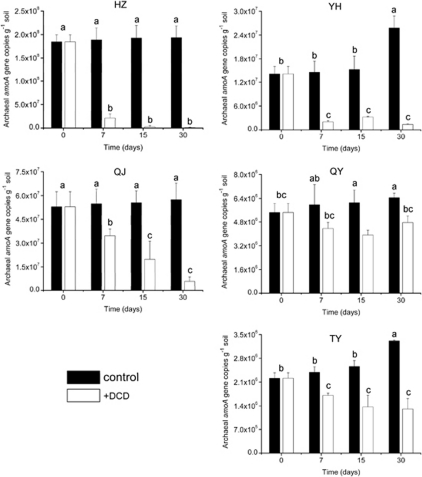
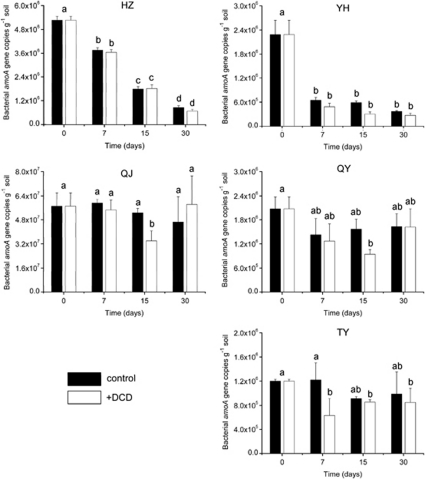
The thaumarchaeal amoA gene copy numbers remained relatively stable in control microcosms of HZ and QJ soils, and significantly increased in all other three acidic soils incubated for 30 days ranging from 1.03 × 106 to 1.17 × 107 copies g−1 soil, implying very different efficiencies of AOA growth during incubation (Figure 2). Application of DCD significantly inhibited the growth of AOA, and various sensitivities of thaumarchaeal amoA genes to DCD inhibition were detected throughout the incubation, and thaumarchaeal amoA gene copy numbers ranging from 2.21 × 106 to 1.84 × 108copies g−1 soil in control microcosms were reduced to between 1.32 × 106 and 4.76 × 106 copies g−1 soil in DCD treatments at day 30. In contrast, no significant changes in bacterial amoA genes were observed in control microcosms of QJ, QY and TY soils, but AOB significantly decreased in HZ and YH soils which might be ascribed to the disadvantage of competition for limited ammonia in acidic soils (Figure 3). Growth of AOB was only minimally inhibited by addition of DCD and even no inhibition effect on AOB was found in HZ soil, thus, the inhibition effect of DCD on bacterial amoA genes was less effective than that on thaumarchaeal amoA genes.
Figure 2.
Changes in abundance of thaumarchaeal amoA genes of five acidic soils in the presence or absence of DCD. Error bars represent standard errors of triplicate samples. Different letters above the bars indicate a significant difference (P<0.05).
Figure 3.
Changes in abundance of bacterial amoA genes of five acidic soils in the presence or absence of DCD. Error bars represent standard errors of triplicate samples. Different letters above the bars indicate a significant difference (P<0.05).
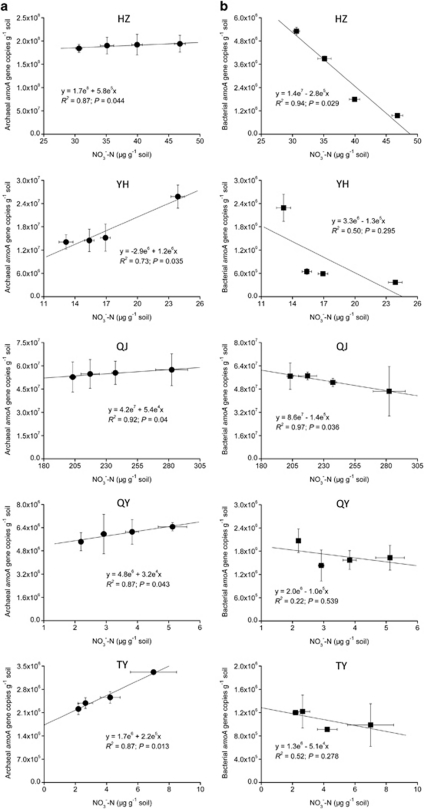
Regression analysis showed a significant positive relationship between nitrate concentration and thaumarchaeal amoA gene abundance in the control microcosms of five acidic soils (Figure 4). However, a significant negative relationship between nitrate concentration and bacterial amoA gene was observed in HZ and QJ soils, and no significant correlation between net nitrification and AOB existed in other three soils.
Figure 4.
Relationship between nitrate concentration and thaumarchaeal (a) or bacterial (b) amoA gene copy numbers in control microcosms. Vertical and horizontal error bars represent standard errors of amoA gene copies and nitrate concentrations from triplicate samples, respectively.
DNA-SIP incubation
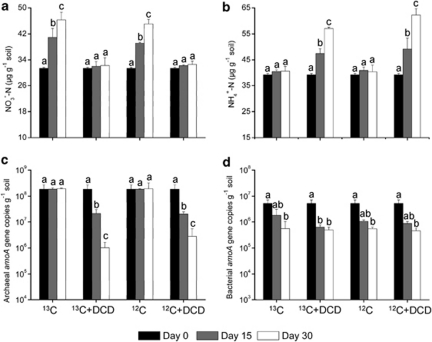
HZ soil sample with a high nitrification rate was typically chosen for 30 days' DNA-SIP microcosm studies. Active nitrification activity was observed in the absence of DCD with nitrate increasing from 31.3 (±0.26) μg NO3−-N g−1 at day 0 to 46.3 (±1.39) and 45.1 (±2.38) μg NO3−-N g−1 in 13C-CO2 and 12C-CO2 treatments at day 30, respectively (Figure 5a). In contrast, addition of DCD significantly hindered the nitrification activity for no apparent nitrate accumulation throughout incubation period. Accordingly, ammonium concentrations in the presence of DCD increased significantly and accumulated from initial 39.2 μg NH4+-N g−1 to 57.1 (±0.46) and 62.2 (±2.47) NH4+-N g−1 in 13C-CO2 + DCD and 12C-CO2 + DCD microcosms at day 30, respectively, indicating a high rate of nitrogen mineralization in this acidic soil (Figure 5b).
Figure 5.
Changes in concentrations of (a) nitrate, (b) ammonium, (c) abundance of thaumarchaeal amoA genes and (d) abundance of bacterial amoA genes during DNA-SIP microcosms of HZ soil for 15 or 30 days. Error bars represent standard errors of triplicate samples. Different letters above the bars indicate a significant difference (P<0.05).
Real-time PCR assays were used to quantify the abundance of thaumarchaeal and bacterial amoA genes in the SIP microcosms. The copy numbers of thaumarchaeal amoA genes in both 13C-CO2 and 12C-CO2 treatments without DCD kept at a high level ranging from 1.84 × 108 to 1.94 × 108 copies g−1 soil (Figure 5c). Addition of DCD significantly reduced the thaumarchaeal amoA gene abundance from 1.84 × 108 to 1.03 × 106 and 2.81 × 106 copies g−1 soil in 13C-CO2 + DCD and 12C-CO2 + DCD incubation after 30 days, respectively (P<0.05), indicating an effective inhibition of DCD on growth of AOA. A significant decrease of bacterial amoA gene abundance was observed in each treatment (P<0.05), showing no significant inhibition effect of DCD on AOB growth at day 15 and day 30 (Figure 5d).
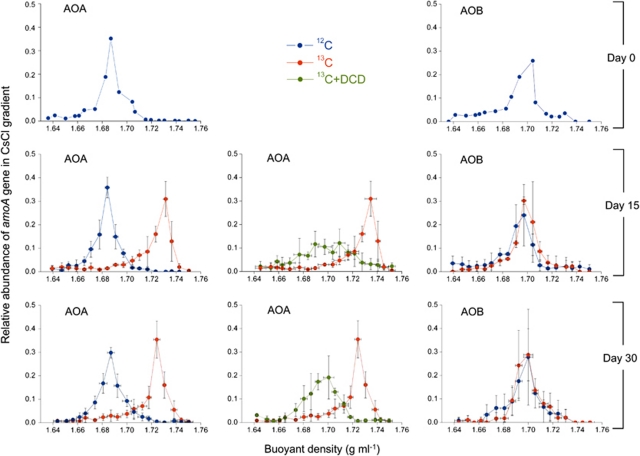
Isopycnic gradient centrifugation was performed with genomic DNA extracted from each treatment to determine which putative autotrophic ammonia oxidizers were labeled by 13C-CO2 during the active nitrification in the SIP incubation. The relative proportion of amoA gene copies in CsCl gradients was calculated from the quantitative PCR of three replicates from each microcosm. The thaumarchaeal amoA genes reached the maximum value for 12C-CO2 control treatment in the light fractions (around a buoyant density of 1.69 g ml−1) throughout the whole incubation (Figure 6). At day 15 and day 30, a clear shift of DNA buoyant density was observed in 13C-CO2 treatment, the relative abundance of thaumarchaeal amoA genes only peaked in the heavy fractions (around a buoyant density of 1.73 g ml−1), which implied the majority of autotrophic AOA assimilated 13C-labeled CO2 during the active nitrification. In 13C-CO2 + DCD treatment, active groups of AOA were effectively inhibited by DCD, which decreased the relative abundance of thaumarchaeal amoA genes in the heavy fractions to lower values at day 15, and even to nearly zero at day 30. With respect to AOB, 15 or 30 day's incubation did not significantly change the distribution of bacterial amoA gene abundance in 13C-CO2 and 12C-CO2 treatments, demonstrating no labeling of 13C-CO2 into bacterial DNA during the active nitrification in the tested soil.
Figure 6.
Distribution of the relative abundance of thaumarchaeal and bacterial amoA gene in CsCl gradient for 13C-CO2, 12C-CO2 and 13C-CO2 + DCD treatments during DNA-SIP microcosms of HZ soil. Twenty fractions of genomic DNA extracted from each 4.9-ml centrifuge tube covered a buoyant density from 1.64 to 1.75 g ml−1. The plotted values are the proportion of thaumarchaeal or bacterial amoA gene copy numbers in each fraction to the total abundance across the gradient. Vertical error bars represent standard errors of the relative abundance from triplicate samples. Horizontal error bars represent standard errors of buoyant density of the same order fraction from triplicate samples.
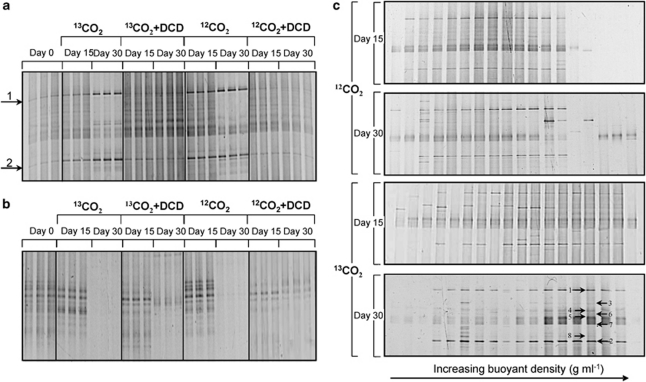
DGGE analysis of the amoA gene PCR products was performed to determine the community changes of thaumarchaeal and bacterial ammonia oxidizers during the incubation (Figure 7). DGGE profiles of thaumarchaeal amoA genes at day 0 were dominated by two bands (bands 1 and 2, Figure 7a), which significantly enhanced their relative intensity in microcosms with 13C-CO2 and 12C-CO2 after 15 or 30 days. However, when active nitrification was blocked by addition of DCD, the two thaumarchaeal amoA bands stayed unchanged in 13C-CO2 + DCD treatment or slightly decreased in their relative intensity in 12C-CO2 + DCD treatment. However, all the bacterial amoA gene band intensity decreased in each of the four treatments after 15 days of incubation (Figure 7b), and even disappeared below the detection limit of DGGE analysis at day 30, which was in accordance with the changes of bacterial amoA gene abundance.
Figure 7.
Denaturing gradient gel electrophoresis analysis of thaumarchaeal (a, c) and bacterial (b) amoA genes during DNA-SIP microcosms of HZ soil incubated with 13C-CO2 or 12C-CO2 in the presence or absence of DCD. Thaumrchaeal amoA gene PCR products of twenty fractions of genomic DNA from individual centrifuge tube were subjected to DGGE for changes of different thaumarchaeal communitys for 15 or 30 days (b). Eight arrows marked the bands in heavy fraction around a buoyant density of 1.73 g ml−1 excised for sequencing.
Thaumarchaeal amoA gene PCR products from twenty fractions of genomic DNA precipitated from each CsCl centrifuge tube were also analyzed by DGGE to separate active communities in microcosms incubated with 13C-CO2 or 12C-CO2 (Figure 7c). During autotrophic growth of specific thaumarchaeal ammonia oxidizers, 13C was incorporated into genomic DNA, which produced a high buoyant density. Apparent transfer of thaumarchaeal amoA genes from light fractions to heavy fractions was observed during incubation with 13C-CO2, with two bands (bands 1 and 2) particularly increasing in their intensity in the high buoyant density region at day 30. The bands intensity of thaumarchaeal amoA gene in the heavy fractions at day 30 was much brighter than that at day 15, indicating a continuous 13C-labeling of AOA during the incubation. Unfortunately, we failed to get successful amplification of bacterial amoA genes with GC-clamped primers after gradient centrifugation, possibly due to the lower DNA concentrations in individual fractions. In fact, the maximal abundance of bacterial amoA gene in the peak fractions ranged from 6.64 × 103 to 1.24 × 104 copies μl−1 CsCl gradient buffer (data not shown).
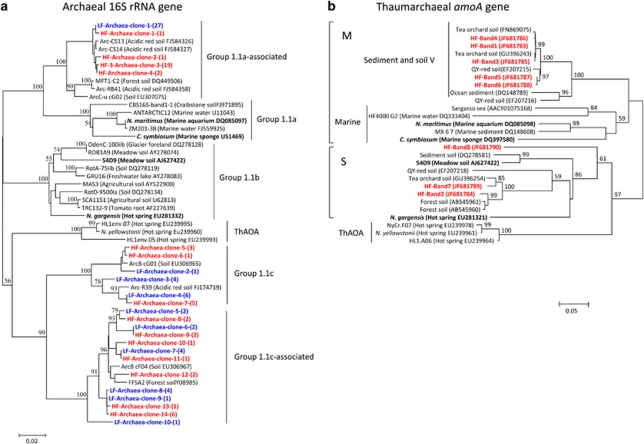
Clone libraries of archaeal 16S rRNA genes were constructed from the light and heavy fractions in the 13CO2 microcosms at day 30, and eight bands representing active thaumarchaeal amoA genes in the heavy fractions around a buoyant density of 1.73 g ml−1 (Figure 7c) were excised and used for clone construction. Phylogenetic analysis revealed that five thaumarchaeal amoA gene band sequences fell within the sediment and soil V cluster in marine lineage previously obtained from marine sequences (Nicol et al., 2008), and are thought presumably to be affiliated with group 1.1a-associated thaumarchaea, while three other DGGE bands belonged to soil and sediment lineage, and are assumed to be related to group 1.1b thaumarchaea (Figure 8b). Many archaeal 16S rRNA sequences (25 out of 52 clones for the light fractions and 24 out of 60 clones for the heavy fractions) were placed within groups 1.1c and 1.1c-associated archaea, which have not yet been observed to participate in ammonia oxidation (Figure 8a).
Figure 8.
Phylogenetic analysis of archaeal 16S rRNA genes (a) retrieved from 13C-CO2 DNA-SIP heavy fractions (HF) and light fractions (LF) of HZ soil microcosms incubated for 30 days, and thaumarchaeal amoA genes (b) derived from eight DGGE bands in the heavy fractions around a buoyant density of 1.73 g ml−1 from the HZ soil incubated for 30 days. The sequences for the HF and LF were highlighted in red and blue, respectively. HF-Archaea-clone-3-(19) indicates that 19 clones showed above 98% similarities to the clone-3. Bootstrap values (>50%) are indicated at branch points. The scale bar represents 2% and 5% nucleic acid sequence divergence for archaeal 16S rRNA genes and thaumarchael amoA genes, respectively. The letters M and S represent Marine Lineage and Soil and Sediment Lineage, respectively.
Discussion
Our studies provide strong evidence for autotrophic ammonia oxidation driven by AOA rather than AOB in the acidic soils tested, which greatly strengthen the hypothesis that AOA and AOB occupy different niches in low-pH environments. First, significant positive relationships between nitrate concentration and thaumarchaeal amoA gene abundance were observed in microcosm incubations with the soil samples (pH<4.50) collected from five distinct sites, indicating a universal phenomenon of AOA dominating the nitrification in acidic soils. Second, assuming that AOA grow chemolithoautotrophically by using inorganic carbon as the sole carbon source via 3-hydroxypropionate/4-hydroxybutyrate pathway (Berg et al., 2007), while AOB use the Calvin-Bassham-Benson cycle to fix CO2 (Schramm et al., 1998), we could use the 13CO2-DNA-SIP approach to identify the active ammonia oxidizers during autotrophic nitrification. By incubating the acidic soil in a headspace of 5% 13C-CO2, incorporation of labeled carbon into genomic DNA was observed for thaumarchaeal amoA genes but not for bacterial genes. In consistence with significant accumulation of nitrate, the intensity of two DGGE bands representing active groups of AOA sharply increased. The abundance of thaumarchaeal amoA genes was maintained at a high level, which would be able to sustain the active nitrification throughout the incubation. By contrast, bacterial amoA gene abundance significantly decreased independently from active nitrification, staying in an inferior position of competing for limited ammonia substrate with thaumarchaea. Therefore, our study presented strong evidence for domination of autotrophic thaumarchaeal ammonia oxidation in the acidic soils.
Ammonia oxidation kinetic study of N. maritimus revealed a half-saturation constant (Km=133 n) for ammonium, by far the lowest Km value of any ammonia oxidizer (Martens-Habbena et al., 2009), and is significantly lower than the Km of known AOB (Stark and Firestone, 1996). Growth of N. maritimus was observed at an ammonium substrate threshold below 10 n at pH 7.5, which is significantly lower than the minimum concentration required by cultivated AOB (above 1 μ near neutral pH; Bollmann et al., 2002), indicating a higher substrate affinity than AOB in ammonia-limited conditions. The recent cultivated thaumarchaeon, N. devanaterra, from an acidic soil could also autotrophically grow at remarkably low ammonia concentration (0.18 n in liquid batch culture at pH 4.5) (Lehtovirta et al., 2011). Assuming ammonia rather than ammonium could serve as the direct substrate for ammonia oxidizers (Suzuki et al., 1974), strongly acidic soils might be perceived as ammonia-limited oligotrophic environments, due to exponential ionization of ammonia to ammonium with decreasing pH (NH3+H+⇔NH4+; pKa=9.25) (De Boer and Kowalchuk, 2001). Although the initial ammonium concentrations seemed relatively high in our five acidic soils ranging from 9.45 to 39.2 mg kg−1 soil, the ammonia concentrations based on the ionization equilibrium in soil water were from 24.6 n to 99.8 n, the same magnitude to the Km value of N. maritimus, but considerably lower than the substrate threshold of AOB. Hence, high levels of ammonium in acidic soils do not necessarily mean a high concentration of ammonia substrate, and upon such low-ammonia energy flux, thaumarchaea outcompete bacteria in the process of ammonia oxidation (Valentine, 2007). Even high rates of organic nitrogen mineralization were observed in SIP microcosms, such ammonia flux would be possibly ionized to ammonium in strongly acidic conditions, which could block the direct delivery of fresh ammonia substrates to ammonia oxidizers. A previous study found that autotrophic ammonia oxidation was dominated by AOA in a pH 7.5 grassland soil without the external nitrogen supply (Zhang et al., 2010), and contrasting findings demonstrated that AOB rather than AOA controlled the nitrification in two agricultural soils with weekly nitrogen fertilization at pH 7.0 (Jia and Conrad, 2009) and 8.0 (Xia et al., 2011), respectively. These three DNA-SIP incubations have a similar soil pH range, but differ in the availability of ammonia substrate. Based on the assumption of 25% water content in these neutral or alkaline soils, the calculated ammonia concentration of the soil tested by Zhang et al. (2010) is reduced from the initial 1.22 μ to 0.48 μ at day 28, which is still lower than the substrate threshold of cultivated AOB. Although ammonium resource was satisfied by weekly supply of 100 μg NH4+-N g−1 soil corresponding to high levels of 120.5 μ and 1.21 × 103 μ ammonia added into the soils tested by Jia and Conrad (2009) and Xia et al. (2011), respectively, such ammonia concentrations might be high enough to stimulate the nitrification of AOB and then suppress the growth of AOA. Therefore, low-ammonia environment is a key factor determining the niche separation of AOA and AOB in both acidic soils and neutral or alkaline soils.
Phylogenetic analyses of archaeal 16S rRNA genes and thaumarchaeal amoA genes retrieved from 13CO2-labeled heavy fractions in DNA-SIP microcosms revealed that active AOA responsible for the ammonia oxidation in the tested strongly acidic soil belonged to groups 1.1a-associated and 1.1b thaumarchaea. Our results are consistent with previous findings that growth of group 1.1a thaumarchaea was associated with autotrophic nitrification in soil microcosms, which seemed to be encouraged by relatively low-level ammonia substrates (Offre et al., 2009; Gubry-Rangin et al., 2010; Zhang et al., 2010). Group 1.1b thaumarchaea comprised only a minority of total archaea in this study (3 out of 60 clones), but 13CO2-incorporating activity demonstrated its active autotrophic ammonia oxidation during incubation. Previous studies suggested that the majority of soil-derived thaumarchaeal sequences were dominated by group 1.1b lineage (Nicol et al., 2008), which had been presumed to possess a heterotrophic or mixotrophic pathway of nitrification until the recent findings that group 1.1b also could autotrophically participate in ammonia oxidation in soil environments (Tourna et al., 2011; Xia et al., 2011). Therefore, both group 1.1a-associated and group 1.1b thaumarchaea are well adapted to low-pH environments, as suggested by the observation that two intensity-increasing DGGE bands of thaumarchaeal amoA genes in our DNA-SIP microcosms were placed within these two distinct lineages. A study performed in an upland acidic soil found considerable AOA community changes in response to long-term fertilization treatments (He et al., 2007), implying a high ability of thaumarchaeal adaptation to low-pH stress. Moreover, many active archaeal 16S rRNA gene sequences derived from both the light and heavy fractions are affiliated with group 1.1c and 1.1c-associated archaea, which is in congruence with previous observations that a high abundance of group 1.1c lineage dominated the archaeal communities in many acidic soil systems (Kemnitz et al., 2007; Lehtovirta et al., 2009). However, currently there is no direct evidence that group 1.1c archaea possess ammonia monooxyenase genes, and the links between thaumarchaeal amoA genes with archaeal 16S rRNA genes seemed less likely in this study. But, we could not entirely rule out the possibility of group 1.1c involving in ammonia oxidation, because the primers used for amoA genes here were designed based on group 1.1a Sargasso sea data and group 1.1b 54d9 (Tourna et al., 2008), which could be biased against targeting group 1.1c archaea, thus the potential ecological function of this lineage in acidic soils requires further investigation in the future researches.
Addition of DCD significantly hindered the nitrification process mainly by inhibiting the AOA in our five acidic soils, and no significant inhibition effect on AOB was found throughout the incubation. Inhibition of soil ammonia oxidation by DCD has been investigated extensively (Di et al., 2009, 2010; O'Callaghan et al., 2010), however, as far as we know, it is the first finding that AOA was significantly inhibited by DCD in a soil microcosm study. DCD has been found to reduce or inhibit the nitrification rate mainly by binding to the active sites of AMO enzyme and impairing the activity of ammonia oxidizers to take up and utilize ammonium resources (Amberger, 1989; Zacherl and Amberger, 1990). As shown in DGGE profiles (Figure 7a), two bands representing active nitrifying groups of AOA were significantly reduced by the nitrification inhibitor DCD. Moreover, addition of DCD effectively impeded the CO2 fixation of thaumarchaea, and decreased the relative proportion of thaumarchaeal amoA genes in the high buoyant density region of CsCl gradients. Hence, DCD might inhibit the nitrification activity in acidic soils by mainly targeting the active groups of AOA in this study. Thus, DCD is not a selective nitrification inhibitor exclusively targeting AOA or AOB, but the dominating ammonia oxidizer group is affected by DCD to a greater extent. This hypothesis was well supported by the similar observations that growth of AOA was inhibited by a nitrification inhibitor acetylene in soil microcosms where AOA dominated ammonia oxidation (Offre et al., 2009; Gubry-Rangin et al., 2010; Zhang et al., 2010).
In conclusion, our studies demonstrate a significant and previously unrecognized contribution of thaumarchaea to mediate the autotrophic nitrification in strongly acidic soils. This result will greatly improve the understanding of the nitrogen-transformation processes in widely distributed acidic soils. The first observation of DCD inhibition on AOA in this study and previous findings of DCD inhibition on AOB suggest that DCD will be a promising nitrification inhibitor to prevent the environmental degradation imposed by intensive fertilization and promote nitrogen fertilization efficiency, which will generate enormous economic impacts in agricultural ecosystems.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (41025004, 40871129, 41020114001, 50921064) and the Chinese Academy of Sciences (KZCX2-YW-JC401). We would like to thank Dr Wenyan Han for access to the tea orchard and Dr Huaiying Yao and Qichun Zhang for assistance in soil sampling.
References
- Amberger A. Research on dicyandiamide as a nitrification inhibitor and future outlook. Commun Soil Sci Plant Anal. 1989;20:1933–1955. [Google Scholar]
- Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in archaea. Science. 2007;318:1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- Bollmann A, Bar-Gilissen MJ, Laanbroek HJ. Growth at low ammonium concentrations and starvation response as potential factors involved in niche differentiation among ammonia-oxidizing bacteria. Appl Environ Microbiol. 2002;68:4751–4757. doi: 10.1128/AEM.68.10.4751-4757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- De Boer W, Kowalchuk GA. Nitrification in acid soils: micro-organisms and mechanisms. Soil Biol Biochem. 2001;33:853–866. [Google Scholar]
- Delong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di HJ, Cameron KC, Shen JP, Winefield CS, O'Callaghan M, Bowatte S, et al. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci. 2009;2:621–624. [Google Scholar]
- Di HJ, Cameron KC, Shen JP, Winefield CS, O'Callaghan M, Bowatte S, et al. Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol Ecol. 2010;72:386–394. doi: 10.1111/j.1574-6941.2010.00861.x. [DOI] [PubMed] [Google Scholar]
- Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag TE, Chang L, Prosser JI. Changes in the community structure and activity of betaproteobacterial ammonia-oxidizing sediment bacteria along a freshwater-marine gradient. Environ Microbiol. 2006;8:684–696. doi: 10.1111/j.1462-2920.2005.00947.x. [DOI] [PubMed] [Google Scholar]
- Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol. 2000;66:5488–5491. doi: 10.1128/aem.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubry-Rangin C, Nicol GW, Prosser JI. Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol. 2010;74:566–574. doi: 10.1111/j.1574-6941.2010.00971.x. [DOI] [PubMed] [Google Scholar]
- Guo JH, Liu XJ, Zhang Y, Shen JL, Han WX, Zhang WF, et al. Significant acidification in major Chinese croplands. Science. 2010;327:1008–1010. doi: 10.1126/science.1182570. [DOI] [PubMed] [Google Scholar]
- Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, et al. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine crenarchaeota. PLoS Biol. 2006;4:520–536. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankinson TR, Schmidt EL. An acidophilic and a neutrophilic nitrobacter strain isolated from the numerically predominant nitrite-oxidizing population of an acid forest soil. Appl Environ Microbiol. 1988;54:1536–1540. doi: 10.1128/aem.54.6.1536-1540.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JZ, Shen JP, Zhang LM, Zhu YG, Zheng YM, Xu MG, et al. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol. 2007;9:2364–2374. doi: 10.1111/j.1462-2920.2007.01358.x. [DOI] [PubMed] [Google Scholar]
- Islam A, Chen D, White RE. Heterotrophic and autotrophic nitrification in two acid pasture soils. Soil Biol Biochem. 2007;39:972–975. [Google Scholar]
- Jia ZJ, Conrad R. Bacteria rather than archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol. 2009;11:1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x. [DOI] [PubMed] [Google Scholar]
- Jiang QQ, Bakken LR. Comparison of nitrosospira strains isolated from terrestrial environments. FEMS Microbiol Ecol. 1999;30:171–186. doi: 10.1111/j.1574-6941.1999.tb00646.x. [DOI] [PubMed] [Google Scholar]
- Jordan FL, Cantera JJL, Fenn ME, Stein LY. Autotrophic ammonia-oxidizing bacteria contribute minimally to nitrification in a nitrogen-impacted forested ecosystem. Appl Environ Microbiol. 2005;71:197–206. doi: 10.1128/AEM.71.1.197-206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemnitz D, Kolb S, Conrad R. High abundance of crenarchaeota in a temperate acidic forest soil. FEMS Microbiol Ecol. 2007;60:442–448. doi: 10.1111/j.1574-6941.2007.00310.x. [DOI] [PubMed] [Google Scholar]
- Kettler TA, Doran JW, Gilbert TL. Simplified method for soil particle-size determination to accompany soil-quality analyses. Soil Sci Soc Am J. 2001;65:849–852. [Google Scholar]
- Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Lehtovirta LE, Prosser JI, Nicol GW. Soil pH regulates the abundance and diversity of group 1.1c crenarchaeota. FEMS Microbiol Ecol. 2009;70:367–376. doi: 10.1111/j.1574-6941.2009.00748.x. [DOI] [PubMed] [Google Scholar]
- Lehtovirta LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci USA. 2011;108:15892–15897. doi: 10.1073/pnas.1107196108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying archaea and bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- Mintie AT, Heichen RS, Cromack K, Myrold DD, Bottomley PJ. Ammonia-oxidizing bacteria along meadow-to-forest transects in the oregon cascade mountains. Appl Environ Microbiol. 2003;69:3129–3136. doi: 10.1128/AEM.69.6.3129-3136.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- Nicol GW, Schleper C. Ammonia-oxidising crenarchaeota: important players in the nitrogen cycle. Trends Microbiol. 2006;14:207–212. doi: 10.1016/j.tim.2006.03.004. [DOI] [PubMed] [Google Scholar]
- O'Callaghan M, Gerard EM, Carter PE, Lardner R, Sarathchandra U, Burch G, et al. Effect of the nitrification inhibitor dicyandiamide (DCD) on microbial communities in a pasture soil amended with bovine urine. Soil Biol Biochem. 2010;42:1425–1436. [Google Scholar]
- Offre P, Prosser JI, Nicol GW. Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol Ecol. 2009;70:99–108. doi: 10.1111/j.1574-6941.2009.00725.x. [DOI] [PubMed] [Google Scholar]
- Pratscher J, Dumont MG, Conrad R. Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. Proc Natl Acad Sci USA. 2011;108:4170–4175. doi: 10.1073/pnas.1010981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotthauwe JH, Witzel KP, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CS, Hultman KA, Robinson D, Killham K, Prosser JI. PCR profiling of ammonia-oxidizer communities in acidic soils subjected to nitrogen and sulphur deposition. FEMS Microbiol Ecol. 2007;61:305–316. doi: 10.1111/j.1574-6941.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- Schramm A, de Beer D, Wagner M, Amann R. Identification and activities in situ of nitrosospira and nitrospira spp. As dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol. 1998;64:3480–3485. doi: 10.1128/aem.64.9.3480-3485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, Spieck E, et al. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Stark JM, Firestone MK. Kinetic characteristics of ammonium-oxidizer communities in a california oak woodland-annual grassland. Soil Biol Biochem. 1996;28:1307–1317. [Google Scholar]
- Stopnisek N, Gubry-Rangin C, Hofferle S, Nicol GW, Mandic-Mulec I, Prosser JI. Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl Environ Microbiol. 2010;76:7626–7634. doi: 10.1128/AEM.00595-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki I, Dular U, Kwok SC. Ammonia or ammonium ion as substrate for oxidation by nitrosomonas-europaea cells and extracts. J Bacteriol. 1974;120:556–558. doi: 10.1128/jb.120.1.556-558.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tourna M, Freitag TE, Nicol GW, Prosser JI. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ Microbiol. 2008;10:1357–1364. doi: 10.1111/j.1462-2920.2007.01563.x. [DOI] [PubMed] [Google Scholar]
- Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA. 2011;108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine DL. Adaptations to energy stress dictate the ecology and evolution of the archaea. Nat Rev Microbiol. 2007;5:316–323. doi: 10.1038/nrmicro1619. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the sargasso sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Verhamme DT, Prosser JI, Nicol GW. Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J. 2011;5:1067–1071. doi: 10.1038/ismej.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitousek PM, Aber JD, Howarth RW, Likens GE, Matson PA, Schindler DW, et al. Human alteration of the global nitrogen cycle: sources and consequences. Ecol Appl. 1997;7:737–750. [Google Scholar]
- Vonuexkull HR, Mutert E. Global extent, development and economic-impact of acid soils. Plant Soil. 1995;171:1–15. [Google Scholar]
- Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace A. Soil acidification from use of too much fertilizer. Commun Soil Sci Plan Anal. 1994;25:87–92. [Google Scholar]
- Xia W, Zhang C, Zeng X, Feng Y, Weng J, Lin X, et al. Autotrophic growth of nitrifying community in an agricultural soil. ISME J. 2011;5:1226–1236. doi: 10.1038/ismej.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D, Yao HY, Huang CY. Microbial biomass, N mineralization and nitrification, enzyme activities, and microbial community diversity in tea orchard soils. Plant Soil. 2006;288:319–331. [Google Scholar]
- Yao HY, Gao YM, Nicol GW, Campbell CD, Prosser JI, Zhang LM, et al. Links between ammonia oxidizer community structure, abundance and nitrification potential in acidic soils. Appl Environ Microbiol. 2011;77:4618–4625. doi: 10.1128/AEM.00136-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacherl B, Amberger A. Effect of the nitrification inhibitors dicyandiamide, nitrapyrin and thiourea on nitrosomonas-europaea. Fert Res. 1990;22:37–44. [Google Scholar]
- Zaman M, Blennerhassett JD. Effects of the different rates of urease and nitrification inhibitors on gaseous emissions of ammonia and nitrous oxide, nitrate leaching and pasture production from urine patches in an intensive grazed pasture system. Agr Ecosyst Environ. 2010;136:236–246. [Google Scholar]
- Zhang LM, Offre PR, He JZ, Verhamme DT, Nicol GW, Prosser JI. Autotrophic ammonia oxidation by soil thaumarchaea. Proc Natl Acad Sci USA. 2010;107:17240–17245. doi: 10.1073/pnas.1004947107. [DOI] [PMC free article] [PubMed] [Google Scholar]