Abstract
There is an increasing appreciation of the polymicrobial nature of many bacterial infections such as those associated with cystic fibrosis (CF) and of the potentially important role for interspecies interactions in influencing both bacterial virulence and response to therapy. Patients with CF are often co-infected with Pseudomonas aeruginosa and other pathogens including Burkholderia cenocepacia and Stenotrophomonas maltophilia. These latter bacteria produce signal molecules of the diffusible signal factor (DSF) family, which are cis-2-unsaturated fatty acids. We have previously shown by in vitro studies that DSF from S. maltophilia leads to altered biofilm formation and increased resistance to antibiotics by P. aeruginosa; these responses of P. aeruginosa require the sensor kinase PA1396. Here we show that DSF signals are present in sputum taken from patients with CF. Presence of these DSF signals was correlated with patient colonization by S. maltophilia and/or B. cenocepacia. Analysis of 50 clinical isolates of P. aeruginosa showed that each responded to the presence of synthetic DSF by increased antibiotic resistance and these strains demonstrated little sequence variation in the PA1396 gene. In animal experiments using CF transmembrane conductance regulator knockout mice, the presence of DSF promoted P. aeruginosa persistence. Furthermore, antibiotic resistance of P. aeruginosa biofilms grown on human airway epithelial cells was enhanced in the presence of DSF. Taken together, these data provide substantial evidence that interspecies DSF-mediated bacterial interactions occur in the CF lung and may influence the efficacy of antibiotic treatment, particularly for chronic infections involving persistence of bacteria.
Keywords: interspecies signaling, diffusible signal factor, biofilm formation, virulence, cystic fibrosis, next-generation sequencing
Introduction
Bacterial infections of the lung are the leading cause of morbidity and mortality in cystic fibrosis (CF) patients. There is an increasing appreciation of the polymicrobial nature of these infections and the potential importance of interspecies interactions in influencing infection status, clinical outcomes or response to therapy (Ryan and Dow, 2008; Sibley et al., 2008, 2009; Ng and Bassler 2009; Shank and Kolter, 2009; Rogers et al., 2010). In many bacteria, the production and perception of diffusible signal molecules allows the organisms to monitor aspects of their environment, such as population density or confinement. These diffusible signal molecules, of which there are a range of structural classes, can also mediate interspecies signaling (Ryan and Dow, 2008; Shank and Kolter, 2009). In addition, it is clear that bacteria can sense signal molecules that they do not produce, thereby eavesdropping on signaling by other organisms in their immediate environment (Ryan and Dow, 2008; Shank and Kolter, 2009).
Chronic colonization and infection by Pseudomonas aeruginosa occurs in 80% of CF patients by 18 years of age, these patients can also be co-infected by other pathogens such as Burkholderia cenocepacia and Stenotrophomonas maltophilia. These latter pathogens produce signal molecules of the diffusible signal factor (DSF) family, which are cis-2-unsaturated fatty acids (Deng et al., 2011; Ryan and Dow, 2011). This class of signal molecule was first described in the plant pathogen Xanthomonas campestris pv. campestris (Xcc) (Barber et al., 1997). DSF from Xcc has been characterized as cis-11-methyl-2-dodecenoic acid (Wang et al., 2004). This molecule is also produced by S. maltophilia, whereas B. cenocepacia produces cis-2-dodecenoic acid (known as BDSF) (Huang and Wong, 2007; Boon et al., 2008). The synthesis of DSF in Xcc is dependent on RpfF, which has some amino-acid sequence similarity to enoyl CoA hydratases, whereas the two-component system comprising the sensor kinase RpfC and regulator RpfG is implicated in DSF perception (Ryan and Dow 2010, 2011; Ryan et al., 2010).
We have previously provided evidence that P. aeruginosa responds to DSF leading to changes in biofilm architecture and increased resistance to cationic antimicrobial peptides (Ryan et al., 2008). This response requires PA1396, a sensor kinase with an input domain of related amino-acid sequence to the sensory input domain of RpfC, which is responsible for DSF perception in Xanthomonads (Ryan et al., 2008). Domain-swapping experiments indicate that PA1396 participates directly in DSF recognition (Ryan et al., 2008) and can also sense BDSF but not cis-2-decenoic acid, a related molecule produced by P. aeruginosa (Davies and Marques 2009; McCarthy et al., 2010). Mutation of PA1396 or addition of DSF to P. aeruginosa leads to increased levels of a number of proteins with roles in bacterial stress tolerance and polymyxin resistance (Ryan et al., 2008). These findings have led to the suggestion that PA1396 acts to negatively regulate the expression of the genes involved in stress and polymyxin tolerance, and that binding of DSF to PA1396 reverses this negative regulatory effect.
Although interspecies signaling mediated by DSF appears to be important for biofilm formation and antibiotic resistance by P. aeruginosa in vitro, its role during infections is poorly understood. Here we have addressed this issue for polymicrobial infections of the CF lung. We have used high-performance liquid chromatography and mass spectrometry to detect DSF family signals in sputum from CF patients at physiologically relevant levels. We present evidence that the presence of these DSF signal molecules is correlated with polymicrobial infection involving Burkholderia and/or Stenotrophomonas species together with P. aeruginosa. We show that clinical P. aeruginosa isolates retain the ability to sense and respond to DSF and have a functional sensor kinase PA1396. Furthermore, we show that the presence of DSF promotes P. aeruginosa persistence in CF transmembrane conductance regulator knockout mice and enhances cationic antimicrobial peptide tolerance of P. aeruginosa biofilms grown on human airway epithelial cells. Taken together, these findings indicate that DSF-mediated interspecies interactions occur in the CF lung where they may influence the efficacy of antibiotic treatment for chronic P. aeruginosa infections.
Materials and methods
Bacterial strains, growth conditions and media
The bacterial strains used in this study are listed in Supplementary Table S1. Cultures were routinely grown at 37 °C in Luria broth with shaking, or on 1.5% Luria broth agar plates. Cultures were also grown in artificial sputum medium which comprises: 5 g mucin from pig stomach mucosa (Sigma, Dublin, Ireland), 4 g DNA (Fluka, Dublin, Ireland), 5.9 mg diethylene triamine pentaacetic acid (Sigma), 5 g NaCl, 2.2 g KCl, 5 ml egg yolk emulsion (Oxoid, Dublin, Ireland) and 5 g amino acids per 1 l water (pH 7.0) (Sriramulu et al., 2005; Fouhy et al., 2007).
Enrolment of subjects, sputum sample collection and P. aeruginosa isolates
Sputum samples from CF patients (January 2010–September 2010) were collected from the Adult Cystic Fibrosis Treatment center in the Cork University Hospital. Sputa from patients with bronchiectasis were also collected for control analysis. All sputa were analyzed and frozen at −70 °C for further study. The protocol was approved by the Animal Ethics Committees of University College Cork and Cork Teaching Hospitals.
Overall, 50 isolates from CF patients with chronic P. aeruginosa lung infection were isolated during this study. These isolates have been collected from sputum of CF patients and stored in nutrient broth containing 5% glycerol at −80 °C. The selection criteria were phenotypic (mucoid and non-mucoid) and different patterns of resistance to antibiotics, as determined on the basis of direct sensitivity testing of isolates from plated sputum.
DSF extraction, DSF bioassay and detection of DSF from sputum samples
Sputum samples were diluted to 5 ml with sterile phosphate-buffered saline (PBS) pH 7.4, centrifuged and DSF was extracted into ethyl acetate, as described previously (Barber et al., 1997). Full details are described in the Supplementary Information.
DNA extraction, 16S rRNA universal PCR amplication and 454 sequencing
DNA extraction from sputa was performed using a high-performance PCR template kit (Roche, Dublin, Ireland). In short, 100 μl of each sample was treated with 200 μl of lysis buffer and 45 μl of proteinaseK and was incubated at 55 °C for 1 h as recommended by manufacturer. The following universal 16S rRNA primers were used for the PCR reaction: KTF (5′-CCAGACTCCTACGGGAGGCAGC-3′) and KTR (5′-CCGTCAATTCCTTTGAGTTT-3′) for the V3–V5 region. Barcode sequences for the V3–V5 samples of either AGCAGAGC or AGCAGATG were attached between the 454 adaptor sequence and the forward primers. Standard PCR reaction conditions were employed for reactions with Taq polymerase—2 m MgCl2, 200 n of each primer, 200 μ dNTPs. The PCR conditions established were 94 °C for 50 s (initialization and denaturing) followed by 40 °C for 30 s (annealing), 72 °C for 60 s in 40 cycles (extension), and a final elongation step at 72 °C for 5 min. Two negative control reactions containing all components, but water instead of template, were performed alongside all test reactions, and were routinely free of PCR product, demonstrating lack of contamination with post-PCR product. The optimal annealing temperature for the primers, which included 454 adapters and barcode sequences, was empirically determined by gradient PCR using control reactions with initially purified bacterial genomic DNA, and validated on Pseudomonas gDNA. The 16S rRNA V3–V5 amplicons were subsequently sequenced on a 454 Genome Sequencer FLX platform (The Genome Analysis Centre, Norwich, UK) according to 454 protocols, one plate each for the V3–V5 region amplicons of 50 samples. Sequence analysis and phylogenetic classification was carried out as described in the Supplementary Information.
Infection and treatment of animals
Mouse infection was performed using a variation on the mouse model described by McCarthy et al. (2010). Briefly, P. aeruginosa strains were grown in Luria broth at 37 °C overnight with shaking, after which bacteria were collected by centrifugation and resuspended in PBS. The exact number of bacteria was determined by plating serial dilutions of each inoculum on Luria broth agar plates. Female C57BL/6 mice (approximately 8-weeks old) or CF transmembrane conductance regulator (CFTR) knockout (CF) mice (approximately 10-weeks old) were anesthetized and infected by the intratracheal route with 20 μl of culture of PAO1, PAO1/DSF, PAO1/trans-11-methyl dodecenoic or PA1396 mutant to have a final inoculum of 5 × 106 colony forming unit (CFU) per mouse. After every 24 h period, bacterial-infected animals were treated with 100 μl of PBS or PBS containing DSF (50 μ) or trans-11-methyl dodecenoic (50 μ). Mice were killed at 1, 3, 5 and 7 days post-infection by intraperitoneal injection of 0.3 ml of 30% pentobarbital. Lungs and spleens were harvested aseptically and homogenized in sterile PBS. A 10-fold serial dilution of lung homogenates was plated on Pseudomonas isolation agar. The results (means±s.e.) are expressed as CFU/organ. All animal experiments were approved by the Animal Ethics Committees of University College Cork or Cork University Hospital.
Epithelial cell co-culture biofilm and cytotoxicity assays
P. aeruginosa biofilms were grown on cystic fibrosis bronchial cells (CFBE) cells using a co-culture model system as previously described (Anderson et al., 2008, Moreau-Marquis et al., 2008). The cytotoxicity of PAO1 (or mutant strains) was assessed by measuring lactate dehydrogenase (LDH) released from epithelial cells in co-culture with the bacteria (Anderson et al., 2008; Moreau-Marquis et al., 2008).
Polymyxin-killing curves
Polymyxin-killing curves were carried out at 37 °C as previously described (Ryan et al., 2008).
Gene expression-profiling experiments
Over-night cultures of P. aeruginosa PAO1 were used to inoculate 200 ml of artificial sputum medium media with a starting OD (600 nm) of 0.05. At an OD of 0.3, 50 μ DSF or 50 μ trans-11-methyl dodecenoic acid was applied to the cultures and left to grow till an OD of 0.8. A detailed description of total RNA extraction, cDNA synthesis and hybridization to Affymetrix GeneChip P. aeruginosa genome arrays and data set analysis can be found in the Supplementary Information.
Quantative real-time PCR assays
The effect of PA1396 mutation or the addition of 50 μ DSF-like signal molecules to PAO1 on expression of genes was monitored by RT-PCR, as described previously (Ryan et al., 2008; McCarthy et al., 2010). The primers used to amplify the genes are listed in Supplementary Table S1.
Results
Signals from the DSF family are present in sputum from CF patients infected with Burkholderia and Stenotrophomonas
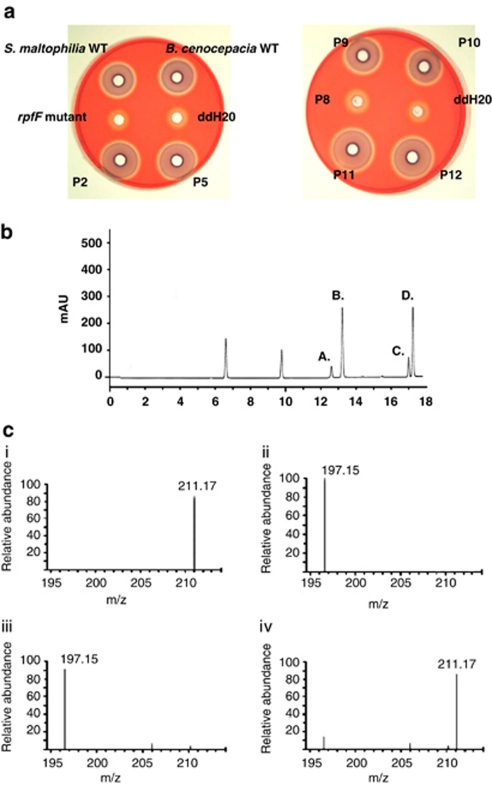
In initial experiments, solvent extracts of 50 sputum samples (from 40 patients with CF and 10 with viral-induced bronchitis) were assessed for the presence of DSF or BDSF using the Xcc rpfF bio-reporter (Barber et al., 1997). Note that this bioassay does not detect cis-decenoic acid (McCarthy et al., 2010). Overall, 15 of the 40 samples from CF patients tested were positive for the presence of DSF family signals (Figure 1; Table 1), although none of the sputum obtained from the 10 bronchitis patients activated the bio-reporter, which can detect DSF down to a concentration of 2.5 n (Barber et al., 1997; McCarthy et al., 2010). Therefore, sputum samples from these bronchitis patients acted as appropriate negative controls in this study.
Figure 1.
Detection of DSF-like signal molecules in the sputum of CF patients. (a) Biossay for DSF activity in extracts from sputum of CF patients (designated ‘P' followed by the sample number, that is, ‘P2') or in culture supernatants of wild-type strains of S. maltophilia or B. cenocepacia. The bioassay depends upon the restoration of endoglucanase activity to an rpfF mutant (DSF minus) strain of Xanthomonas campestris pv. campestris. DSF activity was detected in culture supernatants (as expected) and in some of the sputum samples. Endoglucanase activity in the rpfF mutant and negative control (ddH20) are also shown. (b). A representative high-performance liquid chromatography trace showing analysis of DSF extracts from the CF patient sputum. The compounds in fractions a, b, c and d showed strong activity in the DSF bioassay (c). Identification of DSF and BDSF in sputum from CF patients by liquid chromatography–mass spectrometry. The figure shows MS profiles for (i) synthetic DSF, (ii) synthetic BDSF, (iii) the compound in fraction d identified as DSF and (iv) the compound in fraction c identified as BDSF. The m/z of DSF is 211.17 and BDSF is 197.15.
Table 1. Summary of bacteria identified by culturing or 16S rRNA gene barcoded 454 sequencing and the detection DSF activity in the sputum from CF patients.
| Sample no. | Patient designation | Microbes identified by culture-based methodsa | Bacteria identified by 16S rRNA barcoding 454 sequencingb | DSF bioassay activityc | DSF detected by HPLC/LC-MSd |
|---|---|---|---|---|---|
| 1 | 16A | can, sa | sa, pa | − | − |
| 2 | 27A | sa, can | steno, pa, sa | + | DSF, BDSF |
| 3 | 124A | pa | pa, sa | − | − |
| 4 | 92A | pa | pa, sa | − | − |
| 5 | 10A | steno, can | steno, pa, sa | + | DSF |
| 6 | 74A | sa | pa, sa | − | − |
| 7 | 43A | sa | pa, sa | − | − |
| 8 | 78A | pa | pa, sa | − | − |
| 9 | 35 | burk | pa, sa, steno, burk | ++ | DSF, BDSF |
| 10 | 100A | pa | pa, sa, steno | ++ | DSF, BDSF |
| 11 | 10B | steno, asp, can | pa, sa, steno, burk | ++ | DSF, BDSF |
| 12 | 63B | pa, can | pa, sa, steno | ++ | BDSF |
| 13 | 113B | sa, pa | pa, sa | − | − |
| 14 | 106A | pa, sa | pa, sa | − | − |
| 15 | 62A | pa, can | pa | − | − |
| 16 | 97B | pa | pa, sa | − | − |
| 17 | 117A | can, asp | sa | − | − |
| 18 | 57C | steno, can, sphino | steno, pa, sa | ++ | DSF |
| 19 | 130A | pa | pa, sa, strep | − | − |
| 20 | 102B | pa | pa, sa, strep | − | NA |
| 21 | 19C | pa | pa, sa | − | NA |
| 22 | 47A | sa, pa | pa, sa, strep | − | NA |
| 23 | 45A | pa, can | pa, sa | ++ | DSF, BDSF |
| 24 | 57A | steno, can | pa, sa, steno, burk | ++ | DSF, BDSF |
| 25 | 57B | can, steno | pa, sa, steno | ++ | DSF, BDSF |
| 26 | 54B | pa | pa, sa | − | NA |
| 27 | 41A | steno, can, pa, asp | pa, sa, steno, burk | ++ | DSF, BDSF |
| 28 | 58A | burk | pa, sa, steno, burk | ++ | DSF, BDSF |
| 29 | 126A | sa, pa, strep | pa, strep, steno, burk | − | NA |
| 30 | 107C | pa, strep, can, sa | pa, strep, steno, burk | ++ | DSF, BDSF |
| 31 | 85A | sa, pa | pa, strep, sa | − | NA |
| 32 | 17A | pa | pa, strep, steno, sa | − | NA |
| 33 | 93B | pa, can | − | NA | |
| 34 | 123A | pa, sa | pa, strep, sa | − | NA |
| 35 | 2A | pa, sa | pa, strep, steno, sa | − | NA |
| 36 | 34A | sa | pa, sa | ++ | DSF, BDSF |
| 37 | 72A | pa, can | pa, sa | − | NA |
| 38 | 40A | pa, sa | pa, sa | − | NA |
| 39 | 101A | pa, sa | pa, sa | ++ | DSF, BDSF |
| 40 | 131 | Normal resp. flora only | Normal resp. flora only | − | NA |
| 41 | 132 | Normal resp. flora only | Normal resp. flora only | − | NA |
| 42 | 133 | Normal resp. flora only | Normal resp. flora only | − | NA |
| 43 | 134 | Normal resp. flora only | Normal resp. flora only | − | NA |
| 44 | 135 | Normal resp. flora only | Normal resp. flora only | − | NA |
| 45 | 136 | Normal resp. flora only | Normal resp. flora only | − | NA |
| 46 | 131 | Normal resp. flora only | Normal resp. flora only | − | − |
| 47 | 132 | Normal resp. flora only | Normal resp. flora only | − | − |
| 48 | 133 | Normal resp. flora only | Normal resp. flora only | − | − |
| 49 | 134 | Normal resp. flora only | Normal resp. flora only | − | − |
| 50 | 135 | Normal resp. flora only | Normal resp. flora only | − | − |
Abbreviations: BDSF, cis-2-dodecenoic acid; CF, cystic fibrosis; DSF, diffusible signal factor; HPLC, high-performance liquid chromatography; LC, liquid chromatography; MS, mass spectrometry; NA, not applicable.
Abbreviations of microbes identified by culture-based methods: asp (Aspergillus species); burk (Burkholderia species); man (Candida species); pa (Pseudomonas aeruginosa); sa (Staphylococcus aureus); sphino (Sphingomonas species); steno (Stenotrophomonas species); strep (Streptococcus species).
Abbreviations of bacteria identified based solely on 454 sequence data obtained from the amplification of the V3-V5 region of the universal 16S rRNA gene
Detection of DSF activity using the bioassay (+, activation of the bioassay activity; −, no activation of the bioassay)
DSF signal molecules which were identified in extracts from patient sputa using HPLC and MS. DSF refers to the molecule cis-11-methyl-2-dodecenoic acid, whereas BDSF refers to the molecule cis-2-dodecenoic acid.
The activation of DSF bio-reporter suggested the presence of DSF family signal molecules in the sputum samples. In order to obtain direct evidence that these signals were present, the sputum extracts were separated by reverse-phase high-performance liquid chromatography and fractions tested for DSF activity (Figure 1). Several peaks of activity were detected for each sample. Two of these peaks had the same elution time as synthetic DSF and BDSF standards, and mass spectrometry allowed these to be tentatively identified as DSF (m/z of 211.17) and BDSF (m/z of 197.15), respectively (Figure 1). The compounds in the other peaks could not be identified. The DSF bio-reporter detects concentrations of DSF in the range of 2.5 to 2500 n; the concentration of DSF was estimated to be approximately 250 n in each of the positive sputum samples tested.
To explore the relationship between the presence of DSF family signals and bacterial community composition, we performed a combination of culture-dependent and culture-independent analyses on sputum samples. For culture-independent analysis, bacterial DNA was extracted from sputum samples and the V3–V5 region of the 16S rRNA gene was amplified. These barcoded and pooled amplicons were subsequently sequenced on a 454 Genome Sequencer FLX platform. The 16S rRNA gene-based analyses revealed complex bacterial communities associated with CF infection (see Table 1; Supplementary Table S2; Supplementary Figure S1), as has also been seen in recent studies using similar techniques (Harris et al., 2007; Klepac-Ceraj et al., 2010; Rogers et al., 2010; Guss et al., 2011). In summary, the analysis detected a total of 53,225 PCR amplicons that were <350 bp in length, with an average 325 reads for each sample (Supplementary Table S2). Using a 98% similarity threshold value, we identified a wide range of operational taxonomic units for each sample. The number of sequences from each genus was determined and the 10 most abundant genera were observed to be Pseudomonas, Streptococcus, Rothia, Staphylococcus, Stenotrophomonas, Fusobacterium, Chryseomonas, Actinomyces, Porphyromonas and Neisseria (see Supplementary Figure S1; Supplementary Table S2).
For culture-dependent analysis, samples were plated on standard selective and non-selective media. As expected, the culture-based approach isolated and identified P. aeruginosa, S. aureus, Burkholderia species and S. maltophilia, which are pathogens typically associated with CF infection. Atypical CF species were also isolated by the culture-based approach. These included Streptococcus species, which were also detected using the 454 sequencing of 16S-rRNA amplicons (Supplementary Figure S1).
Comparison of these findings with the analysis of signal molecules in CF sputum indicated a direct correlation between occurrence of DSF-family signals and the presence of Burkholderia species and/or S. maltophilia, but no correlation with the presence of P. aeruginosa or S. aureus (Table 1).
Isolates of Pseudomonas aeruginosa from CF patients retain the ability to respond to DSF
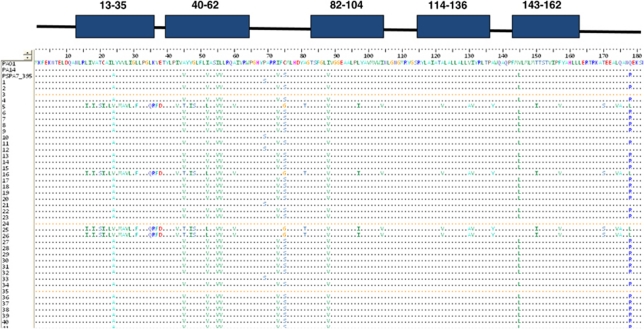
Analysis of P. aeruginosa isolates from the airways of multiple CF patients has revealed a number of genetic adaptations to chronic infection (Smith et al., 2006; D'Argenio et al., 2007). In some cases, adaptation involves mutation of genes involved in intraspecies signaling. In order to assess whether adaptation might influence DSF-dependent interspecies signaling, we assayed a panel of 50 P. aeruginosa isolates from 40 different adult CF patients for the presence of the PA1396 gene that encodes the DSF sensor (Ryan et al., 2008). The PA1396 gene was amplified by PCR from all 50 isolates and amplicons were sequenced. Bioinformatic analysis using ClustalW showed that PA1396 from each of the clinical isolates had a very high degree of sequence similarity to PA1396 from the sequenced strains of P. aeruginosa (Figure 2; Supplementary Figure S2), with very little amino-acid sequence diversity in the sensory input, histidine kinase or receiver domains.
Figure 2.
Comparison of amino-acid sequence of the sensory input domain of the DSF sensor PA1396 from different CF isolates of P. aeruginosa. The amino-acid sequences shown represent residues 1–187 of PA1396. The top panel describes the predicted transmembrane segments found in the input of PA1396. The sequences were obtained by PCR, cloning and sequencing of full-length PA1396 genes from 40 clinical isolates of P. aeruginosa extracted from the sputum from CF patients. Sequences of PA1396 from the model P. aeruginosa strains PAO1, PA14 and PA7 are included for comparative purposes. Sequences were aligned using ClustalW and presented using BIOEDIT. The majority of alterations at different positions appear to be of a conservative nature.
In parallel, the ability of these 50 P. aeruginosa isolates to respond to exogenous synthetic DSF was assessed. We have previously shown that perception of DSF by P. aeruginosa leads to increased expression of pmrAB operon and increases in the level of resistance to the cationic antimicrobial peptides polymyxin B and E (Ryan et al., 2008). The effects of DSF addition to a subset of these clinical isolates was monitored by measurement of the expression of pmrAB (PA4774-4778) by quantitative RT-PCR and of the tolerance to polymyxin B and E. Addition of synthetic DSF to the P. aeruginosa CF isolates lead to the upregulation of pmrAB (Supplementary Figure S3), which was mirrored by an increased tolerance to polymyxin B (Supplementary Figure S4).
Our previous work has demonstrated that mutation of PA1396 in the model strain PAO1 led to increased tolerance to polymyxins B and E (Ryan et al., 2008). To determine whether PA1396 has a similar influence in clinical isolates of P. aeruginosa, the gene was disrupted in five clinical isolates and mutants were assessed for alterations in phenotype. In four of the five strains, mutation of PA1396 led to an enhanced tolerance to polymyxins B and E under high Mg2+ conditions (Supplementary Figure S5). The remaining strain CF23 already had high intrinsic tolerance to polymyxin B and E (1 mg ml–1) and mutation of PA1396 gave no further alteration. The basis of the enhanced polymyxin tolerance of this strain is unclear; it was not associated with enhanced expression of pmrAB and the strain responded to addition of DSF by alteration in pmrAB expression (data not shown). Taken together, the findings indicate that the P. aeruginosa CF isolates retain the ability to respond to DSF and have a functional sensor kinase protein PA1396.
DSF signaling promotes P. aeruginosa persistence in CFTR knockout mice
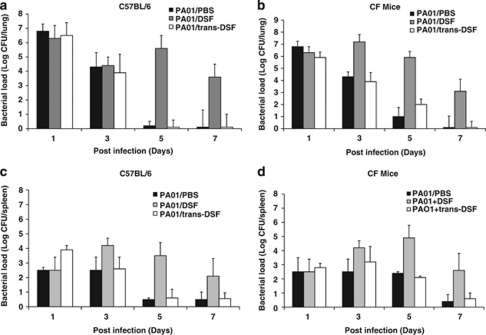
To determine whether DSF can modulate P. aeruginosa infections in a mammalian model, the effects of supplementation of P. aeruginosa PAO1 with DSF on the bacterial load in the lung was investigated in both CFTR knockout (CF) mice or the isogenic parental C57BL/6 mice. As controls for these experiments, bacterial inocula were supplemented with PBS or with synthetic trans-11-methyl dodecenoic acid, which is not recognized by PA1396 and does not influence pmrAB gene expression (Ryan et al., 2008). The lung bacterial load was determined at 1, 3, 5 or 7 days post infection (see Materials and methods).
C57BL/6 mice infected with P. aeruginosa PAO1 with either PBS or trans-11-methyl dodecenoic acid cleared all bacteria within 3 days (Figure 3). With addition of DSF, however, bacteria were still present at 7 days. CF mice infected with PAO1 showed persistence of bacteria for 5 days (Figure 3). CF mice infected with PAO1 with DSF showed persistence of bacteria for up to 7 days and bacteria were also detected in the spleen at 7 days post-infection, indicating that there was dissemination (Figure 3). In contrast to these effects of DSF, addition of trans-11-methyl dodecenoic acid had no effect on bacterial persistence or on dissemination (Figure 3).
Figure 3.
Administration of DSF enhances the persistence P. aeruginosa PAO1 in mice. C57BL/6 or CF mice were infected with PAO1 in PBS with or without supplementation with DSF (designated PAO1/DSF) or trans-11-methyl dodecenoic (designated PAO1/ trans-DSF). Animals were killed at 1, 3, 5, 7 days after infection and 10-fold serial dilutions of the lung (a, b) and spleen (c, d) homogenates were plated to determine the bacterial load. The means and s.d. of triplicate measurements are shown.
The role of PA1396 in P. aeruginosa persistence during CF infection was also investigated by examination of the clearance of the PA1396 mutant from C57BL/6 mice or CF mice (Supplementary Figure S6). The PA1396 mutant showed similar persistence in both C57BL/6 mice or CF mice (for up to 7 days) as P. aeruginosa PAO1 with added DSF (Supplementary Figure S5). Addition of synthetic DSF or trans-11-methyl dodecenoic acid to the PA1396 mutant had no effect in either C57BL/6 mice or CF mice (Supplementary Figure S6). These results suggest that the presence of DSF increases the capacity of P. aeruginosa to persist in both CF and normal mice, and that this persistence effect is dependent on PA1396.
DSF influences polymyxin tolerance in P. aeruginosa biofilms on CF-derived airway epithelial cells
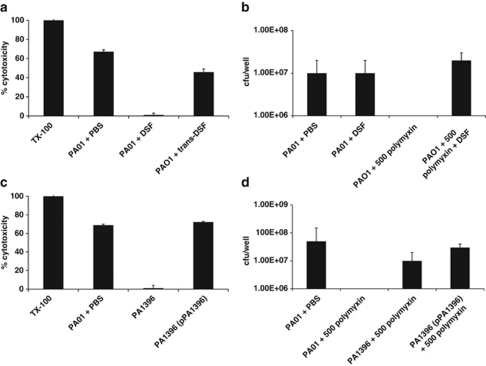
As an approach to understanding the influence of DSF on P. aeruginosa infection in the context of the CF airway, we used an in vitro tissue culture-based system in which P. aeruginosa biofilms were allowed to form on human CF-derived lung epithelial cells. We grew CFBE cells, a human bronchial epithelial cell line with a CFTR F508/F508 genotype, in standard multiwell (24-well) tissue-culture plates, in which they formed confluent monolayers (Anderson et al., 2008; Moreau-Marquis et al., 2008). These monolayer cultures were inoculated with P. aeruginosa strain PAO1, and biofilm formation in this static system was monitored over time by phase-contrast microscopy and CFU count of attached bacteria. In parallel, LDH release from the epithelial cells was measured to assess the cytotoxicity of the bacteria.
In the absence of DSF, after 4 to 6 h of exposure to P. aeruginosa PAO1, most epithelial cells had detached from the plastic, leaving only small patches of isolated cells (Figure 4). The measurements of LDH release suggested that bacteria were cytotoxic (Figure 4). The addition of synthetic DSF to the tissue-culture medium resulted in the preservation of monolayer integrity for up to 12 h after introduction of P. aeruginosa PAO1. There was a concomitant reduction of LDH released from the CFBE epithelial cell monolayer (Figure 4). However, addition of DSF did not alter the numbers of bacteria bound to the epithelial cells after 1 h of incubation. In contrast, the addition of synthetic trans-11-methyl dodecenoic acid did not alter the timing of the effects of P. aeruginosa of monolayer integrity, which was comparable to that seen with no addition (Figure 4).
Figure 4.
Addition of DSF or mutation of PA1396 affects virulence and polymyxin tolerance of P. aeruginosa in co-culture with CFBE epithelial cells. (a) Cytotoxicity of P. aeruginosa to CFBE cells as measured by LDH release at 6 h after inoculation with PAO1 wild type in the absence or presence of DSF. The values, which are expressed as percentage of the response seen after addition of triton X100 (TX-100) detergent, are the means and s.d. of triplicate measurements. (b) Mutation of PA1396 significantly reduces the cytotoxicity of P. aeruginosa PAO1 to CFBE epithelial cells. Cytotoxicity was rescued by complementation in trans with the full-length gene (pPA1396). (c, d) Addition of DSF and mutation of PA1396 affect polymyxin tolerance of P. aeruginosa biofilms on CFBE epithelial cells. For these experiments, 500 μg ml−1 polymyxin was added to the co-culture at 1 h, which prevents the cytotoxic effects of P. aeruginosa (see Materials and methods). After 24 h in the absence of DSF, all bacteria were eradicated. However, in the presence of DSF, 500 μg ml−1 polymyxin had no apparent effect. (d) Mutation of PA1396 increases the polymyxin tolerance of P. aeruginosa in co-culture with CFBE epithelial cells and can be complemented towards wild type by in trans expression of the full-length gene (pPA1396).
Previous studies have shown that bacterial biofilms cultured on CFBE cells are extremely resistant to antibiotic treatment (Anderson et al., 2008; Moreau-Marquis et al., 2008). We treated static CFBE/PAO1 biofilms for 24 h with 500 μg ml–1 polymyxin B, a concentration measured in the lungs of CF patients (McAllister et al., 1999). CFU counts before the antibiotic treatment were approximately 108 CFU/well. After antibiotic treatment, <103 CFU/well remained on the cells suggesting that the minimum bactericidal concentration of polymyxin B was >500 μg ml–1 under these conditions (Figure 4). The presence of synthetic DSF increased level of polymyxin B resistance of P. aeruginosa PAO1 biofilms developed on CFBE cells as 107 CFU/well remained after antibiotic treatment (Figure 4). The data suggest that DSF treatment of P. aeruginosa biofilms on CFBE cells results in an increase in the tolerance to polymyxin B, but decrease in cytotoxicity of the bacteria to the epithelial cells. Mutation of PA1396 had similar effects to DSF addition The PA1396 mutant showed decreased virulence to epithelial cells as measured by LDH release and increased tolerance to polymyxin B (Figure 4). In trans expression of the full-length gene, PA1396, restored cytotoxicity and antibiotic sensitivity to wild-type levels (Figure 4).
DSF modulates expression of genes implicated in a broad range of functions in P. aeruginosa
The broader effects of DSF on regulation of gene expression in P. aeruginosa were investigated by comparative transcriptome profiling using Affymetrix chips. For these experiments, bacteria were grown in artificial CF sputum medium in the absence and presence of DSF or trans-11-methyl dodecenoic. All cultures were harvested at an OD600 of 0.6. A total of 52 genes were differentially expressed by at least five-fold during growth of P. aeruginosa in artificial CF sputum in the presence or absence of DSF (Table 2). Of these, 28 genes were upregulated, whereas 24 genes were downregulated. Those genes whose expression was increased by DSF included those whose products are involved in flagellar synthesis (PA1467), iron uptake (PA4358, PA4359) and biofilm formation (PA4781), as well as antibiotic resistance (PA4599, PA4774–PA4777). Those genes that were downregulated in response to DSF included those encoding subunits of cytochrome c oxidase as well as those involved in type III secretion. For those genes that were tested, these changes were confirmed by RT-PCR (Supplementary Figure S7). Alteration in expression of the majority of these genes is not seen as a response to trans-11-methyl dodecenoic (Table 2), indicating that they were specific responses to DSF. The findings suggest that DSF can influence a broad range of functions in P. aeruginosa that may underpin the enhanced persistence and reduced virulence seen as a response to the signal.
Table 2. P. aeruginosa genes differentially regulated during growth in artificial CF sputum in the presence of DSF.
| ORFa | Gene namea | Category or classa |
Fold changesb |
|
|---|---|---|---|---|
| WT+DSF versus WT | WT+trans-11-methyl-2-dodecenoic acid versus WT | |||
| PA0744 | Enoyl-CoA hydratase | 11 | ||
| PA0745 | Enoyl-CoA hydratase | 21 | ||
| PA0746 | Acyl-CoA dehydrogenase | 23 | ||
| PA0747 | Aldehyde dehydrogenase | 7 | ||
| PA0806 | Hypothetical protein | 10 | 7 | |
| PA1325 | Hypothetical protein | 9 | ||
| PA1326 | ilvA2 | Threonine dehydratase | 17 | |
| PA1327 | Protease | 6 | ||
| PA1467 | Hypothetical protein | 6 | ||
| PA1559 | Hypothetical protein | 10 | 5 | |
| PA1560 | Hypothetical protein | 11 | 10 | |
| PA1797 | Hypothetical protein | 54 | 5 | |
| PA1889 | Hypothetical protein | 5 | ||
| PA2339 | Mannitol transport protein | 9 | ||
| PA2966 | acpP | Acyl carrier protein | 3 | 7 |
| PA3790 | oprC | Copper transport outer membrane porin OprC precursor | 5 | |
| PA4138 | tyrS | Tyrosyl-tRNA synthetase | 12 | |
| PA4357 | Hypothetical protein | 6 | ||
| PA4358 | Ferrous iron transport protein B | 6 | 5 | |
| PA4359 | Ferrous iron transport protein A | 8 | 5 | |
| PA4599 | mexC | Resistance-nodulation-cell division (RND) multidrug efflux | 5 | |
| PA4773 | Hypothetical protein | 7 | ||
| PA4774 | Hypothetical protein | 9 | ||
| PA4775 | Hypothetical protein | 8 | ||
| PA4776 | pmrA | Two-component regulator system response regulator | 7 | |
| PA4777 | pmrB | Two-component regulator system signal sensor kinase | 6 | |
| PA4781 | Cyclic di-GMP phosphodiesterase | 7 | ||
| PA4782 | Hypothetical protein | 6 | ||
| PA4898 | opdK | Vanillate porin | 5 | 5 |
| PA4908 | Ornithine cyclodeaminase | 5 | 9 | |
| PA5101 | Hypothetical protein | 5 | 11 | |
| PA0901 | Succinylglutamate desuccinylase | −8 | ||
| PA0910 | Hypothetical protein | −6 | ||
| PA1421 | gbuA | Guanidinobutyrase | −23 | |
| PA1546 | hemN | Coproporphyrinogen III oxidase | −8 | |
| PA1556 | ccoO | cbb3-Type cytochrome c oxidase subunit II | −7 | |
| PA1557 | ccoN | cbb3-Type cytochrome c oxidase subunit I | −8 | |
| PA1561 | Aerotaxis receptor | −7 | ||
| PA1699 | Hypothetical protein in type III secretion | −7 | ||
| PA1701 | Hypothetical protein in type III secretion | −7 | ||
| PA1708 | popB | Translocator protein | −8 | |
| PA1709 | popD | Translocator outer membrane protein | −6 | |
| PA1710 | exsC | Exoenzyme S synthesis protein | −10 | |
| PA1711 | exsE | −10 | ||
| PA1714 | exsD | −6 | ||
| PA1715 | pscB | Type III export apparatus protein | −6 | |
| PA2222 | Hypothetical protein | −5 | ||
| PA3684 | Hypothetical protein | −8 | −5 | |
| PA3832 | Hypothetical protein | −6 | −9 | |
| PA3842 | Chaperone | −12 | ||
| PA3880 | Hypothetical protein | −7 | ||
| PA3967 | Hypothetical protein | −8 | −6 | |
| PA4000 | Hypothetical protein | −6 | −6 | |
| PA5475 | Hypothetical protein | −7 | −9 | |
| PA5494 | Hypothetical protein | −10 | ||
| PA5505 | TonB-dependent receptor | −6 | ||
Abbreviations: CF, cystic fibrosis; DSF, diffusible signal factor; ORF, open reading frame.
From P. aeruginosa genome website, http://www.pseudomonas.com
Regulation (n-fold) of genes differentially expressed during P. aeruginosa growth in artificial CF sputum with DSF or trans-11-methyl-2-dodecenoic acid compared with wild type; a positive number indicates an upregulation of the gene and a negative number indicates a downregulation of the gene.
Intriguingly, mutation of PA1396 gave rise to altered levels of transcripts for PA4358, PA4359, PA4599, PA4774, PA477, PA4776, PA0901 and PA5505, which are also regulated by DSF; however, transcript levels of other DSF-responsive genes (PA0806, PA1560, PA2966) were not altered in the PA1396 mutant (Supplementary Figure S8). This may suggest the existence of further pathways of DSF sensing in P. aeruginosa that do not involve PA1396.
Discussion
In nature, bacteria are more likely to grow in polymicrobial communities than in monoculture (Sibley et al., 2009; Rogers et al., 2010). The development and maintenance of such multispecies consortia will depend upon interactions between community members (Sibley et al., 2009; Rogers et al., 2010). In vitro studies using model systems indicate the potential importance of interspecies signaling involving diffusible signals in modulation of the behavior of different bacteria in such communities. Here, we have provided evidence to support the contention that interspecies signaling is important in a ‘natural' environment, that of the polymicrobial community associated with the CF lung.
We analyzed sputum taken from patients with CF for the presence of diffusible signal molecules of the DSF family. We detected cis-11-methyl-2-dodecenoic acid (DSF) and cis-2-dodecenoic acid (BDSF) in these sputum samples (Table 1). These signals are produced by the CF-associated pathogens S. maltophilia and B. cenocepacia, and their presence in the sputum samples was strongly correlated with patient colonization by S. maltophilia and/or B. cenocepacia (Table 1). Although P. aeruginosa, the predominant CF pathogen, does not produce DSF or BDSF, it is able to respond to these signals, leading to changes in biofilm architecture and increased resistance to cationic antimicrobial peptides. This response of P. aeruginosa depends upon the sensor kinase PA1396. Each of the 50 clinical (CF) isolates of P. aeruginosa that were tested retained a response to DSF measured in vitro, and each had an intact PA1396 gene based on sequence analysis (Figure 2; Supplementary Figure S2). For a selection of these clinical isolates, we also demonstrated that PA1396 is functional and is involved in the regulation of antibiotic resistance and gene expression. Furthermore, the concentration of signal molecules in the sputum was estimated to be approximately 250 n, which is the physiological range for the P. aeruginosa response. Taken together, these findings indicate the potential for interspecies signaling involving DSF family signals between different bacterial species within the CF lung.
The influence of DSF on P. aeruginosa has been examined thus far only in vitro. Here we have shown that the presence of DSF promotes persistence of P. aeruginosa in CFTR knockout mice, promotes polymyxin tolerance in P. aeruginosa biofilms developed on CF-derived airway epithelial cells, but inhibits the cytotoxicity of P. aeruginosa to cultured cells (Figure 3). Although extrapolations from these model systems should be made cautiously, these findings indicate the possible influence that DSF signaling might have on the behavior of P. aeruginosa in the CF lung. Intriguingly, the co-ordination of the reduction in cytotoxicity and development of an antibiotic resistant biofilm has been associated with the transition between acute and chronic infections of CF lung by P. aeruginosa.
Interestingly, we detected two additional molecules with DSF activity in the bioassay in the sputum samples (Figure 1). Although we were unable to identify them, we speculate that these molecules may be also members of the DSF family. The presence of DSF and BDSF was correlated with the presence of S. maltophilia and/or B. cenocepacia, as determined collectively by culture-based methods and 16S rRNA sequencing. However, for some patients, the occurrence of S. maltophilia and/or B. cenocepacia was indicated only by 16S rRNA sequencing (Table 1). This raises the question of whether the detection of DSF-family signals might find a use as a marker for the presence of certain bacteria within the microbial flora of the CF respiratory tract. Our analysis of the microbial community demonstrated a diverse population of microorganisms within eight bacterial phyla, comprising >50 genera, including facultative and obligate anaerobes, oral bacteria and opportunistic pathogens including previously recognized CF pathogens (Supplementary Figure S1). It is not known how many of these other bacteria produce DSF or BDSF. Bioinformatic analysis of bacterial genome sequences suggest that the synthesis of these molecules is not restricted to the xanthomonads or Burkholderia spp. as unrelated organisms such as Thiobacillus denitrificans, Methylobacillus flagellatus, Sideroxydans lithotrophicus carry an rpf gene cluster (unpublished data). It is noteworthy that BDSF and DSF were not detected in sputum samples from viral bronchitis patients who also showed diverse population of bacteria comprising >40 genera consistent with normal respiratory flora with an absence of bacterial opportunistic pathogens. Detection of DSF family signal molecules may not afford a definitive test for the presence of particular bacterial genera, but may nevertheless allow monitoring of changes in infection status that inform the therapeutic regime.
The sensing of DSF by P. aeruginosa leads to alteration in expression of genes encoding a wide range of functions to include biofilm formation and increased tolerance to polymyxins. These effects, which require the sensor kinase PA1396, could impinge on pathogen persistence and response to antibiotic treatment (Figure 4; Supplementary Figures S5 and S6). Molecules that block key signal sensing or transduction steps in pathogens could represent lead compounds for new drugs (Rasko and Sperandio 2009, 2010). It remains to be seen whether interference with the action of PA1396 or associated downstream signal transduction pathways may render P. aeruginosa more susceptible to antibiotic treatment.
In conclusion, understanding the microbial flora of the CF respiratory tract is of considerable importance, as interactions between community members can potentially affect virulence and persistence of pathogens such as P. aeruginosa; we are beginning to understand the role that the diffusible signals may have in these interactions. In vitro experiments have shown that bacterial signals such as AI-2 and DSF can exert different influences on the behavior of P. aeruginosa. In addition, bacterial signals such as alkyl quinolones and N-acyl homoserine lactones have been detected in the sputum of CF patients. Our data provide substantial evidence to support the contention that interspecies DSF-mediated interactions occur in the CF lung and may influence the efficacy of antibiotic treatment, particularly for chronic infections involving persistence of bacteria.
Acknowledgments
We thank Steve P Bernier for help and guidance in performing co-culture biofilm experiments. We are also indebted to Melanie Febrer and Mark Alston at The Genome Analysis Centre for advice and assistance with the bioinformatic and statistical analyses of next-generation sequencing data. KBT, YM, JMD. and RPR were supported in part by grants awarded by the Science Foundation of Ireland (SFI 07/IN.1/B955 to JMD and SFI 09/SIRG/B1654 to RPR) and through a Royal Irish Academy fellowship (to RRP). This work was also supported by NIH grant R01AI083256 to GAO.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anderson GG, Moreau-Marquis S, Stanton BA, O'Toole GA. In vitro analysis of tobramycin-treated Pseudomonas aeruginosa Biofilms on cystic fibrosis-derived airway epithelial cells. Infect Immun. 2008;76:1423–1433. doi: 10.1128/IAI.01373-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber CE, Tang JL, Feng JX, Pan MQ, Wilson TJG, Slater H, et al. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol Microbiol. 1997;24:555–566. doi: 10.1046/j.1365-2958.1997.3721736.x. [DOI] [PubMed] [Google Scholar]
- Boon C, Deng YY, Wang LH, He YW, Xu JL, Fan Y, et al. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J. 2008;2:27–36. doi: 10.1038/ismej.2007.76. [DOI] [PubMed] [Google Scholar]
- D'Argenio DA, Wu MH, Hoffman LR, Kulasekara HD, Deziel E, Smith EE, et al. Growth phenotypes of Pseudomonas aeruginosalasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol. 2007;64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Marques CNH. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol. 2009;191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng YY, Wu JE, Tao F, Zhang LH. Listening to a new language: DSF-based quorum sensing in gram-negative bacteria. Chem Rev. 2011;111:160–173. doi: 10.1021/cr100354f. [DOI] [PubMed] [Google Scholar]
- Fouhy Y, Scanlon K, Schouest K, Spillane C, Crossman L, Avison MB, et al. Diffusible signal factor-dependent cell-cell signaling and virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J Bacteriol. 2007;189:4964–4968. doi: 10.1128/JB.00310-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Guss AM, Roeselers G, Newton ILG, Young CR, Klepac-Ceraj V, Lory S, et al. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J. 2011;5:20–29. doi: 10.1038/ismej.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JK, De Groote MA, Sagel SD, Zemanick ET, Kapsner R, Penvari C, et al. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci USA. 2007;104:20529–20533. doi: 10.1073/pnas.0709804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TP, Wong ACL. A cyclic AMP receptor protein-regulated cell-cell communication system mediates expression of a FecA homologue in Stenotrophomonas maltophilia. App Environ Microbiol. 2007;73:5034–5040. doi: 10.1128/AEM.00366-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepac-Ceraj V, Lemon KP, Martin TR, Allgaier M, Kembel SW, Knapp AA, et al. Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ Microbiol. 2010;12:1293–1303. doi: 10.1111/j.1462-2920.2010.02173.x. [DOI] [PubMed] [Google Scholar]
- McAllister SM, Alpar HO, Brown MRW. Antimicrobial properties of liposomal polymyxin B. J Antimicrobial Chemo. 1999;43:203–210. doi: 10.1093/jac/43.2.203. [DOI] [PubMed] [Google Scholar]
- McCarthy Y, Yang LA, Twomey KB, Sass A, Tolker-Nielsen T, Mahenthiralingam E, et al. A sensor kinase recognizing the cell-cell signal BDSF (cis-2-dodecenoic acid) regulates virulence in Burkholderia cenocepacia. Mol Microbiol. 2010;77:1220–1236. doi: 10.1111/j.1365-2958.2010.07285.x. [DOI] [PubMed] [Google Scholar]
- Moreau-Marquis S, Bomberger JM, Anderson GG, Swiatecka-Urban A, Ye SY, O'Toole GA, et al. The Delta F508-CFTR mutation results in increased biofilm formation by Pseudomonas aeruginosa by increasing iron availability. Am J Physiol Lung Cell Mol Physiol. 2008;295:L25–L37. doi: 10.1152/ajplung.00391.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Ann Rev Gen. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko DA, Sperandio V.2009Novel approaches to bacterial infection therapy by interfering with cell-to-cell signaling Curr Protoc MicrobiolChapter 17: Unit 17.13. [DOI] [PubMed]
- Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Dis. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- Rogers GB, Hoffman LR, Whiteley M, Daniels TWV, Carroll MP, Bruce KD. Revealing the dynamics of polymicrobial infections: implications for antibiotic therapy. Trends Microbiol. 2010;18:357–364. doi: 10.1016/j.tim.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RP, Dow JM. Diffusible signals and interspecies communication in bacteria. Microbiol. 2008;154:1845–1858. doi: 10.1099/mic.0.2008/017871-0. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Dow JM. Intermolecular interactions between HD-GYP and GGDEF domain proteins mediate virulence-related signal transduction in Xanthomonas campestris. Virulence. 2010;1:404–408. doi: 10.4161/viru.1.5.12704. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Dow JM. Communication with a growing family: diffusible signal factor (DSF) signaling in bacteria. Trends Microbiol. 2011;19:145–152. doi: 10.1016/j.tim.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Fouhy Y, Garcia BF, Watt SA, Niehaus K, Yang L, et al. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol Microbiol. 2008;68:75–86. doi: 10.1111/j.1365-2958.2008.06132.x. [DOI] [PubMed] [Google Scholar]
- Ryan RP, McCarthy Y, Andrade M, Farah CS, Armitage JP, Dow JM. Cell-cell signal-dependent dynamic interactions between HD-GYP and GGDEF domain proteins mediate virulence in Xanthomonas campestris. Proc Natl Acad Sci USA. 2010;107:5989–5994. doi: 10.1073/pnas.0912839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank EA, Kolter R. New developments in microbial interspecies signaling. Curr Opin Microbiol. 2009;12:205–214. doi: 10.1016/j.mib.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CD, Parkins MD, Rabin HR, Duan K, Norgaard JC, Surette MG. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci USA. 2008;105:15070–15075. doi: 10.1073/pnas.0804326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CD, Parkins MD, Rabin HR, Surette MG. The relevance of the polymicrobial nature of airway infection in the acute and chronic management of patients with cystic fibrosis. Curr Opin Invest Drugs. 2009;10:787–794. [PubMed] [Google Scholar]
- Smith EE, Buckley DG, Wu ZN, Saenphimmachak C, Hoffman LR, D'Argenio DA, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriramulu DD, Lunsdorf H, Lam JS, Romling U. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J Med Microbiol. 2005;54:667–676. doi: 10.1099/jmm.0.45969-0. [DOI] [PubMed] [Google Scholar]
- Wang LH, He YW, Gao YF, Wu JE, Dong YH, He CZ, et al. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol Microbiol. 2004;51:903–912. doi: 10.1046/j.1365-2958.2003.03883.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.