Abstract
Recent applications of molecular genetics to edaphic microbial communities of the McMurdo Dry Valleys and elsewhere have rejected a long-held belief that Antarctic soils contain extremely limited microbial diversity. The Inter-Valley Soil Comparative Survey aims to elucidate the factors shaping these unique microbial communities and their biogeography by integrating molecular genetic approaches with biogeochemical analyses. Although the microbial communities of Dry Valley soils may be complex, there is little doubt that the ecosystem's food web is relatively simple, and evidence suggests that physicochemical conditions may have the dominant role in shaping microbial communities. To examine this hypothesis, bacterial communities from representative soil samples collected in four geographically disparate Dry Valleys were analyzed using molecular genetic tools, including pyrosequencing of 16S rRNA gene PCR amplicons. Results show that the four communities are structurally and phylogenetically distinct, and possess significantly different levels of diversity. Strikingly, only 2 of 214 phylotypes were found in all four valleys, challenging a widespread assumption that the microbiota of the Dry Valleys is composed of a few cosmopolitan species. Analysis of soil geochemical properties indicated that salt content, alongside altitude and Cu2+, was significantly correlated with differences in microbial communities. Our results indicate that the microbial ecology of Dry Valley soils is highly localized and that physicochemical factors potentially have major roles in shaping the microbiology of ice-free areas of Antarctica. These findings hint at links between Dry Valley glacial geomorphology and microbial ecology, and raise previously unrecognized issues related to environmental management of this unique ecosystem.
Keywords: Antarctica, Dry Valleys, geochemistry, mineral soil
Introduction
Composed of glacially carved valleys separated from the Polar Plateau by the Transantarctic Mountains, the McMurdo Dry Valleys compose the largest inland ice-free area of the Antarctic continent (Bockheim and McLeod, 2008). Widely considered the coldest, driest desert on Earth (Hopkins et al., 2006) and used by NASA as an analog for Mars, the Dry Valleys often receive <10 cm per year water-equivalent precipitation, most of which arrives in the form of snow that sublimes almost instantly (Marchant and Head, 2007; Cary et al., 2010). High salt content in Dry Valley soils further reduces the amount of available water and imposes additional osmotic barriers on Dry Valley biota (Wynn-Williams, 2000). The diverse glacial geomorphology (that is, the history of geological processes that shaped the environment) of the Dry Valleys is manifested in high degrees of physicochemical heterogeneity in the region, including altitude, surface geology, soil types and weathering stages, and soil salt content (Bockheim, 2002; Barrett et al., 2004; Marchant and Head, 2007; Bockheim and McLeod, 2008; Block et al., 2009).
Because of the absence of vascular plants and macrofauna, the Dry Valleys has long been considered highly hostile to life (Scott, 1905; Wynn-Williams and Edwards, 1990; Vishniac, 1993). Furthermore, the limitation of culture-based techniques (Rappé and Giovannoni, 2003; Janssen, 2006), the lack of direct data on bacterial and archaeal diversity (Adams et al., 2006), and the belief that aeolian transport is the prevailing mechanism for microbiota distribution within the region (Moorhead et al., 1999; Hopkins et al., 2006; Nkem et al., 2006) collectively contributed to a prevailing notion that the microbial ecology of Dry Valley mineral soils is dominated by a few cosmopolitan species homogeneously distributed across the Dry Valley landscape (Horowitz et al., 1972; Johnson et al., 1978; Friedmann et al., 1993; Vishniac, 1993). Recent studies utilizing molecular genetic methods have challenged this view and showed that Dry Valley soils can harbor bacterial communities far more active and diverse than previously expected (La Torre de et al., 2003; Smith et al., 2006; Niederberger et al., 2008; Wood et al., 2008a; Babalola et al., 2009; Cary et al., 2010), but the forces driving this apparent microbial diversity remain unknown (Adams et al., 2006; Cary et al., 2010).
It has been shown that abiotic elements are the dominant factors determining biodiversity in extremely simplified food webs (Convey, 1996; Doran et al., 2002; Hogg et al., 2006; Poage et al., 2008). The lack of clear biotic interactions (Barrett et al., 2006a) and diverse abiotic conditions make the Dry Valley soil ecosystem a highly, if not the most, tractable model for resolving the roles of abiotic factors in structuring soil microbiota (Wall and Virginia, 1999; Hogg et al., 2006; Hopkins et al., 2006). Furthermore, the importance of abiotic factors as the dominant driver of biodiversity in the Dry Valleys has long been recognized for microfauna and microflora (Adams et al., 2006; Convey et al., 2009), and although some reports have hinted at heterogeneous microbial communities in disparate parts of the Dry Valleys (Smith et al., 2006; Niederberger et al., 2008; Smith et al., 2010), no systematic survey of Dry Valley habitats across well-defined physicochemical heterogeneities have been undertaken (Adams et al., 2006; Cary et al., 2010).
Here we present findings from a coordinated comparative survey of soil bacterial communities from four topographically and geomorphologically diverse Dry Valleys, coupling molecular genetic techniques with biogeochemical analyses. In addition to describing the microbial communities associated with four distinct habitats and establishing an ecological framework of Dry Valley soils, we aim to uncover potential links between microbial ecology and environmental variables, which are potentially reflective of historical and ongoing geological processes (Virginia and Wall, 1999).
Materials and methods
Sample collection
Mineral soils representative of local soil types were collected from four study sites in the McMurdo Dry Valleys: Miers Valley (78°05.486′S, 163°48.539′E, elevation 171 m), Beacon Valley (77°52.321′S, 160°29.725′E, elevation 1376 m), Upper Wright Valley (77°31.125′S, 160°45.813′E, elevation 947 m) and Battleship Promontory (76°54.694′S, 160°55.676′E, elevation 1028 m). Detailed descriptions of each valley are available in Supplementary Information. Samples were collected in December 2006 (Miers Valley and Beacon Valley) and January 2008 (Battleship Promontory and Upper Wright Valley). The sampling sites were all located on a south-facing, 0–20° slope and consisted of two 50 m transects crossing in the middle, with the intersection being the central sampling point (X) and four sampling points (A–D) at the end point of the transects (A being the southernmost point and the remaining points in an anti-clockwise order). At each point, a 1-m2 sampling area was identified, and from each corner one scoop of soil was collected from the top 2 cm, after soil pavement pebbles were removed. All reasonable precautions were taken to avoid anthropogenic and intersite contamination, and samples were stored in a sterile Whirl-Pak (Nasco International, Fort Atkinson, WI, USA). Samples were stored at −20 °C at the earliest opportunity and transported back to our laboratory in New Zealand for further analysis.
DNA extraction and community fingerprinting analysis
DNA was extracted using the bead-beating method (Coyne et al., 2001), which has been shown to be highly effective for Dry Valley soil samples (Niederberger et al., 2008) (see also Supplementary Information). Molecular genetic analysis was carried out using automated ribosomal intergenic spacer analysis (ARISA), which resolves community diversity through the hyper-variability of the 16S–23S internal transcribed spacer (ITS). PCR for ARISA was carried out as previously described (Wood et al., 2008a) using both bacterial and cyanobacterial universal primers. The bacterial primers were ITSF (5′-HEX-GTCGTAACAAGGTAGCCGTA-3′) and ITSReub (5′-GCCAAGGCAATCCCACC-3′) (Jones et al., 2007), and the cyanobacterial primers were CY-ARISA-F (5′-FAM-GYCAYRCCCGAAGTCRTTAC-3′) and 23S30R (5′-CHTCGCCTCTGTGTGCCWAGGT-3′) (Wood et al., 2008a). PCR components and conditions, and quality control procedure for PCR amplicons are described in Supplementary Information. For ARISA fragment length (AFL) analysis, 2 μl of diluted PCR product was mixed with 0.25 μl of ROX-labeled genotyping internal size standard (ETR900-R, GE Healthcare, Buckinghamshire, UK) and diluted to 10 μl final volume using 20% Tween-20. AFLs were determined by capillary electrophoresis using the MegaBACE 500 DNA Analysis system (GE Healthcare). ARISA was performed twice for each sample.
Soil physicochemical analysis
pH and conductivity of soil samples were measured on a CyberScan PC 510 Bench Meter (Eutech Instruments Pte Ltd, Singapore) using the slurry technique, which consists of mixing 2 g of soil with 5 ml of deionized water and allowing the samples to settle. Soil gravimetric water content was determined by gravimetrically drying 40 g of air-dried soil at 105 °C until the sample reached a constant weight.
For total percentage carbon and nitrogen measurements, air-dried soil samples were ground to fine powders using a ball grinder and precisely weighed out to 0.2 g aliquots. The samples were then analyzed using a LECO Truspec Elemental Determinator (LECO Corporation, St Joseph, MI, USA). The ground soil samples were also acid digested following the United States EPA 200-2 analysis method for elemental analysis (see Supplementary Information) using inductively coupled plasma mass spectrometry (ICP/MS).
454 pyrosequencing of PCR amplicons
Four sets of barcoded PCR primers were designed to allow direct 454 tag sequencing of four different samples within a single region of a 454 GS FLX Standard (that is, non-titanium) PicoTiterPlate (454 Life Sciences, Branford, CT, USA). The PCR primers consisted of three components: 5′-[454 GS FLX adapter A/B]+[4 nt barcode (aka MID)]+[forward/reverse gene-specific PCR primer]-3′. The 454 GS FLX adapter sequences were 5′-GCCTCCCTCGCGCCATCAG-3′ (adapter A) and 5′-GCCTTGCCAGCCCGCTCAG-3′ (adapter B). The barcode sequences were 5′-AGTC-3′ (Battleship Promontory), 5′-ACAT-3′ (Beacon Valley), 5′-AGAC-3′ (Miers Valley) and 5′-ACGT-3′ (Upper Wright Valley). The forward PCR primer was 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and the reverse PCR primer was 806R (5′-GGACTACCAGGGTATCTAAT-3′) (Huws et al., 2007). Adapter A was always coupled with 338F, and adapter B with 806R. The predicted amplicon size for these primers was 511 bp.
Genomic DNA samples extracted from individual sampling points at a study site (that is, valley) were pooled at equal weight ratio and used as PCR template. PCR components and conditions, and quality control procedure for PCR amplicons are described in Supplementary Information. The prepared PCR amplicons were sent to the University of Otago High-Throughput DNA Sequencing Unit (Dunedin, New Zealand), where they were pooled at equal weight ratios and used to generate two emPCR libraries (for sequencing from adapter A and adapter B, respectively). The resulting emPCR products were pooled at equal weight ratios and sequenced using 1/16th region of a 454 GS FLX Standard PicoTiterPlate.
Sequence analysis
454 PCR amplicon pyrosequencing data was analyzed using AmpliconNoise v1.0. (Quince et al., 2011). Briefly, raw flowgrams (sff files) with perfectly matching primer and barcode sequences were filtered for a minimum flowgram length of 360 cycles (including primer and barcode sequences) before the first noisy signal (that is, 0.5–0.7 or no signal in all four nucleotides). All flowgrams were then truncated at 360 cycles and clustered to remove sequencing noise using PyroNoise (Quince et al., 2009, 2011). PCR noise was removed using SeqNoise (Quince et al., 2011), and PCR chimeras were removed using Perseus (Quince et al., 2011). The resulting dereplicated sequences were aligned using ClustalW 2.0.12 (Thompson et al., 1994), NAST (DeSantis et al., 2006b) and PRANK+F v.090707 (Löytynoja and Goldman, 2008). Default parameters were used for ClustalW, and the +F (for robustness against indel errors) and -uselogs (to enable alignment of large numbers of sequences) options were used for PRANK+F. NAST alignment was performed using the Greengenes reference alignment and default parameters (DeSantis et al., 2006a, 2006b). Mothur 1.10.2 (Schloss et al., 2009) was used to calculate sequence distance matrices and cluster sequences into operational taxonomic units (OTUs) defined at the furthest neighbor Jukes–Cantor distance of 0.03 (OTU0.03), the most stringent OTU definition allowing for intragenomic 16S rRNA variation and PCR/sequencing errors (Schloss and Handelsman, 2006; Kunin et al., 2009). Rank-abundance data were generated for each study site and groups of study sites, and rarefaction curves, collector's curves, and other population diversity indices (for example, ChaoI and ACE) were calculated. Community similarity trees, Venn diagrams and taxonomic heat maps were also generated using Mothur, and libshuff tests were performed. For phylogenetic assignments, representative sequences of all identified OTU0.03 were analyzed using the Classifier function provided by the Ribosomal Database Project Release 10, Update 15 (Wang et al., 2007). Taxonomic assignment threshold was set at 80%.
Statistical analysis
A one-way analysis of variance was performed on the soil physicochemical data using Statistica (Version 7, StatSoft Inc., Tulsa, AZ, USA) to identify significant differences in soil chemistry between sampling sites. Soil physicochemical data were also imported into the PRIMER 6 package (PRIMER-E Ltd, Ivybridge, UK), where the data were first transformed using a square-root transformation, followed by a log (X+1) transformation, and normalized. A distance matrix was generated based on Euclidean distances, with which hierarchical clustering (group average), non-metric multidimensional scaling (MDS) and one-way analysis of similarities (ANOSIM) analyses were performed.
ARISA run data were processed using Genetic Profiler (Version 2, GE Healthcare) to yield all detectable peaks without applying any arbitrary cutoff. The resulting data were processed using a modified version of a previously described program (Abdo et al., 2006) (see Supplementary Information for detailed parameters), and AFL profiles were then imported into PRIMER 6 and converted into presence/absence data. Data from multiple ARISA runs were averaged for each sample, and a similarity matrix was generated based on Bray–Curtis similarity, on which hierarchical clustering, MDS and ANOSIM analyses were performed. Biota-Environmental STepwise and LINKTREE analyses (Clarke et al., 2008) were performed using the transformed and normalized soil physicochemical data as environmental variables, and the ARISA similarity matrices as biotic factors. For Biota-Environmental STepwise analysis, the BIOENV method with Spearman ranking was used, and 99 permutation tests were run (none were ⩾ρ).
Results
Soil geochemistry
Overall, soil physicochemical properties varied significantly among valleys (Table 1). One-way ANOSIM reported a global R-value of 0.771 (R-value of one indicates that the most similar samples are all within the same study site) and an associated P-value of 0.001, indicating significant physicochemical differences between the study sites. When analyzed separately, the ICP/MS profiles for all four valleys were also statistically different from each other (ANOSIM, global R-value=0.691, P-value=0.001). Results from ANOSIM pairwise tests show that Miers Valley was most different from all other valleys (Supplementary Table 1). Copper concentration in Beacon Valley was considerably higher than all other valleys, corroborating previous reports (Wood et al., 2008a).
Table 1. Soil geochemical properties.
| Beacon Valley | Upper Wright Valley | Battleship Promontory | Miers Valley | |
|---|---|---|---|---|
| pH | 7.10 (0.28) | 6.96 (0.09) | 7.68 (0.51) | 8.62 (0.31) |
| Conductivity (μS) | 3920 | 6130 | 107 | 300 |
| Gravimetric water content (%) | 2.36 (0.16) | 1.07 (0.03) | 1.14 (0.05) | 0.53 (0) |
| % C | 0.14 (0.04) | 0.11 (0.01) | 0.1 (0) | 0.46 (0.16) |
| % N | 0.08 (0.01) | 0.12 (0.02) | 0.04 (0.01) | 0.05 (0.02) |
| C/N | 1.80 (0.46) | 0.98 (0.23) | 2.25 (0.25) | 18.22 (20.07) |
| Ag | 0.02 (0.03) | 0.00 (0.00) | 0.00 (0.00) | 0.01 (0.02) |
| Al | 13636 (3633) | 10016 (2463) | 12345 (1555) | 11721 (2426) |
| As | 1.10 (0.25) | 0.98 (0.11) | 0.58 (0.14) | 0.93 (0.71) |
| B | 507 (28) | 514 (16) | 519 (21) | 505 (24) |
| Ba | 27.5 (19.8) | 20.0 (3.9) | 12.3 (2.0) | 75.2 (19.8) |
| Bi | 0.04 (0.01) | 0.03 (0.01) | 0.01 (0.00) | 0.47 (0.97) |
| Ca | 8094 (2208) | 2747 (984) | 4906 (573) | 12337 (2625) |
| Cd | 0.10 (0.04) | 0.09 (0.04) | 0.07 (0.02) | 0.13 (0.02) |
| Co | 11.2 (1.5) | 6.3 (1.2) | 8.1 (1.2) | 15.4 (3.0) |
| Cr | 5.5 (1.8) | 4.3 (0.7) | 3.7 (0.3) | 25.9 (5.0) |
| Cu | 73.7 (12.6) | 33.1 (5.6) | 49.9 (8.0) | 11.9 (2.7) |
| Fe | 24386 (5435) | 13879 (2020) | 15972 (1896) | 22165 (4260) |
| Hf | 0.20 (0.07) | 0.19 (0.05) | 0.14 (0.03) | 0.30 (0.10) |
| Hg | 0.02 (0.02) | 0.01 (0.01) | 0.01 (0.00) | 0.02 (0.01) |
| In | 0.01 (0.00) | 0.01 (0.00) | 0.01 (0.00) | 0.02 (0.01) |
| K | 1049 (442) | 1876 (1058) | 513 (67) | 2579 (572) |
| Li | 7.1 (1.9) | 6.7 (1.2) | 4.2 (0.6) | 8.5 (1.7) |
| Mg | 5281 (1071) | 3941 (697) | 2762 (372) | 20345 (3408) |
| Mn | 213 (49) | 150 (46) | 159 (23) | 367 (68) |
| Na | 2213 (727) | 3515 (1773) | 1400 (215) | 3884 (692) |
| Ni | 14.5 (2.8) | 9.1 (1.3) | 11.3 (1.6) | 84.8 (15.7) |
| P | 487 (88) | 259 (52) | 360 (57) | 875 (167) |
| Pb | 4.42 (0.98) | 4.57 (0.86) | 1.81 (0.25) | 2.11 (0.63) |
| Se | 0.79 (0.19) | 0.56 (0.08) | 0.58 (0.11) | 0.48 (0.13) |
| Si | 943 (38) | 944 (108) | 881 (62) | 6194 (1863) |
| Sr | 35.2 (10.1) | 16.2 (3.9) | 18.6 (2.4) | 160.6 (41.1) |
| Ti | 417 (166) | 230 (48) | 431 (39) | 2556 (622) |
| Tl | 0.04 (0.02) | 0.04 (0.01) | 0.02 (0.01) | 0.04 (0.02) |
| U | 0.55 (0.10) | 0.52 (0.23) | 0.45 (0.06) | 0.58 (0.15) |
| V | 77.9 (28.5) | 32.3 (6.0) | 40.4 (4.6) | 39.2 (7.9) |
| Zn | 52.1 (11.0) | 32.3 (4.9) | 31.6 (4.7) | 52.2 (3.5) |
| Zr | 7.2 (2.6) | 9.0 (3.5) | 5.1 (1.0) | 15.4 (4.7) |
Unless noted, values (including C/N ratio) represent averages and s.d. (in parentheses) calculated from five sampling points from each study site. Conductivity measurements were taken using samples pooled from all five sampling points, and gravimetric water content was measured for sampling points A and C for each study site. Elemental concentrations are in ppm.
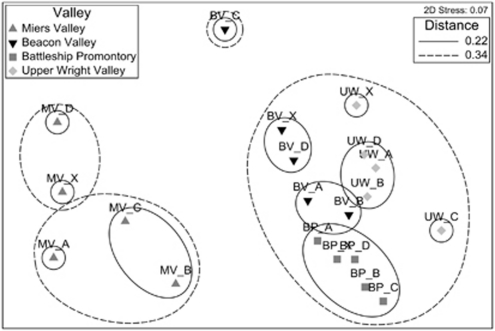
The relationship between physicochemical profiles of all individual samples from four valleys is represented in a non-metric MDS plot (Figure 1), in which Miers Valley is clearly an outlier to the remaining valleys. This observation is supported by the pairwise ANOSIM results (Supplementary Table 1). Soil physicochemical properties are largely consistent within individual study sites, with the exception of sampling point C from Beacon Valley (BV_C) (Figure 1). This is likely because of its high Ag and Hg content (3–5-fold higher than average, Supplementary Table 2).
Figure 1.
MDS plot of soil physicochemical profiles. An Euclidean distance matrix of soil physicochemical profiles was calculated using soil physicochemical properties that have been square-root and log (X+1) transformed and normalized. A MDS plot was then generated using the distance matrix to represent the relative distances between individual samples. The samples are represented using the following symbols: Miers Valley (MV), gray triangles ( ); Beacon Valley (BV), upturned black triangles (▾); Battleship Promontory (BP), gray squares (
); Beacon Valley (BV), upturned black triangles (▾); Battleship Promontory (BP), gray squares ( ); Upper Wright Valley (UW), gray diamonds (
); Upper Wright Valley (UW), gray diamonds ( ). Contour lines enclose samples with distances below certain thresholds (0.22, solid line; 0.34, dashed line).
). Contour lines enclose samples with distances below certain thresholds (0.22, solid line; 0.34, dashed line).
Community profiling using ARISA
Significantly more bacterial AFL groups (AFLs, each representing at least one phylotype) were identified in Miers Valley and Battleship Promontory samples than Beacon Valley and Upper Wright Valley (Supplementary Table 3), an observation supported by one-way global analysis of variance performed on the number of bacterial AFLs (F-statistic=9.4, P-value <0.001). More bacterial AFLs were identified than cyanobacterial ones for all the study sites (Supplementary Table 3) and Miers Valley appears to harbor a moderately higher level of cyanobacterial diversity (one-way global analysis of variance, F-statistic=3.6, P-value=0.037) relative to other study sites (Supplementary Table 3).
Global ANOSIM analysis of the ARISA data revealed that the cyanobacterial communities (R-value=0.775, P-value=0.001) and overall bacterial communities (R-value=0.709, P-value=0.001) of the four study sites were significantly different from each other, suggesting localized microbial communities. MDS plots of ARISA data (Figures 2a and b) show that Miers Valley and Battleship Promontory samples strongly group within respective valleys. Meanwhile, samples from Beacon Valley and Upper Wright Valley appear to be more poorly resolved, an observation reflected in the pairwise ANOSIM data (Supplementary Table 4). The MDS plot of bacterial ARISA data shows that bacterial communities in the four study sites fall into two groups: one consisting of Miers Valley and Battleship Promontory, and the other containing Beacon Valley and Upper Wright Valley. This is different from the relationship shown in the MDS plot of cyanobacterial ARISA data, in which the cyanobacterial community of Miers Valley can be seen as an outlier to the other three study sites.
Figure 2.
MDS plots of bacterial (a) and cyanobacterial (b) ARISA profiles. Bray–Curtis similarity matrices of bacterial and cyanobacterial ARISA profiles were calculated based on presence/absence of AFLs. MDS plots were then generated using the distance matrices to represent the relative distances between individual samples. The samples are represented using the following symbols: Miers Valley (MV), gray triangles ( ); Beacon Valley (BV), upturned black triangles (▾); Battleship Promontory (BP), gray squares (
); Beacon Valley (BV), upturned black triangles (▾); Battleship Promontory (BP), gray squares ( ); Upper Wright Valley (UW), gray diamonds (
); Upper Wright Valley (UW), gray diamonds ( ). Contour lines enclose samples with similarities above certain thresholds (bacterial ARISA: 0.20, solid line; 0.40, dashed line) (cyanobacterial ARISA: 0.20, solid line).
). Contour lines enclose samples with similarities above certain thresholds (bacterial ARISA: 0.20, solid line; 0.40, dashed line) (cyanobacterial ARISA: 0.20, solid line).
Sequencing results
To identify dominant bacteria in the four study sites, PCR amplicons of partial bacterial 16S rRNA gene (hypervariable regions V3–V4) were bidirectionally sequenced, and 4092 raw pyrosequencing reads were obtained. The flowgrams were processed using AmpliconNoise (Quince et al., 2011) for quality filtering, denoising and chimera removal. In total, 2108 reads from adapter B (that is, the reverse PCR primer) passed the quality control and chimera check pipeline, and no forward reads were obtained. Of the 999 reads that contained incorrect barcode or primer sequences and failed the quality control, 831 were attributed to forward reads (81%). Only reverse reads (hereafter referred to as high quality reads, see Materials and methods for details) were used for downstream analysis, and the average read length of the high quality reads was 259 nt (including PCR primer but not barcode sequences), slightly above the 250 nt average length typical of 454 GS FLX runs (Mardis, 2008). The lower-than-expected read counts were likely the result of a strong secondary structure formed between the 454 forward fusion primer and the 5′ end of the amplicon, which interfered with the emPCR and sequencing processes.
Biodiversity and richness measures
A non-redundant set of high quality reads was aligned using ClustalW, PRANK+F and NAST, and the resulting alignments were independently analyzed using Mothur. OTUs were defined as furthest neighbor clusters within a Jukes–Cantor distance of 0.03 (OTU0.03). Alpha diversity indices for the three different multiple sequence alignments showed that the results are reproducible across alignment algorithms (Supplementary Figure 1). All further analyses were based on the PRANK+F alignment.
The terminal slopes of rarefaction curves (that is, the increase in number of OTUs per sequence added) indicate that Beacon Valley and Upper Wright Valley have been sampled comprehensively (Supplementary Figure 2). For Miers Valley and Battleship Promontory, the slopes are 0.13 and 0.18, respectively, much higher than those for Beacon Valley (0.02) and Upper Wright Valley (0.03). These findings validate the higher levels of observed diversity and AFLs seen in Miers Valley and Battleship Promontory's (Supplementary Figure 1 and Table 3, respectively)
Between 33 and 126 OTUs0.03 were obtained for the four study sites from between 262 and 838 reads (Table 2). Miers Valley, which is the most diverse site (126 OTUs0.03), had 542 reads, whereas only 33 OTUs0.03 were observed for Beacon Valley, which had 838 reads. Singleton and duplet OTUs0.03 account for large portions of the observed OTUs0.03, and the proportion of ‘rare' OTUs0.03 (that is, singletons and duplets) appears to increase with overall diversity (Table 2). With the exception of Miers Valley, which is dominated by one OTU0.03, all other study sites contain several dominant OTUs0.03 (Supplementary Figure 3). Although there is considerable variation in sampling depths among the samples, the observations above are not dependent on sampling depth, and we therefore forwent resampling the more comprehensively covered samples to maximize our ability to observe rare OTUs.
Table 2. Distribution and composition of OTUs0.03.
| Study site | Reads | Total OTUs0.03 | Singleton OTUs0.03 | Duplet OTUs0.03 | Ratio of rare OTUs0.03 (%) |
|---|---|---|---|---|---|
| Miers Valley | 542 | 126 | 66 | 25 | 72.2 |
| Battleship Promontory | 262 | 83 | 44 | 12 | 67.5 |
| Beacon Valley | 838 | 41 | 18 | 6 | 58.5 |
| Upper Wright Valley | 466 | 33 | 13 | 3 | 48.5 |
Singleton OTUs0.03 contain only one read and duplet OTUs0.03 contain two reads.
Phylogeny and distribution of diversity
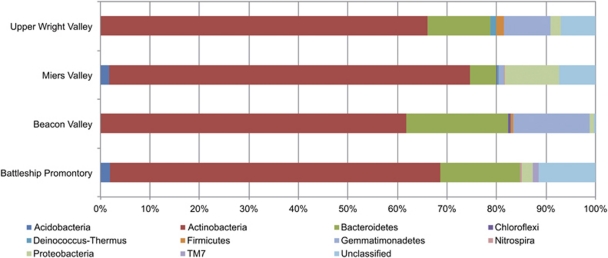
To assess the phylogenetic distribution and diversity present in the sequences, high quality sequences were classified using the Ribosomal Database Project Classifier at 80% confidence threshold (Wang et al., 2007). The overall phylum-level compositions of all four sites are highly similar (Figure 3), as confirmed by repeated analysis of variance test measures of phylum-level abundances (P-value ≈1). Sequences affiliated with Actinobacteria account for 62–73% of sequences in all sites, followed by Bacteroidetes-affiliated sequences at 5–21% (Figure 3). With the exception of Miers Valley, Proteobacteria (mostly α-Proteobacteria) account for <3% of sequences in the collections. Gemmatimonadetes are absent in Battleship Promontory and range from 1% to 15% in other valleys. Acidobacteria account for <2% of sequences in the Miers Valley and Battleship Promontory collections and are completely absent in the other two communities. The remaining phyla were only present at <1% abundance except TM7, which was only found in Battleship Promontory. Between 0.2% and 11.5% of the sequences could not be confidently classified, and no Archaeal sequences were found. Surprisingly, no cyanobacterial sequences were identified despite the cyanobacterial ARISA results and several reports of well-known cyanobacterial species in the Dry Valleys (Friedmann et al., 1993; La Torre de et al., 2003; Smith et al., 2006; Wood et al., 2008a; Pointing et al., 2009).
Figure 3.
Phylum-level distribution of bacterial OTUs0.03. Representative sequences of OTUs0.03 for each study site were phylogenetically assigned using the Ribosomal Database Project Classifier, and phylum-level assignments with >80% confidence threshold were retrieved and plotted.
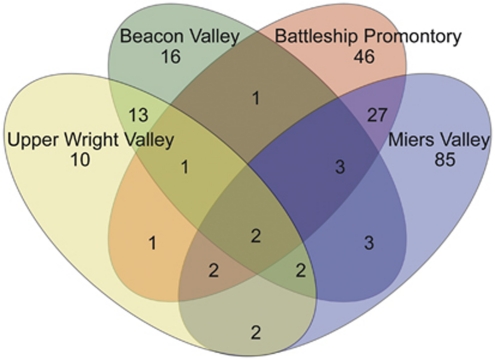
A Venn diagram illustrating the distribution of unique OTUs0.03 among the valleys (Figure 4) shows a highly localized distribution pattern. Battleship Promontory and Miers Valley appeared to share a large number of OTUs0.03, whereas Beacon Valley and Upper Wright Valley share a very different collection of OTUs0.03. This supports the grouping of microbial communities demonstrated by the bacterial ARISA data (Figure 2a). Surprisingly, the number of OTUs0.03 shared by all four valleys (that is, cosmopolitan OTUs0.03) is exceptionally low (2 out of 214 OTUs0.03), and despite the universal dominance of Actinobacteria (Figure 3), both cosmopolitan OTUs0.03 were only dominant in Beacon Valley and Upper Wright Valley. Conversely, OTUs0.03 affiliated with Actinobacteria and dominant in Battleship Promontory and Miers Valley tend to be absent or in very low abundance in Beacon Valley and Upper Wright Valley (Table 3).
Figure 4.
Venn diagram for bacterial OTUs0.03 found in the four Dry Valleys.
Table 3. Phylogenetic assignment and sequence distribution of select OTUs0.03.
| OTU ID |
Number of sequences |
RDP assigned taxonomy | Taxonomic level and confidence | |||
|---|---|---|---|---|---|---|
| BP | BV | MV | UW | |||
| 76 | 17 | 89 | 9 | 136 | Actinomycetales | Order/100% |
| 38 | 20 | 0 | 178 | 1 | Actinomycetales | Order/100% |
| 47 | 0 | 129 | 0 | 44 | Gemmatimonadetes | Phylum/80% |
| 56 | 0 | 89 | 0 | 80 | Rubrobacteridae | Subclass/97% |
| 104 | 0 | 119 | 0 | 37 | Rubrobacterineae | Family/99% |
| 146 | 0 | 98 | 1 | 34 | Actinomycetales | Order/100% |
| 36 | 0 | 120 | 0 | 10 | Flavobacteriaceae | Family/100% |
| 35 | 0 | 42 | 0 | 15 | Bacteroidetes | Phylum/100% |
| 180 | 1 | 29 | 3 | 9 | Rubrobacterineae | Family/92% |
| 121 | 0 | 8 | 0 | 32 | Porphyromonadaceae | Family/88% |
| 145 | 2 | 0 | 27 | 0 | Rubrobacteridae | Subclass/98% |
| 75 | 11 | 0 | 14 | 0 | Actinobacteria | Phylum/88% |
| 95 | 2 | 20 | 0 | 1 | Actinomycetaceae | Family/100% |
| 33 | 0 | 14 | 0 | 6 | Actinomycetales | Order/100% |
| 52 | 4 | 0 | 16 | 0 | Rubrobacteridae | Subclass/100% |
| 57 | 18 | 0 | 2 | 0 | Solirubrobacterales | Order/88% |
The four most abundant OTUs0.03 in each valley are included, with the two OTUs0.03 found in all four valleys in bold. The listed taxonomic assignments are the finest level that passed the RDP Classifier's default 80% confidence threshold. Apart from Flavobacteriaceae and Porphyromonadaceae, which are affiliated with the phylum Bacteroidetes, all other OTUs0.03 with taxonomic assignments finer than phylum belong to the phylum Actinobacteria.
Biogeochemical analysis
Using Biota-Environmental STepwise analysis in the PRIMER 6 package, the bacterial and cyanobacterial ARISA data (as multivariate biotic patterns) were linked to physicochemical data (all 38 variables listed in Table 1 plus altitude) to investigate potential relationships between microbial community structure and the underlying soil geochemistry (Supplementary Table 5). Variables most consistently explanatory of both bacterial and cyanobacterial ARISA patterns were altitude and conductivity. The amount of copper also appears to be significantly correlated with cyanobacterial distribution.
LINKTREE analysis (Clarke et al., 2008) was also performed to examine how physicochemical factors affect specific ARISA patterns (Supplementary Figure 4 and Supplementary Table 6). For the bacterial ARISA data, conductivity and Pb separate Miers Valley and Battleship Promontory from Beacon Valley and Upper Wright Valley, and have the same role for the cyanobacterial ARISA data. Multiple factors, including gravimetric water content and altitude (Supplementary Table 6), signify the split of Miers Valley and Battleship Promontory sites. The factors Ca, Sr and Mn clearly separate Beacon Valley and Upper Wright Valley samples, and may reflect differences in the underlying soil geochemistry.
Discussion
The Dry Valleys encompass a high degree of physicochemical heterogeneity, which is reflected in the four valleys chosen for this study: Beacon Valley, Upper Wright Valley, Battleship Promontory and Miers Valley. In addition to well-characterized differences in altitude (a proxy for annual mean temperature (Körner, 2007)) and soil salt content (a proxy for water availability (Lamsal and Paudyal, 1999) and age of the soil (Bockheim, 1979)), the study sites all possess distinct soil elemental compositions, reflecting the variability in local soil mineralogy (Table 1 and Supplementary Table 1). Overall, geochemical measurements of multiple samples taken from each individual 2500 m2 sampling area in four valleys formed robust clusters on the MDS plot, establishing a framework for inter-valley comparisons (Figure 1). Miers Valley is a clear outlier because of its high C/N ratio, total percentage C content and low gravimetric water content (Table 1). Miers Valley soils are also substantially more alkaline than all other valleys, which may have resulted from greater influence from salts of marine origin (Campbell and Claridge, 1987). Furthermore, the elevated soil pH and high C/N ratio of Miers Valley are potentially reflective of photosynthetic activity by cyanobacteria in the system (Wood et al., 2008a, 2008b). However, Battleship Promontory's average C/N ratio is similar to drier valleys (Table 1) despite considerable cyanobacterial presence in lithic habitats (Johnston and Vestal, 1991; Friedmann et al., 1993; Wynn-Williams, 2000; La Torre de et al., 2003). Miers Valley and Battleship Promontory samples have much lower conductivity values than the other two valleys, reflecting higher precipitation levels and the lack of reverse leaching of ground salts (McLeod et al., 2009).
Community DNA profiling of bacteria and cyanobacteria using ARISA revealed four very distinct communities with varied levels of diversity (Supplementary Tables 3 and 4). Miers Valley contains the most cyanobacterial AFLs (Supplementary Table 3) and is identified as an outlier to the remaining three valleys by ANOSIM (Supplementary Table 4) and MDS plot (Figure 2b), an outcome likely linked to the presence of a permanent lake that supports significant cyanobacterial biomass (Wood et al., 2008a). Despite having the highest copper concentration of all four sampling sites, Beacon Valley contains the second most cyanobacterial AFLs (Supplementary Table 3), which is unexpected given that previous studies have suggested that Miers Valley contains markedly higher cyanobacterial diversity than Beacon Valley (Wood et al., 2008a). This may be due to the more sensitive AFL identification algorithm used in this study, but the absence of cyanobacterial sequences in the pyrosequencing results limits the scope of discussion on the relationship between copper concentration and cyanobacterial diversity in Dry Valley soils. Bacterial ARISA data revealed significantly higher levels of diversity in Battleship Promontory and Miers Valley (Supplementary Table 3) and tight grouping between these two sampling sites, in contrast with looser grouping between Beacon Valley and Upper Wright Valley (Figure 2a and Supplementary Table 4) samples.
Pyrosequencing analysis of partial 16S rRNA gene PCR amplicons (V3–V4 regions), generated from pooled DNA from each valley using a different set of PCR primers, corroborated findings from the bacterial ARISA. Beacon Valley and Upper Wright Valley were found to harbor considerably lower levels of microbial diversity (Supplementary Figure 1) and share a number of OTUs0.03 different from those shared by Battleship Promontory and Miers Valley (Figure 4). Meanwhile, only 2 of the 214 OTUs0.03 identified in this study were found in all four valleys, questioning the idea that Dry Valley microbial ecology is dominated by ubiquitously abundant cosmopolitan species (Cameron et al., 1970; Wynn-Williams and Edwards, 1990; Vishniac, 1993). The discovery of genetically localized microbial communities in Dry Valley soils calls into the question the importance of aeolian transport in determining Dry Valley microbiota distribution (Nkem et al., 2006; Wood et al., 2008a) and suggests that Dry Valley soil bacterial populations may be subject to very limited inter-valley redistribution or that selection by local physicochemical conditions may be much stronger than previously expected (Moorhead et al., 1999; Barrett et al., 2006b).
Given the difference in physicochemical conditions at the four study sites, the similarity between bacterial community phyla structures is surprising (Figure 3). It has been shown in a wide range of soil habitats that phylum-level community structure is highly site-specific even in homogeneous landscapes (Herrera et al., 2007; Labbé et al., 2007; Lamarche et al., 2007; Fulthorpe et al., 2008), and the conservation of phyla structures seen here may be reflective of the strong selective pressure imposed by the Dry Valley environment. On the other hand, diversity at a finer phylogenetic resolution is very high across the four sites. For example, only 9 out of 81 OTUs0.03 affiliated with Actinobacteria and none of the 28 OTUs0.03 affiliated with Bacteroidetes were found in more than two valleys. These observations point to localized, genetically disparate bacterial populations in the Dry Valleys, and suggest potential endemism in Dry Valley soil microbiota. These ideas challenge the notion that aeolian transport has a major role in inter-valley biota distribution (Adams et al., 2007). Although it is tempting to compare the level of spatial heterogeneity observed here with those of more temperate ecosystems, differences in sampling strategy and analysis methodology make such comparisons unreliable.
The distribution pattern described here (Figure 4) is unlikely to be a product of PCR/sequencing artifacts because of two pieces of evidence. Firstly, the number of OTUs0.03 identified for each study site was unrelated to the number of sequences obtained for that site (Supplementary Figure 1) (Pearson's correlation analysis, R=−0.305, P-value=0.695), which indicates that the flowgram clustering approach employed by AmpliconNoise (Quince et al., 2011) and the Perseus chimera checker (Quince et al., 2011) have sufficiently removed ‘noisy reads' that artificially increase the size of the rare biosphere as the number of reads increases (Kunin et al., 2009; Huse et al., 2010; Lee et al., 2011). Secondly, the two ‘cosmopolitan' OTUs0.03 were not universally dominant, and other OTUs0.03 at much higher total abundances were completely absent in one or more of the valleys (Table 3). This observation is not necessarily due to the modest sampling depth, as two of the study sites (Beacon Valley and Upper Wright Valley) were in fact comprehensively sampled according to terminal slopes of their rarefaction curves. Furthermore, although more OTUs0.03 will likely to be found in all four valleys by a more extensive sequencing effort, it will not alter the observation that no OTUs0.03 are dominant in all four study sites, and that dominant microbial species appear highly localized within the Dry Valleys.
Surprisingly, cyanobacterial sequences were completely absent from the pyrosequencing data despite significant cyanobacterial diversity reported in cyanobacterial ARISA. Further examination of the PCR primers used to generate partial 16S rRNA gene amplicons (Probe Match, http://rdp.cme.msu.edu/probematch/search.jsp) revealed that the reverse PCR primer (806R) targets cyanobacteria very poorly (matching <1% known cyanobacterial sequences). Although PCR primer-related biases are well known, some reports suggested that it is largely limited to ‘rare' taxa (Huse et al., 2008; Hamp et al., 2009). Here we demonstrate a case where PCR primer bias can influence observed diversity at the phylum level. It should be noted, however, that a previous report of microbial communities in the McKelvey Valley based on both PCR clone libraries and qPCR reported very low or non-existent cyanobacterial signals in soils (Pointing et al., 2009), and it is possible that PCR primer bias and low abundance conspired in the lack of cyanobacterial sequences in our results. Although Acidobacteria have been reported as a dominant group in many earlier reports of Dry Valley microbial ecology (Smith et al., 2006; Pointing et al., 2009; Cary et al., 2010), they represent <2% of the sequences in the Miers Valley and Battleship Promontory, and are completely absent in the other valleys. Results from Probe Match suggest that this was not due to primer bias (data not shown), and thus the absence of Acidobacteria in Beacon Valley and Upper Wright Valley may have ecological significance. We did not attempt to investigate archaeal diversity in our samples, and the PCR primers used do not target archaeal sequence. However, archaeal diversity in the Dry Valleys had previously been reported as extremely low and limited to the globally ubiquitous Group II low-temperature Crenarchaeotes (Aislabie et al., 2006; Hogg et al., 2006), and archaea-specific PCR assays failed to detect any signal in the McKelvey Valley (Pointing et al., 2009).
By combining biological and physicochemical data, Biota-Environmental STepwise and LINKTREE analyses both identified altitude and conductivity as primary drivers for the biological differences seen in bacterial and cyanobacterial ARISA data (Supplementary Table 5). Altitude can be considered a proxy for average temperature, which generally drops by 5.5 °C for every 1000 m increase in elevation (Körner, 2007). This temperature gradient can be exacerbated by the low air relative humidity in the Dry Valleys and may directly influence the presence of liquid water in the austral summer. Conductivity values (that is, soil salt content) for samples from Miers Valley and Battleship Promontory are remarkably low by Dry Valley standards (Aislabie et al., 2006), reflecting seasonal surface water availability in these two valleys. Conversely, the high conductivity of Beacon Valley and Upper Wright Valley samples suggests that those surface soils are relatively ancient and experience little leaching from precipitation (McLeod et al., 2009). Soil conductivity has been shown to negatively correlate with micro-invertebrate diversity (Freckman and Virginia, 1997; Barrett et al., 2004) within a single valley, and an earlier report comparing Dry Valley soils with various lithic habitats suggested that conductivity might have a role in determining overall biota distribution (Pointing et al., 2009). Our results corroborate previous findings and suggest that soil salt content is instrumental in structuring biota at all levels. Elemental Pb content and gravimetric water content, both of which are linked to the mineralogy of the soil, were also identified as influential in determining bacterial and cyanobacterial community compositions (Supplementary Table 5 and Supplementary Figure 4).
Although our modest sequencing effort was intended to identify dominant bacterial species, the results have shown that Dry Valley soils harbor far more than a few cosmopolitan species (Horowitz et al., 1972; Johnson et al., 1978; Vishniac, 1993), and that Dry Valley microbial communities appear to be highly localized and composed of potentially endemic populations. Moreover, our integrated approach, utilizing two types of molecular genetic techniques, geochemical measurements and statistical analyses, showed that the distribution of Dry Valley microbiota significantly correlates with physicochemical conditions of the environment. These discoveries not only challenge existing paradigms for Dry Valley microbiota distribution and refocus drivers of microbial diversity in the Dry Valley ecosystem, they also support the hypothesis that Dry Valley ecology is largely determined by abiotic factors in this ultraoligotrophic environment (Barrett et al., 2004; Cary et al., 2010). Our findings, based on a systematic survey of habitats across a wide physicochemical gradient, provide statistically robust data showing that a complex set of abiotic factors determine the microbial components of the Dry Valleys. These results also pose novel challenges to the biosecurity management of this unique and fragile habitat as existing policies have been designed under the assumption that Dry Valley soil bacterial populations are homogeneous across the landscape and detached from contemporary environmental conditions (Moorhead et al., 1999; Barrett et al., 2006b). Lastly, the discovery of highly localized, potentially endemic microbial populations in the Dry Valleys lends credence to the theory of potential links between Dry Valley glacial geomorphology and soil microbial ecology, which in turn lead to questions such as colonization opportunities and selection mechanisms; the results presented here therefore has implications for Antarctic research at large.
Acknowledgments
This research was supported by Foundation for Research, Science and Technology of New Zealand through an IPY grant to SCC and a FRST Postdoctoral Research Fellowship to CKL (UOWX0715). The New Zealand Marsden Fund provided financial support for CKL (UOW1003). Logistic support for field research was provided by Antarctica New Zealand. We thank Dr Jo-Ann Stanton of University of Otago for assistance with pyrosequencing of PCR amplicons, and we acknowledge Dr Susanna Wood of Cawthron Institute for useful discussions on the analysis of community genetic profiling results. We are grateful to Dr Megan Balks of University of Waikato for her comments on the geology of study sites.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abdo Z, Schütte UME, Bent SJ, Williams CJ, Forney LJ, Joyce P. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ Microbiol. 2006;8:929–938. doi: 10.1111/j.1462-2920.2005.00959.x. [DOI] [PubMed] [Google Scholar]
- Adams B, Bardgett R, Ayres E, Wall D, Aislabie J, Bamforth S, et al. Diversity and distribution of Victoria Land biota. Soil Biol Biochem. 2006;38:3003–3018. [Google Scholar]
- Adams B, Wall D, Gozel U, Dillman, Chaston, Hogg I. The southernmost worm, Scottnema lindsayae (Nematoda): diversity, dispersal and ecological stability. Polar Biol. 2007;30:809–815. [Google Scholar]
- Aislabie J, Chhour K-L, Saul D, Miyauchi S, Ayton J, Paetzold R, et al. Dominant bacteria in soils of Marble Point and Wright Valley, Victoria Land, Antarctica. Soil Biol Biochem. 2006;38:3041–3056. [Google Scholar]
- Babalola O, Kirby B, Le Roes-Hill M, Cook A, Cary SC, Burton S, et al. Phylogenetic analysis of actinobacterial populations associated with Antarctic Dry Valley mineral soils. Environ Microbiol. 2009;11:566–576. doi: 10.1111/j.1462-2920.2008.01809.x. [DOI] [PubMed] [Google Scholar]
- Barrett JE, Virginia R, Hopkins D, Aislabie J, Bargagli R, Bockheim JG, et al. Terrestrial ecosystem processes of Victoria Land, Antarctica. Soil Biol Biochem. 2006a;38:3019–3034. [Google Scholar]
- Barrett JE, Virginia R, Wall D, Cary SC, Adams B, Hacker AL, et al. Co-variation in soil biodiversity and biogeochemistry in northern and southern Victoria Land, Antarctica. Antarct Sci. 2006b;18:535–548. [Google Scholar]
- Barrett JE, Virginia R, Wall D, Parsons A, Powers L, Burkins M. Variation in biogeochemistry and soil biodiversity across spatial scales in a polar desert ecosystem. Ecology. 2004;85:3105–3118. [Google Scholar]
- Block W, Smith L, Kennedy AD. Strategies of survival and resource exploitation in the Antarctic fellfield ecosystem. Biol Rev Camb Philos Soc. 2009;84:449–484. doi: 10.1111/j.1469-185X.2009.00084.x. [DOI] [PubMed] [Google Scholar]
- Bockheim JG. Relative age and origin of soils in Eastern Wright Valley, Antarctica. Soil Sci. 1979;128:142–152. [Google Scholar]
- Bockheim JG. Landform and soil development in the McMurdo Dry Valleys, Antarctica: a regional synthesis. Arctic Antarct Alpine Res. 2002;34:308–317. [Google Scholar]
- Bockheim JG, McLeod M. Soil distribution in the McMurdo Dry Valleys, Antarctica. Geoderma. 2008;144:43–49. [Google Scholar]
- Cameron RE, King J, David CN. Microbiology, ecology and microclimatology of soil sites in Dry Valleys of South Victoria Land, Antarctica. Antarctic Ecol. 1970;2:702–716. [Google Scholar]
- Campbell IB, Claridge GGC.1987Salts in Antarctic Soils Anarctica: Soils, Weathering Processes and Environment. Development in Soil ScienceVol. 16. Elsevier: New York; 239–273. [Google Scholar]
- Cary SC, McDonald IR, Barrett JE, Cowan D. On the rocks: the microbiology of Antarctic Dry Valley soils. Nat Rev Microbiol. 2010;8:129–138. doi: 10.1038/nrmicro2281. [DOI] [PubMed] [Google Scholar]
- Clarke KR, Somerfield PJ, Gorley RN. Testing of null hypotheses in exploratory community analyses: similarity profiles and biota-environment linkage. J Exp Mar Biol Ecol. 2008;366:56–69. [Google Scholar]
- Convey P. The influence of environmental characteristics on life history attributes of Antarctic terrestrial biota. Biol Rev. 1996;71:191–225. [Google Scholar]
- Coyne K, Hutchins D, Hare C, Cary SC. Assessing temporal and spatial variability in Pfiesteria piscicida distributions using molecular probing techniques. Aquat Microb Ecol. 2001;24:275–285. [Google Scholar]
- Convey P, Stevens M, Hodgson D, Smellie J, Hillenbrand C-D, Barnes D, et al. Exploring biological constraints on the glacial history of Antarctica. Quaternary Sci Rev. 2009;28:3035–3048. [Google Scholar]
- DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006a;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006b;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran PT, Priscu JC, Lyons WB, Walsh JE, Fountain AG, McKnight D, et al. Antarctic climate cooling and terrestrial ecosystem response. Nature. 2002;415:517–520. doi: 10.1038/nature710. [DOI] [PubMed] [Google Scholar]
- Freckman D, Virginia R. Low-diversity Antarctic soil nematode communities: distribution and response to disturbance. Ecology. 1997;78:363–369. [Google Scholar]
- Friedmann EI, Kappen L, Meyer MA, Nienow JA. Long-term productivity in the cryptoendolithic microbial community of the Ross Desert, Antarctica. Microb Ecol. 1993;25:51–69. doi: 10.1007/BF00182129. [DOI] [PubMed] [Google Scholar]
- Fulthorpe R, Roesch L, Riva A, Triplett E. Distantly sampled soils carry few species in common. ISME J. 2008;2:901–910. doi: 10.1038/ismej.2008.55. [DOI] [PubMed] [Google Scholar]
- Hamp T, Jones J, Fodor A. Effects of experimental choices and analysis noise on surveys of the ‘rare biosphere'. Appl Environ Microbiol. 2009;75:3263–3270. doi: 10.1128/AEM.01931-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera A, Héry M, Stach JEM, Jaffré T, Normand P, Navarro E. Species richness and phylogenetic diversity comparisons of soil microbial communities affected by nickel-mining and revegetation efforts in New Caledonia. Eur J Soil Biol. 2007;43:130–139. [Google Scholar]
- Hogg I, Cary SC, Convey P, Newsham K, O Donnell A, Adams B, et al. Biotic interactions in Antarctic terrestrial ecosystems: Are they a factor. Soil Biol Biochem. 2006;38:3035–3040. [Google Scholar]
- Hopkins D, Sparrow A, Novis P, Gregorich EG, Elberling B, Greenfield LG. Controls on the distribution of productivity and organic resources in Antarctic Dry Valley soils. Proc R Soc B. 2006;273:2687–2695. doi: 10.1098/rspb.2006.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz NH, Cameron R, Hubbard J. Microbiology of the Dry Valleys of Antarctica. Science. 1972;176:242–245. doi: 10.1126/science.176.4032.242. [DOI] [PubMed] [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Welch DBM, Relman DA, Sogin M. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huws SA, Edwards JE, Kim EJ, Scollan ND. Specificity and sensitivity of eubacterial primers utilized for molecular profiling of bacteria within complex microbial ecosystems. J Microbiol Methods. 2007;70:565–569. doi: 10.1016/j.mimet.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Janssen P. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol. 2006;72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RM, Madden JM, Swafford JA.1978Taxonomy of Antarctic bacteria from soils and air primarily of the McMurdo Station and Victoria Land Dry Valleys regionIn: Packer BC (ed.).Terrestrial Biology III, Antarctic Research Series The William Byrd Press: Richmond, VA; 35–64. [Google Scholar]
- Johnston C, Vestal J. Photosynthetic carbon incorporation and turnover in Antarctic cryptoendolithic microbial communities: are they the slowest-growing communities on Earth. Appl Environ Microbiol. 1991;57:2308–2311. doi: 10.1128/aem.57.8.2308-2311.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Shade A, McMahon K, Kent A. Comparison of primer sets for use in automated ribosomal intergenic spacer analysis of aquatic bacterial communities: an ecological perspective. Appl Environ Microbiol. 2007;73:659–662. doi: 10.1128/AEM.02130-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner C. The use of ‘altitude' in ecological research. Trends Ecol Evol. 2007;22:569–574. doi: 10.1016/j.tree.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2009;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- La Torre de JR, Goebel BM, Friedmann EI, Pace NR. Microbial diversity of cryptoendolithic communities from the McMurdo Dry Valleys, Antarctica. Appl Environ Microbiol. 2003;69:3858–3867. doi: 10.1128/AEM.69.7.3858-3867.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé D, Margesin R, Schinner F, Whyte LG, Greer CW. Comparative phylogenetic analysis of microbial communities in pristine and hydrocarbon-contaminated Alpine soils. FEMS Microbiol Ecol. 2007;59:466–475. doi: 10.1111/j.1574-6941.2006.00250.x. [DOI] [PubMed] [Google Scholar]
- Lamarche J, Bradley RL, Hooper E, Shipley B, Simao Beaunoir A-M, Beaulieu C. Forest floor bacterial community composition and catabolic profiles in relation to landscape features in Québec's southern boreal forest. Microb Ecol. 2007;54:10–20. doi: 10.1007/s00248-006-9156-3. [DOI] [PubMed] [Google Scholar]
- Lamsal K, Paudyal G. Model for assessing impact of salinity on soil water availability and crop yield. Agric Water Manage. 1999;41:57–70. [Google Scholar]
- Lee CK, Herbold CW, Polson SW, Wommack KE, Williamson SJ, Cary SC.2011Groundtruthing Next-Gen Microbial Ecology—Biases and Errors in Community Structure Estimates Obtained from 454 Titanium Pyrosequencing of PCR AmpliconsIn Review. [DOI] [PMC free article] [PubMed]
- Löytynoja A, Goldman N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. 2008;320:1632–1635. doi: 10.1126/science.1158395. [DOI] [PubMed] [Google Scholar]
- Marchant D, Head JW. Antarctic Dry Valleys: microclimate zonation, variable geomorphic processes, and implications for assessing climate change on Mars. Icarus. 2007;192:187–222. [Google Scholar]
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- McLeod M, Bockheim JG, Balks M, Aislabie J. Soils of western Wright Valley, Antarctica. Antarct Sci. 2009;21:355–365. [Google Scholar]
- Moorhead DL, Doran PT, Fountain AG, Lyons WB, McKnight D, Priscu JC, et al. Ecological legacies: impacts on ecosystems of the McMurdo Dry Valleys. Bioscience. 1999;49:1009–1019. [Google Scholar]
- Niederberger TD, McDonald IR, Hacker AL, Soo RM, Barrett JE, Wall D, et al. Microbial community composition in soils of Northern Victoria Land, Antarctica. Environ Microbiol. 2008;10:1713–1724. doi: 10.1111/j.1462-2920.2008.01593.x. [DOI] [PubMed] [Google Scholar]
- Nkem J, Wall D, Virginia R, Barrett JE, Broos E, Porazinska D, et al. Wind dispersal of soil invertebrates in the McMurdo Dry Valleys, Antarctica. Polar Biol. 2006;29:346–352. [Google Scholar]
- Poage M, Barrett JE, Virginia R, Wall D. The influence of soil geochemistry on nematode distribution, McMurdo Dry Valleys, Antarctica. Arctic Antarct Alpine Res. 2008;40:119–128. [Google Scholar]
- Pointing SB, Chan Y, Lacap DC, Lau MCY, Jurgens JA, Farrell RL. Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci USA. 2009;106:19964–19969. doi: 10.1073/pnas.0908274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzén A, Davenport RJ, Turnbaugh PJ. Removing Noise From Pyrosequenced Amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Toward a census of bacteria in soil. PLoS Comput Biol. 2006;2:e92. doi: 10.1371/journal.pcbi.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RF. Vol. II. Cooper Square Press: New York (2001 Reprint), Annex II; 1905. The Voyage of the Discovery. [Google Scholar]
- Smith JJ, Tow LA, Stafford W, Cary SC, Cowan D. Bacterial diversity in three different Antarctic cold desert mineral soils. Microb Ecol. 2006;51:413–421. doi: 10.1007/s00248-006-9022-3. [DOI] [PubMed] [Google Scholar]
- Smith JL, Barrett JE, Tusnády G, Rejtö L, Cary SC. Resolving environmental drivers of microbial community structure in Antarctic soils. Antarct Sci. 2010;22:673–680. [Google Scholar]
- Thompson J, Higgins D, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virginia R, Wall D. How soils structure communities in the Antarctic Dry Valleys. Bioscience. 1999;49:973–983. [Google Scholar]
- Vishniac HS.1993The microbiology of Antarctic soilsIn: Friedmann EI (ed.).Antarctica Microbiology Wiley-Liss, Inc.: New York; 297–341. [Google Scholar]
- Wall D, Virginia R. Controls on soil biodiversity: insights from extreme environments. Appl Soil Ecol. 1999;13:137–150. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood SA, Rueckert A, Cowan D, Cary SC. Sources of edaphic cyanobacterial diversity in the Dry Valleys of Eastern Antarctica. ISME J. 2008a;2:308–320. doi: 10.1038/ismej.2007.104. [DOI] [PubMed] [Google Scholar]
- Wood SA, Mountfort D, Selwood AI, Holland PT, Puddick J, Cary SC. Widespread distribution and identification of eight novel microcystins in antarctic cyanobacterial mats. Appl Environ Microbiol. 2008b;74:7243–7251. doi: 10.1128/AEM.01243-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn-Williams DD.2000Cyanobacteria in desertsIn: Whitton BA, Potts M (eds).The Ecology of Cyanobacteria Kluwer Academic Publishers: Dordrecht, The Netherlands; 341–366. [Google Scholar]
- Wynn-Williams DD, Edwards HGM.1990Ecological aspects of Antarctic microbiologyIn: Marshall KC (ed.).Advances in Microbial EcologyVol. 11. Springer: New York; 71–146. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.